Abstract
If properly translated to clinical use, our knowledge about biomarkers may lead to a more effective way of combating colorectal cancer (CRC). Biomarkers are biomolecular, genetic, or cytogenetic attributes indicative of the disease’s progression, predisposition, prognosis, or therapeutic options. For CRC, these include chromosomal instability, mutations in KRAS and TP53, loss of 18q, and elevated level of carcinoembryonic antigen (CEA), which are all associated with poor prognosis. The prognostic significance of 18q loss can be attributed to reduced expression of SMAD4, or DCC, although the chromosomal arm is actually heavily populated by genes whose downregulation correlate to worse survival. Potentially, identification of prognostic biomarkers can help the oncologist decide whether adjuvant chemotherapy is necessary after surgery. Testing for therapeutic biomarkers can be necessary if targeted therapeutics are being considered. The identification of highly penetrant predisposition markers (such as mutations in APC and MLH1) can be a lifesaver for carrier individuals, who would then have to undergo colonoscopy at an earlier age. Even sporadic CRCs may have some hereditary components, according to recent studies. Genome-wide association studies (using SNP arrays) showed that polymorphisms of certain genes can have subtle influence on CRC predisposition. Our own SNP array-based analysis suggested that long stretches of germline homozygosity (autozygosity), indicative of consanguinity, may also factor in CRC predisposition.
COLORECTAL CANCER
In 2002, there were about a million new cases of colorectal cancer (CRC) worldwide.1 The incidence was highest in the developed world, and the Western-style diet is suspected to be the primary contributing factor.2 Genetic predisposition, alcohol consumption, and smoking may also increase an individual’s chance of acquiring CRC. Approximately 70% of CRC cases lack clear genetic basis and are generally classified as sporadic. Less than 10% are due to known inherited, highly penetrant mutations (hereditary), while the rest may be due to low penetrant, though not-yet-clearly-defined genetic predisposition (familial). The 2 most common hereditary CRCs are familial adenomatous polyposis (FAP) and hereditary nonpolyposis colorectal cancer (HNPCC) caused by mutations in APC and mismatch repair (MMR) genes respectively.3
PROBING THE MOLECULAR CHANGES THAT OCCUR DURING CRC PROGRESSION
Over the years, the extensive molecular and cytogenetic characterizations of CRC tissues and cell lines, along with linkage studies on CRC-afflicted families, have paved the way to our deeper understanding of the biology behind CRC progression. Recent data generated using modern genomic tools (expression/SNP/CGH arrays, genome-wide sequencing) now remind us that CRC biology is even more complex and diverse. Nonetheless, we have solid understanding that there are 2 major genomic pathways in CRC progression: CIN (chromosomal instability), and MIN (microsatellite instability [MSI]). CIN tumors are often characterized by gross chromosomal aberrations, while MIN tumors usually retain the normal diploid state, have high mutation rate, and have CpG-island methylator phenotype. 4–6 FAP and HNPCC tumors have CIN and MIN phenotypes, respectively. Initially described was a model in which sequential accumulation of mutations, chromosomal aberrations, and gene expression dysregulation drive the progression of spontaneous CIN CRCs.7 It is now widely recognized that in CIN cases, inactivation of APC leads to accumulation of β catenin in the nucleus, elevating the transcription of genes involved in cell proliferation.8 APC is deactivated primarily by truncating mutations within the protein’s mutation cluster region (MCR) and also by copy loss and promoter hypermethylation.9 The most common mutations in sporadic CIN (but not MIN) CRCs are those of APC, TP53, and the oncogene KRAS.10,11 For sporadic MIN CRCs, the epigenetic silencing of MLH1 can result in defective MMR, leading to frequent mutations in genes such as TGFBR2, BRAF, and BAX.12–14
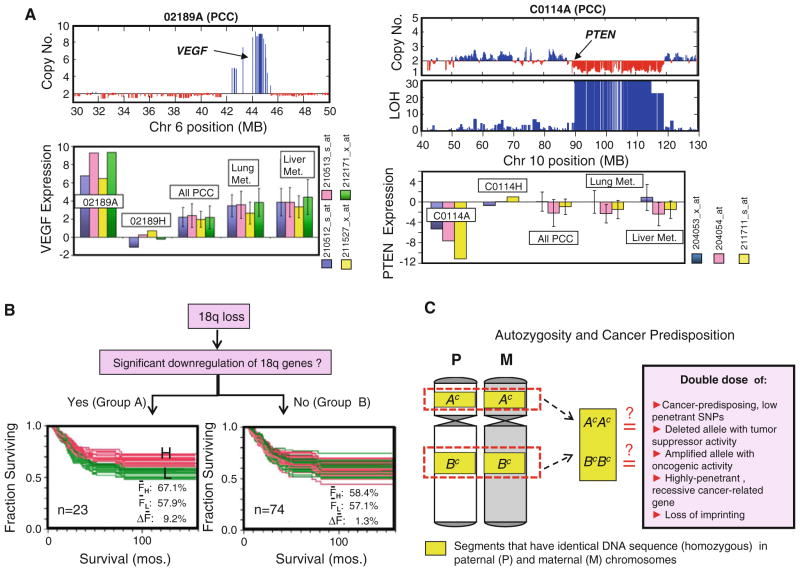
