Abstract
Lassa virus infection elicits distinctive changes in host gene expression and metabolism. We focus on changes in host gene expression that may be biomarkers that discriminate individual pathogens or may help to provide a prognosis for disease. In addition to assessing mRNA changes, functional studies are also needed to discriminate causes of disease from mechanisms of host resistance. Host responses that drive pathogenesis are likely to be targets for prevention or therapy. Host responses to Lassa or its related arenaviruses have been monitored in cell culture, in animal models of hemorrhagic fever, in Lassa-infected nonhuman primates and, to a limited extent, in infected human beings. Here, we describe results from those studies and discuss potential targets for reducing virus replication and mitigating disease.
Keywords: hemorrhagic fever viruses, Lassa fever, transcriptome profiling
Background
History
Lassa fever (LF) is an acute viral disease endemic to West Africa. It was first described in 1969 when two missionary nurses died in the town of Lassa, Nigeria that gave its name to the etiological agent, Lassa virus (LASV) [1–5]. Soon after LASV was described, outbreaks in several hospitals showed the virus was circulating widely in Sierra Leone, Liberia and Guinea [6–11]. Although LASV was identified in 1969, records of hemorrhagic fever (HF) since the 1920s indicate this virus had been circulating for at least 50 years [12,13].
Several studies [10,14–15] have shown LASV seroprevalence up to 50% in the endemic regions. However, since severe LF appears sporadically despite high rates of infection, disease onset is rare. Of more than 300,000 LF cases occurring every year, there are around 5000 human deaths [16,17] but the lack of systematic surveillance and absence of convenient diagnostics obscure the true picture of LF in West Africa. As of 2014, LF outbreaks were reported in Nigeria, Liberia, Sierra Leone, Guinea and the Central African Republic, but the virus likely circulates in the Democratic Republic of the Congo, Mali and Senegal as well [18,19].
The rates for disease are impacted by LASV strain variation with individual isolates differing by up to 27% in nucleotide sequence [20]. We also know that large areas of central and southern Africa have Mopeia-like viruses that are related to LASV but do not cause disease [21]. Disease also depends on variations in host susceptibility due to concurrent infections or genetic differences. For example, west Africans have alleles of the genes LARGE and IL-21 that reduce LASV replication [22,23].
Clinical manifestations
Current knowledge is based on meager data from people, combined with data extrapolated from LF animal models [5]. Even though person-to-person transmission appears to be inefficient, the aerosol transmissibility of LASV and the post-9/11 fears [24], caused LASV to be classified as a category A biothreat agent [25]. Around 80% of LASV infections are mild or asymptomatic; the remaining cases evolve to LF or multisystem organ failure and death. Overall, the mortality rates are between 1 and 2%, and among hospital cases mortality can reach 20% [26].
LF pathogenesis and the host immune response are poorly characterized. There is a strong correlation between viremia and disease, but tissue damage is not caused by direct viral-cell lysis and seems due to the host response [27–29]. Initial flu-like symptoms are poorly differentiated from other diseases and the incubation period is long (between 1 and 2 weeks). Those two situations make the initial diagnosis difficult and delay the initiation of treatment. Twenty percent of the infected patients develop symptoms including: muscle fatigue, facial edema and sore throat. A small number of these patients progress to systemic disease with mucosal, conjunctival, gastrointestinal or genital bleeding. Platelet dysfunction and endothelial damage are important for vascular leakage in the most severe cases.
Causes of death include myocarditis, pulmonary edema, acute respiratory distress and hypo-volemic shock. Liver AST/ALT changes, uncontrolled viremia and high levels of IL-6 in plasma [28] are contributors to fatal disease. Massive viral replication in liver and spleen leads to progressive hemorrhage and increased mortality. It is common to find hepatocellular necrosis, but it is insufficient to cause death [9,30,31]. Platelet counts are usually normal, while platelet aggregation is impaired [32,33]. Other coagulations factors remain in the normal range and there are few signs of disseminated intravascular coagulation [33–36].
CNS manifestations, such as disorientation, motor problems or hiccups, are associated with poor LF prognosis. Approximately 30% of LF patients develop hearing problems and 17% of LF survivors suffer permanent hearing loss [5,9,37–39].
LASV infection during pregnancy increases the risk of death for mothers from 7% during the first two trimesters to 30% during the third trimester [13,40]. High titers of LASV are found in placenta and fetal organs, possibly due to the relatively immunosuppressed state of pregnancy [40].
