SUMMARY
Cancer is driven by genetic and epigenetic alterations that allow cells to overproliferate and escape mechanisms that normally control their survival and migration. Many of these alterations map to signaling pathways that control cell growth and division, cell death, cell fate, and cell motility, and can be placed in the context of distortions of wider signaling networks that fuel cancer progression, such as changes in the tumor microenvironment, angiogenesis, and inflammation. Mutations that convert cellular proto-oncogenes to oncogenes can cause hyperactivation of these signaling pathways, whereas inactivation of tumor suppressors eliminates critical negative regulators of signaling. An examination of the PI3K-Akt and Ras-ERK pathways illustrates how such alterations dysregulate signaling in cancer and produce many of the characteristic features of tumor cells.
Many of the genetic and epigenetic alterations that drive cancer affect signaling molecules. An examination of the PI3K-Akt and Ras-ERK pathways illustrates how this produces the characteristic features of tumor cells.
1. INTRODUCTION
The development of cancer involves successive genetic and epigenetic alterations that allow cells to escape homeostatic controls that ordinarily suppress inappropriate proliferation and inhibit the survival of aberrantly proliferating cells outside their normal niches. Most cancers arise in epithelial cells, manifesting as carcinomas in organs such as the lung, skin, breast, liver, and pancreas. Sarcomas, in contrast, arise from mesenchymal tissues, occurring in fibroblasts, myocytes, adipocytes, and osteoblasts. Nonepithelial tumors can also develop in cells of the nervous system (e.g., gliomas, neuroblastomas, and medulloblastomas) and hematopoietic tissues (leukemia and lymphoma).
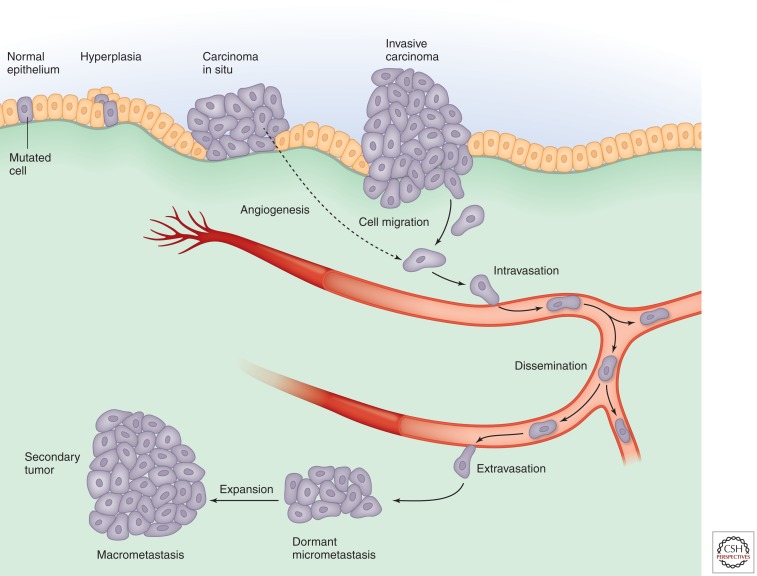
In solid tumors, these alterations typically promote progression from a relatively benign group of proliferating cells (hyperplasias) to a mass of cells with abnormal morphology, cytological appearance, and cellular organization. After a tumor expands, the tumor core loses access to oxygen and nutrients, often leading to the growth of new blood vessels (angiogenesis), which restores access to nutrients and oxygen. Subsequently, tumor cells can develop the ability to invade the tissue beyond their normal boundaries, enter the circulation, and seed new tumors at other locations (metastasis), the defining feature of malignancy (Fig. 1). This linear sequence of events is clearly an oversimplification of complex cancer-associated events that proceed in distinct ways in individual tumors and between tumor sites; however, it provides a useful framework in which to highlight the critical role of dysregulated signaling in processes associated with the initiation and progression of cancer.
Figure 1.
Cancer progression.
The root cause of cancer is usually genetic or epigenetic alterations in the tumor cells (see below). Progression of the cancer, however, is associated with a complex interplay between the tumor cells and surrounding non-neoplastic cells and the extracellular matrix (ECM). Moreover, the tumor cells develop several well-defined features (Hanahan and Weinberg 2000; Solimini et al. 2007). In addition to increased cell proliferation, these include resistance to apoptosis and other forms of cell death, metabolic changes, genetic instability, induction of angiogenesis, and increased migratory capacity. Dysregulation of cellular signal transduction pathways underlies most of these characteristics.
Here, we describe how tumor cells co-opt signaling pathways to allow them to proliferate, survive, and invade other tissues. To cover all of the signaling molecules involved and their myriad contributions to cancer would require an entire textbook (Weinberg 2013). We therefore focus primarily on two pathways—Ras-ERK (Morrison 2012) and PI3K-Akt signaling (Hemmings and Restuccia 2012)—that play central roles in multiple processes associated with cancer, while highlighting the involvement of some other key signaling molecules.
2. MUTATIONS AS THE CAUSE OF CANCER
Most tumors arise as a consequence of genetic alterations to cellular genes, which may be inherited or arise spontaneously—for example, as a result of DNA damage induced by environmental carcinogens or mutations arising from replication errors. These alterations confer a selective advantage to the cells, which together with changes in the microenvironment, promote tumor growth and progression. Some are gain-of-function mutations, producing so-called oncogenes that drive tumor formation. Others inactivate tumor suppressor genes that normally ensure that cells do not proliferate inappropriately or survive outside their normal niche.
Tumors can possess tens to hundreds or even thousands of mutations, but many of these are merely so-called “passengers.” Typically only two to eight are the “driver mutations” that cause progression of the cancer (Vogelstein et al. 2013). These may be point mutations (such as G12V Ras), deletions (as seen with PTEN), inversions, or amplifications (as seen with Myc). Large-scale rearrangements also occur—for example, the BCR-ABL fusions involving chromosomes 9 and 22, which are associated with several leukemias and generate an oncogenic version of the tyrosine kinase Abl. Loss of heterozygosity due to gene conversion or mitotic recombination between normal and mutant parental alleles is another source of genetic alterations that drive cancer. This often affects tumor suppressors such as the retinoblastoma protein (pRB) and p53 (encoded by the TP53 gene in humans).
Changes in the methylation state of promoters of genes that impact cancer can also play an important role in oncogenesis (Sandoval and Esteller 2012; Suva et al. 2013). Indeed, epigenetic silencing is more common than mutational silencing for some genes—for example, the cyclin-dependent kinase (CDK) inhibitor (CKI) p16 (also known as CDKN2A or INK4a) and the mismatch repair (MMR) enzyme MLH1. Silencing of MMR enzymes can lead to additional genetic changes because it affects proteins that prevent errors by repairing DNA. Conversely, several mutations associated with cancer affect epigenetic regulators that influence multiple cellular programs—for example, DNMT1 and TET1, which control DNA methylation, and the histone-modifying enzymes EZH2, SETD2, and KDM6A are deleted or mutated in cancer (Delhommeau et al. 2009; Ley et al. 2010; Wu et al. 2012). Interestingly, mutations in the metabolic enzymes isocitrate dehydrogenase (IDH) 1 and IDH2 may promote cancer by generating an “oncometabolite” not present in normal cells that inhibits certain chromatin-modifying enzymes (see below) (Ward and Thompson 2012a).
Finally, in a minority of cancers, infectious agents are the triggers. A few human cancers are triggered by viruses that encode genes that promote tumorigenesis through activation of oncogene pathways or inactivation of tumor suppressors. The human papilloma virus, which is associated with cervical and head and neck cancers, encodes a protein, E6, that promotes degradation of p53, while another viral protein, E7, inactivates pRB and CKIs, among other effects (Munger and Howley 2002). In hepatocellular carcinoma caused by hepatitis B virus, by contrast, it is not clear whether viral proteins themselves are oncogenic, viral integration promotes expression of nearby cellular oncogenes, or cancer is simply a consequence of persistent liver injury and inflammation (Seeger et al. 2013). Epstein–Barr virus (also known as human herpesvirus 4) produces a protein called LMP1 that acts as a constitutively active tumor necrosis factor (TNF) receptor, engaging a plethora of signaling pathways, including NF-κB, JNK/p38, PI3K, and ERK (Morris et al. 2009). An extreme case of a transmissible cancer is that affecting the Tasmanian devil. All tumors are derived from a founder tumor and are transmitted as allografts from devil to devil during intraspecies facial biting (Murchison et al. 2012; Hamede et al. 2013).
2.1. Cancer-Causing Mutations Affect Signaling Pathways
We can connect the genetic alterations in cancer cells with signaling pathways that control processes associated with tumorigenesis and place these in the context of distortions of wider signaling networks that fuel cancer progression. In each case, the result is dysregulated signaling that is not subject to the normal control mechanisms.
Oncogenic mutations can cause the affected genes to be overexpressed (e.g., gene amplification) or produce mutated proteins whose activity is dysregulated (e.g., point mutations, truncations, and fusions). Examples include proteins involved in signaling pathways that are commonly activated in many physiological responses, such as growth factor receptor tyrosine kinases (RTKs; e.g., the epidermal growth factor receptor, EGFR), small GTPases (e.g., Ras), serine/threonine kinases (e.g., Raf and Akt), cytoplasmic tyrosine kinases (e.g., Src and Abl), lipid kinases (e.g., phosphoinositide 3-kinases, PI3Ks), as well as nuclear receptors (e.g., the estrogen receptor, ER). Components of developmental signaling pathways, such as Wnt, Hedgehog (Hh), Hippo, and Notch can also be affected, as can downstream nuclear targets of signaling pathways—for example, transcription factors (e.g., Myc and NF-κB), chromatin remodelers (e.g., EZH2), and cell cycle effectors (e.g., cyclins).
Alternatively, deletions and other mutations can inactivate negative regulators that normally function as tumor suppressors. Indeed, one of the most commonly mutated genes in cancer is the tumor suppressor p53, the so-called “guardian of the genome.” p53 is a critical hub that controls cell proliferation and stress signals such as apoptosis and DNA damage responses (see below). pRB and CKIs such as p16 are other tumor suppressors whose mutation deregulates the cell cycle. Many tumor suppressors function as negative regulators of cytoplasmic signaling—for example, the adenomatous polyposis coli protein (APC) is a negative regulator of the Wnt pathway, and the lipid phosphatase PTEN is a negative regulator of the PI3K-Akt pathway.
It is worth noting that hyperactivated oncogene pathways can also induce a state of irreversible cell cycle arrest termed senescence (Gorgoulis and Halazonetis 2010; Vargas et al. 2012). This is believed to represent a fail-safe mechanism to inhibit proliferation caused by aberrant activation of oncoproteins in normal cells and is accompanied by changes in cellular structure, chromatin organization, DNA damage, cytokine secretion, and gene expression. Oncogenic transformation requires alterations that abrogate senescence, such as loss of p53 or PTEN.