A decade ago, CRC investigators started using genome-wide expression profiling techniques to look for definitive molecular signatures that define each stage in the tumor’s progression or distinguish between MIN and CIN tumors.15,16 Finding a consensus among these studies has been difficult because of the discrepancy in technical and analytical approaches, or just the natural variability of tumors at the molecular level. In contrast, results from cytogenetic analyses using CGH, CGH arrays, as well as our own SNP array analyses have consistently pictured CIN CRCs as having frequent gains in chromosomal arms 7, 8, 13, and 20q, and losses in 4, 8p, 14q, 17p, and 18.17–20 The higher-resolution SNP and CGH arrays made possible the detection of narrower regions of aberrations that can harbor important oncogenes (e.g., VEGF in an amplified region in the 6p arm), or tumor-suppressor genes (e.g., PTEN in an LOH region in the 10q arm) (see Fig. 1a). Using SNP and expression array analyses, we were able to demonstrate that upregulated expression is common among genes in the often gained chromosomal arms (e.g., 13q, 20q), while a good percentage of genes exhibit downregulated expression in the often lost regions (e.g., 18q, 17p, 8p).19,20 Very likely, within these aberrant regions are genes whose upregulation (for oncogene) or downregulation (for tumor-suppressor gene) (see Fig. 1a) may be advantageous to the proliferating cancer cell. A copy number loss can also completely deactivate a tumor-suppressor gene (such as APC) that had previously acquired a mutation in one allele.21 Owing to its ability to simultaneously read copy number and SNP calls, SNP arrays can identify chromosomal regions with loss of heterozygosity (LOH), but with neutral copy number.22 How such regions of gene conversion/somatic uniparental disomy (UPD) can confer advantage to cancer cell is still unclear. Our hypothesis is that gene conversion is a process that can simultaneously increase the tumor-promoting activities of both oncogenes and tumor-suppressor genes within that chromosomal region.21
FIG. 1.
a SNP array analysis (Affymetrix 50 K Xba) reveals a highly amplified region in the 6p arm (which includes VEGF locus) and a deleted region in the 10q arm (which includes PTEN locus) of primary colon cancer (PCC) samples 02189A and C0114A, respectively. Genome-wide expression analysis (Affymetrix U133A) indicates that 02189A had a very highly upregulated expression of VEGF (as detected by all 4 probe sets), and C0114A had highly downregulated PTEN (3 probe sets), compared with their matching normal mucosa samples (02189H, C0114H), the rest of PCC samples (n = 182), liver metastasis samples (n = 39), and lung metastasis samples (n = 19). Gene expression level is calculated as: zt = (It − Īn)/σn; where It is the normalized, log transformed intensity value (I) of each probe set for the tumor sample, while Īn and σn are the average and standard deviation (respectively) of the I values for 53 normal mucosa samples. b The loss of 18q occurs often in CRC and has been linked to poorer prognosis. Genes within the 18q arm are arbitrarily divided into 2 groups. Group A genes (n = 23) are those with significant expression downregulation, i.e., zt ≤ −3 in at least 10% of PCC samples run in both SNP and expression arrays (n = 71), while the rest of the genes (n = 74) outside this category are classified into group B. CRC patients with expression data for PCC samples are divided into low expression group (L, green) and high expression group (H, red) for every 18q gene, prior to Kaplan–Meir (KM) analysis. Noticeable is the distinct H/L clustering pattern in the overlaid KM plots of Group A (but not in Group B) genes, suggesting that lower expression level of a number of genes (attributed to 18q loss) also correlates to poorer prognosis. The ΔF values (FH – FL; where FH and FL are the 5-year survival rates for H and L patient groups, respectively) are significantly higher in A gene group compared to B gene group (t test, P<0.001). c Possible explanations on why autozygosity contributes to cancer predisposition
BIOMARKERS FOR COLORECTAL CANCER AND THEIR POTENTIAL CLINICAL IMPORTANCE
One of the foremost goals of CRC research is to translate our knowledge about biomarkers into diagnostic tools that would aid in the clinical management of the disease. These biomarkers may be genetic mutations, SNPs, RNA levels, protein levels, and chromosomal features indicative of the extent of the disease’s progression and prognosis, or a person’s predisposition to the disease. Moreover, these markers may be known or potential therapeutic targets themselves or may influence a drug’s efficacy (e.g., drug resistance factors). Not all these biomarkers will find practical use in the clinics. Indeed, the American Society of Clinical Oncology (ASCO) in 2006 concluded that there are still insufficient data to support the clinical use of most of the CRC biomarkers described in literature.23 The difficulty of finding universal biomarkers is probably rooted in the inherent complexity of cancer: that almost every individual will have a distinct path to cancer progression, perhaps necessitating an individualized therapeutic plan (i.e., personalized medicine).
