Abstract
Importance
Use of aspirin and other non-steroidal anti-inflammatory drugs (NSAIDs) is associated with lower risk of colorectal cancer. Prior studies examining a potential differential relationship of aspirin and NSAIDs with colorectal cancer risk according to genetic factors have been limited to analyses of candidate genes or pathways.
Objective
To comprehensively identify common genetic markers that characterize individuals who may obtain differential benefit from aspirin and/or NSAID chemoprevention, we tested gene by environment (G X E) interactions between regular use of aspirin and/or NSAIDs and single nucleotide polymorphisms (SNPs) across the genome in relation to risk of colorectal cancer.
Design
Case-control study using the Colon Cancer Family Registry (CCFR) and the Genetics and Epidemiology of Colorectal Cancer Consortium (GECCO) that enrolled cases of colorectal cancer ascertained between 1976 and 2011 and matched controls. Odds ratios (ORs) of colorectal cancer and 95% confidence intervals (95% CIs) were estimated using conventional logistic regression analysis and case-only interaction analysis, after adjusting for age, sex, center, the first three principal components to account for population structure, and known colorectal cancer risk factors. For all genome-wide analyses, a two-sided p-value<5.0×10-8, which yields a genome-wide significance level of 0.05, was considered statistically significant.
Setting
10 observational studies (5 case-control and 5 cohort studies) that were initiated between 1976 and 2003 across the U.S., Canada, Australia and Germany.
Participants
8,634 colorectal cancer cases and 8,553 controls of European descent.
Exposures
Genome-wide SNP data generated from genome-wide association scans and imputation to HapMap II, as well as information on regular use of aspirin and/or NSAIDs and other colorectal cancer risk factors collected using in-person interviews and/or structured questionnaires.
Main Outcomes and Measures
Colorectal cancer
Results
Regular use of aspirin and/or NSAIDs was associated with lower risk of colorectal cancer (OR=0.69; 95% CI=0.64-0.74; P=6.2×10-28) compared to non-regular use. In the conventional logistic regression analysis, the SNP rs2965667 at chromosome 12p12.3 near the microsomal glutathione S-transferase 1 (MGST1) gene showed a genome-wide significant interaction with aspirin and/or NSAID use (P for interaction=4.6×10-9). Compared to non-regular use, regular use of aspirin and/or NSAIDs was associated with a lower risk of colorectal cancer among individuals with rs2965667-TT genotype (OR=0.66; 95% CI=0.61-0.70; P=7.7×10-33), but a higher risk among those with much less common (4%) TA or AA genotypes (OR=1.89; 95% CI=1.27-2.81; P=0.002). In case-only interaction analysis, the SNP rs16973225 at chromosome 15q25.2 near the interleukin 16 (IL16) gene showed a genome-wide significant interaction with aspirin and/or NSAID use (P for interaction=8.2×10-9). Compared to non-regular use, regular use of aspirin and/or NSAIDs was associated with a lower risk of colorectal cancer among individuals with rs16973225-AA genotype (OR=0.66; 95% CI=0.62-0.71; P=1.9×10-30), but was not associated with risk of colorectal cancer among those with less common (9%) AC or CC genotypes (OR=0.97; 95% CI=0.78-1.20; P=0.76).
CONCLUSIONS AND RELEVANCE
In this genome-wide investigation of G X E interactions, use of aspirin and/or NSAIDs was associated with lower risk of colorectal cancer, and the association of these medications with colorectal cancer risk differed according to genetic variation at two SNPs at chromosomes 12 and 15. Validation of these findings in additional populations may facilitate targeted colorectal cancer prevention strategies.
Introduction
Considerable evidence demonstrates that aspirin and other non-steroidal anti-inflammatory drugs (NSAIDs) are associated with lower risk of colorectal neoplasms.1-5 However, the mechanisms behind this association are not well understood. Routine use of aspirin and/or NSAIDs for chemoprevention of cancer is not currently recommended due to uncertainty about its risk-benefit profile. Hence, understanding the interrelationship between genetic markers and use of aspirin and NSAIDs, also known as gene by environment (G X E) interactions, can help to identify population subgroups defined by genetic background that may preferentially benefit from chemopreventive use of these agents and offer novel insights into underlying mechanisms of carcinogenesis.
Previous genetic studies have examined the association of aspirin and/or NSAIDs with colorectal cancer according to a limited number of candidate genes or pathways.6-10 Thus, to comprehensively identify common genetic markers that characterize individuals who may obtain differential benefit from aspirin and NSAIDs, we conducted a discovery-based, genome-wide analysis of G X E interactions between regular use of aspirin and/or NSAIDs and single nucleotide polymorphisms (SNPs) in relation to risk of colorectal cancer.
Methods
Study population and harmonization of environmental data
We included individual-level data pooled from a case-control study from the Colon Cancer Family Registry (CCFR) and nine studies from the Genetics and Epidemiology of Colorectal Cancer Consortium (GECCO) that were initiated between 1976 and 2003, and enrolled cases of colorectal cancer diagnosed between 1976 and 2011, and matched controls across the U.S., Canada, Australia and Germany (Table 1). The cohorts are described in Supplementary Material. All cases were defined as invasive colorectal adenocarcinoma and confirmed by medical record, pathology report, or death certificate. For prospective cohorts, nested case-control sets were constructed by fixing the cohort at a timepoint upon which risk set sampling was employed to select cases and controls. For other case-control studies, population-based controls were used. For all studies, controls were matched on age, sex, race/ethnicity and in some studies on additional factors.