2.1.1. The PI3K-Akt and Ras-ERK Pathways as Examples of Oncogenic Signaling Pathways
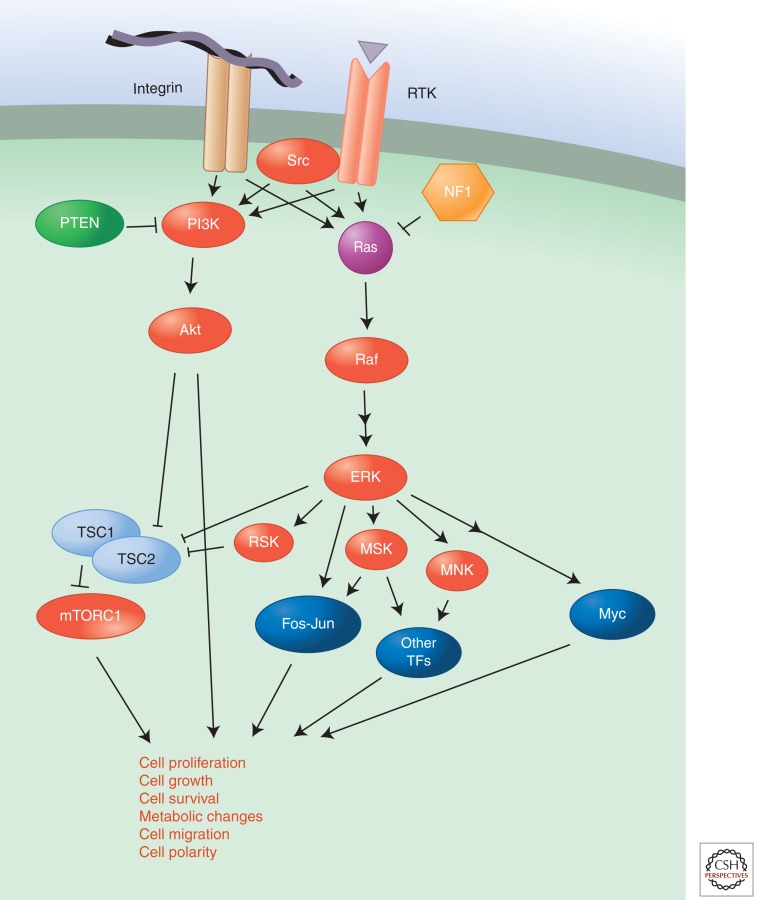
Many of the genes commonly mutated in cancer encode components or targets of the PI3K-Akt and Ras-ERK pathways (Fig. 2). Ordinarily these pathways are transiently activated in response to growth factor or cytokine signaling and ligand occupancy of integrin adhesion receptors, but genetic alterations can lead to constitutive signaling even in the absence of growth factors. The PI3K-Akt pathway can be activated through amplification or activating mutations affecting several PI3K-Akt-pathway proteins—the type I PI3K isoform PIK3CA (p110a), Akt, and the adaptor protein PIK3R1—or through deletion or inactivating mutations in the phosphatases that hydrolyze PI3K products such as phosphatidylinositol 3,4,5-trisphosphate (p1p3)—the PTEN and INPP4B tumor suppressors. Further downstream, mutations in the tumor suppressors TSC1 and TSC2 hyperactivate signaling by mTORC1 (Laplante and Sabatini 2012), an important target of PI3K-Akt signaling. Similarly, the Ras-ERK pathway is activated by mutations in Ras, or its downstream target Raf, that cause constitutive activation of these proteins or by inactivation of GTPase-activating proteins (GAPs), such as NF1 (Cichowski and Jacks 2001), DAB2IP (Min et al. 2010), and RASAL2 (McLaughlin et al. 2013), that stimulate the hydrolysis of GTP bound to Ras, which leads to its inactivation. The transcription factor Myc is an important downstream target of Ras-ERK signaling and many other pathways. It is frequently amplified or overexpressed in cancer; interestingly, Myc can not only bind to promoter regions of genes but also enhance transcriptional elongation of polymerase II, thus extending its effects beyond genes with Myc-binding sites in their promoters. Myc can thus serve as a universal amplifier of expressed genes rather than merely binding to promoters and initiating transcription de novo (Rahl et al. 2010; Lin et al. 2012; Nie et al. 2012).
Figure 2.
The Ras-ERK and PI3K pathways.
Oncogenic mutations, amplification, or gene fusions involving upstream tyrosine kinases lead to constitutive signaling through both the Ras-ERK and PI3K-Akt pathways. RTKs including EGFR, ErbB2, fibroblast growth factor receptor (FGFR), and platelet-derived growth factor receptor (PDGFR) are mutated or amplified in a variety of cancers. Similarly, oncogenic mutations in G-protein-coupled receptors (GPCRs) can also activate these pathways.
Finally, it is important to recognize that deregulated synthesis of growth factors themselves plays an important role in many cancers. Inappropriate synthesis of growth factors by cells expressing the appropriate receptor can generate an autocrine loop driving signaling. This can also be achieved through cleavage and release of anchored soluble growth factors by surface ADAM proteases, which are activated downstream from oncogenic signaling pathways (Turner et al. 2009). Alternatively, the growth factor may be synthesized by a neighboring cell (paracrine stimulation). In both cases, signaling via the Ras-ERK and PI3K-Akt pathways may be increased.
3. DYSREGULATION OF CELLULAR PROCESSES BY ONCOGENIC SIGNALING
How, then, does dysregulation of cellular signaling drive cancer progression and produce the characteristic features of tumor cells mentioned above? Below we discuss the role of signal transduction in cancer-associated processes, surveying the major signals involved and focusing on Ras-ERK and PI3K-Akt signaling to illustrate how their targets influence the behavior of the tumor cells.
4. CELL PROLIFERATION
Excessive cell proliferation is a feature of most cancers. Limited availability of growth factors or nutrients, contact inhibition, and other feedback mechanisms ensure that the pathways that regulate proliferation (see Fig. 3) are normally tightly controlled. As outlined above, however, mutations in proto-oncogenes and tumor suppressors or inappropriate synthesis of ligands/receptors can hyperactivate these pathways, leading to activation of the cell cycle machinery. Note that signaling targets that represent critical components of cell cycle control mechanisms can also undergo genetic alterations in cancer; for example, the genes encoding cyclin D, cyclin E, and CDK4 are amplified in certain cancers and the G1 restriction point inhibitor pRB and p16 can be deleted or mutated as well.
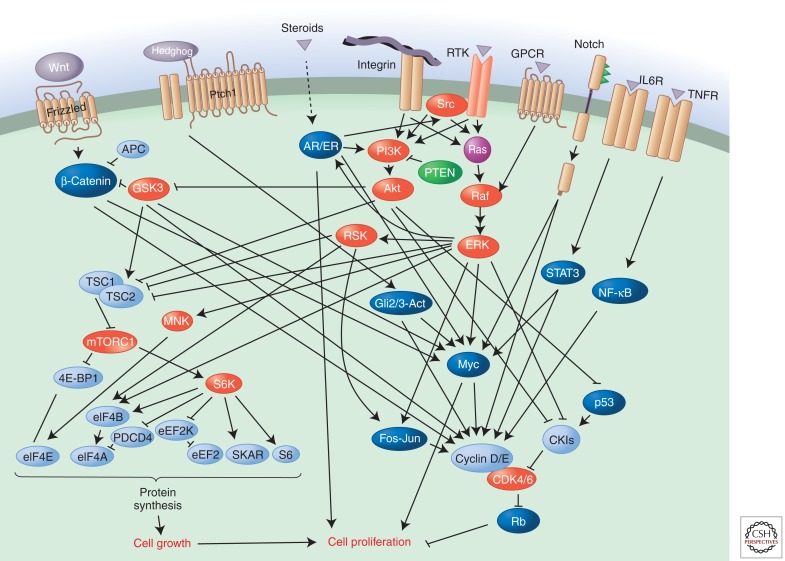
Figure 3.
Regulation of cell proliferation by the Ras-ERK and PI3K-Akt pathways.
The Ras-ERK and PI3K-Akt pathways are important regulators of normal cell proliferation and thus their constitutive hyperactivation can lead to excessive proliferation. One important target of the Ras-ERK pathway is Myc, which is phosphorylated by ERK; this leads to its stabilization by suppression of ubiquitylation (Sears et al. 2000). Myc stimulates cell proliferation by inducing numerous genes that promote cell proliferation, including those encoding G1/S cyclins, CDKs, and the E2F-family transcription factors that drive the cell cycle (Duronio and Xiong 2013). In addition, it represses expression of various cell cycle inhibitors (e.g., CKIs), blocks the activity of transcription factors that promote differentiation (see below), induces genes that enhance translation, and shifts cells to anabolic metabolism. ERK also phosphorylates numerous other transcription factors important for cell proliferation. Elk1, for example, in combination with the SRF transcription factor, induces the immediate early gene FOS, whose product is also stabilized by ERK phosphorylation (Murphy et al. 2002). FOS, also an oncogene, encodes a component of the transcription factor AP1, which regulates many genes involved in cell proliferation.
Multiple kinases in the ribosomal S6 kinase (RSK), mitogen- and stress-activated kinase (MSK), and mitogen-activated protein kinase (MAPK)-interacting kinase (MNK) families are also phosphorylated by ERK, and these kinases, in turn, phosphorylate transcription factors that regulate cell cycle progression—for example, Fos and CREB (Roux and Blenis 2004). MSKs represent the predominant kinases responsible for the nucleosomal response involving phosphorylation of histone H3 at S10, which is commonly induced by mitotic stimuli (Soloaga et al. 2003). MNKs play an important role regulating translation following mitogenic stimulation by phosphorylating the translation initiation factor eIF4E, and loss of the MNK phosphorylation site completely abrogates its ability to transform cell lines or promote tumors in animal models (Soloaga et al. 2003). Activation of RSK family members by ERK also leads to activation of the mTORC pathway through TSC2 phosphorylation and relief of mTORC inhibition. In addition, RSK regulates translation by phosphorylating eIF4B, which increases its interaction with the translation initiation factor eIF3. The promotion of translation by these mechanisms is important for cell growth and, consequently, cell proliferation.
PI3K-Akt signaling controls cell proliferation at various levels. Akt regulates cell growth during cell cycle progression by controlling mTORC1. It inhibits the GAP activity of the TSC1–TSC2 complex toward Rheb, thus allowing GTP-bound Rheb to activate mTORC1. This then phosphorylates eIF4-binding protein, releasing the eIF4E cap-binding factor and allowing it to bind mRNAs, and p70 RSK. This promotes increased protein synthesis, which is critical for enhanced cell growth during cell cycle progression (Richardson et al. 2004). Akt also phosphorylates the kinase GSK3, inhibiting its catalytic activity. Phosphorylation of cyclin D and Myc by GSK3 targets them for degradation; thus, inhibition of this kinase by Akt causes stabilization of these important cell cycle regulators (Diehl et al. 1997; Sears et al. 2000).
In addition, Akt inhibits several cell cycle inhibitors, such as the CKIs p27 (also known as KIP1) and p21 (also known as CIP1); phosphorylation leads to their sequestration in the cytoplasm by 14-3-3 proteins. In the case of p27, phosphorylation also targets it for degradation. Akt-mediated phosphorylation of p21 prevents it from forming a complex with proliferating cell nuclear antigen (PCNA) to inhibit DNA replication, reduces its binding to CDK2/CDK4, and attenuates its inhibitory activity toward CDK2 (Rossig et al. 2001). Furthermore, Akt blocks FoxO-dependent transcription of cell cycle inhibitors such as p27 and RBL2 (retinoblastoma-like protein 2) (Burgering and Medema 2003). It also phosphorylates and activates MDM2 (Ogawara et al. 2002), a ubiquitin ligase that promotes degradation of p53, thereby releasing a key brake on the cell cycle. Later on in the cell cycle, Akt can regulate several enzymes involved in the G2/M transition (Xu et al. 2012b).
Phosphorylation and consequent inhibition of GSK3 by Akt may, in certain contexts, lead to stabilization and nuclear translocation of the Wnt target β-catenin (Haq et al. 2003; Korkaya et al. 2009; Ma et al. 2013), a transcriptional regulator whose degradation would otherwise be promoted by GSK3 (Polakis 2001; Korkaya et al. 2009). This leads to induction of β-catenin target genes that regulate proliferation, including those encoding Myc and cyclin D. Akt can also phosphorylate β-catenin directly, causing its dissociation from cadherin cell–cell adhesion complexes (see below), thus increasing the pool of β-catenin available and its transcriptional activity (Fang et al. 2007).
Numerous other signaling pathways can of course drive cell proliferation in cancer. Cytokine and RTK signaling, for example, activate STAT3, which stimulates synthesis of Myc and cyclin D (Harrison 2012). Notch, Wnt/β-catenin, and Hedgehog, all of which have been implicated in cancer, also induce Myc and cyclin D (see below). Similarly, the transcription factor NF-κB, which can be activated by TNF and various other signals, also targets cyclin D expression. Cyclin E is induced by several of these signals. Estrogen signaling (see Sever and Glass 2013) stimulates cell proliferation via activation of the ERα subtype, which induces cyclin D and Myc. Disruption of the balance between ERα and ERβ or mutations in ERα that yield truncated proteins or activated proteins can dysregulate this pathway (Thomas and Gustafsson 2011; Li et al. 2013; Robinson et al. 2013; Toy et al. 2013). Note that signaling through ERs and the androgen receptor (AR) is coupled to and enhanced by Ras-ERK and PI3K-Akt signaling (Castoria et al. 2004; Renoir et al. 2013). Growth factor stimulation (e.g., EGF and insulin-like growth factor, IGF) and mutations that activate these pathways increase proliferation of ER/AR-dependent tumors. In addition, these steroid receptors form cytoplasmic complexes with Src and PI3K, which leads to activation of their downstream effectors, and ERK can phosphorylate ERα, which causes its activation in the absence of ligand and stimulation of cell proliferation.
