SUMMARY
Spinocerebellar ataxia type 1 (SCA1) is a paradigmatic neurodegenerative proteinopathy, in which a mutant protein (in this case, ATAXIN1) accumulates in neurons and exerts toxicity; in SCA1 this process causes progressive deterioration of motor coordination. Seeking to understand how post-translational modification of ATAXIN1 levels influences disease, we discovered that the RNA-binding protein PUMILIO1 (PUM1) not only directly regulates ATAXIN1 but that it also plays an unexpectedly important role in neuronal function. Loss of Pum1 caused progressive motor dysfunction and SCA1-like neurodegeneration with motor impairment, primarily by increasing Ataxin1 levels. Breeding Pum1+/− mice to SCA1 mice (Atxn1154Q/+) exacerbated disease progression, whereas breeding them to Atxn1+/− mice normalized Ataxin1 levels and largely rescued the Pum1+/− phenotype. Thus, both increased wild-type ATAXIN1 levels and PUM1 haploinsufficiency could contribute to human neurodegeneration. These results demonstrate the importance of studying post-transcriptional regulation of disease-driving proteins to reveal factors underlying neurodegenerative disease.
INTRODUCTION
Misfolded proteins underlie the pathogenesis of a number of neurodegenerative conditions, collectively known as proteinopathies. Alzheimer disease (AD), Parkinson disease (PD), amyotrophic lateral sclerosis (ALS), and polyglutamine diseases such as Huntington disease all fall into this category (Ross and Poirier, 2004; Soto, 2003). Despite the heterogeneity of their pathogenic mechanisms, in each of these diseases the misfolded protein accumulates in neurons and exerts toxicity. Somewhat surprisingly, the brain can also be sensitive to elevated levels of wild-type protein: duplication of the amyloid precursor protein (APP) locus causes autosomal dominant early-onset AD (Rovelet-Lecrux et al., 2006; Rumble et al., 1989), and duplications or triplications of α-synuclein (SNCA) are associated with familial PD (Chartier-Harlin et al., 2004; Ibanez et al., 2004; Singleton et al., 2003). Along similar lines, it has been shown recently that leucine-rich repeat kinase 2 (LRRK2) mutations, the most common cause of inherited PD, increase overall protein synthesis in Drosophila, and that reduction in dLRRK levels is protective (Martin et al., 2014).
Spinocerebellar ataxia type 1 (SCA1) is paradigmatic of the subgroup of polyglutamine (polyQ) proteinopathies caused by expansion of an unstable CAG repeat in the coding region of the relevant disease gene, in this case ATAXIN1 (ATXN1) (Orr et al., 1993). The onset of SCA1 is usually in mid-life, when motor coordination begins to deteriorate because of cerebellar degeneration; patients eventually die of bulbar dysfunction that renders them unable to clear their airway (Zoghbi and Orr, 2009). There is clear evidence that the expanded polyQ tract stabilizes ATXN1 and causes it to resist being cleared by the ubiquitin-proteasome pathway, in effect increasing its abundance in neurons (Cummings et al., 1999). Notably, the severity of neurodegeneration in fly and mouse models of SCA1 correlates directly with levels of mutant ATXN1 protein (Burright et al., 1995; Fernandez-Funez et al., 2000), and massive overexpression of even wild-type ATXN1 under the Purkinje-cell-specific promoter can produce a mild SCA1-like phenotype in mice (Fernandez-Funez et al., 2000).
Although the artificiality of transgenic models limits their relevance to the human disease, these results from SCA1 transgenic mice, along with the evidence from familial AD and PD patients, led us to ask whether there were post-translational modifications that might increase the levels of wild-type ATXN1 in a more physiologically relevant way and shed further light on the role of protein levels in neurodegeneration. The extraordinarily long 3′ UTR, approximately 7kb in ATXN1 messenger RNA (mRNA), seemed to promise a rich source of key brain-enriched post-transcriptional regulatory elements. To our surprise, we found that ATXN1 is regulated directly by an RNA-binding protein, Pumilio1, and that a brain-wide increase in wild-type Atxn1 levels of only ~50%, caused by Pum1 haploinsufficiency, is sufficient to cause marked neurodegeneration in mice.
RESULTS
The RNA-binding protein PUMILIO1 regulates ATAXIN1 levels in cells
Two types of molecules are known to modulate protein levels by binding to the corresponding mRNA: RNA-binding proteins (RBPs) and microRNAs (miRNAs). RBPs bind to specific sequence motifs or secondary structures in mRNAs and regulate multiple steps in RNA metabolism such as splicing, nucleus-cytoplasm transport and translation (Lukong et al., 2008). On the other hand, miRNAs are small non-coding RNAs that control various developmental and physiological processes by suppressing the expression of their target genes via binding of a short (6–8 nucleotide) complementary seed region in the 3′UTR of mRNAs (Bartel, 2009).
We first scanned the ~7kb-long ATXN1 3′UTR for potential binding sites for miRNAs by using the TargetScan (Friedman et al., 2009), CoMeTa (Gennarino et al., 2012) and HOCTARdb (Gennarino et al., 2011) prediction tools. As expected, scanning identified dozens of potential miRNA binding sites (data not shown). Since RNA folding mediates miRNA-RNA interactions by masking or exposing specific binding site sequences, we analyzed the secondary structure of the ATXN1-3′UTR (Wan et al., 2014) to prioritize the best candidate ATXN1-modulating miRNAs. This revealed a complicated secondary structure that masks the binding sites for almost all of the putative miRNAs that might target the ATXN1-3′UTR (Figure S1). For miRNAs to act on ATXN1 mRNA, they would likely require the help of RNA-binding proteins (RBPs) to unfold such a structure.
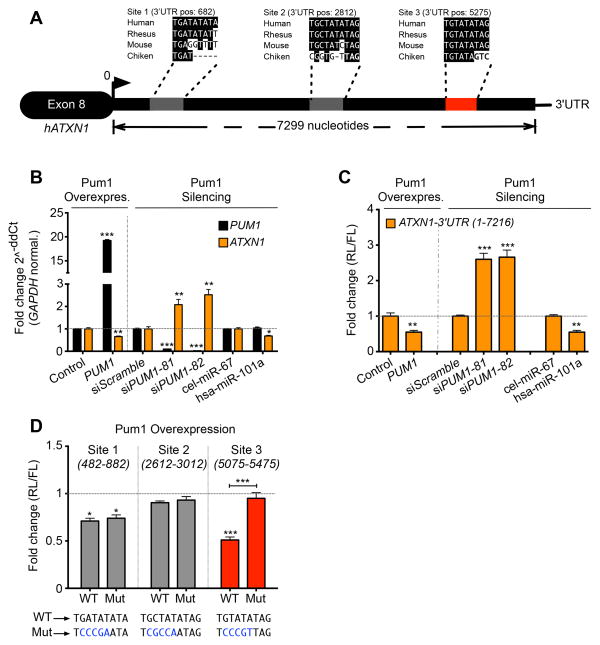
Scan analysis of the human ATXN1-3′UTR revealed three putative Pumilio1 (PUM1) binding motifs (Wang et al., 2002) at positions 682, 2812, and 5275 from the beginning of the UTR (Figure 1A). The RNA-binding protein PUM1 regulates its target genes by inducing a conformational switch in the 3′UTR that unmasks specific miRNA binding sites (Kedde et al., 2010; Li et al., 2010; Miles et al., 2012). Interestingly, the motif in position 5275 (Figure 1A, red box) is highly conserved across several species and represents the canonical PUM1 binding motif (5′-UGUAXAUA-3′) (Galgano et al., 2008; Wang et al., 2002). Overexpressing PUM1 in HEK293T cells reduced ATXN1 mRNA levels, whereas decreasing PUM1 by two different RNA-interferences (RNAi) increased ATXN1 mRNA levels (Figure 1B). In vitro overexpression of PUM1 consistently decreased the luciferase activity of a reporter construct expressing the full-length ATXN1-3′UTR (Figure 1C). Mutation of each PUM1 binding motif within the ATXN1-3′UTR revealed that only the most conserved site, containing the canonical motif, is functional; when mutated, it abolished the effect of PUM1 overexpression on luciferase activity (Figure 1D).
Figure 1. PUM1 regulates ATXN1 levels via a highly conserved binding motif.