CRC Prognosis and Therapeutics
After the tumor is removed by the surgeon, the pathologist will examine the tissue specimen for pathologic staging. The latest proposal by the American Joint Committee on Cancer (AJCC) would classify CRC into either stage 0, I, II (subdivided into IIA, IIB, IIC), III (subdivided into IIIA, IIIB, IIIC), or IV.24,25 Stage I tumors (initial tumor invasion) have excellent prognosis (>90% 5-year survival rate) and do not need a follow-up chemotherapy. In contrast, chemotherapy will be administered to a great majority of stage III tumors (evidence of lymph node involvement).26 The MOSAIC clinical trial demonstrated that the addition of oxaliplatin to LV5FU2 regimen [bolus plus continuous infusion of 5-fluorouracil (5FU) and leucovorin (LV)] can improve the 6-year overall survival (OS) of stage III patients to 73% (compared with 69% for those treated with just LV5FU2).27 However, stage II patients treated with oxaliplatin plus LV5FU2 (also known as FOLFOX-4) had 6-year OS of 87%, which was not significantly different from those in the LV5FU2 group. Treatment of metastatic CRCs (stage IV) remains a great challenge (with 5-year survival rate of only about 10% at best), but the addition of the anti-EGFR cetuximab or the topoisomerase I inhibitor irinotecan to FOLFOX has produced improved outcome.28,29 Although histopathology has contributed immensely to clinical management of CRC, it is also clear that basing the clinical decision entirely on tumor pathological stage may have its drawbacks. There may be scenarios wherein a stage II patient did not receive chemotherapy, but could have benefited from it, or a stage III patient whose condition deteriorated because of drugs’ toxicity. Therefore, there is a real need to find prognostic markers that can complement, or serve as alternatives to, histopathological staging. Among these biomarkers are somatic mutations in TP53 and KRAS, elevated serum level of carcinoembryonic antigen (CEA), and tumors having CIN, all of which were found to correlate to poorer CRC prognosis.23,26
The possibility that CRC prognosis is determined by a set of gene expression dysregulated genes has been explored using genome-wide expression analyses. A number of these studies examined tumor samples from stage II or III CRC patients who have undergone surgical resection without follow-up chemotherapy. Statistical analyses were employed to identify sets of genes whose expression levels are predictive of recurrence risk. What these studies advocate is that stage II or III cases can be further stratified based on their expression profile-determined recurrence risk, with high recurrence risk patients needing postoperative chemotherapy, while those from low recurrence risk group are better off without it. Recurrence predictor sets consisting of 30, 218, and 23 genes have been reported from the analysis of 50 Stage II, 25 Dukes’ C (equivalent to stage III), and 74 Dukes’ B (equivalent to stage II) CRC samples respectively.30–32 These predictor sets, which are markedly different from each other (partly explained by the discrepancies in experimental designs and analytical approaches employed), clearly need further validations before they can be used in clinics. More recently, O’Connell and colleagues reported the analysis of CRC samples from four independent studies (1035 had only surgery, and 816 had surgery plus 5FU/LV treatment) and found that 48 genes were significantly associated with recurrence risk, and 66 genes with benefit to 5FU/LV treatment.33 Using the expression levels of 7 (BGN, FAP, INHBA, GADD45B, Ki-67, C-MYC, MYBL2) out of the 48 genes, plus 5 reference genes (ATP5E, GPX1, PGK1, UBB, VDAC2), a recurrence score (RS) was calculated as a measure of the likelihood a stage II (or III) tumor recurring after its surgical removal. This is the basis of Oncotype Dx colon cancer diagnostic test, reported to have been validated using samples from QUASAR (Quick and Simple and Reliable) clinical trial and is already being marketed as an in-house test for Stage II CRC.34,35
There is also solid evidence that microsatellite instability (oftentimes easily distinguishable from CIN phenotype) is associated with favorable clinical outcome in CRC.36,37 Among stage II/III patients whose tumors were either microsatellite stable (MSS; MIN is negative in all 5 microsatellite markers) or MSI-L (i.e., MIN is positive in only 1 of 5 microsatellite markers), the benefit from 5FU-based chemotherapy was significant.38,39 However, chemotherapy (5FU + LV, or 5FU +levamisole) did not improve the overall survival of stage II/III MSI-H (i.e., MIN is positive in at least 2 of 5 microsatellite markers) patients.38 A more recent report actually indicated that adjuvant 5FU-treatmentmay even reduce the survival rate of MSIH cases.39 At any rate, these studies suggest that 5FU’s effectiveness is limited to tumors with MMR proficiency. A recent report also indicated that the addition of oxaliplatin in the regimen (FOLFOX) can overcome this problem.40,41
At least 17 retrospective studies between 1994 and 2009 have shown that 18q loss is an indicator of poor prognosis among CIN CRCs.42–44 However, the American Society of Surgical Oncology (ASCO) Tumor Marker Expert Panel is not yet endorsing 18q loss as an independent prognostic or predictive (response to 5FU-based chemotherapy) marker in CRC because there are only a few studies in support of it (that was back in 2006).23 What is probably needed is a clinical trial that would divide stage II/III MSS patients into 18q loss and 18q intact groups, with members of each group being randomly assigned to chemotherapy and observational (surgery only) arms. Nevertheless, an ongoing clinical trial (E5202) is already trying to address the importance of the 18q arm in CRC chemotherapy.45 E5202 aims to compare the response of stage II patients considered as high risk (MSS/18q loss, or MSI-L/18q loss) to either FOLFOX or FOLFOX/bevacizumab combination.
