Inhibition of protein–protein interactions (PPIs) with designed molecules represents a key challenge in modern bio-organic chemistry.[1–3] In contrast to competitive enzyme inhibition,[4] in which small molecules are optimised to masquerade as a substrate (or transition-state analogue) and fit a well defined concave pocket or cleft, competitive inhibition of PPIs requires a molecule that must make discontinuous, noncovalent contacts over a much larger (>800 Å2), surface-lacking, defined shape. Whilst high-throughput screening has identified inhibitors of some PPIs,[5,6]—these are generally regarded as ‘low-hanging fruit’[2]—there remains a need to develop our basic understanding of how to design molecules that possess the features needed for protein-surface recognition. Building on fundamental studies of short peptide recognition,[7–9] a number of approaches in which a scaffold is used to project groups capable of making multivalent hydrophobic,[10] ion-pairing[11–29] and metal–ligand interactions[30–33] with proteins have been described. In the current manuscript we illustrate that functionalised RuII tris-bipyridine complexes can be used as selective and low nanomolar sensors for cytochrome c (cyt c).[34] Receptors for cyt c have been described[11–13,16–18,20–26] in addition to inhibitors of its PPIs.[35–37] The current system, however, offers significant advantages for fundamental studies of protein-surface recognition. Binding to metalloproteins and nonmetalloproteins can be detected by using simple fluorescence quenching or anisotropy changes, respectively, whilst structure–affinity studies and screening against a panel of proteins point to specific interactions with the target. Importantly, the highest affinity receptor binds cyt c with 1:1 stoichiometry and an affinity of 2 nm.
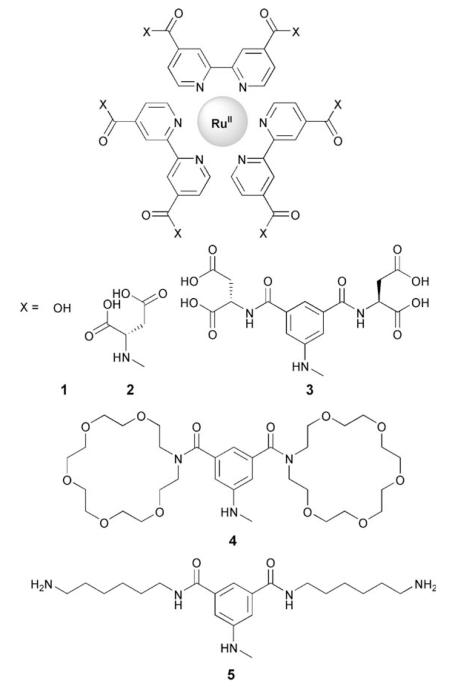
We selected RuII tris-(5,5′-biscarboxybipyridine) as a core to which could be appended functional groups capable of making a diverse array of noncovalent interactions with our target protein. We then synthesised a series of RuII tris-(5,5′-biscarboxamidobipyridine) derivatives, 1–5, (as described in the Supporting Information) which present functional domains of different size and composition that are suitable for matching to the diverse topology of different proteins.
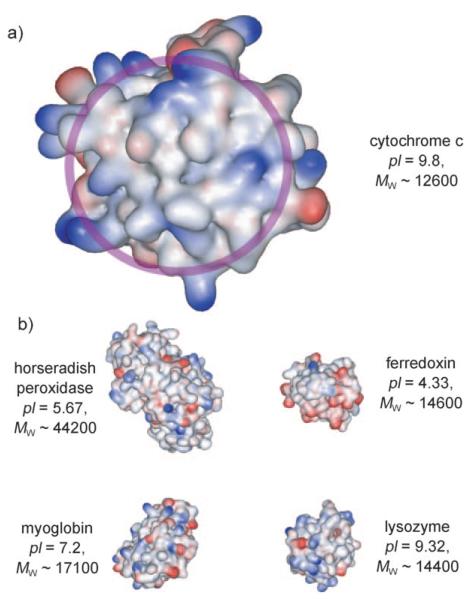
The recognition surface of cyt c centres around a solvent-exposed hydrophobic haem group surrounded by basic residues (Figure 1a).[38] As such, we anticipated compounds 2–4, which have functional groups capable of cation recognition, to be potent receptors for this protein. We also selected a range of additional metallo- and nonmetalloproteins to test against for selective recognition and sensing (Figure 1b). These represent proteins of different sizes, and more significantly, different surface compositions, but in certain cases very similar charge (for example, lysozyme and cyt c).