(A) Schematic representation of Human ATXN1-3′UTR showing three putative PUM1 binding motifs (gray and red boxes) and their conservation in different species. The numbers indicate positions of PUM1 motifs in the human ATXN1-3′UTR. (B) ATXN1 mRNA quantification by qRT-PCR in HEK293T cells upon overexpression (left panel) or knockdown (siPUM1-81 and -82) (right panel) of PUM1. The destination-cloning vector (Control), scrambled siRNAs (siScr.) and cel-miR-67 were used as negative controls. The housekeeping gene GAPDH was used to normalize the expression of genes in all the qRT-PCR experiments. (C) Luciferase assay in HEK293 cells overexpressing the reporter construct harboring the full length ATXN1-3′UTR. In these conditions, we overexpressed (left panel) or decreased expression (right panel) of PUM1. (D) Luciferase assay in HEK293T cells transfected with single WT and mutant (MUT) putative PUM1 binding site on the ATXN1-3′UTR. The position of cloned regions for each PUM1 binding motif are indicated. The mutagenized nucleotides are highlighted in blue. In panels C and D the destination-vector (control), RNAi scramble (siScramble) and cel-miR-67 were used as negative controls; miR-101a was used as positive control. (RL) Renilla, (FL) Firefly luciferase. All the experiments were performed in triplicate (data represent mean ± S.E.M.). P values were calculated by Student’s t-test. Statistical significance is indicated as follows: *P<0.05, **P<0.01, ***P<0.0001. See also Figure S1.
Pum1 is widely expressed in mouse brain and regulates Atxn1 levels in vivo
To examine the endogenous expression pattern of Pum1 in mice, we performed in situ hybridization assays (ISH) and western blot on 3-week-old mouse brain sections. Pum1 was expressed in all major brain regions in wild-type mice, almost completely absent in the brain of null mice, and reduced in heterozygous (Pum1+/−) brains (Figure S2A). We also confirmed that Pum1 protein is widely expressed in the brain at 5 weeks of age (Figure S2B).
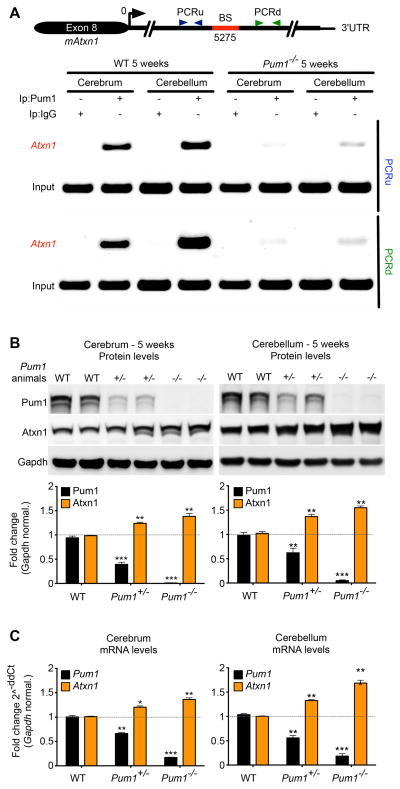
To determine whether Pum1 binds Atxn1 mRNA in vivo, we performed an RNA cross-linking and immunoprecipitation assay (RNA-Clip) on cerebrum and cerebellum from 5-week-old WT animals, using Pum1 knockout mice (Pum1−/−) as negative controls (Figure S2C). We found that Pum1 physically interacts with the conserved binding site of the Atxn1-3′UTR in WT mice (Figure 2A). Consistent with the finding that Pum1 negatively regulates Atxn1, Pum1 heterozygous (Pum1+/−) mice showed increased levels of both Atxn1 protein (Figure 2B) and mRNA (Figure 2C)—approximately 30% in the cerebrum and 50% in the cerebellum—and Pum1−/− mice showed even more pronounced increases (Figure 2B, C). These data demonstrate that Pum1 directly regulates Atxn1 levels in the mouse brain.
Figure 2. Pum1 directly binds the 3′UTR of Atxn1 to regulate its levels in mouse cerebrum and cerebellum.
(A) RNA-Clip for the conserved Pum1 binding site in mouse cerebrum and cerebellum. PCRu and PCRd highlight the PCR fragments upstream and downstream of the conserved Atxn1-3′UTR Pum1 binding site (BS). IP with IgG as well Pum1 null mice were used as negative controls. Isolated RNA from a fraction (10%) of pre-cleared lysate was used as input. The experiment was performed in triplicate. Quantification of Atxn1 protein (B) and mRNA levels (C) in WT and Pum1−/− mice in cerebrum and cerebellum (n=8 per genotype). Data represent mean ± S.E.M. and normalized to Gapdh. See Experimental Procedures for more details. P values were calculated by Student’s t-test; *P<0.05, **P<0.01, ***P<0.0001. See also Figure S2.
PUM1 controls ATXN1 levels by affecting RNA stability and not through the miRNA machinery
Several mRNA subsets contain target sites for both RBPs and miRNAs, and cooperation between these two types of post-transcriptional regulators has been described (Bhattacharyya et al., 2006; Fabian and Sonenberg, 2012; Glorian et al., 2011; Kim et al., 2009; Kundu et al., 2012). This may be particularly relevant for PUM1, as studies have indicated extensive interaction between PUM1 and the miRNA regulatory system (Kedde et al., 2010; Li et al., 2010).
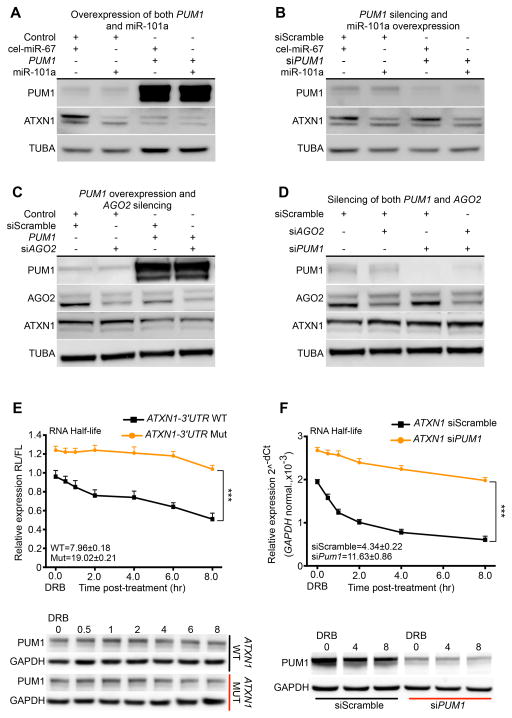
To determine whether PUM1 regulates ATXN1 through miRNA by inducing a conformational switch in its 3′UTR, we overexpressed PUM1 in HEK293T cells along with miR-101a, a miRNA known to modulate ATXN1 levels (Lee et al., 2008). These conditions significantly reduced levels of ATXN1 protein (Figure 3A and S3A) and mRNA (Figure S3E), but no more than overexpressing miR-101a or PUM1 separately. In fact, overexpression of miR-101a along with PUM1 knockdown consistently decreased levels of ATXN1 protein (Figure 3B and S3B) and mRNA (Figure S3F) to a degree comparable to that of miR-101a overexpression alone. These results suggest that PUM1 regulates ATXN1 in a miR-101a-independent fashion but do not exclude the possibility that other miRNAs bind the ATXN1 3′UTR. To obviate testing the effect of PUM1 on all possible miRNAs regulating ATXN1, we knocked down the catalytic engine of the RNA-induced silencing complex (RISC), Argonaute-2 (AGO2), to globally inhibit miRNA binding and retested PUM1’s ability to regulate ATXN1. We found that PUM1 overexpression in the context of AGO2 knockdown still reduced levels of both ATXN1 protein (Figure 3C and S3C) and mRNA (Figure S3G). Conversely, simultaneous RNAi of PUM1 and AGO2 increased levels of both ATXN1 protein (Figure 3D and S3D) and mRNA (Figure S3H), but no more than silencing PUM1 alone. These data establish that PUM1 modulates ATXN1 levels directly by binding its 3′UTR, without the assistance of the miRNA machinery.
Figure 3. PUM1 modulates the levels of wild-type ATXN1 independently of miRNAs.
