The selective enrichment of tagged molecules from complex biological mixtures is of primary importance for methods in chemical biology and proteomics. Affinity purification on (strept)avidin beads using biotin as an affinity tag is one of the most widely applied methods to achieve this, fully utilizing the high affinity of biotin for (strept)avidin (Ka ≈ 1.7 × 1015 M−1)[1]. However, the method is limited by the harsh, denaturing conditions required for elution, such as an SDS boil or treatment with 8 M Guanidine (pH 1.5). Protein structure and function are lost and target-proteins may be contaminated with proteins nonspecifically bound to the beads. Two strategies have been explored to elute under mild conditions: i) weakening of the biotin-(strept)avidin interaction by modulating the Ka[2] and ii) introduction of a proteolytically[3] or chemically[4] cleavable linker. While the first strategy does improve the release of biotinylated proteins from (strept)avidin beads, it adversely affects the stringency of the immobilization. The second strategy enables site-specific cleavage, but premature cleavage has been reported and cleavage conditions have no demonstrated compatibility with active biomolecules. In addition, the general applicability of the cleavable linkers is often limited by the need for multistep organic synthesis before implementation.
Hydrazones have been explored for the reversible conjugation[5] and labeling[6-8] of biomolecules. However, their application has been limited as most hydrazones are prone to hydrolysis and premature cleavage, while hydrazones (and oximes) that are fully stable under biological and mildly acidic conditions hydrolyze slower and are difficult to exchange or cleave. In principle a molecular catalyst could be used to accelerate hydrolysis and enable an effective transition between a stable state for work-up and a dynamic state for cleavage and exchange.
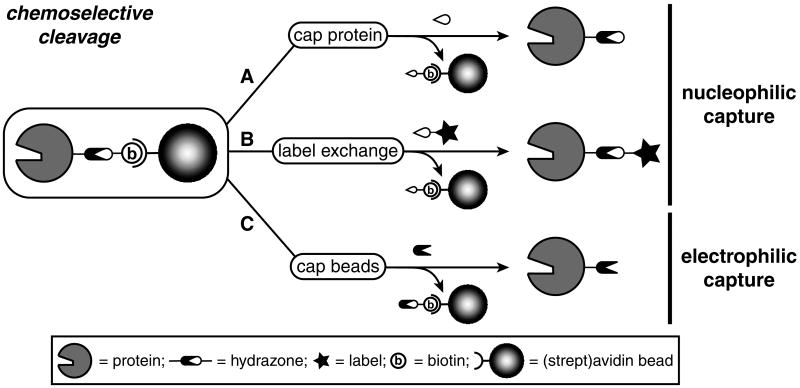
Here, we report that stable, yet exchangeable, bisarylhydrazone linkers can be cleaved under mild conditions by catalytic transimination (Scheme 1). The mechanism of transimination[9] allows for the introduction of a new label to trace the originally biotinylated protein, or a new affinity tag for further purification (Scheme 1, B). Alternatively, the biotin fragment bound to the beads can be capped with a benzaldehyde derivative to elute a protein with a synthetic handle that can be modified at a later time (Scheme 1, C).
Scheme 1.
Three distinct strategies of cleaving a hydrazone linker: A by capping the protein, B by label exchange for further analysis or purification and C by capping the beads, leaving a synthetic handle on the protein for further modification.
The bisarylhydrazone formed between a benzaldehyde and a hydrazinopyridine group has been successfully applied as a stable linker in bioconjugation and biomolecular labeling.[3a,10,11d] Notably, it has been included as a stable component of a protease cleavable biotinylation strategy[3a] and as a stable chromogenic biotinylation agent.[10] The nucleophilic catalyst aniline accelerates effectively hydrazone formation and hydrolysis[11] and a 2 orders of magnitude enhancement in the rate constants can be achieved without changing pH.[11d] As a result, the stable bisarylhydrazone will become dynamic upon addition of the catalyst and make the bond prone to exchange and cleavage.
Indeed, a mixture of two bisarylhydrazones (100 μM each) is stable at pH 7.0 (> 22 hours), but rapidly reequilibrates in the presence of 100 mM aniline at pH 7.0 (< 8 hours) to give four hydrazones (50 μM each) (SI Fig 1). Reequilibration proceeds with the catalyzed rate constant of hydrolysis, suggesting that scrambling occurs through direct reaction of trace free hydrazine and free aldehyde present in solution at equilibrium. Surprisingly, despite considerable analysis of hydrazones in dynamic covalent chemistry[12], this is the first example of hydrazone reequilibration under physiological conditions in the absence of excess monomers.