Table 1. Descriptive characteristics of study populations.
| Study | Design | Country | Years of Inception/Recruitment | Years of Diagnosis | Cases | Controls | Mean Age (range, yrs) | Female No. (%) | Covariates Used in Base Model Analysisa | Definition of Regular Use of Aspirin and/or NSAIDsb |
|---|---|---|---|---|---|---|---|---|---|---|
| CCFR | case-control | U.S., Canada, Australia | 1998-2006 | 1998-2006c | 1163 | 978 | 54.3 (17-81) | 1067 (49.8) | age, gender, 3 PCs, center | At least twice a week for more than a month |
| DACHS | case-control | Germany | 2003-2010 | 2003-2010 | 2339 | 2180 | 68.7 (33-99) | 1801 (39.9) | age, gender, 3 PCs | At least 1 time per month for at least one year |
| DALS | case-control | U.S. | 1991-1994 | 1991-1994 | 1115 | 1173 | 63.8 (28-79) | 1027 (44.9) | age, gender, 3 PCs, center | At least 3 times per week for at least one month |
| HPFS | cohort | U.S. | 1986 | 1986-2008 | 403 | 401 | 65.2 (48-83) | 0 (0) | age, 3 PCs | Currently taking at least 2 times per week |
| NHS | cohort | U.S. | 1976 | 1976-2008 | 553 | 955 | 59.7 (44-69) | 1508 (100) | age, 3 PCs | On average 5 or more days per month |
| OFCCR | case-control | Canada | 2000-2006 | 1998-2003 | 553 | 519 | 62.1 (29-77) | 577 (53.8) | age, gender, 3 PCs | At least twice a week for more than a month |
| PMH-CCFR | case-control | U.S. | 1998-2003 | 1998-2002d | 280 | 122 | 62.8 (48-73) | 402 (100) | age, 3 PCs | At least twice a week for more than a month |
| PLCO | cohort | U.S. | 1993-2001 | 1994-2009 | 485 | 415 | 63.6 (55-75) | 382 (42.4) | age, gender, 3 PCs, center | At least twice a week in the last 12 month |
| VITAL | cohort | U.S. | 2000-2002 | 2000-2009 | 277 | 279 | 66.5 (50-76) | 268 (48.2) | age, gender, 3 PCs | At least four or more days per week for a year |
| WHI | cohort | U.S. | 1993-1998 | 1993-2011 | 1466 | 1531 | 66.3 (50-79) | 2997 (100) | age, 3 PCs, region | At least once a week for at least the last 2 weeks |
PC: principal component
CCFR: Colon Cancer Family Registry; DACHS: Darmkrebs: Chancen der Verhütung durch Screening study; DALS: Diet, Activity and Lifestyle Study;
HPFS: Health Professionals Follow-up Study; NHS: Nurses’ Health Study; OFCCR: Ontario Familial Colorectal Cancer Registry;
PMH-CCFR: Postmenopausal Hormone study-Colon Cancer Family Registry; PLCO: Prostate, Lung, Colorectal and Ovarian Cancer Screening Trial;
VITAL: VITamins And Lifestyle; WHI: Women's Health Initiative
In the Multivariable-Adjusted Model, in addition to the covariates adjusted in the Base Model, those colorectal cancer-related factors, including smoking status (never, former, or current smoker), BMI, alcohol consumption and red meat consumptions, are also adjusted.
Includes regular use of aspirin-only, NSAIDs-only, or both aspirin and NSAIDs.
All cases diagnosed between 1998-2006, except two cases for which the year of diagnosis is 1985 and 1995
All cases diagnosed between 1998-2002, except four cases for which the year of diagnosis is 1983, 1984, 1990, 1991
Study-specific eligibility and our multi-step data harmonization procedure are described in Supplementary Material. Briefly, within each study, all exposure information, including aspirin and/or NSAID use, was collected by in-person interviews and/or structured questionnaires with the reference time for cohort studies as the time of enrollment (WHI, PLCO, and VITAL) or blood draw (HPFS and NHS). Individuals with missing aspirin and/or NSAIDs data were excluded. The precise definition of regular use of aspirin and/or NSAIDs, which was determined individually by each study cohort, is provided in Table 1. All participants provided written or verbal informed consent and studies were reviewed and approved by their respective Institutional Review Boards or ethics committees.
Statistical methods
A detailed description for genotyping, quality assurance/quality control, and imputation is provided in Supplementary Material. Average sample and SNP call rates, and concordance rates for blinded duplicates are listed in Supplementary Table 1. In brief, genotyped SNPs were excluded based on call rate (< 98%), lack of Hardy-Weinberg Equilibrium in controls (HWE, P < 1×10-4), and minor allele frequency (MAF < 5% for WHI Set 1, DALS Set 1, and OFCCR; MAF < 5 / # of samples for each other study). As imputation of genotypes is standard practice in genetic association analysis, all autosomal SNPs of each study were imputed to the CEPH collection (CEU) population in HapMap II using IMPUTE (CCFR), BEAGLE (OFCCR) and MACH (all other studies). After imputation and quality control analyses, a total of about 2.7 million SNPs were used in the analysis. To reduce heterogeneity, all analyses were restricted to samples self-reported as of European descent and clustering with Utah residents with Northern/Western European ancestry from the CEU population in principal component analysis, including the HapMap II populations as reference.
Statistical analyses were conducted centrally on individual-level data. We adjusted for age at reference time, sex, center, and racial composition using the first three principal components from EIGENSTRAT to account for population substructure. Each directly genotyped SNP was coded as 0, 1, or 2 copies of the variant allele. For imputed SNPs, we used the expected number of copies of the variant allele which provides unbiased test statistics.11 Both genotyped and imputed SNPs were examined as continuous variables (i.e., assuming log-additive effects). We analyzed each study separately using logistic regression models and combined study-specific results using fixed effect to obtain summary odds ratios (ORs) and 95% confidence intervals (95% CIs). We calculated p-values for heterogeneity using Cochran's Q test.12 Fixed effect meta-analysis is routinely used in GWAS because it is the most powerful approach for identifying disease associated variants.13,14 Furthermore, in our study fixed effect was more appropriate than random effects since the Q-Q plots and the p-value distributions indicated minimal heterogeneity across studies. Moreover, the effects may not fit a Gaussian distribution as required by the random effects model and the limited number of included studies may lead to an imprecise estimate of heterogeneity.15
To test for G X E interactions between SNPs and the regular use of aspirin and/or NSAIDs (including use of aspirin-only, NSAIDs-only, or both aspirin and NSAIDs) or the regular use of aspirin-only, we used conventional case-control logistic regression and case-only interaction analyses. Equations for the models used in the interaction analyses are provided in Supplementary Material. We examined genome-wide correlations between SNPs and use of aspirin and/or NSAIDs using linear regression analysis, and did not observe deviation from independence. For all genome-wide G X E interaction analyses, a two-sided p-value<5.0×10-8, which yields a genome-wide significance level of 0.05, was considered statistically significant.