Representative western blot (upper panel) of protein lysates from HEK293T cells upon: (A) Overexpression of both PUM1 and miR-101a; (B) RNAi PUM1 (siPUM1) followed by overexpression of miR-101a; (C) overexpression of PUM1 followed by RNAi AGO2 (siAGO2); and (D) RNAi of both PUM1 and AGO2. The negative controls were destination-cloning vector (Control), RNAi scramble (siScramble) and cel-miR-67. All data were normalized to α-Tubulin (TUBA). (E) mRNA half-life quantification of wild-type (WT) and Mutated (Mut) PUM1 ATXN1-3′UTR binding sites in HEK293T cells at different time points upon DRB treatment (time zero). The numeric values within the panel given the extrapolated half-life for WT and Mut RNA, P=3.6×10−06. Firefly (FL) RNA levels were quantified and normalized to Renilla (RL). Bottom panel: representative western blot of PUM1 in HEK293T cells at different time points. Data were normalized to GAPDH. (F) ATXN1 mRNA half-life quantification in HEK293T cells at different time points, from zero (DRB treatment) to 8 hours total upon RNAi of PUM1 (siPUM1) or RNAi of scramble (siScramble) transfection. The numeric values within the panel given the extrapolated half-life for siPUM1 and siScramble RNA, P=0.012. Bottom panel: representative western blot of PUM1 in HEK293T cells at different time points. Data were normalized to GAPDH mRNA (top panel) or protein (bottom panel). All experiments were performed in triplicate (data represent mean ± S.E.M.); ***P<0.0001. See also Figure S3.
To further explore the mechanism by which PUM1 regulates ATXN1 levels, we tested whether PUM1 influences the stability or the translation of ATXN1 mRNA. We transfected HEK293T with a luciferase reporter encoding an ATXN1-3′UTR harboring either the conserved wild-type (WT) or mutated (Mut) PUM1 binding site. Later, we used treatment with DRB (5,6-dichloro-1-β-D-ribofuranosylbenzimidazole), a drug that inhibits RNA translation by blocking RNA polymerase II in the early elongation stage, to assess the levels of the reporter transcript. Upon the addition of DRB (time-point zero), the relative expression of reporter transcripts containing the ATXN1-3′UTR Mut binding site is considerably higher than that of transcripts containing the ATXN1-3′UTR WT binding site (Figure 3E top panel). This difference remains stable over time until 8 hours after DRB addition. Remarkably, ATXN1-3′UTR with Mut binding site reached its half-life after 19 hours, whereas ATXN1-3′UTR with WT binding site decreased linearly over time, reaching its half-life at nearly 8 hours (Figure 3E top panel). Given that the promoter sequences of the ATXN1-3′UTR constructs carrying either WT or Mut binding sites are exactly the same and that transfection of neither construct affected PUM1 protein levels, we conclude that PUM1 promotes degradation of ATXN1 by binding its 3′UTR (Figure 3E bottom panel and S3I).
To investigate physiological changes in ATXN1 mRNA, we decided to knock down PUM1 in HEK293T cells and measure the half-life of endogenous ATXN1 mRNA at different time points after DRB treatment. Knockdown of PUM1 (siPUM1) was associated with a significant increase of ATXN1 mRNA from time zero and remained upregulated up to 8 hours after translation inhibition (Figure 3F top panel). Our calculation consistently showed that the half-life of ATXN1 mRNA was much longer (nearly 12 hours) after siPUM1 than after siScramble transfection (~4 hours) (Figure 3F top panel). We confirmed PUM1 downregulation by quantifying mRNA at time zero (Figure S3J) and protein levels at different time points (Figure 3F bottom panel and S3K). PUM1 thus increases ATXN1 levels by directly regulating the stability of ATXN1 mRNA.
Pum1 mutant mice develop progressive motor dysfunction and neurodegeneration
Recent studies have shown that Pum1 is an essential regulator of spermatogenesis in mice and promotes differentiation of embryonic stem cells (Chen et al., 2012; Leeb et al., 2014), but its role in the mammalian nervous system has not been investigated. We therefore characterized the brain structure and behaviour of Pum1 knockout mice (Chen et al., 2012).
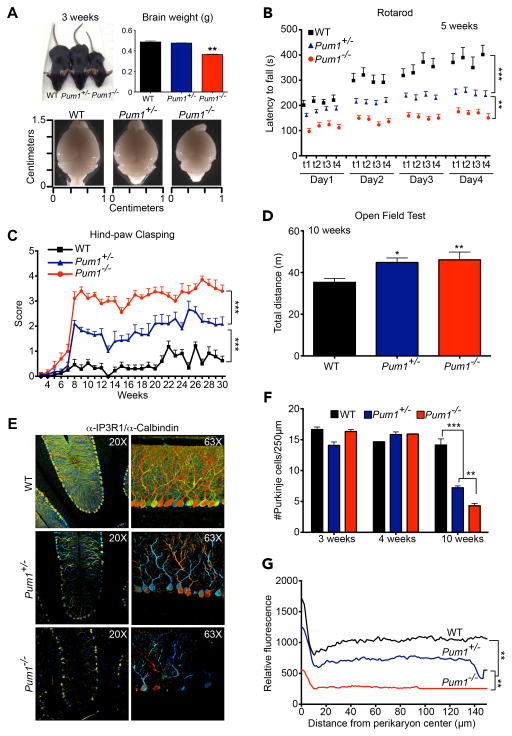
The Pum1 null allele tends to be transmitted with an altered Mendelian ratio (Figure S4A). Compared to WT and Pum1+/− littermates, Pum1−/− mice were significantly smaller in body length, weight, and brain weight and size (Figure 4A, Figure S4B). Surprisingly, the loss of one copy of Pum1 was sufficient to cause impaired performance on the accelerating rotarod assay in 5-week-old mice (Figure 4B): the motor deficit had progressed in severity by 12 weeks (Figure S4C). This motor incoordination was even more dramatic in Pum1−/− age-matched mice (Figure 4B, S4C), which performed equally poorly in the dowel-walking test (Figure S4D, E)—as poorly, in fact, as SCA1 mice at this age in both assays (Watase et al., 2002).
Figure 4. Pum1 mutant mice develop progressive motor deficits and cerebellar degeneration.
(A) Representative pictures of 3-week-old mice. Body size, brain weight, and brain size are reduced in Pum1−/− animals. (B) Accelerating rotarod analysis. Mice were trained over 4 days with four trials (t) per day. The null mice were significantly different from wild-type from Day 1; by Day 2 the difference between wild-type and both Pum1 null and heterozygotes was statistically significant, as was the difference between the two mutants. (C) Hind-paw clasping analysis in mice: a higher score indicates a more severe phenotype (See Figure S4M bottom panel for scoring details). By 6 weeks of age the null mice were statistically different from wild-type; by 8 weeks, both mutant lines were statistically significantly different from wild-type. (D) Open field test measuring the total distance travelled of the Pum1 null mice relative to wild-type. (E) Representative images of immunofluorescence (IF) confocal microscopy in 3D depth-coding (see Experimental Procedures). Co-staining with α-IP3R1 and -Calbindin antibodies was used to label Purkinje cells and to reveal their arborization. (F) Purkinje cell counts at 3-, 4- and 10-week-old for all examined genotypes. (G) IF for Calbindin and IP3R1 were quantified and averaged in selected rectangular cerebellar subsections. All experiments were performed in WT, Pum1+/− and Pum1−/− mice. More than 18 mice per genotype were considered in (A), (B) (C) and (D) and 6 per genotype in (E), (F) and (G). Data in (A), (D) and (F) represent mean ± S.E.M.; *P<0.05, **P<0.01, ***P<0.0001. See also Figure S4.
Beginning at 8 weeks of age, both Pum1+/− and Pum1−/− mice exhibited hind-paw clasping when suspended by the tail (Figure 4C and Figure S4F), a sign of neurological dysfunction. At 10 weeks of age, Pum1−/− mice displayed significantly less vertical activity in an open field chamber (Figure S4G) and spent less time in its center (Figure S4H) but travelled greater distances over 30 minutes in the chamber than wild-type (Figure 4D). Interestingly, Pum1−/− mice covered a greater distance at 18 weeks than at 10 weeks of age (Figure S4I). Using the DigiGait assay, we found that 12-week-old Pum1−/− mice had a wider stance (Figure S4J), shorter stride length (Figure S4K), and greater stride frequency (Figure S4L). At the same age, Pum1+/− and Pum1−/− mice were poor nest builders (Figure S4M). Pum1 deficiency thus causes progressive loss of motor coordination that appears to be cerebellar in origin.