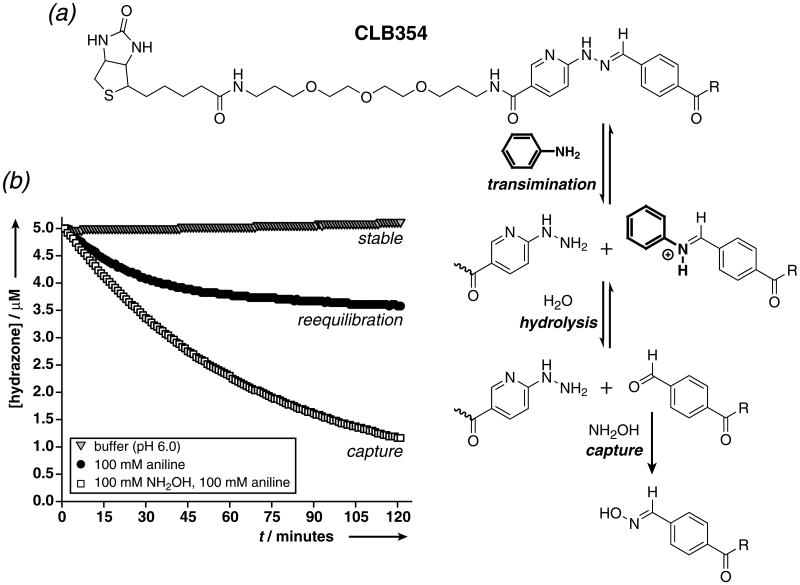
The ability to convert the hydrazone bond effectively between stable and dynamic states is key for its use as an exchangeable linker in pull-down assays for the enrichment and purification of proteins. Biotinylation agent CLB354 (Fig 1a) is commercially available and its hydrazone linker has a distinct absorption in the visible region.[10] The reversibility and stability of CLB354 were first validated in solution at pH 6.0 to facilitate both reequilibration kinetics and protein stability. The CLB354 hydrazone (5 μM) is fully stable overnight, yet upon treatment with 100 mM aniline ∼25% of the hydrazone is rapidly hydrolyzed to reach a new equilibrium (Fig 1b). As the hydrazone has a Keq of ∼106 M−1 in aqueous buffer,[11d] it would need to be diluted significantly to shift its equilibrium toward the starting materials and disrupt the hydrazone bond. Yet, the equilibrium can be pushed toward cleavage by trapping the free aldehyde with an aminooxy compound, such as hydroxylamine (NH2OH), to give a more stable oxime (Fig 1b).[11c,13] The cleavage kinetics depend on the concentration of NH2OH (SI Fig 4) and a significant, but limited enhancement is achieved by the addition of 100 mM aniline (SI Fig 4). This suggests that at high NH2OH concentration (> 10 mM) direct nucleophilic attack on the CLB354 hydrazone competes with aniline-catalyzed transimination. At 100 mM NH2OH and 100 mM aniline, cleavage is quantitative within 8 hours (SI Fig 5).
Figure 1.
(a) Biotinylation agent CLB354.[10] (b) CLB354 (5 μM) in 0.1 M Na phosphate (pH 6.0), in the absence of amines (stable), in presence of 100 mM aniline (reequilibration), and in the presence of 100 mM NH2OH and 100 mM aniline (capture).
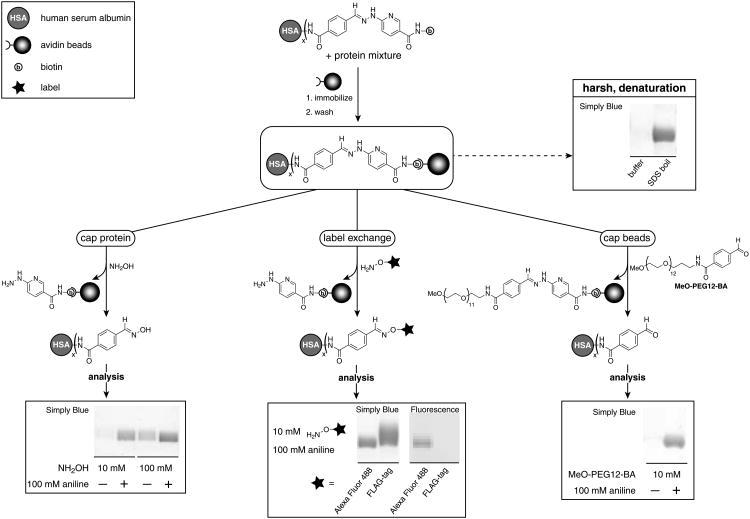
The mechanism of cleavage is expected to be the same in solution and on (strept)avidin beads. However, the kinetics of the transimination reaction may be affected by the accumulation of free hydrazine groups on the beads as the reaction proceeds and by the local environment created by the proteins. To gain insight in the cleavage efficiency on beads, a model protein study was performed in which biotinylated human serum albumin (HSA-CLB354x; x is the average number of CLB354 per HSA) (8 μM) was retrieved from a well-defined protein mixture consisting of aprotinin, cytochrome C, myoglobin, and aldolase (10 μM each) in 50 mM sodium phosphate, 150 mM NaCl (pH 7.4). HSA-CLB354x (x = 1.3) was pulled down from the protein mixture with avidin beads. The beads were washed and incubated for 2 hours at pH 6.0 with varying amounts of NH2OH and aniline (Scheme 2) to elute HSA by capping the protein (Scheme 1, A). As reflected in the SDS-PAGE analysis, the cleavage efficiency is improved, in accordance with the solution study, by increasing the concentration of NH2OH as well as by the addition of aniline.