The tumor suppressors that normally hold proliferative signaling in check are obviously also critical. Furthest downstream, pRB normally directly inhibits the transcriptional activity of the E2F proteins until it is deactivated through phosphorylation by CDKs. p53, in contrast, normally blocks cell proliferation in response to stress signals such as DNA damage by inhibiting CDK activity via induction of CKIs. Consequently, mutations in this tumor suppressor deregulate cell proliferation under potentially dangerous, cancer-promoting conditions. The CKIs themselves directly inhibit CDKs and are also inactivated by mutation in many cancers, p16 being the most common example. Further upstream are pathway-specific tumor suppressors, such as the Ras-GAP NF1 and APC, which block Wnt/β-catenin signaling (by promoting GSK3 phosphorylation and, consequently, ubiquitin-dependent destruction of β-catenin). In each case, mutation of the tumor suppressor removes an important brake, allowing cells to proliferate despite signals that would ordinarily restrain them. The Hippo pathway plays a critical role in regulating contact inhibition of proliferation (Harvey and Hariharan 2012), and disruption of this pathway, which suppresses the transcriptional coactivator YAP, is emerging as a key tumor suppressor pathway in many cancers (Harvey et al. 2013; Lin et al. 2013; Yu and Guan 2013). The Ras-ERK and PI3K-Akt pathways intersect with Hippo pathway components to inactivate its tumor suppressive activity (O’Neill and Kolch 2005; Kim et al. 2010; Collak et al. 2012).
5. CELL SURVIVAL
Cell death functions as a homeostatic mechanism that normally controls cell number. It is also a built-in cancer-protection mechanism that is activated during initial stages of oncogenesis because of stresses associated with unbalanced proliferative signals, excessive cell proliferation, loss of anchorage to natural niches, etc. Mutations that disable cell-death signaling can thus play an important role in cancer. Overexpression of the antiapoptotic protein Bcl2, for example, can occur as a consequence of chromosomal rearrangements in B lymphocytes, and this contributes to follicular lymphoma by preventing cells from undergoing apoptosis. p53 also regulates apoptosis, both by inducing transcription of proapoptotic regulators and binding directly to the proapoptotic protein Bax (Green and Llambi 2014). Loss of this tumor suppressor through mutation can therefore contribute to cancer by reducing cell death, as well as disabling normal cell cycle control. Other cell death regulators that are mutated in cancer include the proapo-ptotic proteins Puma and Bok (which are frequently deleted) and the antiapoptotic proteins Mcl1 and Bcl-xL (whose genes are amplified).
Control of proapoptotic regulators (e.g., Bim and Bad) and antiapoptotic regulators (e.g., Bcl2 and Mcl1) in normal cells ensures that cells undergo apoptosis in the absence of appropriate signals supplied by growth factors or the tissue microenvironment. Hyperactivation of signaling by oncogenic mutations in the Ras-ERK and PI3K-Akt pathways, however, disrupts the balance in favor of antiapoptotic signals, thus contributing to tumor cell survival and abnormal expansion of the cells beyond normal tissue boundaries.
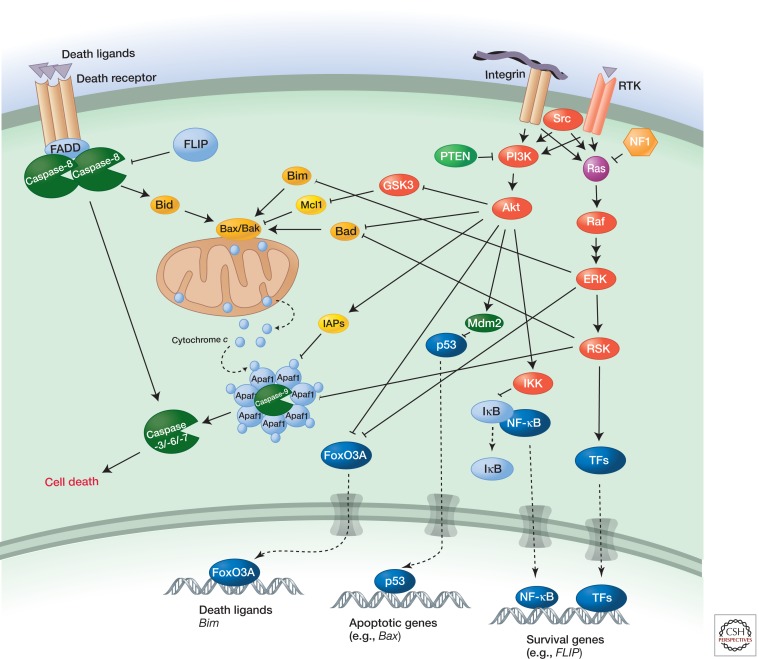
The PI3K-Akt and Ras-ERK pathways regulate cell death in multiple ways (Fig. 4) (review Cagnol and Chambard 2010; Zhang et al. 2011). Akt itself intervenes at several steps in apoptotic signaling from death receptors. It phosphorylates forkhead-family transcription factors such as FoxO3A, which leads to their cytoplasmic sequestration by 14-3-3 proteins, thereby blocking induction of death ligands (e.g., FasL and TRAIL) and the proapoptotic Bcl2-family member Bim. Akt and the ERK-regulated kinase RSK also phosphorylate the proapoptotic Bcl2-family protein Bad, another target for sequestration by 14-3-3 proteins. In addition, Akt phosphorylates and thereby activates the apoptosis inhibitor XIAP. Akt also activates NF-κB, which regulates multiple survival factors, including antiapoptotic proteins (Bcl2, BCLxl, and Mcl1) and the intracellular death receptor inhibitor FLIP (Shen and Tergaonkar 2009). Last, Akt-induced ubiquitylation and degradation of p53 suppresses p53-induced apoptosis (Ogawara et al. 2002).
Figure 4.
Regulation of cell death by Ras-ERK and PI3K-Akt pathways.
ERK phosphorylates Bim and the NF-κB inhibitor IκBα (Ghoda et al. 1997), which targets them for degradation. In addition, RSK phosphorylates the caspase-9 scaffolding protein APAF, which impedes the ability of cytochrome c to nucleate apoptosome formation and activate the downstream caspases that drive apoptosis (Kim et al. 2012).
6. CELL METABOLISM
Cell growth needs to be coordinated with metabolic processes involved in the synthesis of macromolecules. Thus, growth factor pathways that regulate both normal and tumor cells impinge on metabolic pathways to program cells to meet the increased need for synthesis of macromolecules to produce new daughter cells (Ward and Thompson 2012b). Activation of oncogenes and loss of tumor suppressors can directly regulate components of metabolic pathways even in the absence of growth factors and, thereby, produce similar metabolic alterations (Fig. 5).
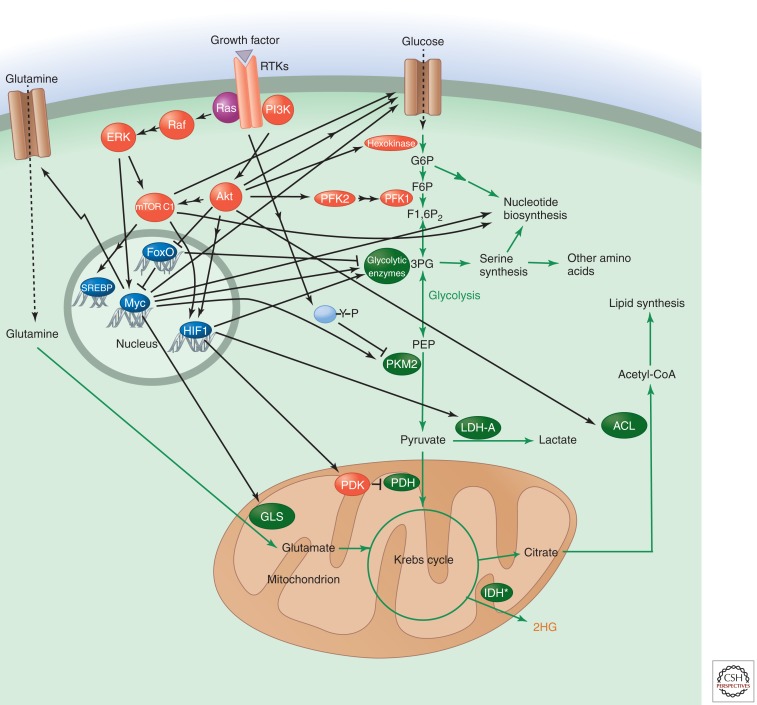
Figure 5.
Regulation of metabolism by Ras-ERK and PI3K-Akt signaling. IDH*, mutated IDH.
The most common metabolic alteration in cancer cells is increased glucose uptake and glycolysis. At first glance, this might appear a disadvantage because glycolysis generates less ATP than oxidative phosphorylation; however, it allows cells to redirect carbon skeletons from glycolysis to anabolic reactions, such as the pentose phosphate pathway, which leads to nucleotide synthesis and regulates redox homeostasis. These also include the serine/glycine synthesis pathway, which generates several amino acids and charges tetrahydrofolate with a methyl group that is used in pyrimidine synthesis and leads to generation of S-adenosylmethionine, the methyl donor for multiple cellular methyltransferase reactions and methylation of essential molecules such as DNA, RNA, proteins, phospholipids, creatine, and neurotransmitters. Cancer cells show increased glutamine uptake and glutaminolysis to support oxidative phosphorylation and biosynthesis of proteins, lipids, and nucleic acids. They also up-regulate lipid synthesis by redirecting citrate from the Krebs cycle to fatty acid synthesis.
The PI3K-Akt pathway targets numerous substrates to promote these metabolic changes (Plas and Thompson 2005). Regulation of glucose transport and hexokinase by Akt promotes glycolysis, leading to generation of nucleotides and amino acids necessary for cell growth (Engelman et al. 2006). Akt2 regulates glucose transport through multiple mechanisms. Regulation of the glucose transporter GLUT4 by Akt2 is critical for circulating glucose homeostasis. The Akt substrate AS160 plays an undefined role in insulin-stimulated GLUT4 translocation and glucose transport through its Rab-GTPase-activating domain (Miinea et al. 2005), and phosphorylation of the protein synip by Akt2 triggers its dissociation from the trafficking regulator syntaxin 4 and assembly of a protein complex that mediates translocation of GLUT4 vesicles to the plasma membrane (Yamada et al. 2005). Akt2 also regulates transcription, accumulation (Barthel et al. 1999; Jensen et al. 2010), and trafficking of GLUT1, which is the principle glucose transporter expressed in most cell types (Wieman et al. 2007). Phosphorylation of TSC2 by Akt affects metabolism through mTORC1-mediated regulation of glycolysis; however, the mechanism of regulation is not known. mTORC1 may regulate glycolysis by increasing translation of glycolytic enzymes or their transcriptional regulators, such as Myc (Kim et al. 2004; Sutrias-Grau and Arnosti 2004). Other Akt targets activated by phosphorylation are hexokinase II, whose association with mitochondria is increased (Roberts et al. 2013), and 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase (Novellasdemunt et al. 2013). Both stimulate glycolysis.
mTORC1 signaling leads to increased synthesis of the transcription factor hypoxia-inducible factor (HIF1). HIF1 induces glycolytic enzymes and lactate dehydrogenase (LDH-A), providing another means of stimulating glycolysis. In addition, it induces pyruvate dehydrogenase kinase (PDK), which inhibits pyruvate dehydrogenase (PDH) in the mitochondrion and thereby reduces flux from glycolysis into the Krebs cycle. mTORC1 can also stimulate pyrimidine biosynthesis via S6K1 (Ben-Sahra et al. 2013; Nakashima et al. 2013).
Akt/mTORC1 promotes lipid synthesis by activating the transcription factor sterol-response-element-binding protein 1 (SREBP), a key regulator of lipid synthesis that is required for tumorigenicity (Bakan and Laplante 2012; Jeon and Osborne 2012; Guo et al. 2013). Loss of SREBP uncouples fatty acid synthase activity from stearoyl-CoA-desaturase-1-mediated desaturation. Another direct target of Akt is ATP-citrate lyase (ACL), an enzyme that converts citric acid to acetyl-CoA, which is required for fatty acid, cholesterol, and isoprenoid synthesis. mTORC1 also regulates amino acid uptake by stimulating translocation of amino acid transporters from intracellular vesicles to the plasma membrane (Berwick et al. 2002; Edinger and Thompson 2002).
Another family of Akt targets that affect cellular and organismal metabolism is FoxO transcription factors. These are negatively regulated by Akt phosphorylation, which causes their sequestration in the cytoplasm by 14-3-3 proteins. Programs regulated by FoxO transcription factors that increase the cellular capacity for oxidative metabolism are thus shut off by active Akt.
Ras-ERK signaling exerts many of its effects on metabolism via Myc. Myc regulates glucose uptake, glycolysis, and the pentose phosphate pathway (Ying et al. 2012) and induces synthesis of glutamine transporters and the enzyme glutaminase (GLS), which converts glutamine into glutamate that can be metabolized in mitochondria (Miller et al. 2012; Dang 2013). It also induces enzymes involved in nucleotide and amino acid synthesis.