Figure 1.
Structures of a) cytochrome c (PDB ID: 1HRC)[39] and its rec ognition surface for interaction with other proteins circled in light purple and b) other proteins tested, including horseradish peroxidase (PDB ID: 1W4W),[40] ferredoxin (PDB ID: 1A7O),[41] myoglobin, (PDB ID: 1HRM),[42] and lysozyme (PDB ID: 2LYM).[43]
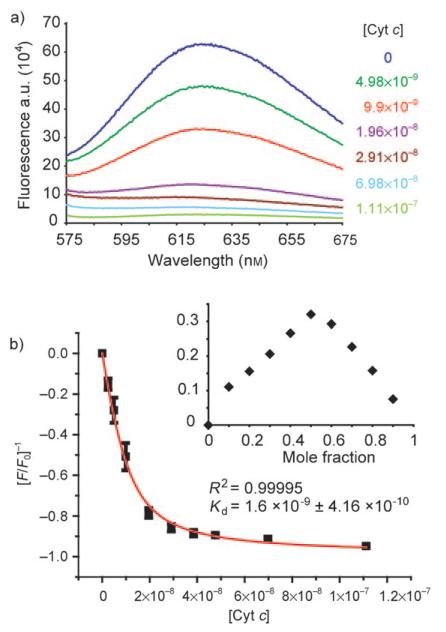
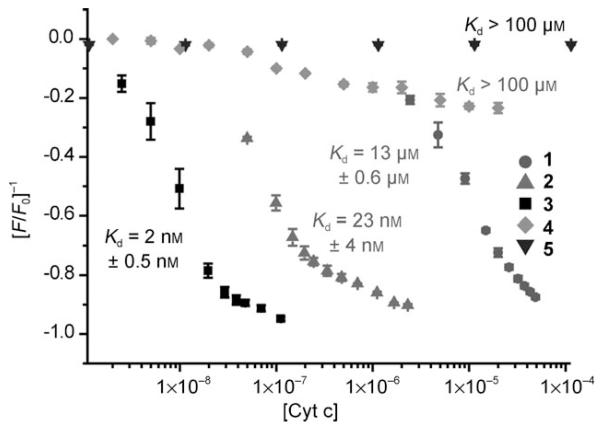
We first tested binding by monitoring the fluorescence response of each receptor upon titration with cyt c. A number of compounds demonstrated almost complete quenching and saturation behaviour during the experiment. Representative data are provided for the most potent compound (3) in Figure 2a and b. The data can be fit to a simple 1:1 binding isotherm by using nonlinear regression (Figure 2b) to afford dissociation constants whilst the Job plot (Figure 2b inset) confirms the expected 1:1 stoichiometry. Shown in Figure 3 are the titration curves for binding of cyt c to each of the metal complexes. Unsurprisingly, the charge-mismatched, amino-functionalised receptor 5 does not bind to the target protein, whilst those compounds with increasing numbers of aspartate residues exhibit progressively stronger binding. Compound 3 binds with a dissociation constant of 2 nm, which is amongst the strongest affinities yet observed for the binding of cyt c by a synthetic receptor[16] and repre sents five orders of magnitude increase in affinity over the core scaffold. This is very dramatic considering that other scaffolds (for example, porphyrins) exhibit significant affinity for the protein,[11,13] whereas the majority of affinity in our case derives from functionalisation. We attempted to evaluate the role of multivalency by increasing the number of carboxylates going from compounds 1–3. The average binding free energy per carboxylate is −4.68 kJmol−1 for 1, −3.66 kJmol−1 for 2 and −2.08 kJmol−1 and for 3; this indicates negative co-operativity. Negatively co-operative binding interactions are commonly observed.[44] This is not unexpected, however, given that our current analysis is somewhat crude in that other noncovalent contacts (for example, hydrophobic) are likely to be important and only a certain number of carboxylates will make contacts with the protein—a phenomenon observed in dendrimer–DNA interactions.[45] We were surprised to find that tris-(5,5′-biscarboxamidobipyridine) (4), which is functionalised with a crown motif, exhibited virtually no binding to cyt c. We had anticipated that the crown ring might recognise lysine residues in preference to arginine residues and thus prove useful for differentiating between basic proteins with different side chains. One reason for this might be that the multivalent contacts that could be made upon cyt c binding are not outweighed by the preferred binding to excess cations present in buffer. Pleasingly, the individual ligands do not appear to bind with high affinity to cyt c. Due to the absence of diagnostic spectroscopic properties, compound 6 (which represents the ligand comprising 3) was tested in a competition assay against the cyt c--1 interaction. Unfortunately, recovery of the fluorescence of 1 upon displacement from the surface of cyt c was not observed—only minor changes in fluorescence at high μm concentrations were seen (see the Supporting Information, Figure 6). This suggests weak binding, however it is not conclusive as there are other interpretations of this result (for example, both species could be binding to different sites).