Scheme 2.
Retrieval of CLB354-labeled human serum albumin (HSA) from a well-defined protein mixture. Cleavage is achieved by capping the aldehyde on HSA with NH2OH (bottom, left) or the hydrazinopyridine group on biotin with PEGylated benzaldehyde (bottom, right). Fluorescently labeled HSA was obtained by cleavage with aminooxyacetyl-Alexa Fluor 488, whereas a FLAG-tag could be introduced by cleavage with aminooxyacetyl-FLAG (bottom, middle). Aniline catalyzes transimination and improves cleavage efficiencies. SDS-PAGE analysis (4-12% Bis-Tris gel, Simply Blue™ stain) shows modified HSA. The hydrazone linker is stable in pH 6.0 buffer (top, right). An SDS boil was performed as a reference (top, right). No detectable levels of the other proteins in the protein mixture are observed after elution. See SI for details.
Cleavage by transimination can be performed with any aminooxy compound and this offers the possibility to refunctionalize HSA by exchanging the biotin tag with a new label or affinity tag (Scheme 1, B). To demonstrate this, the hydrazone of immobilized HSA-CLB354x (x = 1.3) was cleaved in a second experiment by using cleavage buffer containing 10 mM aminooxyacetyl-Alexa Fluor 488 or aminooxyacetyl-FLAG and 100 mM aniline (Scheme 2). Cleavage with 100 mM NH2OH in the presence and absence of 100 mM aniline (see SI), incubation with just cleavage buffer (0.1 M sodium phosphate (pH 6.0)), and an SDS boil were performed as references (Scheme 2). Equal amounts of beads were used per condition. The beads were incubated overnight, except for the SDS boil, and the cleavage efficiencies were determined by densitometry (NIH J Image). In comparison to the SDS boil, 74% of immobilized HSA was recovered with 100 mM NH2OH in the absence of aniline, which is increased to 88% in the presence of aniline. The biotin tag was effectively exchanged with the fluorescent Alexa dye and 63% HSA-Alexa Fluor 488 was retrieved. Exchange with the FLAG tag appeared quantitative. Importantly, no detectable levels of HSA are observed by incubating the beads with buffer alone, confirming that the hydrazone is completely stable at pH 6.0.
All studies utilizing hydrazones as cleavable bonds have used competing amines for cleavage.[5-8] As the use of strongly nucleophilic amines may be incompatible with the function of certain proteins, we explored the alternative strategy by using an aldehyde to capture the hydrazinopyridine group of the biotin fragment (Scheme 1, C). In contrast to cleavage with aminooxy compounds, this strategy requires aniline as a catalyst for transimination, as the aldehyde itself is electrophilic and unable to react directly with the hydrazone bond. A PEGylated benzaldehyde was used to cleave immobilized HSA-CLB354x (x = 1.5) instead of benzaldehyde for solubility. Indeed, in the absence of aniline, 10 mM of PEGylated benzaldehyde does not result in detectable cleavage of the hydrazone overnight (Scheme 2). However, upon addition of 100 mM aniline, transimination occurs and ∼60% of HSA-benzaldehyde is retrieved (Scheme 2), while PEGylated biotin remains on the beads. Importantly, by releasing benzaldehyde-functionalized protein, a new synthetic handle is available for further modification.