The glycolytic enzyme pyruvate kinase is of particular interest in cancer cells. Although glycolysis rates are usually much higher than in noncancer cells, most cancer cells produce an alternative splice form of pyruvate kinase (PKM2) that is less active than the enzyme (PKM1) found in most terminally differentiated cells (Vander Heiden et al. 2009). PKM1 remains active under most physiological conditions, but PKM2 can be turned off by signaling via tyrosine kinases, including the upstream RTKs in the Ras-ERK and PI3K-Akt pathways (Christofk et al. 2008; Hitosugi et al. 2009) and reactive oxygen species (ROS) (Anastasiou et al. 2011). Cancer cells can thus redirect the flux of glycolytic intermediates into anabolic pathways for ribose, serine, and glycine production or production of NADPH and glutathione needed to combat oxidative stress. PKM2 can also enter the nucleus and play a role in gene expression (Luo et al. 2011; Gao et al. 2012). However, deletion of PKM2 accelerates rather than impairs breast tumor formation, which indicates that it is the ability to turn off PKM2 activity that is most critical for tumor growth (Israelsen et al. 2013).
Clearly oncogene activation or loss of tumor suppressors such as PTEN and NF1 can drive these metabolic changes by dysregulating PI3K-Akt and Ras-ERK signaling. Other tumor suppressors also control cell metabolism, however. p53, for example, down-regulates glycolysis by inducing TIGAR, an enzyme that decreases the levels of the glycolytic activator fructose 2,6-bisphosphate. It also stimulates expression of SCO2, which is required for assembly of cytochrome c oxidase and promotes oxidative phosphorylation. Loss of p53 may therefore contribute to the glycolytic phenotype of cancer cells. p53 also regulates glutaminase 2, a metabolic enzyme that controls production of glutamate, which is converted to α-ketoglutarate for mitochondrial respiration and, importantly, glutathione, a critical cellular antioxidant (Hu et al. 2010; Suzuki et al. 2010). Loss of p53 leads to increased levels of ROS and oxidative damage. p53 also regulates the mevalonate pathway that controls cholesterol synthesis and generates intermediates needed for protein geranylgeranylation and farnesylation. Drugs that target this pathway to control cholesterol/cardiovascular disease have been shown to suppress tumor growth (Shibata et al. 2004; Kubatka et al. 2011).
Similarly, loss of the tumor suppressor LKB1 can lead to metabolic alterations. LKB1 activates AMP-activated protein kinase (AMPK), which acts as a cellular energy regulator and inhibits mTORC1 (Hardie 2012). Loss of LKB1 relieves this inhibition, allowing mTORC1 to promote protein synthesis and lipogenesis. Activators of AMPK, such as metformin, are currently being used in diabetes and cancer therapy.
Finally, mutations associated with cancer can lead to the elevation of metabolites uniquely elevated in cancer cells (Kaelin and McKnight 2013). For example, mutations in IDH1 and IDH2 result in the production of 2-hydroxyglutarate (2HG), a metabolite not present at significant levels in normal cells. 2HG inhibits α-ketoglutarate-dependent enzymes such as the TET family, which regulate DNA methylation, and Jumonji C domain histone demethylases. This leads to epigenetic dysregulation that can drive tumorigenesis. Other oncometabolites may include succinate and fumarate, whose levels can increase because of mutations in succinate dehydrogenase and fumarate hydratase. Both can inhibit the activity of prolyl hydroxylases that control HIF levels, leading to induction of PDK and the other glycolytic enzymes mentioned above.
7. CELL POLARITY AND MIGRATION
As tumors progress toward malignancy, the cancer cells frequently become more migratory and develop the capacity to invade surrounding tissue. This is usually accompanied by changes in adhesion, cell polarity, cytoskeletal dynamics, and morphology. Migration is regulated by growth factors, chemokines, adhesion receptors, and other stimuli (Vicente-Manzanares and Horwitz 2011; Devreotes and Horwitz 2014), many of which are targets for dysregulated signaling in cancer. The PI3K-Akt and Ras-ERK pathways regulate migration and invasion through multiple downstream effectors, including the following (Cain and Ridley 2009):
Rho-family GTPases (RhoA, Rac1, Cdc42, ARF6), which control cytoskeletal regulators such as WAVE/WASP-family members, the Arp2/3 complex, formins, the actomyosin contractile machinery, the kinase LIMK, and cofilins (Raftopoulou and Hall 2004);
Integrins and associated matrix adhesion proteins (e.g., FAK, paxillin, and calpains) (Devreotes and Horwitz 2014);
Extracellular proteases, which degrade ECM proteins, facilitating tumor cell invasion by creating space for cells to move and reducing adhesive contacts that may constrain them, and also release various bioactive molecules anchored in the ECM (see below);
Cell–cell adhesion complexes, whose components are regulated through modulation of their stability or protein interactions that affects the strength of adhesion;
Transcription factors such as AP1 and Ets2 that regulate expression of many proteins that control migration/polarity, including matrix metalloproteinases (MMPs), plasminogen activator, cadherins, and actin regulators.
As with other processes regulated by oncoprotein signaling, the outcome of alterations in these pathways is highly context and isoform dependent. For example, Akt1 specifically suppresses migration in many contexts through inhibition of ERK, the transcription factor NFAT, TSC2, or phosphopalladin-induced actin bundling, whereas Akt2 promotes migration through regulation of integrin expression and effects on the epithelial–mesenchymal transition (EMT) (see below; Chin and Toker 2011). Similarly, some isoforms of ERK target RSK to promote cell motility and invasion by altering transcription and integrin activity, whereas others impair cell motility and invasion through effects on the actin cytoskeleton (Sulzmaier and Ramos 2013).
Polarity proteins are critical regulators of tissue architecture. Three protein complexes play central roles in controlling polarity: Scribble, Par, and Crumbs complexes. Through multiple interactions, components of these pathways control signaling pathways that regulate cell polarity and tissue organization. Dysregulation of these pathways is common in tumors and, in some contexts, involves alterations in Ras-ERK and PI3K-Akt signaling. For example, Scribble inhibits ERK activation by functioning as a scaffold to link it with the protein phosphatase PP1γ (Dow et al. 2008; Nagasaka et al. 2013). Loss of Scribble enhances invasion stimulated by H-Ras (Shaikh et al. 1996) and tumor formation promoted by Ras and Myc (Wu et al. 2010). Similarly, loss of the polarity protein Par3 leads to increased invasion in several tumor models (Iden et al. 2012; McCaffrey et al. 2012; Xue et al. 2013) through multiple pathways, including PKC-dependent activation of JAK/STAT3 signaling. This induces expression of a metalloproteinase, MMP9, with subsequent destruction of the ECM and invasion (McCaffrey et al. 2012), and increased Rac activation, leading to decreased cell–cell adhesion (Xue et al. 2013).
Loss of cell polarity is often coupled to cell proliferation because the loss of cell adhesion molecules relieves contact inhibition. One example is the cytoskeletal protein merlin (also known as neurofibromin 2), a tumor suppressor that regulates the Hippo pathway and whose loss is well known to cause increased cell proliferation. Polarity signaling is also coupled to metabolism.
Some subpopulations of epithelial cells in tumors, particularly those at tumor margins, undergo at least a partial EMT. EMTs are associated with various normal physiological processes—for example, wound healing, gastrulation, and branching morphogenesis (Birchmeier and Birchmeier 1995). This developmental process is orchestrated by multiple highly coordinated pathways induced by combinations of different factors, including transforming growth factor β (TGFβ), TNF, Wnt, Notch, and some growth factors. EMT is characterized by a loss of apical-basal polarity, down-regulation of E-cadherin cell–cell adhesion molecules, adoption of a more fibroblast-like appearance, and, in some contexts, acquisition of stem- or progenitor-cell phenotypes and anchorage independence, properties that would enhance the cell’s ability to invade other tissues and initiate tumors at distant sites.
The Ras-ERK and PI3K-Akt pathways drive the EMT in certain contexts, generally under conditions in which these pathways are hyperactivated together with other pathways implicated in EMT (e.g., TGFβ, Wnt, and Notch signaling) (Larue and Bellacosa 2005). Multiple transcription factors, such as Snail, Slug, Twist, and ZEB, play critical roles driving EMT, and these are regulated by ERK and Akt. For example, Akt can phosphorylate the IκB kinases that regulate NF-κB, a transcription factor that induces Snail. Akt also phosphorylates and inactivates GSK3, which normally promotes ubiquitin-dependent degradation of Snail (Doble and Woodgett 2007); Akt activation will therefore increase Snail stability, further promoting EMT. In addition, Akt2 phosphorylates HNRNP E1, a protein that promotes translational elongation on EMT-promoting transcripts such as those encoding interleukin-like EMT inducer and the adaptor protein DAB2 (Hussey et al. 2011). AP1, which is regulated by the Ras-ERK pathway, can also induce transcription factors that promote EMT as well as other gene expression programs that control phenotypic changes associated with EMT. These include up-regulation of specific integrin heterodimers (e.g., α5β1 and αVβ6), vimentin, and fibronectin and down-regulation of cytokeratin, polarity proteins (e.g., Crumbs, PATJ, LGL), and E-cadherin, all of which support cell motility. Interestingly, the polarity protein Scribble maintains cell–cell junctions by suppressing ERK (which stimulates ZEB1) as described above (Elsum et al. 2013; Nagasaka et al. 2013). Dysregulation of both Ras-ERK and PI3K-Akt signaling thus has the potential to play an important role in cancer progression by promoting adoption of an invasive phenotype.
Finally, it is important to note that EMT is not essential for invasion and tumor cell dissemination. Tumor cells can migrate as epithelial sheets within tissues (as occurs during wound healing) or invade by pushing through tissue borders (e.g., basement membrane).
8. CELL FATE AND DIFFERENTIATION
Dysregulation or co-option of developmental signaling pathways is a feature of many cancers. This can disrupt the balance between cell proliferation and differentiation, alter cell fate, and/or inappropriately induce morphogenetic programs such as the EMT (see above) that promote metastasis. Although some oncogenes can directly regulate the developmental state of cells, it is generally believed that cancer progression requires a self-renewing population of “stem-cell-like” cells. These may be induced into a stem-cell-like state by an oncogene(s), or a normal stem/progenitor cell may be the cell-of-origin that sustains the successive mutations that lead to malignancy.
The simplest examples of cancers with dysregulated development are perhaps hematopoietic malignancies in which a differentiation program is stalled before the cells reach their nonproliferative differentiated state. For example, in acute promyelocytic leukemia, a form of acute myelocytic leukemia, myeloblasts fail to differentiate into mature white blood cells because of a translocation that leads to synthesis of a fusion protein combining sequences from a protein called PML and the retinoic acid receptor (RAR). The PML-RAR fusion protein represses RAR-target genes that normally drive differentiation, thereby inactivating the RAR signaling that normally controls this. Subsequently, additional mutations cause overproliferation of the undifferentiated myeloblasts. Inappropriate Wnt signaling has a similar effect in colon cancer. Ordinarily, Wnt signaling via β-catenin (see Nusse 2012) maintains enterocytes in an undifferentiated state in colon crypts but is inactivated by APC-induced degradation of β-catenin as cells move up toward the luminal surface of the intestine. Mutation of the APC tumor suppressor in colon cancer, however, means β-catenin is not destroyed and can maintain cells in an undifferentiated state as they move away. Further mutations can then drive neoplasia.
Developmental signals can also drive cancer progression because they stimulate inappropriate cell proliferation (see above). Mutations that activate Notch, for example, contribute to acute lymphocytic leukemia because Notch signaling (Kopan 2012) can stimulate the cell cycle and also inhibits apoptosis in T cells. Importantly, Notch functions as a tumor suppressor in some other tissues. In others, the concentration of Notch dictates its growth suppressive or stimulatory effects (Mazzone et al. 2010), which illustrates the importance of the signaling context. Activation of the Hedgehog signaling pathway (see Ingham 2012) by mutations in the Patched receptor occurs in basal cell carcinomas and medulloblastomas and again drives cell proliferation. Hedgehog signaling is also hyperactivated via autocrine loops in many tumors that affect tissues derived from the embryonic gut.
Given that the Ras-ERK and PI3K-Akt signaling pathways are activated by growth factors such as EGF, IGF, and fibroblast growth factor (FGF), which play major roles in control of cell fate, they can thus be considered developmental signaling pathways that are hijacked in cancer. Signaling by FGF4/8, for example, activates the Ras-ERK pathway to drive EMT during gastrulation and the Ras-ERK pathway is recapitulated in several cancers (Thiery 2002). The context is important, however; signaling by FGF has the potential to affect cell proliferation, apoptosis, and migration (see above), as well as angiogenesis (see below), but it can also have tumor suppressive effects, maintaining cells in a differentiated, nonproliferative state. For example, whereas FGFR2 is up-regulated in gastric cancers, its expression is reduced in bladder and prostate cancer (Turner and Grose 2010).
