Abstract
Recent advances in molecular and statistical genetics have enabled the reconstruction of human history by studying living humans. The ability to sequence and study DNA by calibrating the rate of accumulation of changes with evolutionary time has enabled robust inferences about how humans have evolved. These data indicate that modern humans evolved in Africa about 150,000 years ago and, consistent with paleontological evidence, migrated out of Africa. And through a series of settlements, demographic expansions, and further migrations, they populated the entire world. One of the first waves of migration from Africa was into India. Subsequent, more recent, waves of migration from other parts of the world have resulted in India being a genetic melting pot. Contemporary India has a rich tapestry of cultures and ecologies. There are about 400 tribal groups and more than 4000 groups of castes and subcastes, speaking dialects of 22 recognized languages belonging to four major language families. The contemporary social structure of Indian populations is characterized by endogamy with different degrees of porosity. The social structure, possibly coupled with large ecological heterogeneity, has resulted in considerable genetic diversity and local genetic differences within India. In this essay, we provide genetic evidence of how India may have been peopled, the nature and extent of its genetic diversity, and genetic structure among the extant populations of India.
One of the first waves of migration from Africa was into India; subsequent waves resulted in a rich tapestry of cultures and diversities. These events are reflected in the genetic structures of contemparary population groups of India.
OUR DNA IS A PALIMPSEST OF OUR HISTORY
Modern biology has provided powerful tools for reconstructing the history of the earth and its inhabitants, including humans. Central to this development has been the ability to study the genetic material of organisms, DNA. Scientists can extract DNA from microbes, plants, animals, and humans, including their fossilized remains. We can sequence DNA and extract valuable information from their sequences about the evolutionary past. Considerable human DNA sequence data have been obtained to date. These have been used to study our diversity and investigate how humans who reside in different geographical regions, or belong to distinct cultural groups, are genetically related. This is zoology and anthropology at the molecular level, and then the biologist turns into an historian.
The data generated by the human genome project indicate that two humans, selected randomly, are genetically ∼99.9% identical in their DNA sequences. Geneticists who study human genomic diversity therefore intensively focus their studies on the tiny fraction (∼0.1%) of our genome in which we differ. This tiny fraction contains very rich information pertaining to our origins and diversity. Because the human genome comprises about three billion nucleotides, this tiny fraction (∼0.1%) corresponds to about three million nucleotide differences.
Normally, only a fraction of the genetic variation of a large population of individuals is represented in a subset of those individuals. In other words, any subset of individuals of a larger set is genetically more homogeneous than the larger set. Residents of a restricted geographical region are usually descendants of a small ancestral group and, therefore, often have limited genetic variation among them. However, genetic variation between two ancestral groups is larger. Thus, descendants of a single ancestral group are genetically more similar than descendants from different ancestral groups. Therefore, by studying the patterns of variation in the genomes of contemporary individuals, resident in one or more geographical regions, it is possible to reconstruct their ancestral affiliations. The differences in DNA sequence in the tiny (0.1%) “variable” fraction of our genome also holds the key to why some individuals are susceptible to, whereas others are protected from, a specific disease. However, these disease-associated sequence differences, like those indicative of our historical origins, are not clustered, but spread across the entire genome.
Inferring human population history rests on a simple reality. New population groups arise or evolve from pre-existing groups. A population splits into subpopulations (population “fission”) because of various cultural and demographic reasons and forces. In the past, when we were predominantly dependent on natural resources for our survival, increase of the numerical size of a population implied that there was pressure on natural resources. This pressure impinged on the survival of members of that group. Therefore, members of an expanding population would possibly have formed subgroups and moved away to new geographical locations in which natural resources were more abundant to form new subpopulations. Because these subgroups may have been small, each subgroup carried with them not the complete catalog, but only a fractional sample of the genomes present in the original population. This creates genomic diversity among the subpopulations. Furthermore, when subgroup sizes are small, demographic bottlenecks are created that would have exacerbated genomic diversity among the subpopulations. With the passage of time, subpopulations of an ancestral population diverge from one another. Despite these pressures and processes that enhance genomic differences among subpopulations of an ancestral population, some core genomic “signatures” are retained in the subpopulations. Thus, by studying the genome of individuals of contemporary subpopulations, it is possible to reconstruct their ancestries and ancestral relationships among the subpopulations.
After subpopulations emerge from an ancestral population, if they remain isolated, that is, if no mate-exchange or admixture takes place among them, they evolve independently. New mutations arise in each subpopulation. These mutations remain confined, because of lack of admixture, to the subpopulations in which they have arisen. Therefore, over time, genomic diversity is even more enhanced among the subpopulations.