To uncover the defects underlying the phenotype, we performed neuropathological studies. At 3 and 4 weeks of age there was no evidence of Purkinje cell pathology in Pum1+/− or Pum1−/− mice (Figure 4F and S4N), but by 10 weeks Pum1 haploinsufficiency had caused loss of Purkinje cells (Figure 4E and F) and dendritic arborization (Figure 4G). Both defects were more dramatic in age-matched Pum1−/− mice (Figure 4E-G). The neuronal loss is thus a result of neurodegeneration and not a developmental defect. Notably, progressive Purkinje cell degeneration and motor dysfunction are a hallmark of SCA1 in both human patients and the SCA1 knock-in mouse model (Atxn1154Q/+) (Watase et al., 2002). Because the progressive defects in Pum1 mutant mice were reminiscent of those observed in SCA1 mice (Watase et al., 2002), we decided to dissect the genetic interaction between Pum1 and SCA1 mice.
Pum1 haploinsufficiency worsens the phenotype of the SCA1 knock-in mouse
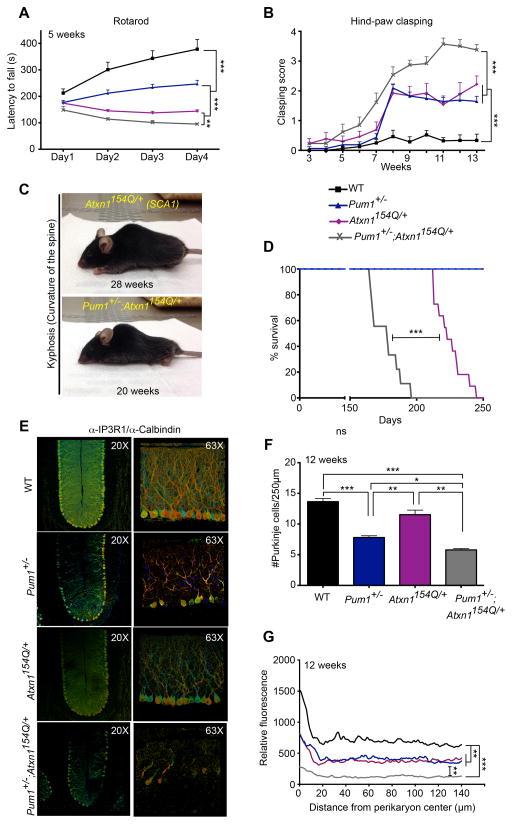
Given that Atxn1 levels were increased in Pum1+/− and in Pum1−/− mice (Figure 2B), we predicted that halving the dosage of Pum1 in the SCA1 knock-in mice (Pum1+/−;Atxn1154Q/+) would exacerbate the SCA1 phenotype, and this proved to be the case. Pum1+/−;Atxn1154Q/+ mice tended to be smaller than Atxn1154Q/+ and the other genotypes (lower body and brain weight, shorter length, smaller brain size; Figure S5A, B) and showed more severe motor incoordination and hind-paw clasping than either Atxn1154Q/+ or Pum1+/− mice (Figure 5A, B). Notably, Pum1+/−;Atxn1154Q/+ mice began to show the hind-paw clasping phenotype at 6 weeks, much earlier than age-matched Atxn1154Q/+ mice or Pum1 single mutants (Figure 5B). At 10 weeks of age Pum1+/−;Atxn1154Q/+ mice travelled greater distances than wild-type and Atxn1154Q/+ but not more than Pum1+/− mice (Figure S5C). Severe kyphosis (curvature of the spine) developed in Pum1+/−;Atxn1154Q/+ mice eight weeks earlier than SCA1 knock-in mice (20 vs. 28 weeks; Figure 5C), confirming the accelerated disease course. In addition, Pum1+/−;Atxn1154Q/+ mice had a significantly shorter life span than their Atxn1154Q/+ littermates (Figure 5D). At 12 weeks of age the Purkinje cell loss (Figure 5E, F) and arborization defects (Figure 5G) were more dramatic in Pum1+/−;Atxn1154Q/+ than all other genotypes. These results suggest a genetic interaction between Pum1 and Atxn1154Q.
Figure 5. Pum1 haploinsufficiency exacerbates SCA1 disease progression.
A 50% reduction of Pum1 in Atxn1154Q/+ mice aggravates (A) motor incoordination on the accelerating rotarod at 5 weeks (n=12 per genotype) and (B) hind-paw clasping when suspended by the tail (n>12 mice per genotype). The double mutants were statistically different from wild-type beginning at week 6. See Experimental Procedures for details. (C) Representative picture of SCA1 and Pum1+/−;Atxn1154Q/+ mice. Pum1+/−;Atxn1154Q/+ showed severe kyphosis (curvature of the spine) at earlier stages than their SCA1 counterparts. (D) Haploinsufficiency of Pum1 reduces life span in SCA1 background mice. P value was calculated by long rank test; **P<0.01, ***P<0.0001. (E) Representative images of immunofluorescence (IF) confocal microscopy in 3D depth-coding (see Experimental Procedures). Co-staining with α-IP3R1 and -Calbindin antibodies was used to label Purkinje cells and to reveal their arborization. (F) Purkinje cell count at 12 weeks for all examined genotypes. (G) IF for Calbindin and IP3R1 were quantified and averaged in selected rectangular cerebellar subsections. Four genotype per mice in (E), (F) and (G) were considered. Data in (F) and (G) represent mean ± S.E.M.; *P<0.05, **P<0.01, ***P<0.0001. See also Figure S5.
Genetic reduction of Atxn1 levels rescued the Pum1 mutant phenotype
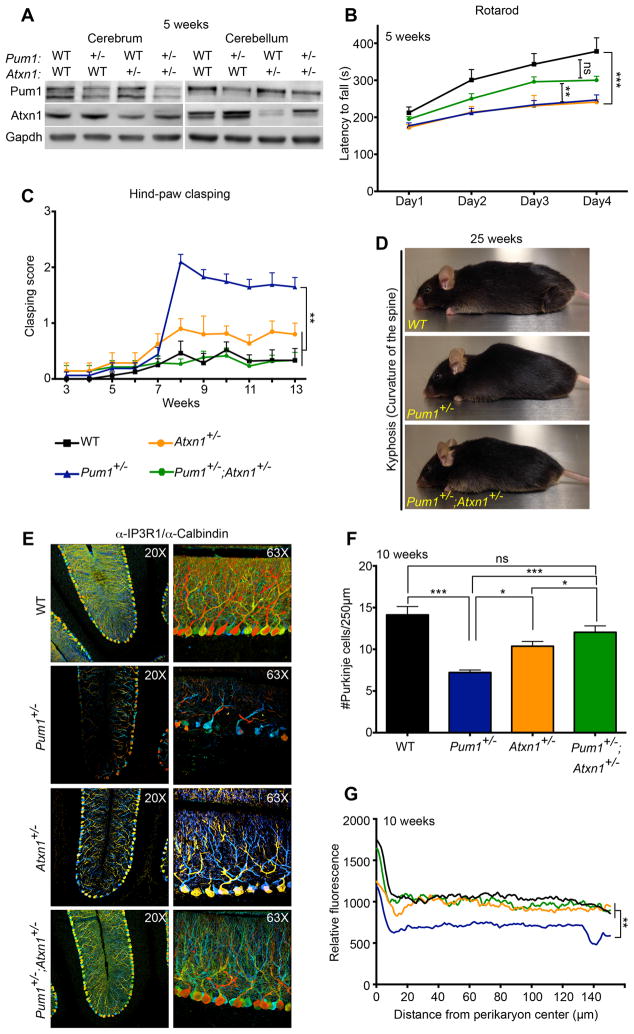
To test our hypothesis that the neurological deficits of Pum1 mutant mice resulted from an increase in Atxn1 levels due to loss of Pum1 regulation, we crossed Pum1+/− with Atxn1+/− mice and characterized the offspring. We first confirmed that Pum1 haploinsufficiency in the Atxn1+/− mice (Pum1+/−;Atxn1+/−) at 5 weeks completely rescued the physiological protein levels of Atxn1 (Figure 6A, S6A and B). At 9 weeks, Pum1+/−;Atxn1+/− mice showed no difference in body weight (Figure S5A), length, brain weight or size relative to any other genotype (Figure S6C). Atxn1 haploinsufficiency in Pum1 mutant mice significantly mitigated the motor deficits observed in Pum1+/− animals at 5 weeks of age (Figure 6B) and completely rescued the hind-paw clasping (Figure 6C) and kyphosis, which occurs at a later age of ~25 weeks (Figure 6D). Interestingly, Pum1+/−;Atxn1+/− mice still travelled farther than other genotypes and not less than Pum1+/− mice (Figure S6D). The Purkinje cell loss (Figure 6E and F) and arborization defects (Figure 6G) typical of 10-week-old Pum1+/− mice were rescued in Pum1+/−;Atxn1+/− cerebellum. This indicates that, whatever the other pathways affected by Pum1 deficiency, the cerebellar degeneration, clasping, and kyphosis are caused primarily by the influence of Pum1 on Atxn1 levels.