9. GENOMIC INSTABILITY
Genomic instability is a common characteristic of cancer cells. Aneuploidy and large-scale DNA rearrangements are frequently observed, and many cancers display elevated mutation rates. Ordinarily, a variety of cellular enzymes repair DNA damage, and checkpoint signaling ensures that DNA replication and cytokinesis are arrested in dividing cells until potentially damaging errors are corrected. Alternatively, checkpoint signaling can induce senescence or apoptosis so that affected cells do not pass on these errors. Whether genomic instability is a cause or a consequence of cancer is still debated, but it clearly reflects a failure of checkpoint signaling and/or DNA repair mechanisms.
DNA damage signals are relayed by the kinases ATM, ATR, Chk1, and Chk2, which stimulate p53, stall the cell cycle, and activate the DNA repair machinery (Rhind and Russell 2012). Downstream of p53, the CKI p21 is induced, and this can halt DNA polymerase if DNA replication has already begun. If the damage cannot be repaired and checkpoint signaling persists, p21 and p53 will induce cells to senesce or undergo apoptosis (see above). Clearly, mutation or epigenetic silencing of these tumor suppressors or upstream kinases can inactivate checkpoint signaling, allowing DNA damage to persist and potentially fuel cancer progression. Indeed, ATM and Chk2 mutations are seen in familial leukemias and colon/breast cancers, respectively, and proteins involved in DNA repair itself are also often mutated—for example, MMR enzymes and BRCA1/2.
The mitotic checkpoint (also known as the spindle assembly checkpoint) ensures that when a cell divides each daughter receives a full complement of chromosomes. A complex containing the proteins Bub1, Bub3, and Mad1-3 monitors attachment of chromosomes to the mitotic spindle, relaying checkpoint signals that block chromosome segregation and subsequent cytokinesis. Once paired, sister chromatids are all attached to microtubules emanating from opposite poles, the signal is switched off, and cells can move from metaphase into anaphase and, ultimately, cytokinesis can proceed (Rhind and Russell 2012). Inactivation of this checkpoint pathway has the potential to lead to aneuploidy, and mutations in Mad1/2 and Bub1 have been observed in cancer (Schvartzman et al. 2010).
Akt has been implicated in multiple aspects of DNA damage responses and genome instability (Xu et al. 2012a). It can inhibit homologous recombinational repair through direct phosphorylation of the checkpoint proteins Chk1 and TopBP1 or indirectly through recruitment of resection factors such as RPA, BRCA1, and Rad51 to sites of double-stranded breaks (DSBs) in DNA. Akt is also activated by DSBs in a DNA-dependent protein-kinase- or ATM/ATR-dependent manner and, in some contexts, can contribute to radioresistance by stimulating DNA repair by nonhomologous end joining. In addition, Akt also inhibits association of BRCA1 with DNA damage foci. As discussed above, dysregulation of the PI3K-Akt pathway suppresses apoptosis through many effectors, thus promoting survival of cells with DNA damage. Because Ras-ERK signaling also inhibits apoptosis, it too could promote survival of damaged cells. Hyperactivation of Ras-ERK signaling has been shown to lead to genomic instability, although the molecular mechanism is unclear (Saavedra et al. 1999). Akt therefore modifies both the response to and repair of genotoxic damage in complex ways that are likely to have important consequences for the therapy of tumors showing deregulation of the PI3K-Akt pathway.
The tumor suppressor PTEN can also regulate chromosome stability, independently of its 3′-phosphatase activity. PTEN regulates the expression of the DNA repair protein RAD51, and loss of PTEN causes extensive centromere breakage and chromosomal translocations (Toda et al. 1993; Liu et al. 2008; Kopan 2012).
Myc overexpression can induce genomic instability. In mammalian cells and Drosophila, overexpression of Myc increases the frequency of chromosomal rearrangements (Prochownik and Li 2007; Greer et al. 2013). Multiple mechanisms have been associated with such genomic rearrangements, including ROS-induced DSBs, suppression of checkpoints that prevent replication of damaged DNA, and telomere clustering.
10. THE TUMOR MICROENVIRONMENT
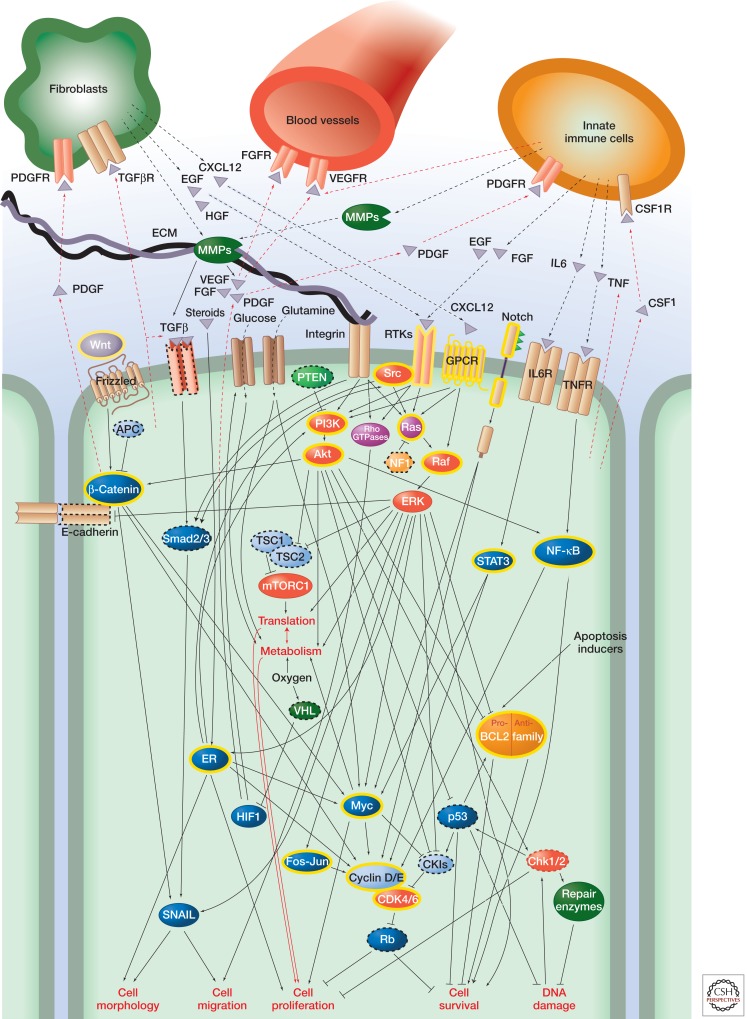
So far, we have primarily considered how signaling within cancer cells themselves is dysregulated in cancer. However, cancer progression (at least in solid tumors) also depends on the ECM, blood vessels, immune cells, and noncancerous cells such as fibroblasts in the tumor microenvironment, all of which communicate with cancer cells by subverted signaling mechanisms (Fig. 6). Many of the changes in the tumor microenvironment during cancer progression mimic changes that occur during wound healing and/or developmental processes. As tumors evolve, the complexity of their “ecosystem” increases; reciprocal paracrine and juxtacrine interactions between populations of neoplastic cells as well as tumor cells and nonneoplastic cells within the microenvironment control cellular signaling pathways in both positive and negative fashions. Dissecting the roles of individual signaling pathways in these ecosystems is complex because it is difficult to distinguish cell-autonomous and non-cell-autonomous activities.
Figure 6.
Cancer signaling networks. The figure illustrates the wide variety of intra- and intercellular signals affected in cancer, focusing on Ras-ERK and PI3K-Akt signaling. It is by no means comprehensive; many more pathways are involved and there are other stromal cells involved in paracrine signaling. Oncoproteins are indicated with yellow highlighting; tumor suppressors are indicated with dashed outlines. Arrows do not necessarily indicate direct interactions in this figure.
10.1. The ECM
The ECM is a scaffold that physically supports tissues and provides a substrate for cell adhesion and migration, as well as a source of bioactive molecules. Far from a static structure, it is constantly being remodeled, and its composition plays a critical role in control of cell behavior. Fibronectin, laminin, collagen, and various other ECM components serve as ligands that activate integrin signaling. Integrin signaling leads to activation of canonical pathways such as Ras-ERK, PI3K-Akt, and Src signaling, as well as other proteins—for example, the tyrosine kinase FAK, a scaffold that links integrins with cytoskeletal proteins, adaptors, and enzymes that transduce signals from matrix adhesion complexes. FAK also regulates p53 and members of the miR-200 family of microRNAs, which control apoptosis and epithelial phenotype (Keely 2011).
Heparin sulfate proteoglycans (HSPGs) in the ECM modulate signaling by associating with various ligands and acting as coreceptors (e.g., for FGF and FGFR). In addition, the ECM actively sequesters a variety of growth factors, including TGFβ, vascular endothelial growth factor (VEGF), and platelet-derived growth factor (PDGF), which can be liberated and/or activated by MMPs. Collagens can also be digested and remodeled by proteinases to enhance tumor cell motility.
The ECM changes as cancer progresses (Lu et al. 2011). It stiffens as large quantities of ECM are deposited by cancer-associated fibroblasts (see below) and collagen fibers become more cross-linked by lysyl oxidases secreted by stromal cells. This increases contractility of the cells, which further fuels stiffening. The changes in stiffness of the ECM promote cell migration and integrin signaling through regulation of Rho family GTPases and other pathways, which synergize with oncogene-activated Ras-ERK and PI3K-Akt pathways to promote invasive growth and cell survival (Keely 2011).
Other changes to the ECM include increased levels of molecules such as tenascin C, a proteoglycan common around developing blood vessels that is induced during inflammation and promotes angiogenesis. MMPs are also up-regulated. These stimulate signaling in various ways and, by degrading the ECM, clear a path for cell migration. Indeed, genes encoding endogenous inhibitors of MMPs such as TIMP3 are known to be targets for hypermethylation in some cancers. HSPGs are also overproduced in cancer and may potentiate oncogenic signaling by FGF, Wnt, and Hedgehog.
10.2. Angiogenesis
Like all tissues, tumors require a blood supply. They acquire this by inducing proliferation and assembly of endothelial cells to form new blood vessels (angiogenesis), co-opting pathways that usually function in wound healing. Central to angiogenesis are signals such as VEGF, PDGF, FGFs, interleukin (IL) 8, and angiopoietin. The PI3K-Akt pathway regulates the induction of angiogenesis as well as vessel integrity (Karar and Maity 2011). Synthesis and secretion of VEGF by cancer cells is induced by HIF1. As mentioned above, HIF1 levels are increased by PI3K-Akt signaling, and hyperactivation of this pathway thus plays an important role in angiogenesis. HIF1 activity can be controlled by the von Hippel–Lindau (VHL) protein, a subunit of an E3 ligase that promotes its ubiquitin-dependent degradation under normoxic conditions when it is proline hydroxylated by proline hydroxylases (see Ward and Thompson 2012b), or through translational control. VHL functions as a tumor suppressor and inactivating VHL mutations occur in a variety of cancers. The PI3K-Akt pathway also modulates the production of other angiogenic factors, such as nitric oxide and angiopoietins. Constitutive endothelial activation of Akt1 has been shown to induce the formation of structurally abnormal blood vessels.
Following its secretion, VEGF is sequestered in the ECM and cannot exert its effects on endothelial cells until it is released by MMPs such as MMP9. These are produced by monocytes and macrophages in the tumor microenvironment (see below), which underscores the importance of immune cells in angiogenesis and existence of the wider signaling network that involves cancer cells, immune cells, and endothelial cells.
Another factor that must be overcome for angiogenesis to occur is inhibitory signals such as thrombospondin 1 (Tsp1). Tsp1 released by various cells normally keeps angiogenesis in check by inducing synthesis of FasL, which causes endothelial cells to undergo apoptosis (see Green and Llambi 2014). Tsp1 is induced by p53, but repressed by Ras, Src, and Myc. It thus represents another control point in angiogenesis that could be activated by dysregulation of the Ras-ERK pathway. Moreover, the gene that encodes Tsp1 is hypermethylated in some cancers.
10.3. Inflammation
Inflammatory cells such as macrophages and neutrophils constitute the first defense against pathogens, but are also involved in tissue remodeling and repair. They are recruited by chemokines secreted by tumor and stromal cells to almost all tumors and secrete various molecules that promote cancer cell proliferation, survival, and migration. In many respects, the contribution of inflammatory cells to tumor progression, like that of angiogenesis, recapitulates their role in wound healing, which also involves these processes.