Figure 2.
Fluorescence emission response of receptor 3 (10 nm, 5 mm sodium phosphate buffer, pH 7.4, λex 467 nm) upon addition of cyt c. a) Emission spectrum with various concentrations of protein. b) Change in response at emission maximum of 625 nm and fit by nonlinear regression (inset: Job Plot). [cyt c] is given in m.
Figure 3.
Change in fluorescence emission response of receptors 1–5 at λem maximum of 625 nm upon titration with cyt c (conditions as for Figure 3). [cyt c] is given in m.
Following these initial binding studies we tested each of the compounds in a functional assay. The reduction kinetics of cyt c by ascorbate have previously been shown to be modified by the binding of receptors to its haem-exposed edge.[12,46,47] Specifically, a retarded rate of reduction is proposed to arise as a result of binding and steric blockage of the reducing agent from approach to the haem group. As expected, at a single concentration, the receptors retarded the rate of reduction in line with their binding affinity determined by fluorescence (see the Supporting Information) that is, receptor 3 exhibited potent inhibition of reduction whilst receptor 5 exhibited no change. These results are therefore indicative of binding to the surface of cyt c as proposed. Furthermore, we were also able to observe the interaction of cyt c and the receptor by using mass spectrometry. ESI mass spectra (see the Supporting Information) of the protein in the presence of receptor 3 clearly showed the presence of noncovalently bound 1:1 and 2:1 products (the observation of two molecules of 3 bound to cyt c in the mass spectrum is not unsurprising given that ESI enhances electrostatic interactions). This technique could be used for rapid screening of this class of receptors against a panel of proteins in future.
We then tested the highest affinity receptor 3 against a range of other proteins. Against all the metalloproteins tested no fluorescence response was observed (Table 1). Whilst a noncovalent interaction cannot be excluded on the basis of the data, this seems likely and the results clearly demonstrate that selective sensing of proteins is possible. Most significantly, acetylated cyt c in which the lysines have been blocked did not respond to receptor 3; this suggests again that molecular recognition derives from interaction of receptor 3 and the basic surface of cyt c. We were also eager to study binding with nonmetalloproteins. Binding, however, does not necessarily result in changes to the fluorescence signal. We therefore monitored changes in both fluorescence intensity and fluorescence anisotropy (in which the signal is dependent upon the molecular weight of the species in solution). Receptor 3 experienced a dose-dependent change in anisotropy signal upon titration with lysozyme, but no changes in emission intensity. The data could be fit to a 1:1 model with a dissociation constant of 273 nm. What is most dramatic about this result is that lysozyme and cyt c have a similar charge state (i.e. pI) and surface composition, yet receptor 3 exhibits two orders of magnitude higher affinity for the latter protein pointing to a structurally well-defined interaction that extends beyond a simple ‘grease and charge’ model of interaction!
Table 1.
Dissociation constants for binding of receptor 3 to different proteins[a]
| Protein | Dissociation constant (Kd) [nm] |
|---|---|
| cytochrome c | 2±0.5 |
| acetylated cytochrome c | >30 000 |
| myoglobin | >30 000 |
| ferredoxin | >30 000 |
| horseradish peroxidase | >30 000 |
| lysozyme | 273±43 |
Conditions for titration as indicated in Figure 2.
In conclusion, we have illustrated that functionalised RuII tris-bipyridine complexes act as selective and high affinity receptors for proteins surfaces. These metal complexes could similarly form the basis of protein sensors for diagnostic purposes.[48–52] Our future studies will focus upon under standing the binding interaction within this model system at a structural and thermodynamic level and inhibiting pharmaceutically relevant PPIs.