Admixture, or the exchange of genes, allows a new genetic variation introduced by mutations to move from one subpopulation to another. Thus, admixture increases genetic similarities or affinities among subpopulations. The primary barriers to admixture are cultural differences, linguistic differences, and geographical distance. In general, unless there has been large-scale admixture on a continued basis, the longer two populations have been separated, the larger the genetic distance between them. Genetic distance, therefore, is a useful “clock” by which we can date evolutionary history. It must, however, be emphasized that various factors, especially natural selection, can, over time, increase or decrease the speed at which the clock ticks, thereby causing significant differences between the actual and estimated times of evolutionary events (Barreiro et al. 2008).
Anatomically, modern humans (Homo sapiens sapiens) evolved in Africa about 150,000 years ago and moved to other geographical regions between 125,000 and 60,000 years ago. There is now abundant evidence that human genomic diversity is greater in Africa than other global regions, indicating that Africa is likely to have been the source of subsequent human dispersals (Campbell and Tishkoff 2008).
Migration is not always symmetric with respect to gender; men appear to be more migratory than women. The genetic consequences of such gender asymmetry of migration can be revealed using genomic variations that are solely transmitted by either fathers or mothers, or transmitted to daughters or sons. Accounting for such gender differences is crucial because the dates of evolutionary events, such as migration to a new geographical area, are dependent on these differential rates. Geneticists have performed these analyses of maternal and paternal lineages using mitochondrial DNA (mtDNA), genetic material that is passed on by a mother to all of her children, therefore marking the genome of the last common female ancestor (female lineage), and, the Y chromosome, possessed only by males and transmitted by father to sons only, marking the genome of the last common male ancestor (male lineage).
The sequences contained in the mtDNA or the Y-chromosomal DNA change over time by mutation and accumulate as differences among individuals. The longer the elapsed time to their common ancestor, the larger the number of accumulated changes. Moreover, earlier changes are embedded in the DNA segment that carries later changes serving as a signature called a haplotype (haploid genotype). Haplotypes can be clustered into groups by the similarities of their DNA profiles called haplogroups and represent branches of maternal or paternal lineages. Consequently, maternal and paternal lineages can be defined by specific genetic variants (markers). Geneticists can estimate the age of a haplogroup; because a haplogroup is defined by the possession of a specific set of DNA variants, additional changes that appear can be used to estimate the age of the haplogroup. A haplogroup with a smaller number of accumulated DNA changes is younger than another with a larger number of accumulated changes. If the rate of accumulation of change per year remains approximately constant, then one can estimate the time of accumulation from the observed number of changes. (Usually, such estimates are quite rough because of random fluctuations in accumulation of changes in different regions of the genome. Therefore, only a broad range of time can usually be ascribed.) The extant variation in our genomes thus carries footprints of the history of our species. Additionally, the pattern with which these mutations and polymorphisms (mutations that increase to high frequencies in a population) accumulate in our genomes is indicative of the dynamics of our population history (Nordborg 1997; Kingman 2000; Rosenberg and Nordborg 2002).
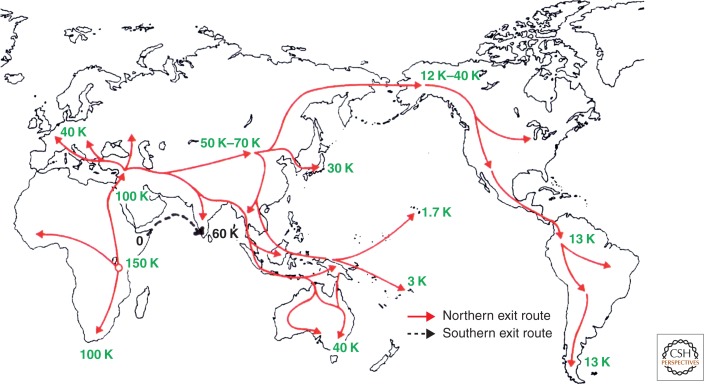
Humans who first migrated from Africa followed a “southern exit route” from the Horn of Africa across the mouth of the Red Sea along the coastline of India to southeastern Asia and Australia (Oppenheimer 2012). The major genetic evidence in support of this route is that the two major branches of the mtDNA L3 haplogroup (the haplogroup that is rooted in Africa), labeled as M and N haplogroups, outside Africa, have a very large number of local lineages in South Asia, and the antiquity of the N lineage found in Europe or the Near East is smaller than that in South Asia (Richards et al. 2006). The probable date of dispersal through the southern exit route was about 70,000–80,000 years ago (Fig. 1) (Lahr and Foley 1998). Archeological evidence in support of this route has been scanty, primarily because the coastlines of that period have become deeply submerged from the subsequent rapid increase in sea levels. There are strong indications, however, that a second and more recent human migration from out of Africa followed a “northern exit route” through the Nile Valley into Central Asia and then beyond, including into India.
Figure 1.