LASV in animal models
At the cellular level, LASV binds α-dystroglycan [41,42] and high-mannose receptors [43] to infect tissue macrophages (MPs) or dendritic cells (DCs). From there, the virus spreads to regional lymph nodes, liver, spleen, kidney, adrenal gland, lung, heart, pancreas, placenta, uterus and breast where it can also infect fibroblastic stromal cells and endothelium [44,45]. Cell culture studies [46–48] showed that LASV inhibits cytochemokine production to decrease the recruitment of immune cells to the sites of infection and dampen the development of a vigorous immune response. At the same time, infection enhances the release of vasoactive mediators that cause edema in a process similar to a septic shock [5,49]. Infection of hepatocytes or adrenal cortical cells contributes to coagulation dysfunction, hypotension, sodium loss with hypovolemia and multi-organ failure [21,50].
Murine models using lymphocytic choriomeningitis virus (LCMV) showed tissue damage resulting from the cytotoxic T lymphocyte response that is preventable with immuno-suppressive drugs [51–53]. However, nonhuman primates (NHPs) or guinea pigs do not respond to these treatments suggesting that the virulence of LASV in man, NHPs and guinea pigs is not a predominantly immune-mediated pathology [52,54].
Our laboratory used the common marmoset (Callithrix jacchus) as a NHP model. After LASV infection, marmosets developed systemic disease resembling LF with weight loss, high viremia, elevated liver enzymes and death 15–20 days after inoculation. Histopathology revealed mild liver inflammation and hepatocyte proliferation with necrotic aggregates composed of MPs with a reduction in the intensity of HLA-DP, DQ and DR staining [55]. These findings are similar to those seen in rhesus macaques [32], cynomolgus macaques [56] and human LF cases [21,57]. Profiling studies in primates will describe specific genes and pathways affected by LASV and how those markers can explain the clinical features of LF.
Diagnosis
Early events in LASV infection are complex and it has been difficult to design early diagnostic tests [57]. Consequently, there is no commercially available diagnostic assay for early laboratory detection of LASV infection. Early diagnosis is important for case classification, implementation of prevention measures, contact-tracing and treatment initiation.
LASV isolation is the ‘gold standard’ for LF diagnosis and must be done in a high containment Biosafety Level-4 (BSL-4) facility, over several days [58]. Virus neutralization tests, reversed passive hemagglutination and inhibition, and complement fixing antibody detections were some of the first techniques tested as LASV diagnostics but detectable antibodies appear late after infection and are only useful for surveillance of the population [59–62]. Indirect immunofluorescence assay (IFA) can be used for LF antibody detection (an increase of more than four-times in IgG-LASV titers, IgG of ~1:256 on admission or IgM-LASV titer of ~1:4 is considered positive) however; specificity is low in populations with small apparent risk of infection and does not distinguish between LASV strains. IFA detects antibodies 7–10 days after the onset of illness, 3–4 days faster than the cytopathic effects (CPE) seen by virus isolation [57,63,64]. ELISA was developed for LASV-IgG, LASV-IgM and antigen detection. This test improved sensitivity and specificity, however, early detection was not improved except when using antigen assays that were positive on the day of patient admission [10,65–71]. Other studies showed increased sensitivity or specificity by combining IgM and antigen assays, but detection has rarely been fast enough for patients to initiate therapy and survive. It is also important to avoid unnecessary ribavirin therapy in case the patients are suffering from other diseases [58].
Molecular methods such as western blot analysis have also been tried for detecting mainly NP and GP2 antibodies [10]. With the use of this technique, it was possible to detect both IgG and IgM against NP in acute samples with very low positive predictive value (53.6%) but high negative predictive value (93%) [72]. Reverse transcription-PCR (RT-PCR) proved useful for LASV detection with 100% sensitivity [73,74]. By using specific primers for a conserved region in the S segment, up to 79% positive predictive value was achieved, compared with 21% for IFA. Testing of three sequential samples after admission increased the positive predictive value to 100% compared with 52% for IFA [75]. Later these protocols were optimized by including a real-time PCR assays [76], second rounds of amplification (nested PCR) [77], targeting several sites on the L segment [78] or combining PCR with sequencing [79]. With PCR there are issues of strain variation (nucleotide mismatching), cross-contamination and high implementation costs in resource-poor countries [79].
A new method called ‘lateral flow immunoassay’ enables NP-specific antigen detection in serum samples and is under preclinical studies by a LF consortium with Corgenix Medical Corporation, CO, USA [71].
All diagnostics described so far detect proteins or nucleic acids of LASV. Host responses to infection might also contribute to accurate diagnosis or the ability to predict clinical disease. Studies in rhesus macaques infected with LCMV [80] or people infected with influenza [81,82] established patterns of virus-specific host responses occurring before viremia. A goal for this work is to develop diagnostic chips that will measure changes in host gene expression that are predictive of disease onset. Unfortunately, despite a great deal of research investment into predictive chips, no such diagnostics are currently available due to the great person-to-person variation in disease susceptibilities and immune responses.