As described in Supplementary Material, for each SNP showing G X E interaction with aspirin and/or NSAID use, we estimated the association of aspirin and/or NSAID use with colorectal cancer risk stratified by SNP genotypes, as well as associations in strata defined by SNP and aspirin and/or NSAID with one common reference group. We also estimated absolute risks associated with aspirin and/or NSAID use among individuals defined by specific genotypes based upon Surveillance, Epidemiology, and End Results (SEER) age-adjusted colorectal cancer incidence rates (Supplementary Material. All analyses were conducted using R 3.1.2.
Results
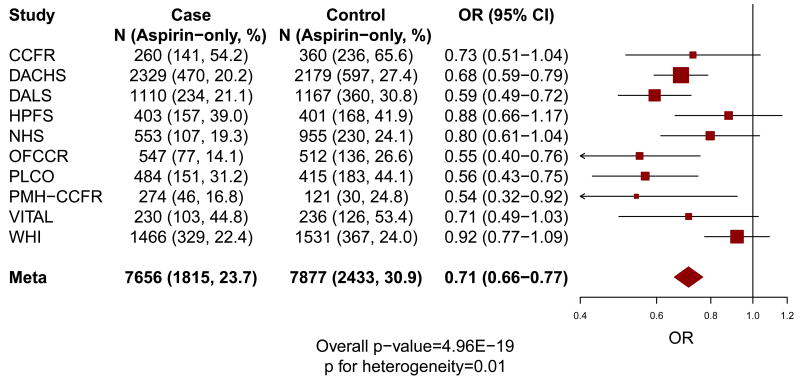
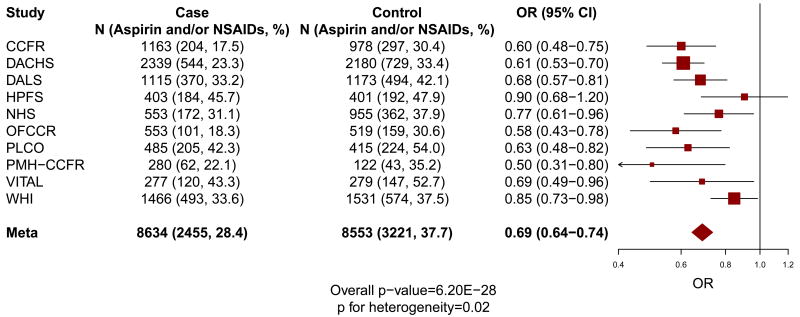
The characteristics of the 8,634 colorectal cancer cases and 8,553 controls of European descent within each cohort from the CCFR and GECCO are provided in Table 1. As shown in Figure 1, compared to non-regular use, regular use of aspirin and/or NSAIDs (OR=0.69; 95% CI=0.64-0.74; P=6.2×10-28; P for heterogeneity=0.02) or aspirin-only (OR=0.71; 95% CI=0.66-0.77; P=5.0×10-19; P for heterogeneity=0.01) was associated with lower risk of colorectal cancer.
Figure 1. Main associations of regular use of aspirin and/or NSAIDs (a) and aspirin-only (b) with the risk of colorectal cancer.
“Aspirin and/or NSAIDs” includes the regular use of aspirin-only, NSAIDs-only, or both aspirin and NSAIDs; and “Aspirin-only” includes the regular use of aspirin-only. The size of the data markers is proportional to the precision of the estimate, which is the inverse of the variance.
For the conventional logistic regression interaction analysis between each SNP and aspirin and/or NSAID use, the p-values are shown in the Manhattan plot and Q-Q plot (Supplementary Figure 1). At chromosome 12p12.3, we observed SNP rs2965667 (minor allele frequency [MAF]=1.7%) showing a genome-wide significant interaction with regular use of aspirin and/or NSAIDs (P for interaction=4.6×10-9). The second top SNP, rs10505806 (MAF=3.8%) was also found in the same locus but it did not reach genome-wide significant interaction (P for interaction=5.5×10-8). These two top SNPs (rs2965667 and rs10505806) were highly correlated (D′=1.0 and r2=0.74 in HapMap CEU). In stratified analysis, compared to non-regular use, regular use of aspirin and/or NSAIDs was statistically significantly associated with lower risk of colorectal cancer among individuals with rs2965667-TT genotype (OR=0.66; 95% CI=0.61-0.70; P=7.7×10-33), which comprised 96% (n=16,465) of the population. In contrast, a higher risk was observed among the 4% (n=722) of the population with TA or AA genotypes (OR=1.89; 95% CI=1.27-2.81; P=0.002). As expected, stratified results for the highly correlated rs10505806 were similar to those for rs2965667. Compared to non-regular use, regular use of aspirin and/or NSAIDs was statistically significantly associated with lower risk of colorectal cancer among individuals with rs10505806-AA genotype (OR=0.66; 95% CI=0.61-0.70; P=8.7×10-33), which comprised 95% (n=16,328) of the population. In contrast, a higher risk was observed among the 5% (n=859) of the population with AT or TT genotypes (OR=1.56; 95% CI=1.12-2.16; P=0.008) (Table 2/Supplementary Figure 2). Rs2965667 also appeared as the SNP with the lowest p-value in the exploratory analyses of aspirin-only, but it did not reach genome-wide significant interaction (P for interaction=8.0×10-7; P for heterogeneity=0.35) (Supplementary Table 2).