The great human exodus: The out-of-Africa journey and dispersal of anatomically modern humans. The numbers indicate the extimated dates in years before present.
PEOPLING OF INDIA, EARLY SETTLERS, AND CONTEMPORARY SOCIAL STRUCTURE
India has served as a major corridor for the migration of modern humans who started to disperse out of Africa about 100,000 (perhaps, sometime between 125,000 and 60,000) years ago (Cann 2001). Nevertheless, the date of entry of modern humans into India remains uncertain. It is quite certain that by the middle of the Paleolithic period (50,000–20,000 years before present [ybp]) humans had spread to many parts of the subcontinent (Misra 1992, 2001). Also, modern human remains dating back to the late Pleistocene (55,000–25,000 ybp) have been found in India (Kennedy et al. 1987). Thus, India has been peopled by contemporary humans at least for the past 55,000 years (Fig. 1).
Molecular genetic evidence, for example, the pattern with which mutations have accumulated on the mtDNA in Indian populations, indicates that a major population expansion of modern humans took place within India (Majumder et al. 1999). Although the period of this demographic expansion remains uncertain, it has been speculated (Mountain et al. 1995) that it took place sometime during the range of 60,000–85,000 ybp. This expansion, followed by subsequent migration, appears to have resulted in the peopling of Southeast Asia and later (50,000–60,000 ybp) of Australia (Crow 1998). An independent expansion of modern humans, ∼60,000 ybp, appears to have taken place in southern China (Ballinger et al. 1992; Crow 1998), which may have resulted in human migration into India through the northeast corridor and also Southeast Asia.
Some recent archaeological finds from India indicate that the major route of dispersal to India from Africa was through the southern route (Mellars 2006). Molecular genetic studies, primarily those based on mtDNA and Y-chromosomal polymorphisms, have also favored a southern dispersal route (Underhill et al. 2001a,b; Forster 2004). The strongest genetic evidence in favor of an early southern exit into India comes from the observation that signatures possessed by Indian and other Asian populations are all derivatives of mitochondrial M and N haplogroups, which themselves derive from the L3 haplogroup now found only in Africa (Quintana-Murci et al. 1999). The southern exit hypothesis is also supported by analyses of mtDNA data from the Andaman Islands (Endicott et al. 2003; Kivisild et al. 2003; Thangaraj et al. 2005) and New Guinea (Forster et al. 2001). Furthermore, the Y-chromosomal haplogroups C and D are found only in the Asian continent and Oceania (Endicott et al. 2003; Kivisild et al. 2003), but not in Eurasia or North Africa. Although available genomic data strongly indicate that India was peopled on one of the earliest waves of human migration from Africa and the dispersal took place via the southern exit route, a major limitation of more direct genetic evidence is the absence of reliable and comparable data from populations that lie on the southern exit route (Stringer 2000).
Contemporary India is a rich tapestry of largely intramarrying ethnic populations. These populations belong to a diverse set of cultures and language groups. There are four distinct language families in India, namely, Austro-Asiatic, Dravidian, Tibeto-Burman, and Indo-European, with a geographical distribution that is largely nonoverlapping within India. The Dravidian-speaking groups inhabit southern India, Indo-European speakers inhabit northern India, and Tibeto-Burman speakers are confined to northeastern India. In contrast, the numerically small group of Austro-Asiatic speakers, who are exclusively tribal, inhabit fragmented geographical areas of eastern and central India. Culturally, the vast majority of the people of India belong to either tribal or caste societies. The tribal populations are characterized by their traditional modes of subsistence: hunting and gathering, unorganized agriculture, slash and burn agriculture, and nomadism (practiced by a limited number of groups). They also have no written form of language and speak a variety of dialects. On the other hand, Hindu society in India (the numerically largest religious group) comprises castes that perform a wide range of occupations and have written forms of language. There is a long-standing debate about the genesis of the caste and tribal populations of India. One model suggests that the tribes and castes share considerable Pleistocene heritage with limited recent gene flow between them (Kivisild et al. 2003), whereas an opposite view concludes that caste and tribes have independent origins (Cordaux et al. 2004). Caste origins, however, appear to be complex. Origins of the same caste in different geographical regions appear to have been different, and genetic contributions to various castes may have also been from different source populations (Basu et al. 2003; Majumder 2010).