Supplementary Material
Acknowledgements
This work was supported by the Wellcome Trust [WT 78112/Z/05/Z].
Footnotes
Supporting information for this article is available on the WWW under http://dx.doi.org/10.1002/chem.200902368.
References
- [1].Yin H, Hamilton AD. Angew. Chem. 2005;117:4200–4235. [Google Scholar]; Angew. Chem. Int. Ed. 2005;44:4130–4163. doi: 10.1002/anie.200461786. [DOI] [PubMed] [Google Scholar]
- [2].Wells JA, McLendon CL. Nature. 2007;450:1001–1009. doi: 10.1038/nature06526. [DOI] [PubMed] [Google Scholar]
- [3].Wilson AJ. Chem. Soc. Rev. 2009 DOI: 10.1039/B807197G. [Google Scholar]
- [4].Babine RE, Bender SL. Chem. Rev. 1997;97:1359–1472. doi: 10.1021/cr960370z. [DOI] [PubMed] [Google Scholar]
- [5].Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, Kong N, Kammlott U, Lukacs C, Klein C, Fotouhi N, Liu EA. Science. 2004;303:844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- [6].Grasberger BL, Lu T, Schubert C, Parks DJ, Carver TE, Koblish HK, Cummings MD, LaFrance LV, Milkiewicz KL, Calvo RR, Maguire D, Lattanze J, Franks CF, Zhao S, Ramachandren K, Bylebyl GR, Zhang M, Manthey CL, Petrella EC, Pantoliano MW, Deckman IC, Spurlino JC, Maroney AC, Tomczuk BE, Molloy CJ, Bone RF. J. Med. Chem. 2005;48:909–912. doi: 10.1021/jm049137g. [DOI] [PubMed] [Google Scholar]
- [7].Maletic M, Wennemers H, McDonald DQ, Breslow R, Still WC. Angew. Chem. 1996;108:1594–1596. [Google Scholar]; Angew. Chem. Int. Ed. Engl. 1996;35:1490–1492. [Google Scholar]
- [8].Schmuck C, Wich P. Angew. Chem. 2006;118:4383–4387. doi: 10.1002/anie.200601046. [DOI] [PubMed] [Google Scholar]; Angew. Chem. Int. Ed. 2006;45:4277–4281. doi: 10.1002/anie.200601046. [DOI] [PubMed] [Google Scholar]
- [9].Peczuh MW, Hamilton AD. Chem. Rev. 2000;100:2479–2493. doi: 10.1021/cr9900026. [DOI] [PubMed] [Google Scholar]
- [10].Leung DK, Yang Z, Breslow R. Proc. Natl. Acad. Sci. USA. 2000;97:5050–5053. doi: 10.1073/pnas.97.10.5050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Clark-Ferris KK, Fisher J. J. Am. Chem. Soc. 1985;107:5007–5008. [Google Scholar]
- [12].Hamuro Y, Calama MC, Park H-S, Hamilton AD. Angew. Chem. 1997;109:2797–2800. [Google Scholar]; Angew. Chem. Int. Ed. Engl. 1997;36:2680–2683. [Google Scholar]
- [13].Jain RK, Hamilton AD. Org. Lett. 2000;2:1721–1723. doi: 10.1021/ol005871s. [DOI] [PubMed] [Google Scholar]
- [14].Park HS, Lin Q, Hamilton AD. Proc. Natl. Acad. Sci. USA. 2002;99:5105–5109. doi: 10.1073/pnas.082675899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Fischer NO, McIntosh CM, Simard JM, Rotello VM. Proc. Natl. Acad. Sci. USA. 2002;99:5018–5023. doi: 10.1073/pnas.082644099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Aya T, Hamilton AD. Bioorg. Med. Chem. Lett. 2003;13:2651–2654. doi: 10.1016/s0960-894x(03)00551-1. [DOI] [PubMed] [Google Scholar]
- [17].Wilson AJ, Groves K, Jain RK, Park HS, Hamilton AD. J. Am. Chem. Soc. 2003;125:4420–4421. doi: 10.1021/ja028574m. [DOI] [PubMed] [Google Scholar]
