Abstract
Molecular analyses of hair follicle formation provide evidence to support the most well-known mathematical model for biological pattern formation.
What are the underlying mechanisms that give rise to complex patterns in biology? Despite recent advances in biotechnology and mathematical modeling, this still remains a largely open question. As reported on page 1447 of this issue, Sick et al. have made a major advance toward answering this question by identifying key molecular players in hair follicle growth and by confirming the validity of perhaps the best-known mathematical model for biological pattern formation (1).
In a seminal paper, Alan Turing proposed that spatial patterns result from a phenomenon he termed “diffusion-driven instability” (2). He showed mathematically that small spatial fluctuations in an otherwise well-mixed system of reacting and diffusing chemicals could become unstable, and that amplification of these fluctuations could lead to a spatial pattern of chemicals that he termed morphogens (i.e., substances that stimulate the development of form or structure in an organism). He proposed that this spatial arrangement could serve as a prepattern for development. Turing's work was ground-breaking because the mathematical nature of the resulting patterns is wholly counterintuitive; since their discovery, they have motivated much mathematical research. However, the model has been the subject of controversy because it has been deemed too simplistic and the search for real biological examples has been neglected. Moreover, although diffusion-driven instability has been shown to be present in chemistry, there is substantial evidence in the fruit fly Drosophila to refute the model for biology (3). The report by Sick et al., by providing the first compelling biological evidence for the Turing model, is thus a landmark publication.
The formation of skin appendages (hairs, feathers, etc.) is an excellent paradigm for patterning because these systems are amenable to experimental manipulation. Nagorcka was the first to propose the Turing model to explain hair pattern formation (4), but at that stage the molecular biology was lagging behind the theory. It was only in 1998 that Jung et al. made the first efforts to link known molecular morphogens with a reaction-diffusion mechanism for feather germ formation (5). They showed how the size, number, and distribution of appendages could be modulated by altering morphogen concentrations (6).
Sick et al. investigated the regulation of hair follicle patterning in developing murine skin. They propose that the protein WNT and its inhibitor DKK are morphogens in the Turing sense. Expression of the protein Dkk1, which inhibits WNT, is actually controlled by secreted WNTs, and both WNTs and DKKs are secreted into the extracellular space where they diffuse, thereby acting over longer distances. Given that the WNT proteins are substantially larger than the DKKs, one would expect a large difference in their rates of diffusion. This makes possible the classical “short-range activation, long-range inhibition” phenomenon that underlies diffusion-driven instability (7).
Because hair follicle patterning occurs in waves, the authors used a reaction-diffusion model to set up an initial pattern of follicles. Then, along the same lines as Mooney and Nagorcka (8), they assumed these follicles to be chemical sources giving rise to a second wave of hair follicle formation on a larger domain (due to the growth of skin).
The model predicts that moderate overexpression of activator (WNT) increases follicular density, whereas moderate overexpression of inhibitor (DKK) during the initial inductive wave increases the interfollicular spacing. Sick et al. have verified these predictions experimentally, providing strong evidence for a genetic underpinning of a Turing reaction-diffusion model.
Together the papers of Jung et al. and Sick et al. show that the skin progenitors are stem cells, in that they are multipotent and may assume appendage or interappendage fates depending on the local chemical environment at the time of specification. In this sense, the molecular components identified by these experiments appear to be acting as morphogens in the true Turing sense.
In principle, a reaction-diffusion model can set up a chemical prepattern before we can visualize changes in cell distribution. That is, it determines sites at which cells will cluster: Regions of high cell density coincide with those of increased morphogen concentration—although the model does not specify how this rearrangement occurs. On the other hand, it is possible for cellular aggregations to form without such a prepattern via simple chemotactic movement in response to gradients in chemical concentration. By way of illustration, the patterns formed by these two different mechanisms are shown in the figure. It is immediately obvious how similar such patterns are.
Biological pattern formation.