Having fully demonstrated the scope of the method in model systems, we implemented CLB354 in our ongoing research program aimed at the discovery of new binding partners of the anticoagulant protein S in addition to its known binding partner, complement factor protein C4bP. Recent studies have suggested that protein S might have several other functions besides its well-known activated protein C (APC) cofactor activity. Owing to its newly discovered functions, there has been renewed interest in the discovery of new binding partners in plasma and on cells.[14] To investigate this, protein S-CLB3543.4 was synthesized and used in a pull-down assay to retrieve its binding partners from human plasma. Protein S-CLB3543.4 and its binding partners were pulled down with avidin beads. The beads were washed and the remaining proteins were eluted overnight with 100 mM NH2OH and 100 mM aniline (pH 6.0). ∼80% of the proteins immobilized on the beads were eluted by this procedure (see SI). The proteins were analyzed by SDS-PAGE and Western blotting. As anticipated, C4bP was enriched and a potential new binding partner of protein S was discovered (see SI).
It is anticipated that the method presented in this paper is compatible with a wide range of biomolecules and the mild cleavage conditions should enable the retrieval of active, structurally intact proteins. Indeed, protein S was found to maintain full APC cofactor activity after incubation overnight with our harshest cleaving condition of 100 mM NH2OH, 100 mM aniline (pH 6.0) (SI Fig 15). Alternatively, proteins that are incompatible with strong nucleophiles can be cleaved with aldehyde in the presence of aniline. The method is highly flexible and can be optimized to meet the conditions and requirements of individual systems. Cleavage times can be reduced significantly by lowering the pH, by increasing temperature, or by increasing the concentration of nucleophiles (aniline, aminooxy groups) in the cleavage cocktail. For example, the bisarylhydrazone can be cleaved quantitatively within 1 hour by using 100 mM NH2OH in 0.1 M anilinium acetate (pH 4.6) (SI Fig 8). The method further distinguishes itself from existing methods by offering the opportunity to introduce a new label or affinity tag, or to preserve a synthetic handle for further modification. These results suggest that detailed analysis of the kinetics and thermodynamics of hydrazones and oximes can be leveraged to expand the utility of these groups in chemical bioapplications.
Supplementary Material
Footnotes
We thank Professor John H. Griffin for providing resources, Phuong M. Nguyen (TSRI) for technical support. This work was supported by NRSA (HL 07695-15 (S.Y.)) and NIH (GM059380 (P.E.D.)).
Supporting information for this article is available on the WWW under http://www.angewandte.org or from the author.
Contributor Information
Dr. Anouk Dirksen, Departments of Cell Biology and Chemistry, The Scripps Research Institute, 10550 N. Torrey Pines Rd, La Jolla, CA 92037 (USA), Fax: (+)1-858-784-7319
Dr. Subramanian Yegneswaran, Department of Molecular & Experimental Medicine, The Scripps Research Institute, 10550 N. Torrey Pines Rd, La Jolla, CA 92037 (USA), Fax: (+)1-858-784-7319
Dr. Philip E. Dawson, Email: dawson@scripps.edu, Departments of Cell Biology and Chemistry, The Scripps Research Institute, 10550 N. Torrey Pines Rd, La Jolla, CA 92037 (USA), Fax: (+)1-858-784-7319.
References
- 1.Green NM. Methods Enzymol. 1990;184:51–67. doi: 10.1016/0076-6879(90)84259-j. [DOI] [PubMed] [Google Scholar]