Considered as an important event in the natural progression of CIN CRCs, 18q loss leads to allelic imbalance of the tumor-suppressor gene SMAD4, or DCC (coding for a netrin-1 receptor), which may help explain the chromosomal arm’s prognostic significance.46,47 Our analysis (integrated SNP/expression array) actually indicated that the 18q arm is populated by numerous genes whose downregulations (consequence of lost chromosomal arm) correlate to worse outcome (see Fig. 1b). These include the gene ATP5A1 (catalytic subunit of mitochondrial H+-ATP synthase), whose lower expression has actually been associated with 5FU-resistance.48
More recent studies have also shown that 8p loss is a marker for poor CRC prognosis.19,49,50 Our analysis also shows that the 8p arm (like18q) harbors a number of genes whose lower expression levels are indicative of poorer clinical outcome.42 One of these is MTUS1, a very likely tumor-suppressor gene coding for a mitotic spindle-associated protein.51–53 Another often downregulated gene within 8p is PPP2CB, which codes for the catalytic subunit of phosphatase 2A, a proven tumor-suppressor gene.54
Bevacizumab (anti-VEGF), cetuximab (anti-EGFR), and panitumumab (anti-EGFR) are now FDA approved for first (in this case bevacizumab) or second line treatments of metastatic CRCs. These monoclonal antibodies can be used alone (in the case of cetuximab and panitumumab) or as components of regimens which include non-targeted therapeutics (e.g., FOLFOX, irinotecan-based FOLFIRI).55 For cost effectiveness, it is very important to identify the characteristics of CRCs that are most (or least) responsive to these targeted therapeutics. Retrospective studies suggest that the effectiveness of anti-EGFR therapeutics is diminished if certain genes downstream of EGFR signaling pathway are altered in metastatic CRCs.56 One of these genes is KRAS, which when mutated can ensure that the MAPK proliferation pathway is still active even after EGFR inhibition. The ASCO Provisional Clinical Opinion states that CRC patients whose metastatic tumors have mutated KRAS (at codons 12 or 13) should not receive anti-EGFR therapy.57 A recent analysis of tumor samples from 172 cetuximab-treated metastatic CRC patients found that the presence of V600E BRAF mutation, or loss in expression of PTEN is associated with shorter overall survival.58
Many ongoing CRC clinical trials involve targeted therapeutics (including those meant to inhibit RAS, mTOR, VEGFR).59 It is therefore expected that in the very near future, screening for certain molecular biomarkers will be necessary prior to the administration of these therapeutics.
CRC Genetic Predisposition
Having a number of immediate family members afflicted with CRC (especially at relatively young age) should be a serious concern. Through genetic tests, members of the family will know if they are indeed carriers of the genetic predisposition and will then recognize how they may pass this down to their children. More important is the understanding that certain genetic predispositions necessitate a different type of clinical management (such as colonoscopic surveillance that is earlier than usual and more frequent). The 2 most common hereditary CRCs are due to FAP/Attenuated FAP (mutations in APC) and HNPCC (mostly mutations in MLH1 and MSH2, but also MSH6 and PMS2).3 Less common are the Hamartomatous Polyposis Syndromes: Peutz-Jeghers (mutations in STK11), Juvenile Polyposis (SMAD4, BMPR1A), Cowden (PTEN), and Bannayan-Riley-Ruvalcaba (PTEN). These aforementioned hereditary CRCs stem from autosomal dominant mutations (with varying penetrance). In contrast, MUTYH-associated polyposis (MAP) originates from a recessively inherited MUTYH mutation. Results from recent genome-wide association studies suggest that polymorphisms at the regulatory or structural regions of certain genes can have subtle effects (odds ratio [OR] = 1.1–1.26) on CRC predisposition.60 These genes include MYC, SMAD7, and CDH1. Investigations on whether copy number variations (CNV) can add to CRC risk are currently being pursued.61 Using SNP array data as well, we reported that the presence of long stretches of homozygosity in the germline genome of an individual may also add to CRC predisposition.62 These genomic regions can reflect the degree of consanguinity in someone’s ancestry and may also be referred to as autozygous. Consanguinity as risk factor in cancer is well documented in literature.21 A simple explanation (though it can be far more complex) is that autozygosity increases the chance of cancer predisposition since within these segments are genes with double doses of low-penetrant cancer-predisposition SNP, CNV, highly penetrant recessive mutation, or loss of imprinting (Fig. 1c).
USE OF BIOMARKERS TOWARD MORE EFFECTIVE MANAGEMENT OF COLORECTAL CANCER
Personalized cancer medicine entails that a patient’s treatment regimen be designed based on the tumor’s molecular signatures. Discussed previously is how retrospective studies were able to demonstrate that certain biomarkers (e.g., 18q loss, MSS) are indicative of poorer prognosis for stage II/III CRCs in the absence of chemotherapy. However, prospective validations may still be needed to convince oncologists that testing for these biomarkers is necessary before designing the most appropriate clinical management plan. For example, 18q’s prognostic value can be tested by dividing stage II/III patients into 18q-intact and 18q-loss groups, with each group being subdivided into “surgery + chemotherapy (i.e., FOLFOX)” and “surgery only” arms. Aside from validating the inherent prognostic value of a biomarker, a prospective study of this design can also examine whether the biomarker is predictive of the tumor’s response to FOLFOX (e.g., whether “18q-loss” group’s response to FOLFOX therapy differs from those of “18q-intact” group).