Signaling via the transcription factor NF-κB (see Newton and Dixit 2012) is important in both cancer cells and tumor-associated inflammatory cells because it can promote cell survival and proliferation and stimulates production of cytokines such as TNF. Oncogenic mutations affecting NF-κB or upstream regulators such as MALT1 and Bcl10 occur in some lymphoid malignancies; however, in most cancers, NF-κB activity is simply increased by cytokine signaling. For example, in colon cancer, TNF produced by macrophages increases NF-κB activity in intestinal epithelial cells, which promotes cell survival; meanwhile, other cytokines such as IL6 and IL11 increase phospho-STAT3 levels (see Harrison 2012), which promote cell proliferation. A similar phenomenon occurs in hepatocytes in hepatocellular carcinoma, the most common form of liver cancer, and prostate cancer (Karin 2009). NF-κB activation also leads to production of more TNF and synthesis of prostaglandin E2, which further fuels cell proliferation and loss of cell polarity.
In addition to cytokines, inflammatory cells secrete growth factors such as EGF and FGF. These are obviously important regulators of Ras-ERK and PI3K-Akt signaling in cancer cells and will, therefore, dysregulate control of cell proliferation, cell death, metabolism, and cell migration, as discussed above. They also lead to production of colony-stimulating factor 1 (CSF1), a key reciprocal signal that stimulates macrophages, causing them to produce more EGF. Immune cells also produce VEGF and MMPs, which promotes angiogenesis, ECM remodeling, and release of other bioactive molecules (see above). Importantly, all these factors participate in paracrine loops involving various immune cells and cancer cells that sustain chronic inflammation and promote tumor growth and progression.
Note that cells of the adaptive immune system can also be involved, producing signals such as IL17 that stimulate both cancer cells and cells of the innate immune system. Resident microbiota and pathogenic bacteria in the tumor microenvironment can also release products that promote inflammation and immune cell infiltration (Grivennikov et al. 2012).
10.4. Cancer-Associated Fibroblasts
Fibroblasts are present in most tissues, helping to shape organs and control the composition of the ECM. They are activated by tissue injury and, in cancer, produce various factors that stimulate proliferation and migration of cancer cells, along with angiogenesis and inflammation. Release of signals such as TGFβ and PDGF by macrophages and cancer cells activates fibroblasts, which, in turn, can release EGF, HGF, IGF, and chemokines such as CXCR12 (also known as stromal-cell-derived factor 1). These can then further activate Ras-ERK and PI3K-Akt signaling and other pathways in the cancer cells and drive feedback loops that amplify this. Cancer-associated fibroblasts are also responsible for the distinct ECM associated with advanced carcinomas, which also affects signaling within the tumor.
The interplay between cancer cells and these different components of the tumor microenvironment is thus incredibly complex and mimics signaling that tissues display both during development and in normal tissue homeostasis and repair. As cancers metastasize, this extends to other organs. Different cancers are known to seed secondary tumors in particular tissues. Successful colonization depends on the cell surface receptors expressed by the cancer cells and target tissue and the suitability of the microenvironment the latter provides.
11. CONCLUDING REMARKS
Cancer cells show a number of defining characteristics. Underlying these is a dysregulation of cellular signal transduction induced by the genetic and epigenetic changes that drive cancer. This affects not only the cancer cells themselves, but the wider signaling network that encompasses other cells, the ECM, blood vessels, and the immune system. Indeed, metastatic cancer can be considered a systemic disease that affects signaling throughout the affected individual, and systemic effects are ultimately what kill patients in cancer.
Pharmacologic and antibody-based inhibitors that target signaling proteins mutated in tumors or proteins downstream from these have had significant impact as cancer treatments. For example, inhibitors of the nonreceptor tyrosine kinase Abl and RTK ErbB2 dramatically reduce patient mortality in chronic myelogenous leukemia and breast cancer. Other inhibitors, such as those that target B-Raf, EGFR, and the kinase ALK induce remarkable reductions of tumor volume and extend survival in patients with melanoma and nonsmall-cell lung carcinomas; however, the rate of recurrence is high because of the development of drug resistance (Gainor and Shaw 2013; Giroux 2013; Holohan et al. 2013; Lito et al. 2013).
The complexity of the cancer signaling network (see Fig. 6) presents a huge challenge for efforts to develop such anticancer drugs because of the redundancy of pathways that control cell proliferation and survival, crosstalk between pathways, and feedback inhibition mechanisms that cause pathway reactivation. The fact that pathways such as Ras-ERK and Akt-PI3K signaling control so many characteristics of cancer cells, and that components of these pathways, or upstream receptors, are so commonly mutated in a variety of cancers gives reason to be optimistic that approaches based on targeting them will be successful. The efficacy of therapies that target these pathways is, however, limited by multiple factors. For example, rewiring of signaling pathways is associated with adaptive responses to inhibition of driver mutations, and this is commonly because of either loss of feedback inhibition or induction of stress pathways (Pratilas and Solit, 2010; Rodrik-Outmezguine et al. 2011; Lito et al. 2013). Moreover, factors from the tumor microenvironment may stimulate alternative pathways that maintain cell viability despite inhibition of the targeted pathways (Castells et al. 2012; Muranen et al. 2012). Alternatively, there can be selection for rare tumor cells that contain drug-resistant variants of the targeted protein or mutations in other pathways that circumvent the dependency on the targeted pathway, and epigenetic or stochastic changes in the state of tumor cells can also activate intrinsic resistance pathways (Holohan et al. 2013; Holzel et al. 2013).
Further complicating matters is the degree of intratumoral genetic heterogeneity. Recent evidence emerging from sequencing of single cells and multiple regions of tumors from individual patients has revealed this is far greater than previously imagined (Navin et al. 2011; Ruiz et al. 2011; Ding et al. 2012; Gerlinger et al. 2012; Xu et al. 2012b; Bashashati et al. 2013) In one study of kidney tumors, only ∼45% of mutations were detected in all tumor regions. This heterogeneity also contributes significantly to intratumoral variation in sensitivity to drugs targeting signaling proteins mutated in cancer, and means that single biopsies may not be sufficient to customize patient treatment.
Overcoming these challenges will require a deeper understanding of the nature of resistance mechanisms and how different cellular signaling programs mediate resistant states in heterogeneous populations of tumor cells. Combination therapies that target these should increase the efficacy of targeted therapies. This is a significant challenge, but one that we feel is not insurmountable.
ACKNOWLEDGMENTS
We would like to thank Lew Cantley, Tony Hunter, and Jeremy Thorner for useful feedback on the manuscript.
Footnotes
Editors: Lewis Cantley, Tony Hunter, Richard Sever, and Jeremy Thorner
Additional Perspectives on Signal Transduction available at www.cshperspectivesinmedicine.org
REFERENCES
*Reference is also in this collection.
- Anastasiou D, Poulogiannis G, Asara JM, Boxer MB, Jiang JK, Shen M, Bellinger G, Sasaki AT, Locasale JW, Auld DS, et al. 2011. Inhibition of pyruvate kinase M2 by reactive oxygen species contributes to cellular antioxidant responses. Science 334: 1278–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakan I, Laplante M 2012. Connecting mTORC1 signaling to SREBP-1 activation. Curr Opin Lipidol 23: 226–234. [DOI] [PubMed] [Google Scholar]
- Barthel A, Okino ST, Liao J, Nakatani K, Li J, Whitlock JP Jr., Roth RA 1999. Regulation of GLUT1 gene transcription by the serine/threonine kinase Akt1. J Biol Chem 274: 20281–20286. [DOI] [PubMed] [Google Scholar]
- Bashashati A, Ha G, Tone A, Ding J, Prentice LM, Roth A, Rosner J, Shumansky K, Kalloger S, Senz J, et al. 2013. Distinct evolutionary trajectories of primary high-grade serous ovarian cancers revealed through spatial mutational profiling. J Pathol 231: 21–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Sahra I, Howell JJ, Asara JM, Manning BD 2013. Stimulation of de novo pyrimidine synthesis by growth signaling through mTOR and S6K1. Science 339: 1323–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berwick DC, Hers I, Heesom KJ, Moule SK, Tavare JM 2002. The identification of ATP-citrate lyase as a protein kinase B (Akt) substrate in primary adipocytes. J Biol Chem 277: 33895–33900. [DOI] [PubMed] [Google Scholar]
- Birchmeier W, Birchmeier C 1995. Epithelial-mesenchymal transitions in development and tumor progression. EXS 74: 1–15. [DOI] [PubMed] [Google Scholar]
- Burgering BM, Medema RH 2003. Decisions on life and death: FOXO Forkhead transcription factors are in command when PKB/Akt is off duty. J Leukoc Biol 73: 689–701. [DOI] [PubMed] [Google Scholar]
- Cagnol S, Chambard JC 2010. ERK and cell death: Mechanisms of ERK-induced cell death—Apoptosis, autophagy and senescence. FEBS J 277: 2–21. [DOI] [PubMed] [Google Scholar]
- Cain RJ, Ridley AJ 2009. Phosphoinositide 3-kinases in cell migration. Biol Cell 101: 13–29. [DOI] [PubMed] [Google Scholar]
- Castells M, Thibault B, Delord JP, Couderc B 2012. Implication of tumor microenvironment in chemoresistance: Tumor-associated stromal cells protect tumor cells from cell death. Int J Mol Sci 13: 9545–9571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castoria G, Lombardi M, Barone MV, Bilancio A, Di Domenico M, De Falco A, Varricchio L, Bottero D, Nanayakkara M, Migliaccio A, et al. 2004. Rapid signalling pathway activation by androgens in epithelial and stromal cells. Steroids 69: 517–522. [DOI] [PubMed] [Google Scholar]
- Chin YR, Toker A 2011. Akt isoform-specific signaling in breast cancer: Uncovering an anti-migratory role for palladin. Cell Adh Migr 5: 211–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christofk HR, Vander Heiden MG, Wu N, Asara JM, Cantley LC 2008. Pyruvate kinase M2 is a phosphotyrosine-binding protein. Nature 452: 181–186. [DOI] [PubMed] [Google Scholar]
- Cichowski K, Jacks T 2001. NF1 tumor suppressor gene function: Narrowing the GAP. Cell 104: 593–604. [DOI] [PubMed] [Google Scholar]
- Collak FK, Yagiz K, Luthringer DJ, Erkaya B, Cinar B 2012. Threonine-120 phosphorylation regulated by phosphoinositide-3-kinase/Akt and mammalian target of rapamycin pathway signaling limits the antitumor activity of mammalian sterile 20-like kinase 1. J Biol Chem 287: 23698–23709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang CV 2013. MYC, metabolism, cell growth, and tumorigenesis. Cold Spring Harb Perspect Med 3: a014217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delhommeau F, Dupont S, Della Valle V, James C, Trannoy S, Masse A, Kosmider O, Le Couedic JP, Robert F, Alberdi A, et al. 2009. Mutation in TET2 in myeloid cancers. N Engl J Med 360: 2289–2301. [DOI] [PubMed] [Google Scholar]
- *.Devreotes P, Horwitz AR 2014. Signaling networks that regulate cell migration. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a005959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehl JA, Zindy F, Sherr CJ 1997. Inhibition of cyclin D1 phosphorylation on threonine-286 prevents its rapid degradation via the ubiquitin-proteasome pathway. Genes Dev 11: 957–972. [DOI] [PubMed] [Google Scholar]
- Ding L, Ley TJ, Larson DE, Miller CA, Koboldt DC, Welch JS, Ritchey JK, Young MA, Lamprecht T, McLellan MD, et al. 2012. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature 481: 506–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doble BW, Woodgett JR 2007. Role of glycogen synthase kinase-3 in cell fate and epithelial-mesenchymal transitions. Cells Tissues Organs 185: 73–84. [DOI] [PubMed] [Google Scholar]
- Dow LE, Elsum IA, King CL, Kinross KM, Richardson HE, Humbert PO 2008. Loss of human Scribble cooperates with H-Ras to promote cell invasion through deregulation of MAPK signalling. Oncogene 27: 5988–6001. [DOI] [PubMed] [Google Scholar]