There are more than 400 tribal groups and greater than 4000 groups of castes and subcastes in India. Although caste and tribe have been administrative social categories in British India, their existence as a social construct probably predates similar social categories found elsewhere in the world. Populations belonging to the caste fold have a ranked social order. There are four broad caste groups; however, commonly, the caste populations are now ranked as “high,” “middle,” and “low.” Although the usage of the ranks as high, middle, and low can appear to have a value judgment attached to them, it is important to take cognizance of this extant hierarchy because, as Kosambi (1965) has pointed out, “stratification of Indian society reflects and explains a great deal of Indian history, if studied in the field without prejudice.” Kosambi emphasized that the social and economic histories of ranked caste groups were different; the same also appears to be true of their genetic histories. There is virtually no evidence of the exchange of genes among tribal populations or between tribal and caste populations. There is also little exchange of genes among castes, primarily because of strict social rules of marriage within the caste system. Social stratification and norms governing mate-exchange among social strata impact on the genetic relationships of populations. Therefore, human geneticists have studied the genetic structures, similarities, and dissimilarities of ranked caste groups in India aiming to shed light on human history. Historical and anthropological studies suggest that, in the establishment of the caste system in India, there have been varying levels of admixture between the tribal people of India and the later immigrants who brought knowledge of agriculture, artisanship, and metallurgy from Central and West Asia. The migrants from Central and West Asia, who likely entered India through the northwestern corridor, spread to most areas of northern but not southern India. There is a distinct gradient of decreasing genetic similarity (representing a cline) of Indian populations with the West- and Central-Asian gene pools as we move eastward or southward from the northwestern corridor (Basu et al. 2003; Sengupta et al. 2006; Indian Genome Variation Consortium 2008; Reich et al. 2009). In other words, South and North India have had differential inputs of genes from Central and West Asia.
Who, then, are the earliest inhabitants of India? The Austro-Asiatic speakers are possibly the earliest settlers of India. They are exclusively tribal and show the highest frequencies of the ancient mtDNA lineage M. They also show the highest frequency (∼20%) of the sublineage M2, which has the highest nucleotide variation within a fast evolving segment of the mitochondrial genome as compared with other sublineages. Recent results on Y-chromosomal markers provide further support for this inference. The Y lineage O-M95, found in high frequency in India, had originated in the Indian Austro-Asiatic populations around 65,000 ybp, very close to the estimated date of entry of the wave of human migration into India on the southern exit route from Africa. These findings are consistent with linguist Colin Renfrew’s (Renfrew 2000) observation that the present distribution of the Austric language group is a result of the initial dispersal from Africa, whereas later agricultural dispersal can account for the distribution of the Elamo-Dravidian or Sino-Tibetan languages (the family to which Tibeto-Burman languages belong). However, recent studies have found that many Dravidian tribal populations also have M2 frequencies comparable to those of Austro-Asiatic tribal people (Kumar et al. 2008; Chandrasekhar et al. 2009). A recent (Chaubey et al. 2011) combined analysis of the uniparentally inherited Y-chromosomal markers with a large number of common single-nucleotide polymorphisms from the nuclear genome have, however, resulted in the proposal that the Austro-Asiatic speakers in India today are derived from dispersal from Southeast Asia, followed by extensive sex-specific admixture with local Indian populations.
Subjugation of an existing population by a relatively small group of highly organized, militarily powerful immigrants, the “elite dominance” model (Renfrew 2000), can obliterate preexisting genomic signatures through strong sexual exploitation by the immigrants, thereby presenting difficulties to our genomic inferences on the antiquities of populations. The antiquity of Dravidian speakers in India, who are not all tribal, but also belong to the organized caste system, has been enigmatic. Some historians have claimed that the Dravidians were widespread over nearly the entire landmass of India (Thapar 2004) and shared areas with Austro-Asiatic speakers. There is some genetic evidence (Basu et al. 2003) in support of this claim, and, furthermore, the Dravidian speakers probably retreated to their current habitat in southern India by the expansion of the more militarily powerful Indo-European speakers who arrived in India from Central Asia through the northwestern corridor. A contrary hypothesis, discussed later, has recently been postulated (Reich et al. 2009).
POPULATION DIVERSITY AND STRUCTURE: INFERENCES FROM mtDNA AND Y-CHROMOSOMAL DNA
Analysis of the genetic structure of Indian populations has shown that Indian ethnic populations, when grouped as tribal versus nontribal by geographical region of habitat or linguistic affiliation, have resulted from admixture of four or five ancestral populations (Indian Genome Variation Consortium 2008; Abdulla et al. 2009). One source of contribution is from Tibeto-Burman-speaking tribals who are distinct from the non-Tibeto-Burman speakers (Basu et al. 2003; Indian Genome Variation Consortium 2008; Abdulla et al. 2009). As ancestral sources of the Indian gene pool, in addition to the original African source population, West and Central Asia have been other major source contributors. However, these migrations from Asia may have taken place only in historical times, perhaps not earlier than 8000 ybp. Migration from West Asia was possibly associated with demic diffusion of agriculture, meaning the actual movement of people in the carriage of an idea or technology. The extent of genetic variation of female lineages (mtDNA) in India is rather restricted (Roychoudhury et al. 2001; Basu et al. 2003), indicating a small founding group of females. In contrast, the variation of male lineages (Y-chromosomal) is very high (Basu et al. 2003; Sengupta et al. 2006). This pattern may be indicative of sex-biased ancient gene flow into India with more male immigrants than female (Bamshad et al. 1998), possibly occurring within the last 5000 years through invasions and wars. This phenomenon obscures ancient genetic signatures and results in the quick introduction of high genetic variability, often mimicking extreme natural selection (Zerjal et al. 2003). The success of some of the Y-chromosomal haplotypes that arose in Central Asia to spread across vast regions of Eurasia (Zerjal et al. 2003), as well as South and Southeast Asia, is indicative of the “success” of the cultural and technological dominance of west Eurasia and Central Asia (Zerjal et al. 2003; Underhill et al. 2009).