As more anti-CRC drugs undergo clinical trials, accompanying prognostic and predictive biomarker analyses for these therapeutics are now being recognized as necessary. For instance, if an anticancer regimen is being tested on stage II CRC patients, the state of certain prognostic biomarkers (e.g., 18q, 8p, MSS/MSI, Oncotype Dx’s 12-gene signature) should be examined and correlated with survival data. The expression levels or mutational status of genes likely to influence the drug’s activity (from preclinical studies) should also be examined. Also, the use of genome-wide expression profiling, chromosomal profiling, as well as high throughput mutational scanning may lead to discoveries of certain biomarkers associated with poor or good response to the drug(s) being tested.
The extensive use of prognostic/predictive marker tests in clinical trials may eventually lead to approval of more drugs targeting only asmall percentage of CRC patients (even if they are just outliers). Comprehensive biomarker test will soon be routinely applied after clinicopathological analysis of CRC tissues taken from surgical resection, with results determining the type of treatment the patient will be subjected to. Certainly, there are still plenty of challenges ahead. Aside from the need for prospective validations of these prognostic biomarkers, the tests have to be simplified and standard set for routine clinical use. Ultimately, a highly cost-effective CRC treatment is what we hope these accompanying prognostic and predictive biomarker tests will result in.
Acknowledgments
We thank our colleagues and collaborators at Cornell (Owen Parker, Yu-Wei Cheng, Hanna Pincas, Sarah Giardina, Richard Shattock, Jianmin Huang), Princeton (Dan Notterman, Gunter Schemmann), MSKCC (Philip Paty, Jinru Shia, Zhaoshi Zeng, Ken Offit), UMDNJ (Emmanuel Zachariah), Weizmann Institute (Eytan Domany, Dafna Tsafrir, Michal Sheffer), Columbia (Shuang Wang), and Rockefeller (Jurg Ott). National Cancer Institute (Grants P01-CA65930 and 263MQ610681), Ludwig Institute for Cancer Research/Conrad N. Hilton Foundation joint Hilton-Ludwig Cancer Metastasis Initiative, and the Gilbert Family Foundation
Contributor Information
Manny D. Bacolod, Email: mbacolod@partners.org.
Francis Barany, Email: barany@med.cornell.edu.
References
- 1.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 2.Ahmed FE. Colon cancer: prevalence, screening, gene expression and mutation, and risk factors and assessment. J Environ Sci Health C Environ Carcinog Ecotoxicol Rev. 2003;21:65–131. doi: 10.1081/GNC-120026233. [DOI] [PubMed] [Google Scholar]
- 3.Lynch HT, Lynch JF, Lynch PM, Attard T. Hereditary colorectal cancer syndromes: molecular genetics, genetic counseling, diagnosis and management. Fam Cancer. 2008;7:27–39. doi: 10.1007/s10689-007-9165-5. [DOI] [PubMed] [Google Scholar]
- 4.Lengauer C, Kinzler KW, Vogelstein B. Genetic instability in colorectal cancers. Nature. 1997;386:623–7. doi: 10.1038/386623a0. [DOI] [PubMed] [Google Scholar]
- 5.Eshleman JR, Markowitz SD. Microsatellite instability in inherited and sporadic neoplasms. Curr Opin Oncol. 1995;7:83–9. [PubMed] [Google Scholar]
- 6.Cheng YW, Pincas H, Bacolod MD, Schemmann G, Giardina SF, Huang J, et al. CpG island methylator phenotype associates with low-degree chromosomal abnormalities in colorectal cancer. Clin Cancer Res. 2008;14:6005–13. doi: 10.1158/1078-0432.CCR-08-0216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–67. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 8.Behrens J. The role of the Wnt signalling pathway in colorectal tumorigenesis. Biochem Soc Trans. 2005;33:672–5. doi: 10.1042/BST0330672. [DOI] [PubMed] [Google Scholar]
- 9.Fearnhead NS, Britton MP, Bodmer WF. The ABC of APC. Hum Mol Genet. 2001;10:721–33. doi: 10.1093/hmg/10.7.721. [DOI] [PubMed] [Google Scholar]
- 10.Sjoblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–74. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 11.Wood LD, Parsons DW, Jones S, Lin J, Sjoblom T, Leary RJ, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–13. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 12.Kane MF, Loda M, Gaida GM, Lipman J, Mishra R, Goldman H, et al. Methylation of the hMLH1 promoter correlates with lack of expression of hMLH1 in sporadic colon tumors and mismatch repair-defective human tumor cell lines. Cancer Res. 1997;57:808–11. [PubMed] [Google Scholar]