- *.Duronio RJ, Xiong Y 2013. Signaling pathways that control cell proliferation. Cold Spring Harb Perspect Biol 5: a008904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edinger AL, Thompson CB 2002. Akt maintains cell size and survival by increasing mTOR-dependent nutrient uptake. Mol Biol Cell 13: 2276–2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsum IA, Martin C, Humbert PO 2013. Scribble regulates an EMT polarity pathway through modulation of MAPK-ERK signaling to mediate junction formation. J Cell Sci 126: 3990–3999. [DOI] [PubMed] [Google Scholar]
- Engelman JA, Luo J, Cantley LC 2006. The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat Rev Genet 7: 606–619. [DOI] [PubMed] [Google Scholar]
- Fang D, Hawke D, Zheng Y, Xia Y, Meisenhelder J, Nika H, Mills GB, Kobayashi R, Hunter T, Lu Z 2007. Phosphorylation of β-catenin by AKT promotes β-catenin transcriptional activity. J Biol Chem 282: 11221–11229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gainor JF, Shaw AT 2013. Emerging paradigms in the development of resistance to tyrosine kinase inhibitors in lung cancer. J Clin Oncol 31: 3987–3996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Wang H, Yang JJ, Liu X, Liu ZR 2012. Pyruvate kinase M2 regulates gene transcription by acting as a protein kinase. Mol Cell 45: 598–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, Martinez P, Matthews N, Stewart A, Tarpey P, et al. 2012. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med 366: 883–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghoda L, Lin X, Greene WC 1997. The 90-kDa ribosomal S6 kinase (pp90rsk) phosphorylates the N-terminal regulatory domain of IκBα and stimulates its degradation in vitro. J Biol Chem 272: 21281–21288. [DOI] [PubMed] [Google Scholar]
- Giroux S 2013. Overcoming acquired resistance to kinase inhibition: The cases of EGFR, ALK and BRAF. Bioorg Med Chem Lett 23: 394–401. [DOI] [PubMed] [Google Scholar]
- Gorgoulis VG, Halazonetis TD 2010. Oncogene-induced senescence: The bright and dark side of the response. Curr Opin Cell Biol 22: 816–827. [DOI] [PubMed] [Google Scholar]
- *.Green DR, Llambi F 2014. Cell death signaling. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a006080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer C, Lee M, Westerhof M, Milholland B, Spokony R, Vijg J, Secombe J 2013. Myc-dependent genome instability and lifespan in Drosophila. PLoS ONE 8: e74641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivennikov SI, Wang K, Mucida D, Stewart CA, Schnabl B, Jauch D, Taniguchi K, Yu G-Y, Österreicher CH, Hung KE, et al. 2012. Adenoma-linked barrier defects and microbial products drive IL-23/IL-17-mediated tumour growth. Nature 491: 254–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo D, Bell EH, Mischel P, Chakravarti A 2013. Targeting SREBP-1-driven lipid metabolism to treat cancer. Curr Pharm Des. 10.2174/13816128113199990486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamede RK, McCallum H, Jones M 2013. Biting injuries and transmission of Tasmanian devil facial tumour disease. J Anim Ecol 82: 182–190. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA 2000. The hallmarks of cancer. Cell 100: 57–70. [DOI] [PubMed] [Google Scholar]
- Haq S, Michael A, Andreucci M, Bhattacharya K, Dotto P, Walters B, Woodgett J, Kilter H, Force T 2003. Stabilization of β-catenin by a Wnt-independent mechanism regulates cardiomyocyte growth. Proc Natl Acad Sci 100: 4610–4615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Hardie DG 2012. Organismal carbohydrate and lipid homeostasis. Cold Spring Harb Perspect Biol 4: a006031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Harrison DA 2012. The Jak/STAT pathway. Cold Spring Harb Perspect Biol 4: a011205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Harvey KF, Hariharan IK 2012. The Hippo pathway. Cold Spring Harb Perspect Biol 4: a011288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey KF, Zhang X, Thomas DM 2013. The Hippo pathway and human cancer. Nat Rev Cancer 13: 246–257. [DOI] [PubMed] [Google Scholar]
- *.Hemmings BA, Restuccia DF 2012. PI3K-PKB/Akt pathway. Cold Spring Harb Perspect Biol 4: a011189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitosugi T, Kang S, Vander Heiden MG, Chung TW, Elf S, Lythgoe K, Dong S, Lonial S, Wang X, Chen GZ, et al. 2009. Tyrosine phosphorylation inhibits PKM2 to promote the Warburg effect and tumor growth. Sci Signal 2: ra73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holohan C, Van Schaeybroeck S, Longley DB, Johnston PG 2013. Cancer drug resistance: An evolving paradigm. Nat Rev Cancer 13: 714–726. [DOI] [PubMed] [Google Scholar]
- Holzel M, Bovier A, Tuting T 2013. Plasticity of tumour and immune cells: A source of heterogeneity and a cause for therapy resistance? Nat Rev Cancer 13: 365–376. [DOI] [PubMed] [Google Scholar]
- Hu W, Zhang C, Wu R, Sun Y, Levine A, Feng Z 2010. Glutaminase 2, a novel p53 target gene regulating energy metabolism and antioxidant function. Proc Natl Acad Sci 107: 7455–7460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussey GS, Chaudhury A, Dawson AE, Lindner DJ, Knudsen CR, Wilce MC, Merrick WC, Howe PH 2011. Identification of an mRNP complex regulating tumorigenesis at the translational elongation step. Mol Cell 41: 419–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iden S, van Riel WE, Schafer R, Song JY, Hirose T, Ohno S, Collard JG 2012. Tumor type-dependent function of the par3 polarity protein in skin tumorigenesis. Cancer Cell 22: 389–403. [DOI] [PubMed] [Google Scholar]
- *.Ingham PW 2012. Hedgehog signaling. Cold Spring Harb Perspect Biol 4: a011221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israelsen WJ, Dayton TL, Davidson SM, Fiske BP, Hosios AM, Bellinger G, Li J, Yu Y, Sasaki M, Horner JW, et al. 2013. PKM2 isoform-specific deletion reveals a differential requirement for pyruvate kinase in tumor cells. Cell 155: 397–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen PJ, Gunter LB, Carayannopoulos MO 2010. Akt2 modulates glucose availability and downstream apoptotic pathways during development. J Biol Chem 285: 17673–17680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon TI, Osborne TF 2012. SREBPs: Metabolic integrators in physiology and metabolism. Trends Endocrinol Metab 23: 65–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaelin WG Jr, McKnight SL 2013. Influence of metabolism on epigenetics and disease. Cell 153: 56–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karar J, Maity A 2011. PI3K/AKT/mTOR pathway in angiogenesis. Front Mol Neurosci 4: 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karin M 2009. NF-κB as a critical link between inflammation and cancer. Cold Spring Harb Perspect Biol 1: a000141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keely PJ 2011. Mechanisms by which the extracellular matrix and integrin signaling act to regulate the switch between tumor suppression and tumor promotion. J Mammary Gland Biol Neoplasia 16: 205–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JW, Zeller KI, Wang Y, Jegga AG, Aronow BJ, O’Donnell KA, Dang CV 2004. Evaluation of myc E-box phylogenetic footprints in glycolytic genes by chromatin immunoprecipitation assays. Mol Cell Biol 24: 5923–5936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Shu S, Coppola MD, Kaneko S, Yuan ZQ, Cheng JQ 2010. Regulation of proapoptotic mammalian ste20-like kinase MST2 by the IGF1-Akt pathway. PLoS ONE 5: e9616. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Kim J, Parrish AB, Kurokawa M, Matsuura K, Freel CD, Andersen JL, Johnson CE, Kornbluth S 2012. Rsk-mediated phosphorylation and 14-3-3ɛ binding of Apaf-1 suppresses cytochrome c-induced apoptosis. EMBO J 31: 1279–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Kopan R 2012. Notch signaling. Cold Spring Harb Perspect Biol 4: a011213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korkaya H, Paulson A, Charafe-Jauffret E, Ginestier C, Brown M, Dutcher J, Clouthier SG, Wicha MS 2009. Regulation of mammary stem/progenitor cells by PTEN/Akt/β-catenin signaling. PLoS Biol 7: e1000121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubatka P, Zihlavnikova K, Kajo K, Pec M, Stollarova N, Bojkova B, Kassayova M, Orendas P 2011. Antineoplastic effects of simvastatin in experimental breast cancer. Klin Onkol 24: 41–45. [PubMed] [Google Scholar]
- *.Laplante M, Sabatini DM 2012. mTOR signaling. Cold Spring Harb Perspect Biol 4: a011593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larue L, Bellacosa A 2005. Epithelial-mesenchymal transition in development and cancer: Role of phosphatidylinositol 3′ kinase/AKT pathways. Oncogene 24: 7443–7454. [DOI] [PubMed] [Google Scholar]
- Ley TJ, Ding L, Walter MJ, McLellan MD, Lamprecht T, Larson DE, Kandoth C, Payton JE, Baty J, Welch J, et al. 2010. DNMT3A mutations in acute myeloid leukemia. N Engl J Med 363: 2424–2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Shen D, Shao J, Crowder R, Liu W, Prat A, He X, Liu S, Hoog J, Lu C, et al. 2013. Endocrine-therapy-resistant ESR1 variants revealed by genomic characterization of breast-cancer-derived xenografts. Cell Rep 4: 1116–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CY, Loven J, Rahl PB, Paranal RM, Burge CB, Bradner JE, Lee TI, Young RA 2012. Transcriptional amplification in tumor cells with elevated c-Myc. Cell 151: 56–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JI, Poon CL, Harvey KF 2013. The Hippo size control pathway-ever expanding. Sci Signal 6: pe4. [DOI] [PubMed] [Google Scholar]
- Lito P, Rosen N, Solit DB 2013. Tumor adaptation and resistance to RAF inhibitors. Nat Med 19: 1401–1409. [DOI] [PubMed] [Google Scholar]
- Liu W, Zhou Y, Reske SN, Shen C 2008. PTEN mutation: Many birds with one stone in tumorigenesis. Anticancer Res 28: 3613–3619. [PubMed] [Google Scholar]
- Lu P, Takai K, Weaver VM, Werb Z 2011. Extracellular matrix degradation and remodeling in development and disease. Cold Spring Harb Perspect Biol 3: a005058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W, Hu H, Chang R, Zhong J, Knabel M, O’Meally R, Cole RN, Pandey A, Semenza GL 2011. Pyruvate kinase M2 is a PHD3-stimulated coactivator for hypoxia-inducible factor 1. Cell 145: 732–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Zhang G, Miao XB, Deng XB, Wu Y, Liu Y, Jin ZR, Li XQ, Liu QZ, Sun DX, et al. 2013. Cancer stem-like cell properties are regulated by EGFR/AKT/β-catenin signaling and preferentially inhibited by gefitinib in nasopharyngeal carcinoma. FEBS J 280: 2027–2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzone M, Selfors LM, Albeck J, Overholtzer M, Sale S, Carroll DL, Pandya D, Lu Y, Mills GB, Aster JC, et al. 2010. Dose-dependent induction of distinct phenotypic responses to Notch pathway activation in mammary epithelial cells. Proc Natl Acad Sci 107: 5012–5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaffrey LM, Montalbano J, Mihai C, Macara IG 2012. Loss of the Par3 polarity protein promotes breast tumorigenesis and metastasis. Cancer Cell 22: 601–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin SK, Olsen SN, Dake B, De Raedt T, Lim E, Bronson RT, Beroukhim R, Polyak K, Brown M, Kuperwasser C, et al. 2013. The RasGAP gene, RASAL2, is a tumor and metastasis suppressor. Cancer Cell 24: 365–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miinea CP, Sano H, Kane S, Sano E, Fukuda M, Peranen J, Lane WS, Lienhard GE 2005. AS160, the Akt substrate regulating GLUT4 translocation, has a functional Rab GTPase-activating protein domain. Biochem J 391: 87–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DM, Thomas SD, Islam A, Muench D, Sedoris K 2012. c-Myc and cancer metabolism. Clin Cancer Res 18: 5546–5553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min J, Zaslavsky A, Fedele G, McLaughlin SK, Reczek EE, De Raedt T, Guney I, Strochlic DE, Macconaill LE, Beroukhim R, et al. 2010. An oncogene-tumor suppressor cascade drives metastatic prostate cancer by coordinately activating Ras and nuclear factor-κB. Nat Med 16: 286–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris MA, Dawson CW, Young LS 2009. Role of the Epstein-Barr virus-encoded latent membrane protein-1, LMP1, in the pathogenesis of nasopharyngeal carcinoma. Future Oncol 5: 811–825. [DOI] [PubMed] [Google Scholar]
- *.Morrison DK 2012. MAP kinase pathways. Cold Spring Harb Perspect Biol 4: a011254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munger K, Howley PM 2002. Human papillomavirus immortalization and transformation functions. Virus Res 89: 213–228. [DOI] [PubMed] [Google Scholar]