We speculate that the problem of human variation could be solved by an intense effort to classify people into transcriptome types. For example, after characterizing the blood transcriptome of 100 healthy people and hierarchical clustering of their profiles, it may be possible to determine whether the clustering follows any external characteristics (such as gender, age, ethnicity, metabolic rate and size, among others). Let us imagine that the clustering allows us to classify the 100 subjects into five ‘groups.’ Since biomarker chip analysis relies on comparison to an appropriate healthy control, then a healthy person from your ‘group’ can serve as your comparator when you get sick. When we profiled NHPs it was possible to use their pre-infection bloods as the ‘healthy control’ and our results were very consistent for a single animal. Based on NHP studies, the next-generation LASV diagnostic would determine whether a suspected Lassa case has a drop in mRNA for NR4A2 or COX2 and a rise in mRNA for CD14, fibronectin1 or chemokine receptor CCR2. The drop or rise would be with respect to the expression of those genes in a healthy comparator. If the sick person fits this profile, they would be likely to be harboring a virulent case of LF disease.
Clinical and laboratory diagnosis of LASV has been improved during the last 20 years by the introduction of new antibody or nucleic acid detection assays. The new technology shortens the time between infection and diagnosis but fails to detect infection during the incubation period. Thus, there are important opportunities for more advanced LF diagnostics that incorporate early patterns of host responses with pathogen detection.
Treatment
To date there are no vaccines or therapies specifically licensed for LF. Patients suspected to have LF are treated based on symptoms and differential diagnosis. Supportive measures such as aggressive rehydration, blood transfusion and continuous monitoring are used when available. If the probability of having a LASV infection is high, patients are isolated [57].
Ribavirin is the only licensed antiviral drug with reported activity against LASV, Junin (JUNV) and Machupo viruses (MACV) [54,83,84]. However, its utility is limited because it must be administrated early in disease, during the time when diagnosis is difficult. Patients treated seven or more days after the onset of fever had a mortality rate of 26% compared with 9% in individuals treated during the first 6 days of fever [54]. Intravenous ribavirin (the optimal route of administration) is costly and not widely available [85], so new antivirals are needed.
Treating people with immune-antiserum showed inconsistent results and animal studies showed that sera often failed to protect against LASV challenge [54,86]. Anecdotal reports of protection with human convalescent plasma or of guinea pigs with high neutralizing titer sera did imply success [87,88]. A combination of ribavirin plus immune serum increased survival in LASV-infected monkeys, suggesting that this combination might be useful for treating human beings with LF [89]. The combination of IFN-alfacon-1 (Escherichia coli-expressed human IFN-α) plus ribavirin had synergistic activity that slowed disease and reduced fatality rates in hamsters inoculated with the arenavirus, Pichindé [90], which serves as a small animal model for LF.
New antiviral compounds against LASV are being developed. Guinea pigs treated with ST-193 had a survival rate of 62.5% in contrast to no survival among controls or ribavirin-treated animals [91]. In the same animal model favipiravir (T-705) allowed full recovery from Pichindé virus (PICV) infection [92]. Several groups are still searching for safe and effective anti-LASV medicines, for example, Zapata et al. published a table of US FDA-approved drugs that would be expected to counteract disease-associated gene-expression changes [93–95].
Lassa vaccine approaches
Both practical and biological obstacles have impeded the search for a LASV vaccine. LASV testing is limited to BSL-4 facilities, which are scarce and inconvenient. Genetic diversity among LASV strains requires vaccine candidates to induce a broad cross-protective immunity [20]. Our lack of knowledge about disease mechanisms and immune correlates also slows progress [96]. Despite these obstacles, some progress has been made.
Early vaccine studies focused on inactivated virus that elicited a strong humoral immune response but failed to protect rhesus macaques from LASV challenge [97], or protected 50% of challenged north African baboons (Papio hamadryas) [98]. These differing results may be due to the differing NHP models, inactivation methods or challenge doses [99]. Viral subparticles [100], peptides (LASV NP conferred around 50% protection in mice but was highly toxic in sensitized mice) [101,102], DNA vaccines [103] or live-attenuated vectors are more promising options due to their capacity for inducing strong cellular immune responses.