- [18].Braun M, Atalick S, Guldi DM, Lanig H, Brettreich M, Burghardt S, Hatzimarinaki M, Ravanelli E, Prato M, Eldik R. v., Hirsch A. Chem. Eur. J. 2003;9:3867–3875. doi: 10.1002/chem.200204680. [DOI] [PubMed] [Google Scholar]
- [19].Salvatella X, Martinell M, Gairf M, Mateu MG, Feliz M, Hamilton AD, de Mendoza J, Giralt E. Angew. Chem. 2004;116:198–200. doi: 10.1002/anie.200352115. [DOI] [PubMed] [Google Scholar]; Angew. Chem. Int. Ed. 2004;43:196–198. [Google Scholar]
- [20].Groves K, Wilson AJ, Hamilton AD. J. Am. Chem. Soc. 2004;126:12833–12842. doi: 10.1021/ja0317731. [DOI] [PubMed] [Google Scholar]
- [21].Tagore DM, Sprinz KI, Fletcher S, Jayawickramarajah J, Hamilton AD. Angew. Chem. 2007;119:227–229. doi: 10.1002/anie.200603479. [DOI] [PubMed] [Google Scholar]; Angew. Chem. Int. Ed. 2007;46:223–225. [Google Scholar]
- [22].Paul D, Miyake H, Shinoda S, Tsukube H. Chem. Eur. J. 2006;12:1328–1338. doi: 10.1002/chem.200501131. [DOI] [PubMed] [Google Scholar]
- [23].Wilson AJ, Hong J, Hamilton AD. Org. Biomol. Chem. 2007;5:276–285. doi: 10.1039/b612975g. [DOI] [PubMed] [Google Scholar]
- [24].Bayraktar H, You C-C, Rotello VM, Knapp MJ. J. Am. Chem. Soc. 2007;129:2732–2733. doi: 10.1021/ja067497i. [DOI] [PubMed] [Google Scholar]
- [25].De M, You C-C, Srivastava S, Rotello VM. J. Am. Chem. Soc. 2007;129:10747–10753. doi: 10.1021/ja071642q. [DOI] [PubMed] [Google Scholar]
- [26].Chiba F, Hu T-C, Twyman LJ, Wagstaff M. Chem. Commun. 2008:4351–4353. doi: 10.1039/b806517a. [DOI] [PubMed] [Google Scholar]
- [27].Hayashida O, Ogawa H, Uchiyama M. J. Am. Chem. Soc. 2007;129:13698–13705. doi: 10.1021/ja074906h. [DOI] [PubMed] [Google Scholar]
- [28].Arendt M, Sun W, Thomann J, Xie X, Schrader T. Chem. Asian J. 2006;1:544–554. doi: 10.1002/asia.200600125. [DOI] [PubMed] [Google Scholar]
- [29].Zadmard R, Schrader T. J. Am. Chem. Soc. 2005;127:904–915. doi: 10.1021/ja045785d. [DOI] [PubMed] [Google Scholar]
- [30].Ojida A, Mito-oka Y, Inoue M.-a., Hamachi I. J. Am. Chem. Soc. 2002;124:6256–6258. doi: 10.1021/ja025761b. [DOI] [PubMed] [Google Scholar]
- [31].Ojida A, Inoue M-A, Mito-oka Y, Hamachi I. J. Am. Chem. Soc. 2003;125:10184–10185. doi: 10.1021/ja036317r. [DOI] [PubMed] [Google Scholar]
- [32].Banerjee AL, Swanson M, Roy BC, Jia X, Haldar MK, Malik S, Srivastava DK. J. Am. Chem. Soc. 2004;126:10875–10883. doi: 10.1021/ja047557p. [DOI] [PubMed] [Google Scholar]
- [33].Grauer A, Riechers A, Ritter S, Kçnig B. Chem. Eur. J. 2008;14:8922–8927. doi: 10.1002/chem.200800432. [DOI] [PubMed] [Google Scholar]
- [34].Hamachi and co-workers previously demonstrated RuII tris-(bipyri-dine)complexes engage in electron transfer reactions selectively with cyt c see: ; Takashima H, Shinkai S, Hamachi I. Chem. Commun. 1999:2345–2346. [Google Scholar]