Table 2. Risk for colorectal cancer according to regular use of aspirin and/or NSAIDs, stratified by the genotypes of rs2965667, rs10505806, and rs16973225.
| rs2965667b | Non-regular aspirin and/or NSAID users | Regular aspirin and/or NSAID usersa | P-value |
|---|---|---|---|
| TT | |||
| Cases/Controls | 5,933/5,088 | 2,325/3,119 | |
| Base Model (OR)d | 1.00 | 0.66 (0.61-0.70) | 7.7×10-33 |
| Multivariable-Adjusted Model (OR)e | 1.00 | 0.63 (0.59-0.68) | 2.3×10-35 |
| TA or AA | |||
| Cases/Controls | 246/244 | 130/102 | |
| Base Model (OR)d | 1.00 | 1.89 (1.27-2.81) | 0.002 |
| Multivariable-Adjusted Model (OR)e | 1.00 | 1.76 (1.16-2.66) | 0.008 |
| P for interactionf | 4.6×10-9 | ||
|
| |||
| rs10505806b | Non-regular aspirin and/or NSAID users | Regular aspirin and/or NSAID usersa | P-value |
|
| |||
| AA | |||
| Cases/Controls | 5,896/5,039 | 2,301/3,092 | |
| Base Model (OR)d | 1.00 | 0.66 (0.61-0.70) | 8.7×10-33 |
| Multivariable-Adjusted Model (OR)e | 1.00 | 0.63 (0.59-0.68) | 4.2×10-35 |
| AT or TT | |||
| Cases/Controls | 283/293 | 154/129 | |
| Base Model (OR)d | 1.00 | 1.56 (1.12-2.16) | 0.008 |
| Multivariable-Adjusted Model (OR)e | 1.00 | 1.42 (1.01-2.00) | 0.045 |
| P for interactionf | 5.5×10-8 | ||
|
| |||
| rs16973225c | Non-regular aspirin and/or NSAID users | Regular aspirin and/or NSAID usersa | P-value |
|
| |||
| AA | |||
| Cases/Controls | 5,686/4,840 | 2,181/2,909 | |
| Base Model (OR)d | 1.00 | 0.66 (0.62-0.71) | 1.9×10-30 |
| Multivariable-Adjusted Model (OR)e | 1.00 | 0.63 (0.59-0.68) | 3.5×10-33 |
| AC or CC | |||
| Cases/Controls | 491/492 | 274/311 | |
| Base Model (OR)d | 1.00 | 0.97 (0.78-1.20) | 0.760 |
| Multivariable-Adjusted Model (OR)e | 1.00 | 0.93 (0.75-1.17) | 0.550 |
| P for interactionf | 8.2×10-9 | ||
The numbers of cases and controls were from the Base Model. For the SNP rs16973225, the total sample size is slightly smaller than in Table 1 due to missing genotype (n=3).
Regular use of aspirin-only, NSAIDs-only, or both aspirin and NSAIDs
SNPs rs2965667 and rs10505806 were identified from conventional logistic regression analysis.
SNP rs16973225 was identified from case-only interaction analysis.
ORs in Base Models are adjusted for age at the reference time, sex, center, and the first three principal components from EIGENSTRAT.
ORs in Multivariable-Adjusted Models are adjusted for age at the reference time, sex, center, the first three principal components, smoking status (never, former, or current smoker), BMI, alcohol consumption, and red meat consumption.
P-values for interactions were calculated after adjusting for age at the reference time, sex, center, and the first three principal components from EIGENSTRAT.
Both of these two highly correlated SNPs (rs2965667 and rs10505806) were imputed across all studies (100% study samples) with a mean imputation R2 of 0.7 for rs2965667 and 0.8 for rs10505806 (Supplementary Table 3). To further validate accuracy of imputation, we conducted direct genotyping of rs10505806 in participants enrolled in the NHS (553 cases and 955 controls) and the HPFS (403 cases and 401 controls). The overall concordance of the SNP rs10505806 between imputed vs. genotyped data was high (Pearson's correlation coefficient r of 0.89). Among the total 956 cases and 1,356 controls within NHS and HPFS whom we also directly genotyped rs10505806, we compared the G X E interaction statistical effect using direct genotype data with the imputed data. We confirmed no material difference in interaction estimates (P for heterogeneity=0.50) between imputed (OR=2.57; 95% CI=1.02-6.43; P for interaction=0.045) and directly genotyped (OR=2.19; 95% CI=1.04-4.59; P for interaction=0.04) data.
In case-only interaction analysis, SNP rs16973225 at chromosome 15q25.2 showed a genome-wide significant interaction with regular use of aspirin and/or NSAIDs (P for interaction=8.2×10-9). In the stratified analysis, compared to non-regular use, regular use of aspirin and/or NSAIDs was statistically significantly associated with lower risk of colorectal cancer among individuals with rs16973225-AA genotype (OR=0.66; 95% CI=0.62-0.71; P=1.9×10-30), which comprised 91% (n=15,616) of the population, but was not associated with risk of colorectal cancer among those with AC or CC genotypes (OR=0.97; 95% CI=0.78-1.20; P=0.76) (Table 2/Supplementary Figure 2), which comprised 9% (n=1,568) of the population.
The SNP rs16973225 was directly genotyped in 9 out of 15 study sets and was imputed with high quality (R2 of 0.9) in the remaining 6 study sets (38% of study samples) (Supplementary Table 3). To validate imputation of rs16973225, we compared the G X E interaction statistical effect with colorectal cancer between imputed vs. genotyped study sets in case-only interaction analysis. We found that the interaction statistical effect size was not different (P for heterogeneity=0.73) within cohorts based on imputed data (OR=1.68; 95% CI=1.30-2.17; P for interaction=4.7×10-5) compared with cohorts based on directly genotyped data (OR=1.59; 95% CI=1.28-1.97; P for interaction=4.2×10-5). In the case-only analysis of aspirin-only, we did not observe genome-wide significant interactions.
The SNP rs2965667 showing a genome-wide significant interaction with aspirin and/or NSAID use in conventional logistic regression case-control analysis also appeared as a notable variant in case-only interaction analysis, although it did not achieve a genome-wide significance level (P for interaction=7.5×10-8). Similarly, the SNP rs16973225 reaching a genome-wide significant interaction with aspirin and/or NSAID use in case-only interaction analysis also showed evidence for G X E interaction in conventional logistic regression analysis (P for interaction=2.2×10-4).