- Muranen T, Selfors LM, Worster DT, Iwanicki MP, Song L, Morales FC, Gao S, Mills GB, Brugge JS 2012. Inhibition of PI3K/mTOR leads to adaptive resistance in matrix-attached cancer cells. Cancer Cell 21: 227–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murchison EP, Schulz-Trieglaff OB, Ning Z, Alexandrov LB, Bauer MJ, Fu B, Hims M, Ding Z, Ivakhno S, Stewart C, et al. 2012. Genome sequencing and analysis of the Tasmanian devil and its transmissible cancer. Cell 148: 780–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy LO, Smith S, Chen RH, Fingar DC, Blenis J 2002. Molecular interpretation of ERK signal duration by immediate early gene products. Nat Cell Biol 4: 556–564. [DOI] [PubMed] [Google Scholar]
- Nagasaka K, Seiki T, Yamashita A, Massimi P, Subbaiah VK, Thomas M, Kranjec C, Kawana K, Nakagawa S, Yano T, et al. 2013. A novel interaction between hScrib and PP1γ downregulates ERK signaling and suppresses oncogene-induced cell transformation. PLoS ONE 8: e53752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima A, Kawanishi I, Eguchi S, Yu EH, Eguchi S, Oshiro N, Yoshino K, Kikkawa U, Yonezawa K 2013. Association of CAD, a multifunctional protein involved in pyrimidine synthesis, with mLST8, a component of the mTOR complexes. J Biomed Sci 20: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navin N, Kendall J, Troge J, Andrews P, Rodgers L, McIndoo J, Cook K, Stepansky A, Levy D, Esposito D, et al. 2011. Tumour evolution inferred by single-cell sequencing. Nature 472: 90–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Newton K, Dixit VM 2012. Signaling in innate immunity and inflammation. Cold Spring Harb Perspect Biol 4: a006049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie Z, Hu G, Wei G, Cui K, Yamane A, Resch W, Wang R, Green DR, Tessarollo L, Casellas R, et al. 2012. c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell 151: 68–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novellasdemunt L, Tato I, Navarro-Sabate A, Ruiz-Meana M, Mendez-Lucas A, Perales JC, Garcia-Dorado D, Ventura F, Bartrons R, Rosa JL 2013. Akt-dependent activation of the heart 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase (PFKFB2) isoenzyme by amino acids. J Biol Chem 288: 10640–10651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Nusse R 2012. Wnt signaling. Cold Spring Harb Perspect Biol 4: a011163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawara Y, Kishishita S, Obata T, Isazawa Y, Suzuki T, Tanaka K, Masuyama N, Gotoh Y 2002. Akt enhances Mdm2-mediated ubiquitination and degradation of p53. J Biol Chem 277: 21843–21850. [DOI] [PubMed] [Google Scholar]
- O’Neill E, Kolch W 2005. Taming the Hippo: Raf-1 controls apoptosis by suppressing MST2/Hippo. Cell Cycle 4: 365–367. [DOI] [PubMed] [Google Scholar]
- Plas DR, Thompson CB 2005. Akt-dependent transformation: There is more to growth than just surviving. Oncogene 24: 7435–7442. [DOI] [PubMed] [Google Scholar]
- Polakis P 2001. More than one way to skin a catenin. Cell 105: 563–566. [DOI] [PubMed] [Google Scholar]
- Pratilas CA, Solit DB 2010. Targeting the mitogen-activated protein kinase pathway: Physiological feedback and drug response. Clin Cancer Res 16: 3329–3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prochownik EV, Li Y 2007. The ever expanding role for c-Myc in promoting genomic instability. Cell Cycle 6: 1024–1029. [DOI] [PubMed] [Google Scholar]
- Raftopoulou M, Hall A 2004. Cell migration: Rho GTPases lead the way. Dev Biol 265: 23–32. [DOI] [PubMed] [Google Scholar]
- Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, Young RA 2010. c-Myc regulates transcriptional pause release. Cell 141: 432–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renoir JM, Marsaud V, Lazennec G 2013. Estrogen receptor signaling as a target for novel breast cancer therapeutics. Biochem Pharmacol 85: 449–465. [DOI] [PubMed] [Google Scholar]
- *.Rhind N, Russell P 2012. Signaling pathways that regulate cell division. Cold Spring Harb Perspect Biol 4: a005942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson CJ, Schalm SS, Blenis J 2004. PI3-kinase and TOR: PIKTORing cell growth. Semin Cell Dev Biol 15: 147–159. [DOI] [PubMed] [Google Scholar]
- Roberts DJ, Tan-Sah VP, Smith JM, Miyamoto S 2013. Akt phosphorylates HK-II at Thr-473 and increases mitochondrial HK-II association to protect cardiomyocytes. J Biol Chem 288: 23798–23806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson DR, Wu YM, Vats P, Su F, Lonigro RJ, Cao X, Kalyana-Sundaram S, Wang R, Ning Y, Hodges L, et al. 2013. Activating ESR1 mutations in hormone-resistant metastatic breast cancer. Nat Genet 45: 1446–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrik-Outmezguine VS, Chandarlapaty S, Pagano NC, Poulikakos PI, Scaltriti M, Moskatel E, Baselga J, Guichard S, Rosen N 2011. mTOR kinase inhibition causes feedback-dependent biphasic regulation of AKT signaling. Cancer Discov 1: 248–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossig L, Jadidi AS, Urbich C, Badorff C, Zeiher AM, Dimmeler S 2001. Akt-dependent phosphorylation of pp21cip1 regulates PCNA binding and proliferation of endothelial cells. Mol Cell Biol 21: 5644–5657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux PP, Blenis J 2004. ERK and p38 MAPK-activated protein kinases: A family of protein kinases with diverse biological functions. Microbiol Mol Biol Rev 68: 320–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz C, Lenkiewicz E, Evers L, Holley T, Robeson A, Kiefer J, Demeure MJ, Hollingsworth MA, Shen M, Prunkard D, et al. 2011. Advancing a clinically relevant perspective of the clonal nature of cancer. Proc Natl Acad Sci 108: 12054–12059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saavedra HI, Fukasawa K, Conn CW, Stambrook PJ 1999. MAPK mediates RAS-induced chromosome instability. J Biol Chem 274: 38083–38090. [DOI] [PubMed] [Google Scholar]
- Sandoval J, Esteller M 2012. Cancer epigenomics: Beyond genomics. Curr Opin Genet Dev 22: 50–55. [DOI] [PubMed] [Google Scholar]
- Schvartzman JM, Sotillo R, Benezra R 2010. Mitotic chromosomal instability and cancer: Mouse modelling of the human disease. Nat Rev Cancer 10: 102–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sears R, Nuckolls F, Haura E, Taya Y, Tamai K, Nevins JR 2000. Multiple Ras-dependent phosphorylation pathways regulate Myc protein stability. Genes Dev 14: 2501–2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeger C, Zoulim F, Mason W 2013. Hepsdna viruses. In Fields virology, 3rd ed. (ed. Knipe DM, Howley P). Lippincott Williams & Wilkins, Philadelphia. [Google Scholar]
- *.Sever R, Glass CK 2013. Signaling by nuclear receptors. Cold Spring Harb Perspect Biol 5: a016709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaikh S, Collier DA, Sham PC, Ball D, Aitchison K, Vallada H, Smith I, Gill M, Kerwin RW 1996. Allelic association between a Ser-9-Gly polymorphism in the dopamine D3 receptor gene and schizophrenia. Hum Genet 97: 714–719. [DOI] [PubMed] [Google Scholar]
- Shen HM, Tergaonkar V 2009. NFκB signaling in carcinogenesis and as a potential molecular target for cancer therapy. Apoptosis 14: 348–363. [DOI] [PubMed] [Google Scholar]
- Solimini NL, Luo J, Elledge SJ 2007. Non-oncogene addiction and the stress phenotype of cancer cells. Cell 130: 986–988. [DOI] [PubMed] [Google Scholar]
- Soloaga A, Thomson S, Wiggin GR, Rampersaud N, Dyson MH, Hazzalin CA, Mahadevan LC, Arthur JS 2003. MSK2 and MSK1 mediate the mitogen- and stress-induced phosphorylation of histone H3 and HMG-14. EMBO J 22: 2788–2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulzmaier FJ, Ramos JW 2013. RSK isoforms in cancer cell invasion and metastasis. Cancer Res 73: 6099–6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutrias-Grau M, Arnosti DN 2004. CtBP contributes quantitatively to Knirps repression activity in an NAD binding-dependent manner. Mol Cell Biol 24: 5953–5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suva ML, Riggi N, Bernstein BE 2013. Epigenetic reprogramming in cancer. Science 339: 1567–1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki S, Tanaka T, Poyurovsky MV, Nagano H, Mayama T, Ohkubo S, Lokshin M, Hosokawa H, Nakayama T, Suzuki Y, et al. 2010. Phosphate-activated glutaminase (GLS2), a p53-inducible regulator of glutamine metabolism and reactive oxygen species. Proc Natl Acad Sci 107: 7461–7466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiery JP 2002. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer 2: 442–454. [DOI] [PubMed] [Google Scholar]
- Thomas C, Gustafsson JA 2011. The different roles of ER subtypes in cancer biology and therapy. Nat Rev Cancer 11: 597–608. [DOI] [PubMed] [Google Scholar]
- Toda M, Shirao T, Minoshima S, Shimizu N, Toya S, Uyemura K 1993. Molecular cloning of cDNA encoding human drebrin E and chromosomal mapping of its gene. Biochem Biophys Res Commun 196: 468–472. [DOI] [PubMed] [Google Scholar]
- Toy W, Shen Y, Won H, Green B, Sakr RA, Will M, Li Z, Gala K, Fanning S, King TA, et al. 2013. ESR1 ligand-binding domain mutations in hormone-resistant breast cancer. Nat Genet 45: 1439–1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner N, Grose R 2010. Fibroblast growth factor signalling: From development to cancer. Nat Rev Cancer 10: 116–129. [DOI] [PubMed] [Google Scholar]
- Turner SL, Blair-Zajdel ME, Bunning RA 2009. ADAMs and ADAMTSs in cancer. Br J Biomed Sci 66: 117–128. [DOI] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB 2009. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science 324: 1029–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas J, Feltes BC, Poloni Jde F, Lenz G, Bonatto D 2012. Senescence: An endogenous anticancer mechanism. Front Biosci (Landmark Ed) 17: 2616–2643. [DOI] [PubMed] [Google Scholar]
- Vicente-Manzanares M, Horwitz AR 2011. Cell migration: An overview. Methods Mol Biol 769: 1–24. [DOI] [PubMed] [Google Scholar]
- Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA Jr, Kinzler KW 2013. Cancer genome landscapes. Science 339: 1546–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward PS, Thompson CB 2012a. Metabolic reprogramming: A cancer hallmark even warburg did not anticipate. Cancer Cell 21: 297–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Ward PS, Thompson CB 2012b. Signaling in control of cell growth and metabolism. Cold Spring Harb Perspect Biol 4: a006783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg R 2013. The biology of cancer. Garland, New York. [Google Scholar]
- Wieman HL, Wofford JA, Rathmell JC 2007. Cytokine stimulation promotes glucose uptake via phosphatidylinositol-3 kinase/Akt regulation of Glut1 activity and trafficking. Mol Biol Cell 18: 1437–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M, Pastor-Pareja JC, Xu T 2010. Interaction between RasV12 and scribbled clones induces tumour growth and invasion. Nature 463: 545–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Broniscer A, McEachron TA, Lu C, Paugh BS, Becksfort J, Qu C, Ding L, Huether R, Parker M, et al. 2012. Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat Genet 44: 251–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu N, Lao Y, Zhang Y, Gillespie DA 2012a. Akt: A double-edged sword in cell proliferation and genome stability. J Oncol 2012: 951724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Hou Y, Yin X, Bao L, Tang A, Song L, Li F, Tsang S, Wu K, Wu H, et al. 2012b. Single-cell exome sequencing reveals single-nucleotide mutation characteristics of a kidney tumor. Cell 148: 886–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue B, Krishnamurthy K, Allred DC, Muthuswamy SK 2013. Loss of Par3 promotes breast cancer metastasis by compromising cell-cell cohesion. Nat Cell Biol 15: 189–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada E, Okada S, Saito T, Ohshima K, Sato M, Tsuchiya T, Uehara Y, Shimizu H, Mori M 2005. Akt2 phosphorylates Synip to regulate docking and fusion of GLUT4-containing vesicles. J Cell Biol 168: 921–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying H, Kimmelman AC, Lyssiotis CA, Hua S, Chu GC, Fletcher-Sananikone E, Locasale JW, Son J, Zhang H, Coloff JL, et al. 2012. Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell 149: 656–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu FX, Guan KL 2013. The Hippo pathway: Regulators and regulations. Genes Dev 27: 355–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Tang N, Hadden TJ, Rishi AK 2011. Akt, FoxO and regulation of apoptosis. Biochim Biophys Acta 1813: 1978–1986. [DOI] [PubMed] [Google Scholar]