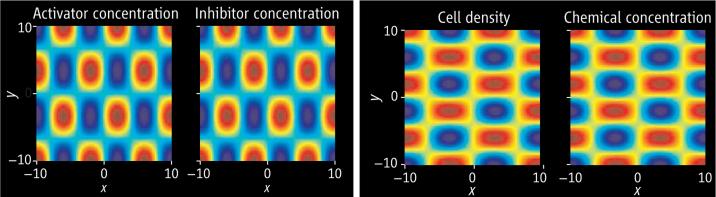
Two mechanisms can show similar results. (Left) Outcome of a reaction-diffusion model (7) in which activator and inhibitor react and diffuse. Small random fluctuations in the initial field lead to coinciding spatial patterns of activator and inhibitor concentration. (Right) Results of a cell chemotaxis model (9) in which cells and chemical both diffuse, with cells also moving up gradients in chemical concentration. Again, small random fluctuations in the initial field lead to coinciding spatial patterns in cell density and chemical concentration. Blue indicates low concentration levels; red indicates high levels.
This highlights one of the difficulties in mathematical modeling: determining which is the “correct” model. Now that WNT and DKK have been identified as possible morphogens, this issue can be addressed experimentally. The key requirement, then, is that the results of such experiments are used to test and refine models, ruling some out if the data allow us to do so. The WNT-DKK interaction does appear to be qualitatively of the form necessary for a Turing-type system, but it is now imperative that we try to overcome the experimental challenges in measuring key parameters (rates of production, decay, diffusion coefficients, etc.) so that quantitative tests can be performed to determine whether the system actually is of Turing type. This would then be the first definitive example of the Turing model in biology.
Turing models have been proposed to describe other types of patterns observed in developmental biology. Two applications currently receiving much attention from experimentalists are pigmentation patterning in fish and skeletal development in the mouse limb. Although the evidence for a Turing diffusion-driven instability in these systems is not as strong as that presented by Sick et al., their report should stimulate further work in biological pattern formation.
Acknowledgments
Supported by Research Councils UK, Lloyds Tercentenary, Microsoft Corporation, and St. Hugh's College, Oxford (R.E.B.) and by NIH (C.-M.C.).
Contributor Information
Philip K. Maini, Center for Mathematical Biology, University of Oxford, Oxford OX1 3LB, UK, and the Oxford Center for Integrative Systems Biology, University of Oxford, Oxford OX1 3QU, UK.
Ruth E. Baker, Center for Mathematical Biology, University of Oxford, Oxford OX1 3LB, UK.
Cheng-Ming Chuong, Department of Pathology, University of Southern California, Los Angeles, CA 90033, USA..
References and Notes
- 1.Sick S, Reinker S, Timmer J, Schlake T. Science. 2006;314:1447. doi: 10.1126/science.1130088. published online 2 November 2006 (10.1126/science.1130088) [DOI] [PubMed] [Google Scholar]
- 2.Turing AM. Philos. Trans. R. Soc. London Ser. B. 1952;237:37. [Google Scholar]
- 3.Akam M. Nature. 1989;341:282. doi: 10.1038/341282a0. [DOI] [PubMed] [Google Scholar]
- 4.Nagorcka BN. Biosystems. 1983–1984;16:323. doi: 10.1016/0303-2647(83)90015-1. [DOI] [PubMed] [Google Scholar]
- 5.Jung H-S, et al. Dev. Biol. 1998;196:11. doi: 10.1006/dbio.1998.8850. [DOI] [PubMed] [Google Scholar]
- 6.Jiang T-X, Jung H-S, Widelitz RB, Chuong C-M. Development. 1999;126:4997. doi: 10.1242/dev.126.22.4997. [DOI] [PubMed] [Google Scholar]
- 7.Gierer A, Meinhardt H. Kybernetik. 1972;12:30. doi: 10.1007/BF00289234. [DOI] [PubMed] [Google Scholar]
- 8.Mooney JR, Nagorcka BN. J. Theor. Biol. 1985;115:299. doi: 10.1016/s0022-5193(85)80102-8. [DOI] [PubMed] [Google Scholar]
- 9.Myerscough MR, Maini PK, Murray JD, Winters KH. In: Dynamics of Complex Interconnected Biological Systems. Vincent TL, Mees AI, Jennings LS, editors. Birkhäuser; Boston, MA: 1990. pp. 65–83. [Google Scholar]