One oral administration of a recombinant aroA-attenuated Salmonella typhimurium expressing LASV-NP induced LASV humoral and cell-mediated immune responses in mice [104]. Immunization with vaccinia virus expressing the complete glycoprotein gene (GPC) or NP from LASV, partially protected guinea pigs from LASV lethal challenge infection but those animals developed viremia and some LF symptoms [105–107]. Vaccinia vectors expressing several combinations of LASV structural proteins (NP, GP1 and GP2) were most successful in NHPs, using constructs expressing the whole GPC or the combined GP1 and GP2. However, immunized monkeys survived LF challenge but were still infected [108]. Similar results were seen in cynomolgus macaques vaccinated with vesicular stomatitis virus (VSV) expressing LASV-GPC or in guinea pigs vaccinated either with Venezuelan equine encephalitis or yellow fever 17D (YF17D) vectors, both expressing LASV-GPC. Despite achieving protection from lethal disease, these vectors often triggered small increases in liver enzymes and detectable LASV viremia indicating only partial protection [109–112]. The VSV- and Vaccinia-vectored Lassa vaccines could confer 100% protection against homologous LASV challenge in animal models, but safety concerns about their use in populations with high HIV-seroprevalence have dampened interest in them. For example, vaccinia vectors have been known to cause pustules in infants and immune-suppressed people, whereas the VSV vectors can be neurotropic and cause encephalitis. Live vectors cannot be confined to the vaccinated person so they pose a danger to bystanders with weak immune systems.
Mopeia virus (MOPV) is closely related to LASV, infecting the same Mastomys species of rodents as LASV. MOPV endemic areas have no reported Lassa-like cases and animals inoculated with this virus survive LASV lethal challenge [26,69,113–117]. This observation suggests that MOPV may be an excellent natural vaccine [99]. Fears that the MOPV genome could re-associate with other viruses to transform into a human pathogen are mitigated by the fact that the attenuated forms of these viruses will be much more abundant in fully immunized populations [69].
Our laboratory developed and characterized a reassortant virus called ML29 that contained the L segment from nonpathogenic MOPV and the S segment from LASV Josiah strain [118,119]. The L segment controls virus replication and pathogenesis; the S segment carries most of the viral antigens. ML29 has proved to be, in vitro or in vivo more attenuated than its parental MOPV [69,120]. So far ML29 is the only vaccine to provide sterilizing immunity after a lethal LASV challenge in multiple animal models and using broadly different LASV strains. It is the only LHF vaccine shown to elicit strong cell-mediated immune responses prior to challenge (allowing the use of surrogate markers for efficacy during vaccine production) [77,118,121]. In guinea pigs, ML29 provided effective protection when delivered up to 2 days after challenge (as a postexposure therapy) [121]. In addition, ML29 was safe and immunogenic in rhesus macaques with AIDS [121,122]. These experiments add value to ML29, since it is most likely to be used in HIV endemic areas and as a LASV postexposure vaccine.
Profiling of host responses is an emerging technology for predicting vaccine efficacy. A landmark paper on YF17D responses showed a combination of six upregulated genes [123] that predicted good antibody responses to YF17D in man. None of these six biomarkers were upregulated after vaccinating monkeys with ML29, the Lassa vaccine candidate, indicating that these markers might be specific for yellow fever or might differ between humans and NHPs. Transcriptome profiling was able to demonstrate the benign nature of ML29 vaccination, even in the context of AIDS, when compared with lethal infection with LCMV-WE [124].
Several vaccine candidates showed protection against LF in animal models. However, only ML29 conferred sterilizing immunity with broad pre- and postexposure protection and no adverse events in healthy or immuno-compromised animals. ML29 succeeded in complying with the ‘two-animal rule’ required by the FDA and, in comparison to other vaccine approaches, can be produced cheaply in West Africa.
Profiling disease progression
Profiling LCMV-infected NHPs as a model for LASV infection
Studies in vivo and in vitro sought to obtain the RNA-expression profiles of LASV infection or the profile of a related arenavirus, LCMV [80,124–126]. Those experiments were aimed at finding early diagnostic biomarkers that might also predict disease outcomes. Virulent and mild infections were compared in order to identify changes specific for severe disease. For practical reasons, most studies concentrated on venous blood sampling. Virus infection of primates progressed from previremic to viremic stages before recovery or death (Figure 1). Rhesus macaques infected with virulent LCMV-WE or the attenuated strain, LCMV-Armstrong (LCMV-ARM) were sampled 1–12 days after infection and then euthanized humanely. Blood and tissue samples were analyzed by RNA extraction and transcriptome-profiling with Affymetrix human microarray chips. Of 54,000 probesets, 12% represented genes that were differentially regulated in periperal blood mononuclear cells (PBMCs) during infection with virulent LCMV-WE or mild LCMV-Armstrong. Approximately 480 of these genes differed in expression between LCMV-ARM and LCMV-WE infections and of these, 91 genes differentiated mild from severe infections during the previremic stage [80]. A caveat for interpreting transcriptome profiles is that changes in gene expression do not always reflect changes in gene product expression that are often subject to post-transcriptional controls; therefore, we present these profiles as work in progress.