Figure 6. Atxn1 haploinsufficiency rescues motor deficits and cerebellar pathology in Pum1+/− mice.
(A) Representative western blot showing that haploinsufficiency of Pum1 restores physiological Atxn1 protein levels. All experiments were performed in triplicate in cerebrum and cerebellum from mice at 5 weeks of age (data represent mean ± SD). All data were normalized to Gapdh. Pum1+/−;Atxn1+/− mice showed (B) significant improvement in motor performance on the accelerating rotarod (n=12 per genotype), (C) reduced hind-paw clasping (n>12 per genotype), and (D) reduced kyphosis (curvature of the spine; photo taken at 25 weeks). Purkinje cells loss (E–F) and loss of dendritic arborization (G) were rescued in Pum1+/−;Atxn1+/− mice (n=6 per genotype). Staining was performed with Calbindin/IP3R1 in 3D-deepth-coding images. Data represent mean ± S.E.M. See Experimental Procedures. P values was calculated by Student’s t-test. ns=not significant; *P<0.05, **P<0.01, ***P<0.0001. See also Figure S6.
Lastly, we analyzed whether Pum1 overexpression could decrease levels of mutant (polyglutamine-expanded) Atxn1 (154Q). We used AAV8 viral injection and found that, indeed, Atxn1154Q/+ mice injected with Pum1/AAV8 showed a reduction of both WT and mutant Atxn1 in the cerebellum, the region most affected in SCA1, compared to controls (Atxn1154Q/+ animals injected with YFP/AAV8; Figure S6E).
DISCUSSION
Accumulation of mutant proteins in the brain has been known for some time to underlie the progression of neurodegenerative disorders such Alzheimer disease (AD), Parkinson disease (PD), Huntington disease (HD), amyotrophic lateral sclerosis (ALS), and the spinocerebellar ataxias. In all these diseases, the mutant proteins form insoluble aggregates (Haass and Selkoe, 2007; Klement et al., 1998; Ross and Poirier, 2004; Zoghbi and Orr, 2000). Considerable attention has been devoted to the question of whether inhibiting aggregate formation or promoting their dissolution would mitigate disease (Arrasate et al., 2004; Bowman et al., 2007; Bowman et al., 2005). The aggregates in all these proteinopathies appear late in the disease course, however, and clearly result from processes that have been taking place for decades. Here we asked how protein levels affect the brain long before aggregates form. Specifically, we sought post-transcriptional regulatory mechanisms that regulate wild-type ATXN1 levels independently of the polyglutamine tract in order to determine whether reducing protein levels might delay disease progression. Our studies have unexpectedly revealed two candidate genes for neurodegenerative conditions in humans: wild-type (but upregulated) ATAXIN1 and PUMILIO1.
We began our investigation by scanning the long 3′UTR of ATXN1 to identify regulatory elements that could be used to modulate ATXN1 levels. We found three PUM1 binding motifs in the 3′UTR of ATXN1, one of which is highly conserved. Using mutagenesis and RNA-Clip we showed that Pum1 regulates Atxn1 levels by binding directly to the highly conserved motif in its 3′UTR.
Pum1 is a member of a well-characterized family of RBPs, known as the PUF family, which are involved in various physiological processes (Spassov and Jurecic, 2003; Wickens et al., 2002). A typical feature of these proteins is the presence of an RNA-binding Pumilio homology domain (PUM-HD) that binds a highly conserved eight-nucleotide motif (Galgano et al., 2008). PUF proteins regulate mRNA stability by several mechanisms leading to mRNA instability or translational repression (Goldstrohm et al., 2006; Suh et al., 2009). One of the most well-studied mechanisms for PUM1 activity, however, involves the miRNA machinery: PUM1 modifies the secondary structure of the 3′UTR of its target mRNAs to allow regulation through specific miRNAs (Fabian and Sonenberg, 2012; Friend et al., 2012; Galgano et al., 2008; Kedde et al., 2010; Li et al., 2010; Miles et al., 2012). It was thus surprising to find that PUM1 directly regulates ATXN1 mRNA stability without harnessing the miRNA regulatory system.
Equally unexpected was the discovery of a role for Pum1 in the maintenance of proper brain structure and neurological function. We found Pum1 expressed in all brain regions, but the deficits we observed in the heterozygous mice—progressive motor incoordination, hind-paw clasping, kyphosis, Purkinje cell and dendritic degeneration—were reminiscent of the SCA1 mouse phenotype. The Pum1 null mice phenocopied the SCA1 knock-in mice but developed even more severe Purkinje cell pathology, showing significant neuronal loss after only two months. We believe this is explained at least in part by constitutive ~50% increase in WT Atxn1 in contrast with the gradual nature of the accumulation of polyglutamine-expanded proteins. Even in the SCA1 knock-in mice, the mutant protein takes time to accumulate to levels that produce symptoms. In the Pum1 mutants, however, the levels of Atxn1 are elevated from the very beginning of life.
The dramatic exacerbation of disease progression in SCA1 knock-in mice lacking a copy of Pum1 indicates a genetic interaction between Pumilio1 and Ataxin1, but the precocity of disease symptoms in the double mutants could conceivably arise through either of two mechanisms: the loss of one copy of Pum1 directly increasing levels of mutant Atxn1, or an additive effect ascribable to the combination of two severe mutations (loss of Pum1 and the CAG expansion in Atxn1). Our results argue for the former possibility. First, the defects observed in Pum1 mutant mice were largely corrected by reducing Atxn1 levels. Second, the Pum1+/−;Atxn1+/− mice were healthier than either Pum1+/− or Atxn1+/− mice. Third, we found that viral overexpression of Pum1 in the mouse brain reduced both the WT and expanded[154Q] forms of the Atxn1 protein (Figure S6E). These three facts support the notion that the neurological deficits exhibited by the Pum1 mutant mice are caused primarily by a rise in wild-type Atxn1 levels, even though other Pum1 targets are undoubtedly affected as well. Without knowing the full set of Pum1 targets we cannot rule out all other pathways, but we were able to evaluate levels of two well-studied PUM1 targets, E2F3 and p27, in Pum1+/−as well as Pum1+/−;Atxn1+/− mice. We did not find any significant change in the levels of these two proteins in the cerebella of either Pum1+/− or Pum1+/−;Atxn1+/− mice compared to other genotypes (data not shown).
It is well established that the severity of neurodegeneration in SCA1 correlates with the levels of expanded (or even wild-type) ATXN1 and that decreasing ATXN1 accumulation can reverse the disease phenotypes in SCA1 models (Fernandez-Funez et al., 2000; Park et al., 2013). It has been only relatively recently, however, that we have understood that in some neurodegenerative diseases, such as AD and PD, too much of the wild-type protein can produce the same phenotype as the mutant protein (Chartier-Harlin et al., 2004; Ibanez et al., 2004; Rovelet-Lecrux et al., 2006; Rumble et al., 1989; Singleton et al., 2003). Our first study of transgenic mice overexpressing wild-type human ATXN1[30Q] under the control of a Purkinje-cell-specific promoter (Pcp2-Atxn1-(CAG)30Q) failed to reveal cerebellar pathology or ataxia (Burright et al., 1995), but our later work monitoring the mice throughout their lifespan revealed that Pcp2-Atxn1-(CAG)30Q mice develop mild Purkinje cell degeneration in later life (Fernandez-Funez et al., 2000). This suggested that only dramatic overexpression of wild-type ATXN1 would be neurotoxic, but the artificiality of the transgenic model—ATXN1 cDNA was massively and postnatally expressed only in cerebellar Purkinje cells and without the 3′UTR—limited its relevance for human patients. Here we introduce the evidence that a moderate (30–60%) increase in the levels of endogenous wild-type Atxn1, expressed in the correct temporal and spatial pattern throughout the brain and preserving all its regulatory elements, is deleterious to neuronal function.
Atxn1 is expressed throughout the brain (Figure S2A) from very early embryonic stages (Banfi et al., 1996; Banfi et al., 1994; Servadio et al., 1995). The present study suggests that mutations in PUM1 or copy number changes in Atxn1 could cause cerebellar neurodegeneration in humans by increasing the levels of ATXN1 throughout development. Variations in Pum1 or other factors that govern ATXN1 levels could also underlie the individual differences in SCA1 onset for the same CAG repeat length.