- 13.Jung B, Smith EJ, Doctolero RT, Gervaz P, Alonso JC, Miyai K, et al. Influence of target gene mutations on survival, stage and histology in sporadic microsatellite unstable colon cancers. Int J Cancer. 2006;118:2509–13. doi: 10.1002/ijc.21710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McGivern A, Wynter CV, Whitehall VL, Kambara T, Spring KJ, Walsh MD, et al. Promoter hypermethylation frequency and BRAF mutations distinguish hereditary non-polyposis colon cancer from sporadic MSI-H colon cancer. Fam Cancer. 2004;3:101–7. doi: 10.1023/B:FAME.0000039861.30651.c8. [DOI] [PubMed] [Google Scholar]
- 15.Nannini M, Pantaleo MA, Maleddu A, Astolfi A, Formica S, Biasco G. Gene expression profiling in colorectal cancer using microarray technologies: results and perspectives. Cancer Treat Rev. 2009;35:201–9. doi: 10.1016/j.ctrv.2008.10.006. [DOI] [PubMed] [Google Scholar]
- 16.Dunican DS, McWilliam P, Tighe O, Parle-McDermott A, Croke DT. Gene expression differences between the microsatellite instability (MIN) and chromosomal instability (CIN) phenotypes in colorectal cancer revealed by high-density cDNA array hybridization. Oncogene. 2002;21:3253–7. doi: 10.1038/sj.onc.1205431. [DOI] [PubMed] [Google Scholar]
- 17.Diep CB, Kleivi K, Ribeiro FR, Teixeira MR, Lindgjaerde OC, Lothe RA. The order of genetic events associated with colorectal cancer progression inferred from meta-analysis of copy number changes. Genes Chromosomes Cancer. 2006;45:31–41. doi: 10.1002/gcc.20261. [DOI] [PubMed] [Google Scholar]
- 18.Nakao K, Mehta KR, Fridlyand J, Moore DH, Jain AN, Lafuente A, et al. High-resolution analysis of DNA copy number alterations in colorectal cancer by array-based comparative genomic hybridization. Carcinogenesis. 2004;25:1345–57. doi: 10.1093/carcin/bgh134. [DOI] [PubMed] [Google Scholar]
- 19.Sheffer M, Bacolod MD, Zuk O, Giardina SF, Pincas H, Barany F, et al. Association of survival and disease progression with chromosomal instability: a genomic exploration of colorectal cancer. Proc Natl Acad Sci USA. 2009;106:7131–6. doi: 10.1073/pnas.0902232106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsafrir D, Bacolod M, Selvanayagam Z, Tsafrir I, Shia J, Zeng Z, et al. Relationship of gene expression and chromosomal abnormalities in colorectal cancer. Cancer Res. 2006;66:2129–37. doi: 10.1158/0008-5472.CAN-05-2569. [DOI] [PubMed] [Google Scholar]
- 21.Bacolod MD, Schemmann GS, Giardina SF, Paty P, Notterman DA, Barany F. Emerging paradigms in cancer genetics: some important findings from high density SNP array studies. Cancer Res. 2009;69:723–7. doi: 10.1158/0008-5472.CAN-08-3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Andersen CL, Wiuf C, Kruhoffer M, Korsgaard M, Laurberg S, Orntoft TF. Frequent occurrence of uniparental disomy in colorectal cancer. Carcinogenesis. 2007;28:38–48. doi: 10.1093/carcin/bgl086. [DOI] [PubMed] [Google Scholar]
- 23.Locker GY, Hamilton S, Harris J, Jessup JM, Kemeny N, Macdonald JS, et al. ASCO 2006 update of recommendations for the use of tumor markers in gastrointestinal cancer. J Clin Oncol. 2006;24:5313–27. doi: 10.1200/JCO.2006.08.2644. [DOI] [PubMed] [Google Scholar]
- 24.Gunderson LL, Jessup JM, Sargent DJ, Greene FL, Stewart AK. Revised TN categorization for colon cancer based on national survival outcomes data. J Clin Oncol. 28:264–71. doi: 10.1200/JCO.2009.24.0952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gunderson LL, Jessup JM, Sargent DJ, Greene FL, Stewart A. Revised tumor and node categorization for rectal cancer based on surveillance, epidemiology, and end results and rectal pooled analysis outcomes. J Clin Oncol. 28:256–63. doi: 10.1200/JCO.2009.23.9194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mutch MG. Molecular profiling and risk stratification of adenocarcinoma of the colon. J Surg Oncol. 2007;96:693–703. doi: 10.1002/jso.20915. [DOI] [PubMed] [Google Scholar]
- 27.Andre T, Boni C, Navarro M, Tabernero J, Hickish T, Topham C, et al. Improved overall survival with oxaliplatin, fluorouracil, and leucovorin as adjuvant treatment in stage II or III colon cancer in the MOSAIC trial. J Clin Oncol. 2009;27:3109–16. doi: 10.1200/JCO.2008.20.6771. [DOI] [PubMed] [Google Scholar]
- 28.Bokemeyer C, Bondarenko I, Makhson A, Hartmann JT, Aparicio J, de Braud F, et al. Fluorouracil, leucovorin, and oxaliplatin with and without cetuximab in the first-line treatment of metastatic colorectal cancer. J Clin Oncol. 2009;27:663–71. doi: 10.1200/JCO.2008.20.8397. [DOI] [PubMed] [Google Scholar]