The results for the three SNPs showing G X E interaction (rs2965667, rs10505806, and rs16973225) did not materially change after adjusting for additional colorectal cancer risk factors, including smoking status, BMI, alcohol consumption, and red meat consumption (Table 2/Supplementary Table 4). For these three SNPs, we show in Supplementary Table 5 the ORs for aspirin and/or NSAID use across genotypes corresponding to 0, 1, or 2 copies of the variant allele; and in Supplementary Table 6 the ORs for each SNP by aspirin and/or NSAID use strata with one common reference group, to fully describe the interaction.
We estimated absolute risks associated with use of aspirin and/or NSAIDs among individuals with specific genotypes defined by each of these three SNPs. Compared with non-use of aspirin and/or NSAIDs, regular use of aspirin and/or NSAIDs was associated with 16.6 fewer colorectal cancer cases per 100,000 individuals with the rs2965667-TT genotype per year; 16.7 fewer colorectal cancer cases per 100,000 individuals with the rs10505806-AA genotype per year; and 16.8 fewer colorectal cancer cases per 100,000 individuals with the rs16973225-AA genotype per year. In contrast, regular use of aspirin and/or NSAIDs was associated with 34.7 additional colorectal cancer cases per 100,000 individuals with rs2965667-TA or AA genotypes per year; 21.1 additional colorectal cancer cases per 100,000 individuals with rs10505806-AT or TT genotypes per year; and only 1.5 fewer colorectal cancer cases per 100,000 with rs16973225-AC or CC genotypes per year.
Discussion
Consistent with the preponderance of experimental, epidemiologic, and clinical trial evidence,1-5 we found that aspirin and/or NSAID use was associated with overall lower risk of colorectal cancer in this large genome-wide investigation of G X E interaction which included 8,634 colorectal cancer cases and 8,553 controls. However, we identified that aspirin and/or NSAID use was differentially associated with colorectal cancer risk according to genetic variation at two highly correlated SNPs at chromosome 12p12.3 (rs2965667 and rs10505806) using a conventional logistic regression analysis.
These SNPs are 927 kb to 971 kb downstream from microsomal glutathione S-transferase 1 (MGST1) (Supplementary Figure 3), a member of the superfamily of Membrane-associated Proteins in Eicosanoid and Glutathione metabolism (MAPEG). MGST1 has high sequence homology to prostaglandin E synthase (MGST1L1), another homologue of the MAPEG family that shares 38% of its DNA sequences with MGST1.16 MGST1 and MGST1L1 are upregulated in several cancers, including colorectal cancer.17,18 MGST1L1 is coexpressed and functionally coupled to prostaglandin-endoperoxide synthase 2 (PTGS2/COX-2), and the combined activity of MGST1L1 and COX-2 increases production of proinflammatory prostaglandin E2 (PGE2), which promotes carcinogenesis through several mechanisms, including stimulation of WNT signaling, an essential oncogenic pathway of colorectal cancer.19-22 An in vitro experiment has demonstrated that NSAIDs can inhibit expression of MGST1L1 and COX-2, thereby blocking COX-2 mediated synthesis of PGE2 in human colon carcinoma cells.23 Taken together, both MGST1L1 and the closely related gene MGST1 may influence NSAID-mediated inhibition of colorectal carcinogenesis partially through involvement in the PGE2-induced WNT signaling pathway. This finding is consistent with strong biologic evidence linking genes in WNT signaling, aspirin and/or NSAIDs, and colorectal cancer.24,25
Another candidate gene in this region is LIM domain only 3 (LMO3), a known oncogene located about 686 kb upstream from rs2965667 (Supplementary Figure 3). Altered expression of LMO3 may contribute to the development of several cancers, such as neuroblastoma and lung cancer.26,27
Rs2965667 is also located about 970 kb upstream from phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 gamma (PIK3C2G) (Supplementary Figure 3). The protein encoded by PIK3C2G gene belongs to the phosphatidylinositol-4,5-bisphosphonate 3-kinase (PI3K) family, which plays a critical role in cancer.28 Experimental evidence suggests that activation of PI3K signaling enhances COX-2/PGE2 production that results in inhibition of apoptosis in colon cancer cell lines that can be restored with NSAID-mediated blockade of PI3K.29 Moreover, our previous study found that regular use of aspirin after diagnosis was associated with longer survival among the 15-30% of colorectal cancer patients with a mutation in PIK3CA, one of the PI3K family genes.30 Markedly improved survival associated with aspirin according to PIK3CA status was also found in an analysis within a separate clinical trial cohort.31 Further investigations for the joint effect of these genes would be helpful to better understand the underlying molecular mechanisms of aspirin/NSAIDs and colorectal cancer.
In the case-only interaction analysis, another SNP rs16973225 at chromosome 15q25.2 was identified with genome-wide significant association. This SNP is about 625 kb upstream of interleukin16 (IL16) (Supplementary Figure 4). As a multifunctional cytokine, IL16 plays a critical role in pro-inflammatory processes, including inflammatory bowel disease, Clostridium difficile-associated colitis, and many cancers including colorectal.32-34 Moreover, IL16 may stimulate monocyte induction of pro-inflammatory cytokines associated with tumorigenesis, including IL6 and TNF (tumor necrosis factor-α),35,36 induction of COX-2 expression, and activation of WNT signaling.36 This evidence suggests the possibility that polymorphisms in or near IL16 gene may regulate the production of inflammatory cytokines that modify the chemopreventive effect of aspirin and/or NSAIDs on colorectal cancer. It is plausible that those GWAS-identified promising loci outside of known coding regions affect more distant genes rather than the closest gene since GWAS loci may be enhancers that can influence gene expression over several hundred kilobases.37
Our study has several strengths. First, our large sample size facilitated detection of genome-wide G X E interactions, even using a conventional logistic regression or case-only interaction analysis and accounting for the stringent threshold for statistical significance. Second, we identified promising variants near genes possessing high functional plausibility given their critical roles in inflammation and prostaglandin synthesis, which have been mechanistically linked to aspirin and/or NSAID use and colorectal carcinogenesis.