In conclusion, we propose that identifying molecules capable of regulating ATXN1 levels provides insight into factors that contribute to cerebellar degeneration We further propose that studying factors that regulate the RNA stability of proteins such as APP, TAU or a-SYN might uncover candidate genes as well as binding sites whose mutation could lead to Alzheimer or Parkinson disease—two diseases for which our understanding of molecular genetic causes is still very limited. For these and the ever-lengthening list of neurodegenerative conditions that do not fit Mendelian categories, it may prove most fruitful to search for factors that elevate the levels of key disease-driving proteins.
EXPERIMENTAL PROCEDURES
Bioinformatic analysis
The ATXN1-3′UTR was downloaded from UTRdb (Grillo et al., 2010). The ATXN1 3′UTR was scanned against CoMeTa (Gennarino et al., 2012), HOCTARdb (Gennarino et al., 2011) and TargetScan (Friedman et al., 2009) to identify all putative miRNAs regulating ATXN1. The secondary structure of ATXN1-3′UTR was calculated with the Vienna RNAfold (Gruber et al., 2008) package by using default parameters in a Minimum Free energy “MFE” (Zuker and Stiegler, 1981) and Boltzmann ensemble “Centroid” (Hofacker and Stadler, 2006). ATXN1-3′UTR was scanned against all know RNA-binding protein motifs downloaded from the database of RNA-binding protein specificities (RBPDB) (Cook et al., 2011).
Cell Culture and Transfection
The Human embryonic kidney immortalized 293 cells (HEK293T) were grown in Dulbecco’s Modified Eagle’s Medium (DMEM, Invitrogen), supplemented with 10% of heat-inactivated Fetal Bovine Serum (FBS) and penicillin/streptomycin. All cells were incubated at 37°C in a humidified chamber supplemented with 5% CO2. Transfection of HEK293T cells was performed using jetPRIME Transfection Reagent (Polyplus transfection) according to the manufacturer’s protocol. Cells were transfected with 50 pmol of either miRIDIAN Dharmacon microRNA Mimics (miR-101a or negative control cel-miR-67) or Ambion small-interfering RNA (siAGO2, siPUM1 or scramble-siRNA control). For overexpression studies, the full cDNA of PUM1 (4635nt) was amplified by Platinum Taq DNA Polymerase High Fidelity (Invitrogen) and cloned into a mammalian expression vector termed pcDNA3.1(+) (Invitrogen). Cells were transfected with 0.5μg of either pcDNA3.1(+)-PUM1 or control pcDNA3.1(+).
RNA Extraction and Quantitative real-time PCR
HEK293T cells were seeded in six-well plates before transfection. After 48 hrs, cells were collected and processed for RNA extraction. Total RNA was obtained using the miRNeasy kit (Quiagen) according to the manufacturer’s instructions. RNA extraction from mouse cerebrum or cerebellum was extracted from 75 mg of tissue. RNA was quantified using the NanoDrop 1000 (Thermo Fisher). Quality of RNA was assessed by gel electrophoresis. cDNA was synthesized using Quantitect Reverse Transcription kit (Quiagen) starting from 1 μg of DNase-treated RNA. Quantitative RT-polymerase chain reaction (qRT-PCR) experiments were performed using the CFX96 Touch Real-Time PCR Detection System (Bio-Rad Laboratories) with PerfeCta SYBR Green FastMix, ROX (Quanta Biosciences). Real-time PCR results were analyzed using the comparative Ct method normalized against the housekeeping gene GAPDH (Vandesompele et al., 2002). The range of expression levels was determined by calculating the standard deviation of the ΔCt (Pfaffl, 2001).
Luciferase Assay
The full-length 3′UTR of human ATXN1 mRNA was subcloned into psiCHECK-2 vector (Promega) by XbaI and NheI restriction enzymes (Lee et al., 2008). The partial 3′UTR, containing binding sites 1 (582-782), 2 (2712-2912) and 3 (5175-5375), was amplified by PCR and cloned into psiCHECK-2 vector (Promega). Mutagenesis reactions were performed using the QuikChange XL Site-Directed Mutagenesis kit (Stratagene). Primers for mutagenesis analysis were automatically designed by QuickChange software (Stratagene). HEK293T cells in 24-well plates were transfected with 30 ng of psiCHECK-2 construct plus: 50 pmol of siPUM1 or control scramble-siRNA and 0.5 μg of pcDNA3.1(+)-PUM1 or control pcDNA3.1(+) using Lipofectamine 2000 (Invitrogen). After 24 hrs, luciferase activity was measured using the Dual Luciferase Reporter Assay System (Promega) according to the manufacturer’s instructions.
Western Blot
HEK293T cells were seeded in six-well plates before transfection. After 72 hrs cells were processed for protein extraction. For mouse tissues, the entire cerebrum and cerebellum were processed for protein extraction. Both pellet or tissues were lysed with RIPA buffer (25 mM Tris-HCL pH 7.6, 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate, 0.1% SDS, and complete protease inhibitor cocktail (Roche)) then placed for 15 minutes on ice followed by centrifugation at 13000 rpm at 4°C for 15 minutes. Proteins were quantified by Pierce BCA Protein Assay Kit (Thermo Scientific) and resolved by high resolution Bolt 4–12% Bis-Tris Plus Gel (Life Technologies) according to the manufacturer’s instruction.
RNA-Cross linking Immunoprecipitation (CLIP)
Brains from wild type or Pum1 knockout mice were dissected out and cerebellum was separated from the rest of the brain. After separation, the tissue was triturated in 8 ml of ice cold HBSS until cells were evenly dissociated with no visible chunks. The cell suspension was layered on a chilled 10 cm sterile tissue culture plate and exposed to 150 mJ/cm2 UVC (Stratagene, model UV Stratalinker 2400) on ice. After one exposure, the cell suspension was gently swirled and exposed again to UVC at 100 mJ/cm2. The cell suspension was pelleted for individual immunoprecipitation (IP). Cells were lysed in lysis buffer (50 mMTris-HCl, pH 7.4, 100 mMNaCl, 1% NP-40, 0.1% SDS, 0.5% sodium deoxycholate, 80 U/ml RNase OUT (Invitrogen) with protease inhibitor (Roche)). Soluble fractions were pre-cleared with Protein A-Sepharose beads, Rabbit control IgG (Sigma), 0.05% BSA and 0.2 μg/ml yeast tRNA (Invitrogen). Pre-cleared lysates was incubated with control IgG or Pum1 (5μg) (Bethyl Laboratories, see Antibodies) together with Protein A-Sepharose beads, and incubated overnight at 4°C with gentle rotation. Next day, beads were washed 5 times with lysis buffer. Beads were treated using 20 units of RNase-free DNase (Roche) for 15 min at room temperature. Followed by 50 ug proteinase K (Roche) treatment for 30 min at 37° C. Immunoprecipated RNA was isolated using miRNeasy kit (Quiagen) and RT-PCR was performed using primers designed to amplify Atxn1 cDNA regions upstream and downstream of the predicted Pum1 binding site (see Primers). Isolated RNA from a fraction (10%) of pre-cleared lysate was used as input.
RNA stability
Total RNA from HEK293T cells, RNA quality, cDNA synthesized and qRT-PCR experiments were obtained as described above in “RNA Extraction and Quantitative real-time PCR”. HEK293T cells were seeded in 24-well plates before transfection for both experiments (Figure 3E, F). For Figure 3E the partial 3′UTR containing the PUM1 WT or mutated (Mut) binding sites 3 (5175-5375) were the same as used for luciferase assay (see Luciferase assay section). HEK293T cells were transfected with 30ng of psiCHECK-2 vectors (Promega) containing ATXN1-3′UTR WT- or Mut binding sitess using Lipofectamine 2000 (Invitrogen). After 36 hours cells were treated with 5,6-dichloro-1-β-D-ribofuranosylbenzimidazole (DRB) at the final concentration of 20μg/ml, and total RNA for qRT-PCR analysis was collected at different time points. Firefly data were normalized to the respective Renilla. For Figure 3F HEK293T cells were transfected with Ambion small-interfering RNA for PUM1 (siPUM1) or Scramble (siScramble) at the final concentration of 40nM. After 48 hours cells were collected and the total RNA were processed for qRT-PCR. All data were normalized to GAPDH. The housekeeping gene GAPDH was used to compare all the qRT-PCR values. All western blot experiments were performed as described in the “Western blot” section.