- [35].Wei Y, McLendon GL, Hamilton AD, Case MA, Purring CB, Lin Q, Park HS, Lee CS, Yu TN. Chem. Commun. 2001:1580–1581. doi: 10.1039/b104142h. [DOI] [PubMed] [Google Scholar]
- [36].Bayraktar H, Ghosh PS, Rotello VM, Knapp MJ. Chem. Commun. 2006:1390–1392. doi: 10.1039/b516096k. [DOI] [PubMed] [Google Scholar]
- [37].Azuma H, Yoshida Y, Paul D, Shinoda S, Tsukube H, Nagasaki T. Org. Biomol. Chem. 2009;7:1700–1704. doi: 10.1039/b900154a. [DOI] [PubMed] [Google Scholar]
- [38].Pelletier H, Kraut J. Science. 1992;258:1748–1755. doi: 10.1126/science.1334573. [DOI] [PubMed] [Google Scholar]
- [39].Bushnell GW, Louie GV, Brayer GD. J. Mol. Biol. 1990;214:585–595. doi: 10.1016/0022-2836(90)90200-6. [DOI] [PubMed] [Google Scholar]
- [40].Carlsson GH, Nicholls P, Svistunenko D, Berglund GI, Hajdu J. Biochemistry. 2005;44:635–642. doi: 10.1021/bi0483211. [DOI] [PubMed] [Google Scholar]
- [41].Binda C, Coda A, Aliverti A, Zanetti G, Mattevi A. Acta Cryst. 1998;D54:1353–1358. doi: 10.1107/s0907444998005137. [DOI] [PubMed] [Google Scholar]
- [42].Hildebrand DP, Burk DL, Maurus R, Ferrer JC, Brayer GD, Mauk AG. Biochemistry. 1995;34:1997–2005. doi: 10.1021/bi00006a021. [DOI] [PubMed] [Google Scholar]
- [43].Kundrot CE, Richards FM. J. Mol. Bio. 1987;193:157–170. doi: 10.1016/0022-2836(87)90634-6. [DOI] [PubMed] [Google Scholar]
- [44].Mammen M, Choi SK, Whitesides GM. Angew. Chem. 1998;110:2908–2953. doi: 10.1002/(SICI)1521-3773(19981102)37:20<2754::AID-ANIE2754>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]; Angew. Chem. Int. Ed. 1998;37:2754–2794. doi: 10.1002/(SICI)1521-3773(19981102)37:20<2754::AID-ANIE2754>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- [45].Pavan GM, Danani A, Pricl S, Smith DK. J. Am. Chem. Soc. 2009;131:9686–9694. doi: 10.1021/ja901174k. [DOI] [PubMed] [Google Scholar]
- [46].Mochan E, Nicholls P. Biochim. Biophys. Acta. 1972;267:309–319. doi: 10.1016/0005-2728(72)90119-3. [DOI] [PubMed] [Google Scholar]
- [47].Petersen LC, Cox RP. Biochem. J. 1980;192:687–693. doi: 10.1042/bj1920687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Schrader T, Koch S. Mol. BioSyst. 2007;3:241–248. doi: 10.1039/b614103j. [DOI] [PubMed] [Google Scholar]
- [49].Baldini L, Wilson AJ, Hong J, Hamilton AD. J. Am. Chem. Soc. 2004;126:5656–5657. doi: 10.1021/ja039562j. [DOI] [PubMed] [Google Scholar]
- [50].Wright AT, Griffin MJ, Zhong Z, McCleskey SC, Anslyn EV, McDevitt JT. Angew. Chem. 2005;117:6533–6536. doi: 10.1002/anie.200501137. [DOI] [PubMed] [Google Scholar]; Angew. Chem. Int. Ed. 2005;44:6375–6378. doi: 10.1002/anie.200501137. [DOI] [PubMed] [Google Scholar]
- [51].Zhou H, Baldini L, Hong J, Wilson AJ, Hamilton AD. J. Am. Chem. Soc. 2006;128:2421–2425. doi: 10.1021/ja056833c. [DOI] [PubMed] [Google Scholar]
- [52].You C-C, Miranda OR, Gider B, Ghosh PS, Kim I-B, Erdogan B, Krovi SA, Bunz UHF, Rotello VM. Nat. Nanotechnol. 2007;2:318–323. doi: 10.1038/nnano.2007.99. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.