We acknowledge some limitations. First, there may be heterogeneity in the definition of regular use of aspirin and/or NSAIDs and the range of time periods encompassed by each study. However, we used a standardized harmonization process on a range of environmental variables, including aspirin and/or NSAID use across 10 cohort and case-control studies. The forest plots (Figure 1) show the consistency of the association between aspirin and/or NSAID use and colorectal cancer on a per-study level and the pooled risk estimate (i.e., OR) is remarkably similar to prior studies.38 Thus, bias due to heterogeneity in the definition and time period of exposure is likely to be minimal. Second, we acknowledge that SNP rs2965667 and the highly correlated rs10505806 are relatively rare and imputed in all studies. However, we directly genotyped rs10505806 in cases and controls within two cohorts included in our study population. The high overall concordance (r=0.89) between imputed and directly genotyped data and the consistent G X E interaction statistical effect using either imputed or directly genotyped data support our assumption that our results are not greatly affected by the amount of imputed data.
Although prior GWAS-based studies have traditionally examined promising findings within a replication cohort, we did not split our data into discovery and replication sets as the most powerful analytical approach is a combined analysis across all studies.39 This approach is increasingly employed as more individual-level GWAS data are becoming available.40 Moreover, the consistency of our findings and lack of heterogeneity across distinct study cohorts provides strong evidence of validation.
Conclusions
In this genome-wide investigation of G X E interactions, use of aspirin and/or NSAIDs was associated with lower risk of colorectal cancer, and the association of these medications with colorectal cancer risk differed according to genetic variation at two SNPs at chromosomes 12 and 15. Validation of these findings in additional populations may facilitate targeted colorectal cancer prevention strategies.
Supplementary Material
Acknowledgments
We thank all study participants for making this work possible. We appreciate the efforts of the GECCO Coordinating Center ensuring the success of this collaboration. For DACHS, we thank all participants and cooperating clinicians, and Ute Handte-Daub, Renate Hettler-Jensen, Utz Benscheid, Muhabbet Celik and Ursula Eilber for excellent technical assistance. For NHS and HPFS, we acknowledge all participants and staff; Patrice Soule and Hardeep Ranu of the Dana Farber Harvard Cancer Center High-Throughput Polymorphism Core who assisted in the genotyping under the supervision of Dr. Immaculata Devivo and Dr. David Hunter; Qun Guo who assisted in programming. We also thank the following state cancer registries: AL, AZ, AR, CA, CO, CT, DE, FL, GA, ID, IL, IN, IA, KY, LA, ME, MD, MA, MI, NE, NH, NJ, NY, NC, ND, OH, OK, OR, PA, RI, SC, TN, TX, VA, WA, WY. The authors assume full responsibility for analyses and interpretation of these data. For PLCO, we thank Drs. Christine Berg and Philip Prorok, Division of Cancer Prevention, National Cancer Institute; the Screening Center investigators and staff for the PLCO Cancer Screening Trial; Mr. Tom Riley and staff, Information Management Services, Inc.; Ms. Barbara O’Brien and staff, Westat, Inc.; and Drs. Bill Kopp, Wen Shao, and staff, SAIC-Frederick. For PMH, we thank the staff of the Hormones and Colon Cancer study. For WHI, we thank the WHI investigators and staff for their dedication, and the study participants for making the program possible. A full listing of WHI investigators can be found on the WHI website. Lastly, we acknowledge COMPASS (Comprehensive Center for the Advancement of Scientific Strategies) at the Fred Hutchinson Cancer Research Center for their work harmonizing the GECCO epidemiological data set.
Grant Support
GECCO: National Cancer Institute, National Institutes of Health, U.S. Department of Health and Human Services (U01 CA137088; R01 CA059045)
CCFR: National Institutes of Health (RFA # CA-95-011) and through cooperative agreements with members of the Colon Cancer Family Registry and P.I.s. This genome wide scan was supported by the National Cancer Institute, National Institutes of Health by U01 CA122839. The content of this manuscript does not necessarily reflect the views or policies of the National Cancer Institute or any of the collaborating centers in the CFRs, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government or the CFR. The following Colon CFR centers contributed data to this manuscript and were supported by National Institutes of Health: Australasian Colorectal Cancer Family Registry (U01 CA097735), Ontario Registry for Studies of Familial Colorectal Cancer (U01 CA074783), and Seattle Colorectal Cancer Family Registry (U01 CA074794).
DACHS: German Research Council (Deutsche Forschungsgemeinschaft, BR 1704/6-1, BR 1704/6-3, BR 1704/6-4 and CH 117/1-1), and the German Federal Ministry of Education and Research (01KH0404 and 01ER0814)
DALS: National Institutes of Health (R01 CA48998 to M.L.S)
HPFS: is supported by the National Institutes of Health (P01 CA 055075, UM1 CA167552, R01 137178, R01 CA151993, and P50 CA 127003) and NHS by the National Institutes of Health (R01 CA137178, P01 CA 087969, R01 CA151993, and P50 CA 127003). Dr. Chan is also supported by K24 DK098311 and is a Damon Runyon Clinical Investigator.
OFCCR: National Institutes of Health, through funding allocated to the Ontario Registry for Studies of Familial Colorectal Cancer (U01 CA074783); see CCFR section above. As subset of ARCTIC, OFCCR is supported by a GL2 grant from the Ontario Research Fund, the Canadian Institutes of Health Research, and the Cancer Risk Evaluation (CaRE) Program grant from the Canadian Cancer Society Research Institute. Thomas J. Hudson and Brent W. Zanke are recipients of Senior Investigator Awards from the Ontario Institute for Cancer Research, through generous support from the Ontario Ministry of Research and Innovation.
PLCO: Intramural Research Program of the Division of Cancer Epidemiology and Genetics and supported by contracts from the Division of Cancer Prevention, National Cancer Institute, NIH, DHHS
PMH-CCFR: National Institutes of Health (R01 CA076366 to P.A. Newcomb)
VITAL: National Institutes of Health (K05 CA154337)
WHI: The WHI program is funded by the National Heart, Lung, and Blood Institute, National Institutes of Health, U.S. Department of Health and Human Services through contracts HHSN268201100046C, HHSN268201100001C, HHSN268201100002C, HHSN268201100003C, HHSN268201100004C, and HHSN271201100004C.