Figure 1. Comparison of progression of viral hemorrhagic fever in lymphocytic choriomeningitis virus WE strain-infected rhesus macaques, in Lassa virus-infected macaques and in surviving people or macaques.
ISN: IFN-stimulated gene; LASV: Lassa virus; LCMV-WE: Lymphocytic choriomeningitis virus WE strain.3 For color figures, please see online at www.futuremedicine.com/doi/full/10.2217/FVL.15.1
Previremic gene expression revealed major disruptions in eicosanoid (COX2), immune response and signaling pathways. The eicosanoids, including prostaglandins and leukotrienes, are potent arachidonic acid derivatives. The activator of ALOX5AP and the gene for COX-2 or PTGS2 were downregulated in virulent infection [80]. Disruption of the eicosanoid pathway could contribute to myocarditis and hemorrhagic disease signs. Immune response genes (early growth response genes, EGR, EGFR and IL-1R) were downregulated early in the previremic phase while IFN-stimulated genes (ISGs) including STAT1, CIG5, IFI44, IFIT4, GBP1, OAS2, OAS-L and MX2 were upregulated and remained high in the viremic phase [80]. The ISGs are upregulated during most viral infections (Table 1) and are known to have antiviral functions like promoting the production of cytokines and innate immunity.
Table 1.
Genes upregulated after exposure to different RNA or DNA viruses.
| Gene name† | Virus‡ | |||||
|---|---|---|---|---|---|---|
| LASV | YF17D | Dengue | Hantavirus | Ebola | Vaccinia | |
| DDX58 (RIG-1) | ✓ | ✓ | ✓ | ✓ | ||
| DHX58 | ✓ | ✓ | ||||
| DSPTI1 | ✓ | ✓ | ✓ | |||
| EIF2AK2 | ✓ | ✓ | ✓ | |||
| EPSTI1 | ✓ | ✓ | ||||
| GBP1 | ✓ | ✓ | ✓ | ✓ | ||
| HERC5 | ✓ | ✓ | ✓ | ✓ | ||
| HERC6 | ✓ | ✓ | ✓ | ✓ | ||
| IFIH1 | ✓ | ✓ | ||||
| IFI6 | ✓ | ✓ | ✓ | ✓ | ||
| IFI44 | ✓ | ✓ | ✓ | ✓ | ✓ | |
| IFI44L | ✓ | ✓ | ✓ | |||
| IFIT1 | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| IFIT2 | ✓ | ✓ | ✓ | |||
| IFIT3 | ✓ | ✓ | ✓ | ✓ | ||
| IFIT5 | ✓ | ✓ | ||||
| IFI35 | ✓ | ✓ | ||||
| IFIH1 (MDA-5) | ✓ | ✓ | ✓ | ✓ | ||
| IFITM1 | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| IFITM3 | ✓ | ✓ | ||||
| IL-1a | ✓ | ✓ | ✓ | ✓ | ||
| IRF7 | ✓ | ✓ | ✓ | ✓ | ✓ | |
| ISG15 | ✓ | ✓ | ✓ | ✓ | ||
| ISG20 | ✓ | ✓ | ✓ | |||
| LY6E | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| MMP1 | ✓ | ✓ | ||||
| MX1 | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| MX2 | ✓ | ✓ | ✓ | ✓ | ✓ | |
| OAS1 | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| OAS2 | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| OAS3 | ✓ | ✓ | ✓ | ✓ | ||
| OASL | ✓ | ✓ | ✓ | |||
| PARP12 | ✓ | ✓ | ✓ | |||
| PLAU | ✓ | ✓ | ||||
| PNPT-1 | ✓ | ✓ | ||||
| RSAD2 | ✓ | ✓ | ✓ | |||
| SP110 | ✓ | ✓ | ✓ | |||
| SCG5 | ✓ | ✓ | ||||
| STAT1 | ✓ | ✓ | ✓ | ✓ | ||
| THBM | ✓ | ✓ | ✓ | |||
| TRIM5 | ✓ | ✓ | ||||
| TRIM22 | ✓ | ✓ | ✓ | ✓ | ||
| TNFS10 | ✓ | ✓ | ✓ | |||
| XAF-1 | ✓ | ✓ | ✓ | |||
These are names of commonly upregulated genes after a cell is exposed to a virus. Most of these genes are ‘IFN-stimulated genes’ or ISGs.