- 29.Falcone A, Ricci S, Brunetti I, Pfanner E, Allegrini G, Barbara C, et al. Phase III trial of infusional fluorouracil, leucovorin, oxaliplatin, and irinotecan (FOLFOXIRI) compared with infusional fluorouracil, leucovorin, and irinotecan (FOLFIRI) as first-line treatment for metastatic colorectal cancer: the Gruppo Oncologico Nord Ovest. J Clin Oncol. 2007;25:1670–6. doi: 10.1200/JCO.2006.09.0928. [DOI] [PubMed] [Google Scholar]
- 30.Barrier A, Boelle PY, Roser F, Gregg J, Tse C, Brault D, et al. Stage II colon cancer prognosis prediction by tumor gene expression profiling. J Clin Oncol. 2006;24:4685–91. doi: 10.1200/JCO.2005.05.0229. [DOI] [PubMed] [Google Scholar]
- 31.Arango D, Laiho P, Kokko A, Alhopuro P, Sammalkorpi H, Salovaara R, et al. Gene-expression profiling predicts recurrence in Dukes’ C colorectal cancer. Gastroenterology. 2005;129:874–84. doi: 10.1053/j.gastro.2005.06.066. [DOI] [PubMed] [Google Scholar]
- 32.Wang Y, Jatkoe T, Zhang Y, Mutch MG, Talantov D, Jiang J, et al. Gene expression profiles and molecular markers to predict recurrence of Dukes’ B colon cancer. J Clin Oncol. 2004;22:1564–71. doi: 10.1200/JCO.2004.08.186. [DOI] [PubMed] [Google Scholar]
- 33.O’Connell MJ, Lavery I, Yothers G, Paik S, Clark-Langone KM, Lopatin M, et al. Relationship between tumor gene expression and recurrence in four independent studies of patients with stage II/III colon cancer treated with surgery alone or surgery plus adjuvant fluorouracil plus leucovorin. J Clin Oncol. 2010;28:3937–44. doi: 10.1200/JCO.2010.28.9538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gray R, Barnwell J, McConkey C, Barnwell J. Adjuvant chemotherapy versus observation in patients with colorectal cancer: a randomised study. Lancet. 2007;370:2020–9. doi: 10.1016/S0140-6736(07)61866-2. [DOI] [PubMed] [Google Scholar]
- 35.Webber EM, Lin JS, Evelyn PW. Oncotype DX tumor gene expression profiling in stage II colon cancer. Application: prognostic, risk prediction. PLoS Curr. 2010:2. doi: 10.1371/currents.RRN1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Popat S, Hubner R, Houlston RS. Systematic review of microsatellite instability and colorectal cancer prognosis. J Clin Oncol. 2005;23:609–18. doi: 10.1200/JCO.2005.01.086. [DOI] [PubMed] [Google Scholar]
- 37.Guastadisegni C, Colafranceschi M, Ottini L, Dogliotti E. Microsatellite instability as a marker of prognosis and response to therapy: a meta-analysis of colorectal cancer survival data. Eur J Cancer. 2010;46:2788–98. doi: 10.1016/j.ejca.2010.05.009. [DOI] [PubMed] [Google Scholar]
- 38.Ribic CM, Sargent DJ, Moore MJ, Thibodeau SN, French AJ, Goldberg RM, et al. Tumor microsatellite-instability status as a predictor of benefit from fluorouracil-based adjuvant chemotherapy for colon cancer. N Engl J Med. 2003;349:247–57. doi: 10.1056/NEJMoa022289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sargent DJ, Marsoni S, Monges G, Thibodeau SN, Labianca R, Hamilton SR, et al. Defective mismatch repair as a predictive marker for lack of efficacy of fluorouracil-based adjuvant therapy in colon cancer. J Clin Oncol. 2010;28:3219–26. doi: 10.1200/JCO.2009.27.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim ST, Lee J, Park SH, Park JO, Lim HY, Kang WK, et al. Clinical impact of microsatellite instability in colon cancer following adjuvant FOLFOX therapy. Cancer Chemother Pharmacol. 2010;66:659–67. doi: 10.1007/s00280-009-1206-3. [DOI] [PubMed] [Google Scholar]
- 41.Ng K, Schrag D. Microsatellite instability and adjuvant fluorouracil chemotherapy: a mismatch? J Clin Oncol. 2010;28:3207–10. doi: 10.1200/JCO.2010.28.9314. [DOI] [PubMed] [Google Scholar]
- 42.Bacolod MD, Barany F. Gene dysregulations driven by somatic copy number aberrations-biological and clinical implications in colon tumors. A paper from the 2009 William Beaumont Hospital Symposium on Molecular Pathology. J Mol Diagn. 2010;12:552–61. doi: 10.2353/jmoldx.2010.100098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lanza G, Matteuzzi M, Gafa R, Orvieto E, Maestri I, Santini A, et al. Chromosome 18q allelic loss and prognosis in stage II and III colon cancer. Int J Cancer. 1998;79:390–5. doi: 10.1002/(sici)1097-0215(19980821)79:4<390::aid-ijc14>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 44.Watanabe T, Wu TT, Catalano PJ, Ueki T, Satriano R, Haller DG, et al. Molecular predictors of survival after adjuvant chemotherapy for colon cancer. N Engl J Med. 2001;344:1196–206. doi: 10.1056/NEJM200104193441603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Benson AB., 3rd New approaches to assessing and treating early-stage colon and rectal cancers: cooperative group strategies for assessing optimal approaches in early-stage disease. Clin Cancer Res. 2007;13:6913s–20s. doi: 10.1158/1078-0432.CCR-07-1188. [DOI] [PubMed] [Google Scholar]