The mtDNA lineage U, which is likely to have arisen in Central Asia, has a high frequency in India, implying that large-scale migration brought with it a large number of copies of this lineage into India. However, this lineage is composed of two deep sublineages, U2i and U2e, with an estimated split ∼50,000 years ago. The sublineage U2i is found in high frequency in India (particularly among tribes; ∼77%), but not in Europe (∼0%), whereas U2e is found in high frequency in Europe (>10%), but not in India (it has very low frequencies among castes, but not among tribals). Thus, a substantial fraction of the U lineage, specifically, the U2i sublineage, may be indigenous to India (Kivisild et al. 1999; Basu et al. 2003; Sengupta et al. 2006). Analysis of the complete mtDNA genome sequence has revealed a large number of sequence variants within major haplogroups within Indian populations, many of which, however, are infrequent (Palanichamy et al. 2004). This indicates a common spread of the root haplotypes of haplogroups M, N, and R between 70,000 and 60,000 years ago along the southern exit route. This analysis has further revealed that entry of the haplogroup U2 postdates the earliest settlement along the southern route.
Central Asian populations are supposed to have been the major contributors to the Indian gene pool, particularly to the North Indian gene pool, and the migrants had supposedly moved into India through what is now Afghanistan and Pakistan. Using mtDNA variation data collated from various studies, we have previously shown (Basu et al. 2003) that populations of Central Asia and Pakistan show the lowest genetic distance with the North Indian populations (FST = 0.017) (see Box 1), higher distances (FST = 0.042) with the South Indian populations, and the highest values (FST = 0.047) with the northeast Indian populations; FST is a standard measure of genetic difference among populations derived from a common source population. Thus, northern Indian populations are genetically closer to Central Asians than populations of other geographical regions of India (Bamshad et al. 2001; Basu et al. 2003).
BOX 1. FST: FIXATION INDEX.
FST is a measure of genetic differentiation among a number (k) of subpopulations. Consider a biallelic locus with alleles A and a whose frequencies in the ith subpopulation are, respectively, pi and qi = 1 − pi (i = 1,2, …, k). Let p and q denote the frequencies of these two alleles, averaged over the k subpopulations. Then, the extent of genetic differentiation among the populations is measured by
in which s2 = variance of the frequency of allele A among the k subpopulations. When k = 2, FST can be used as a measure of genetic distance between two populations. FST = 0 indicates that the two populations are genetically identical, whereas FST = 1 indicates that they are as distinct as they can be.
Although considerable cultural impact on social hierarchy and language in South Asia is attributable to the arrival of nomadic Central Asian pastoralists, studies using Y-chromosomal polymorphisms reveal that the influence of Central Asia on the preexisting gene pool was minor. Y-chromosomal data do not support models that invoke a pronounced recent genetic input from Central Asia to explain the observed genetic variation within South Asia. Genomic variation within Y-haplogroups R1a1 and R2 indicate demographic scenarios that are inconsistent with a recent single history. Deeper statistical analyses of the high-frequency R1a1 haplogroup chromosomes indicate independent recent histories of the Indus Valley and peninsular Indian region. These data are also more consistent with a peninsular origin of Dravidian speakers than a source with proximity to the Indus and significant genetic input resulting from demic diffusion associated with agriculture; rather, it indicates that pre-Holocene and Holocene-era, not Indo-European, expansions have shaped the distinctive South Asian Y-chromosome landscape (Sengupta et al. 2006).