References
- 1.Chan AT, Giovannucci EL, Meyerhardt JA, Schernhammer ES, Curhan GC, Fuchs CS. Long-term use of aspirin and nonsteroidal anti-inflammatory drugs and risk of colorectal cancer. JAMA. 2005 Aug 24;294(8):914–923. doi: 10.1001/jama.294.8.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Flossmann E, Rothwell PM, British Doctors Aspirin T the UKTIAAT. Effect of aspirin on long-term risk of colorectal cancer: consistent evidence from randomised and observational studies. Lancet. 2007 May 12;369(9573):1603–1613. doi: 10.1016/S0140-6736(07)60747-8. [DOI] [PubMed] [Google Scholar]
- 3.Friis S, Poulsen AH, Sorensen HT, et al. Aspirin and other non-steroidal anti-inflammatory drugs and risk of colorectal cancer: a Danish cohort study. Cancer causes & control : CCC. 2009 Jul;20(5):731–740. doi: 10.1007/s10552-008-9286-7. [DOI] [PubMed] [Google Scholar]
- 4.Garcia Rodriguez LA, Huerta-Alvarez C. Reduced incidence of colorectal adenoma among long-term users of nonsteroidal antiinflammatory drugs: a pooled analysis of published studies and a new population-based study. Epidemiology. 2000 Jul;11(4):376–381. doi: 10.1097/00001648-200007000-00003. [DOI] [PubMed] [Google Scholar]
- 5.Rothwell PM, Wilson M, Elwin CE, et al. Long-term effect of aspirin on colorectal cancer incidence and mortality: 20-year follow-up of five randomised trials. Lancet. 2010 Nov 20;376(9754):1741–1750. doi: 10.1016/S0140-6736(10)61543-7. [DOI] [PubMed] [Google Scholar]
- 6.Hutter CM, Chang-Claude J, Slattery ML, et al. Characterization of gene-environment interactions for colorectal cancer susceptibility loci. Cancer Res. 2012 Apr 15;72(8):2036–2044. doi: 10.1158/0008-5472.CAN-11-4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hutter CM, Slattery ML, Duggan DJ, et al. Characterization of the association between 8q24 and colon cancer: gene-environment exploration and meta-analysis. BMC cancer. 2010;10:670. doi: 10.1186/1471-2407-10-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Makar KW, Poole EM, Resler AJ, et al. COX-1 (PTGS1) and COX-2 (PTGS2) polymorphisms, NSAID interactions, and risk of colon and rectal cancers in two independent populations. Cancer causes & control : CCC. 2013 Dec;24(12):2059–2075. doi: 10.1007/s10552-013-0282-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nan H, Morikawa T, Suuriniemi M, et al. Aspirin use, 8q24 single nucleotide polymorphism rs6983267, and colorectal cancer according to CTNNB1 alterations. J Natl Cancer Inst. 2013 Dec 18;105(24):1852–1861. doi: 10.1093/jnci/djt331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seufert BL, Poole EM, Whitton J, et al. IkappaBKbeta and NFkappaB1, NSAID use and risk of colorectal cancer in the Colon Cancer Family Registry. Carcinogenesis. 2013 Jan;34(1):79–85. doi: 10.1093/carcin/bgs296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jiao S, Hsu L, Hutter CM, Peters U. The use of imputed values in the meta-analysis of genome-wide association studies. Genet Epidemiol. 2011 Nov;35(7):597–605. doi: 10.1002/gepi.20608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cochran WG. The combination of estimates from different experiments. Biometrics. 1954;10:101–129. [Google Scholar]
- 13.Pfeiffer RM, G MH, Pee D. On Combining Data From Genome-Wide Association Studies to Discover Disease-Associated SNPs. Statist Sci. 2009;24(4):547–560. [Google Scholar]
- 14.Evangelou E, Ioannidis JP. Meta-analysis methods for genome-wide association studies and beyond. Nature reviews Genetics. 2013 Jun;14(6):379–389. doi: 10.1038/nrg3472. [DOI] [PubMed] [Google Scholar]
- 15.Begum F, Ghosh D, Tseng GC, Feingold E. Comprehensive literature review and statistical considerations for GWAS meta-analysis. Nucleic acids research. 2012 May;40(9):3777–3784. doi: 10.1093/nar/gkr1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prage EB, Pawelzik SC, Busenlehner LS, et al. Location of inhibitor binding sites in the human inducible prostaglandin E synthase, MPGES1. Biochemistry. 2011 Sep 6;50(35):7684–7693. doi: 10.1021/bi2010448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morgenstern R, Zhang J, Johansson K. Microsomal glutathione transferase 1: mechanism and functional roles. Drug metabolism reviews. 2011 May;43(2):300–306. doi: 10.3109/03602532.2011.558511. [DOI] [PubMed] [Google Scholar]
- 18.Nakanishi M, Gokhale V, Meuillet EJ, Rosenberg DW. mPGES-1 as a target for cancer suppression: A comprehensive invited review “Phospholipase A2 and lipid mediators”. Biochimie. 2010 Jun;92(6):660–664. doi: 10.1016/j.biochi.2010.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Castellone MD, Teramoto H, Gutkind JS. Cyclooxygenase-2 and colorectal cancer chemoprevention: the beta-catenin connection. Cancer Res. 2006 Dec 1;66(23):11085–11088. doi: 10.1158/0008-5472.CAN-06-2233. [DOI] [PubMed] [Google Scholar]
- 20.Castellone MD, Teramoto H, Williams BO, Druey KM, Gutkind JS. Prostaglandin E2 promotes colon cancer cell growth through a Gs-axin-beta-catenin signaling axis. Science. 2005 Dec 2;310(5753):1504–1510. doi: 10.1126/science.1116221. [DOI] [PubMed] [Google Scholar]