- 46.Tarafa G, Villanueva A, Farre L, Rodriguez J, Musulen E, Reyes G, et al. DCC and SMAD4 alterations in human colorectal and pancreatic tumor dissemination. Oncogene. 2000;19:546–55. doi: 10.1038/sj.onc.1203353. [DOI] [PubMed] [Google Scholar]
- 47.Fearon ER, Cho KR, Nigro JM, Kern SE, Simons JW, Ruppert JM, et al. Identification of a chromosome 18q gene that is altered in colorectal cancers. Science. 1990;247:49–56. doi: 10.1126/science.2294591. [DOI] [PubMed] [Google Scholar]
- 48.Shin YK, Yoo BC, Chang HJ, Jeon E, Hong SH, Jung MS, et al. Down-regulation of mitochondrial F1F0-ATP synthase in human colon cancer cells with induced 5-fluorouracil resistance. Cancer Res. 2005;65:3162–70. doi: 10.1158/0008-5472.CAN-04-3300. [DOI] [PubMed] [Google Scholar]
- 49.Liu XP, Kawauchi S, Oga A, Sato T, Ikemoto K, Ikeda E, et al. Chromosomal aberrations detected by comparative genomic hybridization predict outcome in patients with colorectal carcinoma. Oncol Rep. 2007;17:261–7. [PubMed] [Google Scholar]
- 50.Al-Mulla F, Behbehani AI, Bitar MS, Varadharaj G, Going JJ. Genetic profiling of stage I and II colorectal cancer may predict metastatic relapse. Mod Pathol. 2006;19:648–58. doi: 10.1038/modpathol.3800564. [DOI] [PubMed] [Google Scholar]
- 51.Zuern C, Heimrich J, Kaufmann R, Richter KK, Settmacher U, Wanner C, et al. Down-regulation of MTUS1 in human colon tumors. Oncol Rep. 2010;23:183–9. [PubMed] [Google Scholar]
- 52.Rodrigues-Ferreira S, Di Tommaso A, Dimitrov A, Cazaubon S, Gruel N, Colasson H, et al. 8p22 MTUS1 gene product ATIP3 is a novel anti-mitotic protein underexpressed in invasive breast carcinoma of poor prognosis. PLoS One. 2009;4:e7239. doi: 10.1371/journal.pone.0007239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Seibold S, Rudroff C, Weber M, Galle J, Wanner C, Marx M. Identification of a new tumor suppressor gene located at chromosome 8p21.3-22. FASEB J. 2003;17:1180–2. doi: 10.1096/fj.02-0934fje. [DOI] [PubMed] [Google Scholar]
- 54.Westermarck J, Hahn WC. Multiple pathways regulated by the tumor suppressor PP2A in transformation. Trends Mol Med. 2008;14:152–60. doi: 10.1016/j.molmed.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 55.Tol J, Punt CJ. Monoclonal antibodies in the treatment of metastatic colorectal cancer: a review. Clin Ther. 2010;32:437–53. doi: 10.1016/j.clinthera.2010.03.012. [DOI] [PubMed] [Google Scholar]
- 56.Modjtahedi H, Essapen S. Epidermal growth factor receptor inhibitors in cancer treatment: advances, challenges and opportunities. Anticancer Drugs. 2009;20:851–5. doi: 10.1097/CAD.0b013e3283330590. [DOI] [PubMed] [Google Scholar]
- 57.Allegra CJ, Jessup JM, Somerfield MR, Hamilton SR, Hammond EH, Hayes DF, et al. American Society of Clinical Oncology provisional clinical opinion: testing for KRAS gene mutations in patients with metastatic colorectal carcinoma to predict response to anti-epidermal growth factor receptor monoclonal antibody therapy. J Clin Oncol. 2009;27:2091–6. doi: 10.1200/JCO.2009.21.9170. [DOI] [PubMed] [Google Scholar]
- 58.Laurent-Puig P, Cayre A, Manceau G, Buc E, Bachet JB, Lecomte T, et al. Analysis of PTEN, BRAF, and EGFR status in determining benefit from cetuximab therapy in wild-type KRAS metastatic colon cancer. J Clin Oncol. 2009;27:5924–30. doi: 10.1200/JCO.2008.21.6796. [DOI] [PubMed] [Google Scholar]
- 59.Waldner MJ, Neurath MF. The molecular therapy of colorectal cancer. Mol Aspects Med. 2010;31:171–8. doi: 10.1016/j.mam.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 60.Tenesa A, Dunlop MG. New insights into the aetiology of colorectal cancer from genome-wide association studies. Nat Rev Genet. 2009;10:353–8. doi: 10.1038/nrg2574. [DOI] [PubMed] [Google Scholar]
- 61.Venkatachalam R, Ligtenberg MJ, Hoogerbrugge N, Geurts van Kessel A, Kuiper RP. Predisposition to colorectal cancer: exploiting copy number variation to identify novel predisposing genes and mechanisms. Cytogenet Genome Res. 2008;123:188–94. doi: 10.1159/000184708. [DOI] [PubMed] [Google Scholar]
- 62.Bacolod MD, Schemmann GS, Wang S, Shattock R, Giardina SF, Zeng Z, et al. The signatures of autozygosity among patients with colorectal cancer. Cancer Res. 2008;68:2610–21. doi: 10.1158/0008-5472.CAN-07-5250. [DOI] [PMC free article] [PubMed] [Google Scholar]