- 2.a) Hirsch JD, Eslamizar L, Filanoski BJ, Malekzadeh N, Haugland RP, Beechem JM, Haugland RP. Anal Biochem. 2002;308:343–357. doi: 10.1016/s0003-2697(02)00201-4. [DOI] [PubMed] [Google Scholar]; b) Zeheb R, Orr GA. Methods Enzymol. 1986;122:87–94. doi: 10.1016/0076-6879(86)22153-9. [DOI] [PubMed] [Google Scholar]; c) Howarth M, Chinnapen DJF, Gerrow K, Dorrestein PC, Grandy MR, Kelleher NL, El-Husseini A, Ting AY. Nat Methods. 2006;3:267–273. doi: 10.1038/NMETHXXX. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Wu SC, Wong SL. J Biol Chem. 2005;280:23225–23231. doi: 10.1074/jbc.M501733200. [DOI] [PubMed] [Google Scholar]; e) Malmstadt N, Hyre DE, Ding Z, Hoffman AS, Stayton PS. Bioconjugate Chem. 2003;14:575–580. doi: 10.1021/bc020055l. [DOI] [PubMed] [Google Scholar]; f) Morag E, Bayer EA, Wilchek M. Biochem J. 1996;316:193–199. doi: 10.1042/bj3160193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.a) Pfander S, Fiammengo R, Kirin SI, Metzler-Nolte N, Jäschke A. Nucleic Acids Res. 2007;35:e25. doi: 10.1093/nar/gkl1110. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Speers AE, Cravatt BF. J Am Chem Soc. 2005;127:10018–10019. doi: 10.1021/ja0532842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.a) Verhelst SHL, Fonović M, Bogyo M. Angew Chem. 2007;119:1306–1308. doi: 10.1002/anie.200603811. Angew. Chem. Int. Ed. 2007, 46, 1284–1286. [DOI] [PubMed] [Google Scholar]; b) Gartner CA, Elias JE, Bakalarski CE, Gygi SP. J Prot Res. 2007;6:1482–1491. doi: 10.1021/pr060605f. [DOI] [PubMed] [Google Scholar]; c) Fauq AH, Kache R, Kahn MA, Vega IE. Bioconjugate Chem. 2006;17:248–254. doi: 10.1021/bc0503059. [DOI] [PubMed] [Google Scholar]; d) van der Veken P, Dirksen EHC, Ruijter R, Elgersma RC, Heck AJR, Rijkers DTS, Slijper M, Liskamp RMJ. ChemBioChem. 2005;6:2271–2280. doi: 10.1002/cbic.200500209. [DOI] [PubMed] [Google Scholar]; e) Shimkus M, Levy J, Herman T. Proc Nat Acad Sci USA. 1985;82:2593–2597. doi: 10.1073/pnas.82.9.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.King TP, Zhao SW, Lam T. Biochemistry. 1986;25:5774–5779. doi: 10.1021/bi00367a064. [DOI] [PubMed] [Google Scholar]
- 6.Carrico IS, Carlson BL, Bertozzi CR. Nat Chem Biol. 2007;3:321–322. doi: 10.1038/nchembio878. [DOI] [PubMed] [Google Scholar]
- 7.Park KD, Liu R, Kohn H. Chem Biol. 2009;16:763–772. doi: 10.1016/j.chembiol.2009.06.005. During the preparation of this manuscript an alternative cleavable acylhydrazone linker was described. Acylhydrazones have a small Keq (∼104 M−1) and hydrolyze appreciably within 2 hours in physiological (∼ 5% at pH 7.4) and mildly acidic (∼40% at pH 5.8) buffer, leading to premature cleavage, consistent with studies that state that acylhydrazones are not stable and should not be used for bioconjugation.[13] In contrast, bisarylhydrazones have a large Keq (∼106 M−1), are fully stable for > 16 hours (also at slightly acidic pH), and only cleaved when cleavage agents are added. [DOI] [PubMed] [Google Scholar]
- 8.Takaoka Y, Tsutsumi H, Kasagi N, Nakata E, Hamachi I. J Am Chem Soc. 2006;128:3273–3280. doi: 10.1021/ja057926x. [DOI] [PubMed] [Google Scholar]
- 9.Cordes EH, Jencks WP. J Am Chem Soc. 1962;84:832–837. [Google Scholar]
- 10.Schwartz DA SoluLink Biosciences Inc. U S Patent. 2004 Oct 5;6,800,728 [Google Scholar]
- 11.a) Cordes EH, Jencks WP. J Am Chem Soc. 1962;84:826–831. [Google Scholar]; b) Dirksen A, Hackeng TM, Dawson PE. Angew Chem. 2006;118:7743–7746. doi: 10.1002/anie.200602877. Angew. Chem. Int. Ed. 2006, 45, 7581–7584. [DOI] [PubMed] [Google Scholar]; c) Dirksen A, Dirksen S, Hackeng TM, Dawson PE. J Am Chem Soc. 2006;128:15602–15603. doi: 10.1021/ja067189k. [DOI] [PubMed] [Google Scholar]; d) Dirksen A, Dawson PE. Bioconjugate Chem. 2008;19:2543–2548. doi: 10.1021/bc800310p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Corbett PT, Leclaire J, Vial L, West KR, Wietor JL, Sanders JKM, Otto S. Chem Rev. 2006;106:3652–3711. doi: 10.1021/cr020452p. [DOI] [PubMed] [Google Scholar]
- 13.a) Kalia J, Raines RT. Angew Chem. 2008;120:7633–7636. Angew. Chem. Int. Ed. 2008, 47, 7523–7526. [Google Scholar]; b) Kalia J, Raines RT. Curr Org Chem. 2010;14:138–147. doi: 10.2174/138527210790069839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.a) Yegneswaran S, Hackeng TM, Dawson PE, Griffin JH. J Biol Chem. 2008;283:33046–33052. doi: 10.1074/jbc.M806527200. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Heeb MJ, Prashun D, Griffin JH, Bouma BN. FASEB J. 2009;23:2244–2253. doi: 10.1096/fj.08-123174. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.