- 21.Kamei D, Murakami M, Nakatani Y, Ishikawa Y, Ishii T, Kudo I. Potential role of microsomal prostaglandin E synthase-1 in tumorigenesis. The Journal of biological chemistry. 2003 May 23;278(21):19396–19405. doi: 10.1074/jbc.M213290200. [DOI] [PubMed] [Google Scholar]
- 22.Murakami M, Naraba H, Tanioka T, et al. Regulation of prostaglandin E2 biosynthesis by inducible membrane-associated prostaglandin E2 synthase that acts in concert with cyclooxygenase-2. The Journal of biological chemistry. 2000 Oct 20;275(42):32783–32792. doi: 10.1074/jbc.M003505200. [DOI] [PubMed] [Google Scholar]
- 23.Sherratt PJ, McLellan LI, Hayes JD. Positive and negative regulation of prostaglandin E2 biosynthesis in human colorectal carcinoma cells by cancer chemopreventive agents. Biochemical pharmacology. 2003 Jul 1;66(1):51–61. doi: 10.1016/s0006-2952(03)00206-5. [DOI] [PubMed] [Google Scholar]
- 24.Dihlmann S, Siermann A, von Knebel Doeberitz M. The nonsteroidal anti-inflammatory drugs aspirin and indomethacin attenuate beta-catenin/TCF-4 signaling. Oncogene. 2001 Feb 1;20(5):645–653. doi: 10.1038/sj.onc.1204123. [DOI] [PubMed] [Google Scholar]
- 25.Gala MK, Chan AT. Molecular Pathways: Aspirin and Wnt Signaling-A Molecularly Targeted Approach to Cancer Prevention and Treatment. Clinical cancer research : an official journal of the American Association for Cancer Research. 2014 Dec 11; doi: 10.1158/1078-0432.CCR-14-0877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kwon YJ, Lee SJ, Koh JS, et al. Genome-wide analysis of DNA methylation and the gene expression change in lung cancer. Journal of thoracic oncology : official publication of the International Association for the Study of Lung Cancer. 2012 Jan;7(1):20–33. doi: 10.1097/JTO.0b013e3182307f62. [DOI] [PubMed] [Google Scholar]
- 27.Aoyama M, Ozaki T, Inuzuka H, et al. LMO3 interacts with neuronal transcription factor, HEN2, and acts as an oncogene in neuroblastoma. Cancer Res. 2005 Jun 1;65(11):4587–4597. doi: 10.1158/0008-5472.CAN-04-4630. [DOI] [PubMed] [Google Scholar]
- 28.Fresno Vara JA, Casado E, de Castro J, Cejas P, Belda-Iniesta C, Gonzalez-Baron M. PI3K/Akt signalling pathway and cancer. Cancer treatment reviews. 2004 Apr;30(2):193–204. doi: 10.1016/j.ctrv.2003.07.007. [DOI] [PubMed] [Google Scholar]
- 29.Kaur J, Sanyal SN. PI3-kinase/Wnt association mediates COX-2/PGE(2) pathway to inhibit apoptosis in early stages of colon carcinogenesis: chemoprevention by diclofenac. Tumour biology : the journal of the International Society for Oncodevelopmental Biology and Medicine. 2010 Dec;31(6):623–631. doi: 10.1007/s13277-010-0078-9. [DOI] [PubMed] [Google Scholar]
- 30.Liao X, Lochhead P, Nishihara R, et al. Aspirin use, tumor PIK3CA mutation, and colorectal-cancer survival. The New England journal of medicine. 2012 Oct 25;367(17):1596–1606. doi: 10.1056/NEJMoa1207756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Domingo E, Church DN, Sieber O, et al. Evaluation of PIK3CA mutation as a predictor of benefit from nonsteroidal anti-inflammatory drug therapy in colorectal cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2013 Dec 1;31(34):4297–4305. doi: 10.1200/JCO.2013.50.0322. [DOI] [PubMed] [Google Scholar]
- 32.Azimzadeh P, Romani S, Mohebbi SR, et al. Interleukin-16 (IL-16) gene polymorphisms in Iranian patients with colorectal cancer. Journal of gastrointestinal and liver diseases : JGLD. 2011 Dec;20(4):371–376. [PubMed] [Google Scholar]
- 33.Cruikshank WW, Kornfeld H, Center DM. Interleukin-16. Journal of leukocyte biology. 2000 Jun;67(6):757–766. doi: 10.1002/jlb.67.6.757. [DOI] [PubMed] [Google Scholar]
- 34.Gerhard R, Queisser S, Tatge H, et al. Down-regulation of interleukin-16 in human mast cells HMC-1 by Clostridium difficile toxins A and B. Naunyn-Schmiedeberg's archives of pharmacology. 2011 Mar;383(3):285–295. doi: 10.1007/s00210-010-0592-8. [DOI] [PubMed] [Google Scholar]
- 35.Grivennikov SI, Karin M. Inflammatory cytokines in cancer: tumour necrosis factor and interleukin 6 take the stage. Annals of the rheumatic diseases. 2011 Mar;70(Suppl 1):i104–108. doi: 10.1136/ard.2010.140145. [DOI] [PubMed] [Google Scholar]
- 36.Klampfer L. Cytokines, inflammation and colon cancer. Current cancer drug targets. 2011 May;11(4):451–464. doi: 10.2174/156800911795538066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Symmons O, Uslu VV, Tsujimura T, et al. Functional and topological characteristics of mammalian regulatory domains. Genome research. 2014 Mar;24(3):390–400. doi: 10.1101/gr.163519.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chan AT, Arber N, Burn J, et al. Aspirin in the chemoprevention of colorectal neoplasia: an overview. Cancer Prev Res (Phila) 2012 Feb;5(2):164–178. doi: 10.1158/1940-6207.CAPR-11-0391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Skol AD, Scott LJ, Abecasis GR, Boehnke M. Joint analysis is more efficient than replication-based analysis for two-stage genome-wide association studies. Nat Genet. 2006 Feb;38(2):209–213. doi: 10.1038/ng1706. [DOI] [PubMed] [Google Scholar]
- 40.Pearce CL, Rossing MA, Lee AW, et al. Combined and Interactive Effects of Environmental and GWAS-Identified Risk Factors in Ovarian Cancer. Cancer Epidemiol Biomarkers Prev. 2013 May;22(5):880–890. doi: 10.1158/1055-9965.EPI-12-1030-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.