Supplementary Material
Acknowledgments
We thank Dr. Haifan Lin (Yale Stem Cell Center and Department of Cell Biology, Yale University School of Medicine) for sharing the Pum1 mutant mice; A. McCall for helpful suggestions on analysis; X. Liu for genotyping; C. Spencer for behavioral training; C. Ljungberg for ISH analysis; D. Yu for confocal microscopy training; and members of the H.Y.Z. laboratory for helpful discussions. We also thank A. Ballabio, T. Klisch, S. Yamamoto, M. Rousseaux, M. Sardiello, C. Schaaf, C. Alcott, and V. Brandt for helpful suggestions and critical reading of the manuscript. This research was supported by NINDS NS27600 (to H.Y.Z.) and the RNA In Situ Hybridization, Confocal and Mouse Behavioral Cores at the BCM Intellectual and Developmental Disabilities Research Center (IDDRC; NIH/NICHHDP30-HD024064) and 1R01NS089664-01 (to R.V.S.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the Eunice Kennedy Shriver National Institute of Child Health & Human Development or the National Institutes of Health.
Footnotes
AUTHOR CONTRIBUTIONS
V.A.G. and H.Y.Z. conceived the study, designed experiments, analyzed and interpreted data and wrote the manuscript. V.A.G. performed molecular and behavioral tests. R.K.S., J.J.W., A.D.M., K.H., J.K., P.J.N., A.D.R. and L.S.S. contributed to molecular work. H.K. performed bioinformatic analyses. T.A.C., H.T.O. and R.V.S. contributed to interpreting data.
The authors declare that they have no conflicts of interest in the research.
References
- Arrasate M, Mitra S, Schweitzer ES, Segal MR, Finkbeiner S. Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature. 2004;431:805–810. doi: 10.1038/nature02998. [DOI] [PubMed] [Google Scholar]
- Banfi S, Servadio A, Chung M, Capozzoli F, Duvick LA, Elde R, Zoghbi HY, Orr HT. Cloning and developmental expression analysis of the murine homolog of the spinocerebellar ataxia type 1 gene (Sca1) Human Molecular Genetics. 1996;5:33–40. doi: 10.1093/hmg/5.1.33. [DOI] [PubMed] [Google Scholar]
- Banfi S, Servadio A, Chung MY, Kwiatkowski TJ, Jr, McCall AE, Duvick LA, Shen Y, Roth EJ, Orr HT, Zoghbi HY. Identification and characterization of the gene causing type 1 spinocerebellar ataxia. Nature Genetics. 1994;7:513–520. doi: 10.1038/ng0894-513. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1111–1124. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- Bowman AB, Lam YC, Jafar-Nejad P, Chen HK, Richman R, Samaco RC, Fryer JD, Kahle JJ, Orr HT, Zoghbi HY. Duplication of Atxn1l suppresses SCA1 neuropathology by decreasing incorporation of polyglutamine-expanded ataxin-1 into native complexes. Nature Genetics. 2007;39:373–379. doi: 10.1038/ng1977. [DOI] [PubMed] [Google Scholar]
- Bowman AB, Yoo SY, Dantuma NP, Zoghbi HY. Neuronal dysfunction in a polyglutamine disease model occurs in the absence of ubiquitin-proteasome system impairment and inversely correlates with the degree of nuclear inclusion formation. Human Molecular Genetics. 2005;14:679–691. doi: 10.1093/hmg/ddi064. [DOI] [PubMed] [Google Scholar]
- Burright EN, Clark HB, Servadio A, Matilla T, Feddersen RM, Yunis WS, Duvick LA, Zoghbi HY, Orr HT. SCA1 transgenic mice: a model for neurodegeneration caused by an expanded CAG trinucleotide repeat. Cell. 1995;82:937–948. doi: 10.1016/0092-8674(95)90273-2. [DOI] [PubMed] [Google Scholar]
- Chartier-Harlin MC, Kachergus J, Roumier C, Mouroux V, Douay X, Lincoln S, Levecque C, Larvor L, Andrieux J, Hulihan M, et al. Alpha-synuclein locus duplication as a cause of familial Parkinson’s disease. Lancet. 2004;364:1167–1169. doi: 10.1016/S0140-6736(04)17103-1. [DOI] [PubMed] [Google Scholar]
- Chen D, Zheng W, Lin A, Uyhazi K, Zhao H, Lin H. Pumilio 1 suppresses multiple activators of p53 to safeguard spermatogenesis. Current Biology: CB. 2012;22:420–425. doi: 10.1016/j.cub.2012.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook KB, Kazan H, Zuberi K, Morris Q, Hughes TR. RBPDB: a database of RNA-binding specificities. Nucleic Acids Research. 2011;39:D301–308. doi: 10.1093/nar/gkq1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings CJ, Reinstein E, Sun Y, Antalffy B, Jiang Y, Ciechanover A, Orr HT, Beaudet AL, Zoghbi HY. Mutation of the E6-AP ubiquitin ligase reduces nuclear inclusion frequency while accelerating polyglutamine-induced pathology in SCA1 mice. Neuron. 1999;24:879–892. doi: 10.1016/s0896-6273(00)81035-1. [DOI] [PubMed] [Google Scholar]
- Fabian MR, Sonenberg N. The mechanics of miRNA-mediated gene silencing: a look under the hood of miRISC. Nature Structural & Molecular biology. 2012;19:586–593. doi: 10.1038/nsmb.2296. [DOI] [PubMed] [Google Scholar]
- Fernandez-Funez P, Nino-Rosales ML, de Gouyon B, She WC, Luchak JM, Martinez P, Turiegano E, Benito J, Capovilla M, Skinner PJ, et al. Identification of genes that modify ataxin-1-induced neurodegeneration. Nature. 2000;408:101–106. doi: 10.1038/35040584. [DOI] [PubMed] [Google Scholar]
- Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Research. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friend K, Campbell ZT, Cooke A, Kroll-Conner P, Wickens MP, Kimble J. A conserved PUF-Ago-eEF1A complex attenuates translation elongation. Nature Structural & Molecular Biology. 2012;19:176–183. doi: 10.1038/nsmb.2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galgano A, Forrer M, Jaskiewicz L, Kanitz A, Zavolan M, Gerber AP. Comparative analysis of mRNA targets for human PUF-family proteins suggests extensive interaction with the miRNA regulatory system. PloS One. 2008;3:e3164. doi: 10.1371/journal.pone.0003164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gennarino VA, D’Angelo G, Dharmalingam G, Fernandez S, Russolillo G, Sanges R, Mutarelli M, Belcastro V, Ballabio A, Verde P, et al. Identification of microRNA-regulated gene networks by expression analysis of target genes. Genome Research. 2012;22:1163–1172. doi: 10.1101/gr.130435.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gennarino VA, Sardiello M, Mutarelli M, Dharmalingam G, Maselli V, Lago G, Banfi S. HOCTAR database: a unique resource for microRNA target prediction. Gene. 2011;480:51–58. doi: 10.1016/j.gene.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glorian V, Maillot G, Poles S, Iacovoni JS, Favre G, Vagner S. HuR-dependent loading of miRNA RISC to the mRNA encoding the Ras-related small GTPase RhoB controls its translation during UV-induced apoptosis. Cell Death and Differentiation. 2011;18:1692–1701. doi: 10.1038/cdd.2011.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstrohm AC, Hook BA, Seay DJ, Wickens M. PUF proteins bind Pop2p to regulate messenger RNAs. Nature Structural & Molecular Biology. 2006;13:533–539. doi: 10.1038/nsmb1100. [DOI] [PubMed] [Google Scholar]
- Grillo G, Turi A, Licciulli F, Mignone F, Liuni S, Banfi S, Gennarino VA, Horner DS, Pavesi G, Picardi E, et al. UTRdb and UTRsite (RELEASE 2010): a collection of sequences and regulatory motifs of the untranslated regions of eukaryotic mRNAs. Nucleic Acids Research. 2010;38:D75–80. doi: 10.1093/nar/gkp902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber AR, Lorenz R, Bernhart SH, Neubock R, Hofacker IL. The Vienna RNA websuite. Nucleic Acids Research. 2008;36:W70–74. doi: 10.1093/nar/gkn188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haass C, Selkoe DJ. Soluble protein oligomers in neurodegeneration: lessons from the Alzheimer’s amyloid beta-peptide. Nature Reviews Molecular Cell Biology. 2007;8:101–112. doi: 10.1038/nrm2101. [DOI] [PubMed] [Google Scholar]