The viruses are LASV [124,127], the vaccine strain Yellow Fever-17D (YF17D) [123,128,129], Dengue virus [130,131], Hantavirus [132], Ebola virus [133,134] and a vaccine strain of poxvirus, Vaccinia [129].
LASV: Lassa virus.
Early downregulation of EGFR and IL-1R during virulent LCMV infection in NHPs was similar in the PICV-infected guinea pig model as shown by protein phosphorylation in guinea pigs infected with the virulent P18 or the attenuated P2 isolates of PICV. The attenuated P2 virus elicited an early pro-inflammatory response while P18 suppressed host cell signaling [135]. P2 increases DNA binding of the activating RelA/p50 dimer of NF-κB, whereas P18 increases DNA binding of the repressive p50/p50 homodimeric form [136]. A similar mechanism was reported for suppression of IL-8 or IL-6 expression in cultured cells by virulent but not attenuated arenaviruses [137]. Meta-analyses of profiling studies in several laboratories identified transcription factors that were shared among some of the virulent infections. For example, the mitogen-activated protein kinase pathway, p38 in particular, seems to control responses to LCMV and Pichindé infections [138].
RNA profiling analyses of liver tissue from monkeys infected with LCMV-WE (virulent), or with LCMV-ARM (attenuated), showed that both viruses alter glucose, amino acid and fatty acid metabolism in addition to the complement and coagulation cascades. The main group of affected genes included ISGs and innate immune response genes (e.g., ISG15, IFIT1 and STAT1) [139]. These genes are also upregulated by several pathogenic viruses (Table 1) as well as hepatitis C virus (HCV) [140].
The trends in gene expression may be similar among several viral infections, but significant differences can be seen in the comparison of attenuated and pathogenic viruses (Table 1) [80,124]. In the comparisons of attenuated and pathogenic arenaviruses, differences in gene expression do not reflect differences in replication potential of the viruses but rather the fact that the pathogenic viruses suppress innate immunity, allowing higher levels of virus production, so the high viremia triggers an inflammatory response that contributes to disease [137,141]. Both the pathogenic and nonpathogenic infections upregulate ISGs, but since viremia is more sustained for the pathogenic virus, sustained ISG expression becomes a biomarker of severe disease. Treatments would include antivirals (since viral components are driving ISG production) and anti-inflammatory drugs such as nucleoside reverse-transcriptase inhibitors [142].
Global gene expression can be used as a predictive marker of disease outcome. Transcriptome analysis of rhesus macaque liver showed that, as with our previous transcriptome analysis for blood [80], prominent expression of inflammatory-response genes during disease. Other transcriptome studies on viral diseases in liver have also identified higher levels of ISGs as a common theme. We observed significant upregulation of STAT1, ISG15 and IFIT1 in LCMV-WE-infected liver compared with nondiseased liver, and similar increases in gene expression were seen in human liver infected with HCV [140]. A recent paper on the Ebola virus transcriptome [133] noted that most of the cells used for profiling are actually uninfected bystanders that are responding to plasma IFN and not infected cells that are modulated by the anti-IFN activities of viral genes [127]. A large drop in hemoglobin gene expression during virulent infection has been attributed to IFN inhibition of erythropoiesis [143]. It is quite possible that in the primate, IFN-α/β initiates platelet dysfunction and treating the primate disease with platelet transfusions, as was done in murine models [139] might help recovery.
An example of early virus-specific gene expression is that Serpin genes were downregulated by HCV, but Serpin C is 30-fold upregulated in the LCMV liver and would be expected to inhibit the intrinsic coagulation pathway [139].
In LCMV-infected monkey liver there were significant decreases among transcripts promoting glycogenolysis (e.g., G6PC and FBP1) in the early stages of infection, and increases in transcripts promoting gluconeogenesis in both viremic and previremic stages of infection. It is likely that gluconeogenesis was driven by hydrolysis of triglycerides since transcripts for β-oxidation enzymes and lipases were upregulated in LCMV-infected livers [139]. Gene-expression changes during LCMV infection predict that glycolysis plus gluconeogenesis constituted a ‘futile cycle’ with increased usage of ketone bodies and fatty acids early, followed by liver functioning in a glucose-producing role, using fatty acids and amino acids as an energy source [139]. These alterations are similar to responses seen during fasting [144].