NEW INFERENCES AND PARADIGMS FROM LARGE SETS OF GENOMIC MARKERS
Analyses of data on 405 single-nucleotide polymorphisms (SNPs) from 75 genes and a 5.2-Mb region on chromosome 22 in 1871 individuals from 55 diverse endogamous Indian populations (Indian Genome Variation Consortium 2008) have revealed that these populations form a genetic link bridging Caucasian and Asian populations. The HUGO Pan Asian SNP Consortium’s study (Abdulla et al. 2009) also showed that most Indian populations share ancestry with European populations, which is consistent with recent observations and our understanding of the expansion of Indo-European-speaking populations. The study also provided strong evidence that the peopling of India (and Southeast Asia) was via a single primary wave of migration out of Africa (Abdulla et al. 2009). In the Indian Genome Variation Consortium (2008) study, genetic distances (FST) between pairs of populations were found to vary from 0.00 to 0.11, with a mean of 0.03, suggesting that the extent of overall differentiation was low. Maximum FST values were observed among the tribal populations of different linguistic lineages. On a pan-India level, when populations were grouped by language or geographical region of habitat, the extent of genetic differentiation among linguistic or geographical groups was not statistically significant. However, grouping by ethnicity (caste and tribe) indicated significant differentiation, possibly caused by antiquity and isolation of the tribal compared with the caste populations. Although no clear geographical grouping of populations was found, ethnicity (tribal/nontribal) and language were the major determinants of genetic affinities among the populations of India.
Using more than 500,000 biallelic autosomal markers, Reich et al. (2009) have also found a north-to-south gradient of genetic proximity of Indian populations to western Eurasians/Central Asians. In general, the Central Asian populations were found to be genetically closer to the higher-ranking caste populations than to the middle- or lower-ranking caste populations. Among the higher-ranking caste populations, those of northern India are, however, genetically much closer to each other (FST = 0.016) than those of southern India (FST = 0.031). Phylogenetic analysis of Y-chromosomal data collated from various sources yields a similar picture.
Reich et al. (2009) have proposed that extant populations of India were “founded” by two hypothetical ancestral populations, one ancestral North Indian (ANI) and another ancestral South Indian (ASI). Presumably, these ancestral populations were derived from ancient humans who entered India via the southern and northern exit routes from out of Africa. All extant Indian populations are derived from admixture between these two putative ancestral populations, with the ANI contribution being higher among extant North Indian populations and that of ASI being higher among extant South Indian populations. In a more recent study (Moorjani et al. 2013), these investigators have shown that between 1900 and 4200 ybp, there was extensive admixture among Indian population groups, followed by a shift to endogamy. Because a large number of unbiasedly selected autosomal markers were used, this study does not suffer from the shortcomings of studies that used data from only one genetic locus (mitochondrial or Y-chromosomal). This model is simplistic, but intuitive and consistent with findings of earlier studies. It is simplistic because the origins of populations in the northeastern region of India cannot be explained by this model, and many past studies (cited earlier in this essay) have indicated genetic inputs into these populations from populations of Southeast Asia. More genome-wide studies with a larger sample of populations from India will provide further clarifications and insights into the population history of India.
ACKNOWLEDGMENTS
We are grateful to our collaborators and coauthors of papers, and also to those Indians who, over many years, have supported our genome diversity studies in many ways, including by donating blood samples. We are grateful to the Department of Biotechnology, Government of India, for providing financial support to our genome diversity studies in India.
Footnotes
Editor: Aravinda Chakravarti
Additional Perspectives on Human Variation available at www.cshperspectives.org
REFERENCES
- Abdulla MA, Ahmed I, Assawamakin A, Bhak J, Brahmachari SK, Calacal GC, Chaurasia A, Chen CH, Chen J, Chen YT, et al. 2009. Mapping human genetic diversity in Asia. Science 326: 1541–1545. [DOI] [PubMed] [Google Scholar]