- Hofacker IL, Stadler PF. Memory efficient folding algorithms for circular RNA secondary structures. Bioinformatics. 2006;22:1172–1176. doi: 10.1093/bioinformatics/btl023. [DOI] [PubMed] [Google Scholar]
- Ibanez P, Bonnet AM, Debarges B, Lohmann E, Tison F, Pollak P, Agid Y, Durr A, Brice A. Causal relation between alpha-synuclein gene duplication and familial Parkinson’s disease. Lancet. 2004;364:1169–1171. doi: 10.1016/S0140-6736(04)17104-3. [DOI] [PubMed] [Google Scholar]
- Kedde M, van Kouwenhove M, Zwart W, Oude Vrielink JA, Elkon R, Agami R. A Pumilio-induced RNA structure switch in p27-3′ UTR controls miR-221 and miR-222 accessibility. Nature Cell Biology. 2010;12:1014–1020. doi: 10.1038/ncb2105. [DOI] [PubMed] [Google Scholar]
- Kim HH, Kuwano Y, Srikantan S, Lee EK, Martindale JL, Gorospe M. HuR recruits let-7/RISC to repress c-Myc expression. Genes & Development. 2009;23:1743–1748. doi: 10.1101/gad.1812509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klement IA, Skinner PJ, Kaytor MD, Yi H, Hersch SM, Clark HB, Zoghbi HY, Orr HT. Ataxin-1 nuclear localization and aggregation: role in polyglutamine-induced disease in SCA1 transgenic mice. Cell. 1998;95:41–53. doi: 10.1016/s0092-8674(00)81781-x. [DOI] [PubMed] [Google Scholar]
- Kundu P, Fabian MR, Sonenberg N, Bhattacharyya SN, Filipowicz W. HuR protein attenuates miRNA-mediated repression by promoting miRISC dissociation from the target RNA. Nucleic Acids Research. 2012;40:5088–5100. doi: 10.1093/nar/gks148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Samaco RC, Gatchel JR, Thaller C, Orr HT, Zoghbi HY. miR-19, miR-101 and miR-130 co-regulate ATXN1 levels to potentially modulate SCA1 pathogenesis. Nature Neuroscience. 2008;11:1137–1139. doi: 10.1038/nn.2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeb M, Dietmann S, Paramor M, Niwa H, Smith A. Genetic exploration of the exit from self-renewal using haploid embryonic stem cells. Cell Stem Cell. 2014;14:385–393. doi: 10.1016/j.stem.2013.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Quon G, Lipshitz HD, Morris Q. Predicting in vivo binding sites of RNA-binding proteins using mRNA secondary structure. Rna. 2010;16:1096–1107. doi: 10.1261/rna.2017210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukong KE, Chang KW, Khandjian EW, Richard S. RNA-binding proteins in human genetic disease. Trends in Genetics: TIG. 2008;24:416–425. doi: 10.1016/j.tig.2008.05.004. [DOI] [PubMed] [Google Scholar]
- Martin I, Kim JW, Lee BD, Kang HC, Xu JC, Jia H, Stankowski J, Kim MS, Zhong J, Kumar M, et al. Ribosomal protein s15 phosphorylation mediates LRRK2 neurodegeneration in Parkinson’s disease. Cell. 2014;157:472–485. doi: 10.1016/j.cell.2014.01.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miles WO, Tschop K, Herr A, Ji JY, Dyson NJ. Pumilio facilitates miRNA regulation of the E2F3 oncogene. Genes & Development. 2012;26:356–368. doi: 10.1101/gad.182568.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr HT, Chung MY, Banfi S, Kwiatkowski TJ, Jr, Servadio A, Beaudet AL, McCall AE, Duvick LA, Ranum LP, Zoghbi HY. Expansion of an unstable trinucleotide CAG repeat in spinocerebellar ataxia type 1. Nature Genetics. 1993;4:221–226. doi: 10.1038/ng0793-221. [DOI] [PubMed] [Google Scholar]
- Park J, Al-Ramahi I, Tan Q, Mollema N, Diaz-Garcia JR, Gallego-Flores T, Lu HC, Lagalwar S, Duvick L, Kang H, et al. RAS-MAPK-MSK1 pathway modulates ataxin 1 protein levels and toxicity in SCA1. Nature. 2013;498:325–331. doi: 10.1038/nature12204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Research. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross CA, Poirier MA. Protein aggregation and neurodegenerative disease. Nature Medicine. 2004;10(Suppl):S10–17. doi: 10.1038/nm1066. [DOI] [PubMed] [Google Scholar]
- Rovelet-Lecrux A, Hannequin D, Raux G, Le Meur N, Laquerriere A, Vital A, Dumanchin C, Feuillette S, Brice A, Vercelletto M, et al. APP locus duplication causes autosomal dominant early-onset Alzheimer disease with cerebral amyloid angiopathy. Nature Genetics. 2006;38:24–26. doi: 10.1038/ng1718. [DOI] [PubMed] [Google Scholar]
- Rumble B, Retallack R, Hilbich C, Simms G, Multhaup G, Martins R, Hockey A, Montgomery P, Beyreuther K, Masters CL. Amyloid A4 protein and its precursor in Down’s syndrome and Alzheimer’s disease. The New England Journal of Medicine. 1989;320:1446–1452. doi: 10.1056/NEJM198906013202203. [DOI] [PubMed] [Google Scholar]
- Servadio A, Koshy B, Armstrong D, Antalffy B, Orr HT, Zoghbi HY. Expression analysis of the ataxin-1 protein in tissues from normal and spinocerebellar ataxia type 1 individuals. Nature Genetics. 1995;10:94–98. doi: 10.1038/ng0595-94. [DOI] [PubMed] [Google Scholar]
- Singleton AB, Farrer M, Johnson J, Singleton A, Hague S, Kachergus J, Hulihan M, Peuralinna T, Dutra A, Nussbaum R, et al. alpha-Synuclein locus triplication causes Parkinson’s disease. Science. 2003;302:841. doi: 10.1126/science.1090278. [DOI] [PubMed] [Google Scholar]
- Soto C. Unfolding the role of protein misfolding in neurodegenerative diseases. Nature Reviews Neuroscience. 2003;4:49–60. doi: 10.1038/nrn1007. [DOI] [PubMed] [Google Scholar]
- Spassov DS, Jurecic R. The PUF family of RNA-binding proteins: does evolutionarily conserved structure equal conserved function? IUBMB Life. 2003;55:359–366. doi: 10.1080/15216540310001603093. [DOI] [PubMed] [Google Scholar]
- Suh N, Crittenden SL, Goldstrohm A, Hook B, Thompson B, Wickens M, Kimble J. FBF and its dual control of gld-1 expression in the Caenorhabditis elegans germline. Genetics. 2009;181:1249–1260. doi: 10.1534/genetics.108.099440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biology. 2002;3:RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y, Qu K, Zhang QC, Flynn RA, Manor O, Ouyang Z, Zhang J, Spitale RC, Snyder MP, Segal E, et al. Landscape and variation of RNA secondary structure across the human transcriptome. Nature. 2014;505:706–709. doi: 10.1038/nature12946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, McLachlan J, Zamore PD, Hall TM. Modular recognition of RNA by a human pumilio-homology domain. Cell. 2002;110:501–512. doi: 10.1016/s0092-8674(02)00873-5. [DOI] [PubMed] [Google Scholar]
- Watase K, Weeber EJ, Xu B, Antalffy B, Yuva-Paylor L, Hashimoto K, Kano M, Atkinson R, Sun Y, Armstrong DL, et al. A long CAG repeat in the mouse Sca1 locus replicates SCA1 features and reveals the impact of protein solubility on selective neurodegeneration. Neuron. 2002;34:905–919. doi: 10.1016/s0896-6273(02)00733-x. [DOI] [PubMed] [Google Scholar]
- Wickens M, Bernstein DS, Kimble J, Parker R. A PUF family portrait: 3′UTR regulation as a way of life. Trends in Genetics: TIG. 2002;18:150–157. doi: 10.1016/s0168-9525(01)02616-6. [DOI] [PubMed] [Google Scholar]
- Zoghbi HY, Orr HT. Glutamine repeats and neurodegeneration. Annual Review of Neuroscience. 2000;23:217–247. doi: 10.1146/annurev.neuro.23.1.217. [DOI] [PubMed] [Google Scholar]
- Zoghbi HY, Orr HT. Pathogenic mechanisms of a polyglutamine-mediated neurodegenerative disease, spinocerebellar ataxia type 1. The Journal of Biological Chemistry. 2009;284:7425–7429. doi: 10.1074/jbc.R800041200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M, Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Research. 1981;9:133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.