Similar to our observations with LCMV, other viral infections also affect liver energy metabolism. HBV transcription requires a regulatory enzyme of gluconeogenesis, encoded by PPARGC1 that is upregulated by fasting, cold temperature or stress and acts through nuclear receptor 4a, forkhead transcription factor and glucocorticoid receptor [145]. Remarkably, PPARGC1 was significantly upregulated in the attenuated LCMV infection but not during virulent infection [139]. Sendai virus transcription initiation requires tubulin and the glycolytic enzyme, phosphoglycerate kinase [146]. Mayaro, an alphavirus, increases glucose consumption in infected cells [147], but this may not be directly related to virus replication. Both phosphoglycerate kinase and the key glycolytic enzyme, glyceraldehyde 3-phosphate dehydrogenase were strongly upregulated in both mild and pathogenic LCMV infections. Similarly, glyceraldehyde 3-phosphate dehydrogenase is needed in the life cycle of parainfluenza virus 3 (HPIV3) [148]. In the LCMV-infected primate, IRS1 expression is decreased and expression of IRS2 is moderated, through a 7.2-fold upregulation of mRNA for SOCS2 [139], similar to the situation described for HCV infection [149].
Severe LCMV disease has characteristics of starvation that resemble both anorexia (appetite loss) and cachexia (TNF-α or IL-6-mediated wasting). High IL-6 was found in plasma of deceased macaques after LASV infection [56] and high IL-6 was found in LCMV-WE-infected macaque livers [150]. IL-6 stimulates protein catabolism that triggers gluconeogenesis [151], while anorexia is driven by IL-1β [152] and mobilizes fat instead of protein stores [151]. Although we did not see increases in IL-1β, we saw upregulation of genes for both protein catabolism and lipid oxidation. We also saw early decreases in blood triglycerides that corroborated our transcriptome result of suppressed acetyl-COA-carboxylase. Low triglycerides differentiated the viral disease from bacterial sepsis where high triglycerides coincide with high liver IL-6 [153]. Although high IL-6 is detected by day 7 [150], high circulating triglycerides are not seen until the day of death for LCMV-WE-infected macaques [139].
Metabolic changes in LCMV-infected liver may not account for the lethal disease resembling LF. Even though the liver transcriptome resembles starvation, possibly due to appetite loss and dehydration, primates can survive weeks of starvation and do not die quickly as they do from LASV [154]. Since most of the transcriptome changes affecting intermediary metabolism were seen in diseased and nondiseased animals, it is unlikely that those changes were the primary cause of death. Importantly, some genes involved in intermediary metabolism (PPARGC1, PCK1, IRS2, G6PC) were considerably more upregulated in mild compared with fatal infections [139] leading to the conclusion that the altered energy metabolism is a self-preservation mechanism and not a pathogenic one.
The transcriptome in liver from LCMV-WE monkeys revealed changes in gene expression related to fatty acid and glucose metabolism, steroid metabolism, complement or coagulation cascades, MAPK signaling pathway and cell adhesion. Virus-induced cytokine production could be responsible for platelet dysfunction and diminished erythropoiesis. Altered gene expression related to the coagulation cascade could be directly responsible for thrombocytopenia and capillary leakage in severe LCMV infection.
Profiling LASV-exposed human cells
Results from transcriptional profiling of cell cultures should be cautiously interpreted, as they may not accurately reflect what is taking place in live tissue. As the liver is a target organ for LASV, one study used a differentiated human hepatoma cell line, HuH-7, for transcriptome profiling. However, in that study insignificant differences were seen between LASV- or LCMV-infected cells and their uninfected controls [125]. Another transcriptome study using human PBMCs exposed to LASV or ML29 (for 4, 8 and 24 h) showed that ISGs comprise the most highly affected group of transcripts (Table 1) as would be expected for RNA viruses. For instance, in influenza-infected animals [155] or in yellow fever-vaccinated people, antiviral ISGs were the most common group of upregulated genes [123]. PBMC mRNA isolated during febrile episodes of dengue HF exhibited upregulation of GBP1, IFI44, IFIH1, IFIT1, MX1, LY6E, as seen in LASV-PBMC exposure. LASV infection in vivo produces high levels of viremia with mild IFN induction, insignificant tissue damage and little macrophage activation. In vitro, LASV is an efficient inhibitor of type I IFN and cytokine production in contrast to the less virulent MOPV [46,47,156].
Among ISGs, IFI6 (encoding γIFN-inducible protein 6) was tenfold upregulated in LASV-exposed PBMCs compared with cells exposed to the vaccine strain ML29. IFI6 encodes a cytokine belonging to the hematopoietic IFN-inducible nuclear antigens with 200 amino acid repeats (HIN-200) family. Its gene product contains domains involved in DNA binding, transcription regulation and protein–protein interactions. IFI6 protein localizes to the nucleus, then interacts with p53 and retinoblastoma-1 to inhibit cell growth via the Ras/Raf signaling pathway. IFI6 functions as a transcriptional repressor and plays a role in the regulation of hematopoietic differentiation that controls cellular proliferation [157].