- Ballinger SW, Schurr TG, Torroni A, Gan YY, Hodge JA, Hassan K, Chen K-H, Wallace DC 1992. Southeast Asian mitochondrial DNA analysis reveals continuity of ancient mongoloid migrations. Genetics 130: 139–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamshad MJ, Watkins WS, Dixon ME, Jorde LB, Rao BB, Naidu JM, Prasad BV, Rasanayagam A, Hammer MF 1998. Female gene flow stratifies Hindu castes. Nature 395: 651–652. [DOI] [PubMed] [Google Scholar]
- Bamshad M, Kivisild T, Watkins WS, Dixon ME, Ricker CE, Rao BB, Naidu JM, Prasad BV, Reddy PG, Rasanayagam A, et al. 2001. Genetic evidence on the origins of Indian caste populations. Genome Res 11: 994–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreiro LB, Laval G, Quach H, Patin E, Quintana-Murci L 2008. Natural selection has driven population differentiation in modern humans. Nat Genet 40: 340–345. [DOI] [PubMed] [Google Scholar]
- Basu A, Mukherjee N, Roy S, Sengupta S, Banerjee S, Chakraborty M, Dey B, Roy M, Roy B, Bhattacharyya NP, et al. 2003. Ethnic India: A genomic view, with special reference to peopling and structure. Genome Res 13: 2277–2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell MC, Tishkoff SA 2008. African genetic diversity: Implications for human demographic history, modern human origins, and complex disease mapping. Annu Rev Genomics Hum Genet 9: 403–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cann RL 2001. Genetic clues to dispersal in human populations: Retracing the past from the present. Science 291: 1742–1748. [DOI] [PubMed] [Google Scholar]
- Chandrasekar A, Kumar S, Sreenath J, Sarkar BN, Urade BP, Mallick S, Bandopadhyay SS, Barua P, Barik SS, Basu D, et al. 2009. Updating phylogeny of mitochondrial DNA macrohaplogroup m in India: Dispersal of modern human in South Asian corridor. PLoS ONE 4: e7447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaubey G, Metspalu M, Choi Y, Mägi R, Romero IG, Soares P, van Oven M, Behar DM, Rootsi S, Hudjashov G, et al. 2011. Population genetic structure in Indian Austroasiatic speakers: The role of landscape barriers and sex-specific admixture. Mol Biol Evol 28: 1013–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordaux R, Aunger R, Bentley G, Nasidze I, Sirajuddin SM, Stoneking M 2004. Independent origins of Indian caste and tribal paternal lineages. Curr Biol 14: 231–235. [DOI] [PubMed] [Google Scholar]
- Crow TJ 1998. Was the speciation event on the Y chromosome? In Abstracts of contributions to the dual congress, p. 109 University of Witwatersrand Medical School, Johannesburg, South Africa. [Google Scholar]
- Endicott P, Gilbert MT, Stringer C, Lalueza-Fox C, Willerslev E, Hansen AJ, Cooper A 2003. The genetic origins of the Andaman Islanders. Am J Hum Genet 72: 178–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster P 2004. Ice ages and the mitochondrial DNA chronology of human dispersals: A review. Philos Trans R Soc Lond B Biol Sci 359: 255–264; discussion 264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster P, Torroni A, Renfrew C, Rohl A 2001. Phylogenetic star contraction applied to Asian and Papuan mtDNA evolution. Mol Biol Evol 18: 1864–1881. [DOI] [PubMed] [Google Scholar]
- Indian Genome Variation Consortium. 2008. Genetic landscape of the people of India: A canvas for disease gene exploration. J Genet 87: 3–20. [DOI] [PubMed] [Google Scholar]
- Kennedy KAR, Deraniyagala SU, Roertgen WJ, Chiment J, Sisotell T 1987. Upper Pleistocene fossil hominids from Sri Lanka. Am J Phys Anthrop 72: 441–461. [DOI] [PubMed] [Google Scholar]
- Kingman JF 2000. Origins of the coalescent: 1974–1982. Genetics 156: 1461–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kivisild T, Bamshad MJ, Kaldma K, Metspalu M, Metspalu E, Reidla M, Laos S, Parik J, Watkins WS, Dixon ME, et al. 1999. Deep common ancestry of Indian and western-Eurasian mitochondrial DNA lineages. Curr Biol 9: 1331–1334. [DOI] [PubMed] [Google Scholar]
- Kivisild T, Rootsi S, Metspalu M, Mastana S, Kaldma K, Parik J, Metspalu E, Adojaan M, Tolk HV, Stepanov V, et al. 2003. The genetic heritage of the earliest settlers persists both in Indian tribal and caste populations. Am J Hum Genet 72: 313–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosambi DD 1965. The culture and civilisation of ancient India in historical outline. Routledge & Kegan Paul, London. [Google Scholar]
- Kumar S, Padmanabham PB, Ravuri RR, Uttaravalli K, Koneru P, Mukherjee PA, Das B, Kotal M, Xaviour D, Saheb SY, et al. 2008. The earliest settlers’ antiquity and evolutionary history of Indian populations: Evidence from M2 mtDNA lineage. BMC Evol Biol 8: 230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahr MM, Foley RA 1998. Towards a theory of modern human origins: Geography, demography, and diversity in recent human evolution. Am J Phys Anthropol 27: 137–176. [DOI] [PubMed] [Google Scholar]
- Majumder PP 2010. The human genetic history of South Asia. Curr Biol 20: R184–R187. [DOI] [PubMed] [Google Scholar]
- Majumder PP, Roy B, Banerjee S, Chakraborty M, Dey B, Mukherjee N, Roy M, Thakurta PG, Sil SK 1999. Human-specific insertion/deletion polymorphisms in Indian populations and their possible evolutionary implications. Eur J Human Genet 7: 435–446. [DOI] [PubMed] [Google Scholar]
- Mellars P 2006. Going east: New genetic and archaeological perspectives on the modern human colonization of Eurasia. Science 313: 796–800. [DOI] [PubMed] [Google Scholar]
- Misra VN 1992. Stone age in India: An ecological perspective. Man Env 14: 17–64. [Google Scholar]
- Misra VN 2001. Prehistoric human colonization of India. J Biosci 26: 491–531. [DOI] [PubMed] [Google Scholar]
- Moorjani P, Thangaraj K, Patterson N, Lipson M, Loh P-R, Govindaraj P, Berger B, Reich D, Singh L 2013. Genetic evidence for recent population mixture in India. Am J Hum Genet 93: 422–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mountain JL, Hebert JM, Bhattacharyya S, Underhill PA, Ottolenghi C, Gadgil M, Cavalli-Sforza LL 1995. Demographic history of India and mtDNA sequence diversity. Am J Hum Genet 56: 979–992. [PMC free article] [PubMed] [Google Scholar]
- Nordborg M 1997. Structured coalescent processes on different time scales. Genetics 146: 1501–1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppenheimer S 2012. Out-of-Africa, the peopling of continents and islands: Tracing uniparental gene trees across the map. Phil Trans R Soc B 367: 770–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palanichamy MG, Sun C, Agrawal S, Bandelt HJ, Kong QP, Khan F, Wang CY, Chaudhuri TK, Palla V, Zhang YP 2004. Phylogeny of mitochondrial DNA macrohaplogroup N in India, based on complete sequencing: Implications for the peopling of South Asia. Am J Hum Genet 75: 966–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintana-Murci L, Semino O, Bandelt HJ, Passarino G, McElreavey K, Santachiara-Benerecetti AS 1999. Genetic evidence of an early exit of Homo sapiens sapiens from Africa through eastern Africa. Nat Genet 23: 437–441. [DOI] [PubMed] [Google Scholar]
- Reich D, Thangaraj K, Patterson N, Price AL, Singh L 2009. Reconstructing Indian population history. Nature 461: 489–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renfrew C 2000. At the edge of knowability: Towards a prehistory of languages. Cambridge Archaeol J 10: 7–34. [Google Scholar]
- Richards M, Bandelt HJ, Kivisild T, Oppenheimer S 2006. A model for the dispersal of modern humans out of Africa. In Human mitochondrial DNA and the evolution of Homo sapiens (ed. Bandelt H-J, Macaulay V, Richards M), pp. 227–257 Springer, Berlin. [Google Scholar]
- Rosenberg NA, Nordborg M 2002. Genealogical trees, coalescent theory and the analysis of genetic polymorphisms. Nat Rev Genet 3: 380–390. [DOI] [PubMed] [Google Scholar]
- Roychoudhury S, Roy S, Basu A, Banerjee R, Vishwanathan H, Usha Rani MV, Sil SK, Mitra M, Majumder PP 2001. Genomic structures and population histories of linguistically distinct tribal groups of India. Hum Genet 109: 339–350. [DOI] [PubMed] [Google Scholar]
- Sengupta S, Zhivotovsky LA, King R, Mehdi SQ, Edmonds CA, Chow CE, Lin AA, Mitra M, Sil SK, Ramesh A, et al. 2006. Polarity and temporality of high-resolution Y-chromosome distributions in India identify both indigenous and exogenous expansions and reveal minor genetic influence of Central Asian pastoralists. Am J Hum Genet 78: 202–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stringer C 2000. Palaeoanthropology. Coasting out of Africa. Nature 405: 24–25, 27. [DOI] [PubMed] [Google Scholar]
- Thangaraj K, Chaubey G, Kivisild T, Reddy AG, Singh VK, Rasalkar AA, Singh L 2005. Reconstructing the origin of Andaman Islanders. Science 308: 996. [DOI] [PubMed] [Google Scholar]
- Thapar R 2004. Early India: From the origins to AD 1300. University of California Press, Oakland, CA. [Google Scholar]
- Underhill PA, Passarino G, Lin AA, Marzuki S, Oefner PJ, Cavalli-Sforza LL, Chambers GK 2001a. Maori origins, Y-chromosome haplotypes and implications for human history in the Pacific. Hum Mutat 17: 271–280. [DOI] [PubMed] [Google Scholar]
- Underhill PA, Passarino G, Lin AA, Shen P, Mirazon LM, Foley RA, Oefner PJ, Cavalli-Sforza LL 2001b. The phylogeography of Y chromosome binary haplotypes and the origins of modern human populations. Ann Hum Genet 65: 43–62. [DOI] [PubMed] [Google Scholar]
- Underhill PA, Myres NM, Rootsi S, Metspalu M, Zhivotovsky LA, King RJ, Lin AA, Chow CE, Semino O, Battaglia V, et al. 2009. Separating the post-glacial coancestry of European and Asian Y chromosomes within haplogroup R1a. Eur J Hum Genet 18: 479–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerjal T, Xue Y, Bertorelle G, Wells RS, Bao W, Zhu S, Qamar R, Ayub Q, Mohyuddin A, Fu S, et al. 2003. The genetic legacy of the Mongols. Am J Hum Genet 72: 717–721. [DOI] [PMC free article] [PubMed] [Google Scholar]