IFITM 1-3 is an IFN-inducible viral restriction factor that differentially restricts the cellular entry of enveloped viruses (influenza-, flavi-, filo-viruses) and modulates cellular tropism independently of viral receptor expression [158–160]. Even though enveloped RNA viruses share some entry mechanisms with arenaviruses, pseudo-particles expressing envelope glycoproteins of LCMV, LASV, MACV, as well as MLV (retrovirus) were not restricted by IFITM expression [158].
The vaccine strain ML29 downregulated genes related to the IFN pathway including INHBA (negative regulator of IFN-γ), IFI44, TNFSF10 (TR AIL), SPP1 and LY6E (RIGE) genes, as well as immune response-related genes such as ITGAM encoding integrin α-M/β-2, RSAD2 (Viperin) and TRIM5 [161–163]; all more expressed after LASV than after ML29-exposure. Interstitial collagenase and MMP-1 were upregulated more than 65-fold in LASV versus ML-29 exposed cells [124]. MMP proteins are involved in the normal breakdown of extracellular matrix during reproduction, tissue remodeling and embryonic development and are also involved in metastasis and arthritis. MMP-1 interacts with the HIV transactivating protein Tat, resulting in the degradation of Tat and downregulation of Tat-mediated transactivation and neurotoxicity [164].
Several cytokines with anti-inflammatory activities were upregulated 20-fold to 25-fold after exposing PBMCs to LASV [124]. The cytokine inhibin β A (INHBA) subunit joins the α subunit to form pituitary follicle-stimulating hormone secretion inhibitor. Inhibin negatively regulates gonadal stromal cell proliferation and has tumor-suppressor activity. Vaccinia virus-induced INHBA expression (mediated by the viral E3 protein) blocked expression of multiple cytokines [165] by antagonizing signaling cascades. Genes for CXCL6 and IL-1A were also upregulated in PBMCs exposed to LASV (and CXCL6 shares functional homology with IL-8). Clinical studies showed that serum IL-8 levels were higher in patients with nonfatal LF and, along with other pro-inflammatory chemokines (IP-10, IL-1β, TNF-α) [166], lower in patients with fatal LF. The clinical data match results with observations in primary human cells in vitro. In animal models of fatal LF [56,122,150,167] it also seems that higher levels of IL-8 are protective against LF in vivo. While cell culture studies [46] show that replication of LASV in monocyte-derived MPs or DCs is associated with suppressing innate immune responses, in vivo studies with patients or experimental animals show that cytokines such as IP-10 and IL-8 (possibly coming from uninfected bystander cells) can recruit immune cells and increase survival. This view contrasts with the early ‘cytokine storm’ observed in filovirus HF [168,169].
In contrast to HF caused by the South American arenaviruses Machupo and Junin, hemorrhage with disseminated intravascular coagulation is rarely seen in patients with LF. Nevertheless, LASV effects on vascular function and coagulation abnormalities are involved in LF pathogenesis [168,170]. Platelets remain either normal or moderately low but there is a strong reduction in platelet function. Plasma from LHF patients has inhibitory activity on platelets from healthy individuals, suggesting the presence of soluble factors causing platelet malfunctions [36]. We showed that in vitro replication of LASV in endothelial cells (HUVEC) produced high virus titers up to 107 plaque-forming units (pfu)/ml without causing CPE. However, some cellular functions were perturbed including reduced levels of IL-8. Infecting HUVEC with nonpathogenic MOPV had no effect on IL-8 production [46] suggesting that LHF disease may not be the direct consequence of CPE but is more likely due to altered expression of soluble factors and dampened immunity.
Six genes related to the coagulation pathway are differently expressed in LASV-exposed versus ML29-exposed PBMCs: thrombomodulin (THBD), HB-EGF, ITGAM, PLAU, ITGB3 and PD-ECGF [124]. THBD is predominantly synthesized by vascular endothelial cells, and is a 60 kDa type I trans-membrane protein that forms a 1:1 molar complex with thrombin and exerts pronounced inhibitory effects on thrombotic, inflammatory and redox-related responses initiated in response to thrombin generation [171]. THBD significantly enhances the rate of thrombin inactivation by ATIII and dramatically accelerates the ability of thrombin to activate protein C. To validate cDNA array results, DCs derived from human PBMCs exposed to LASV versus ML29 and THBD expression was measured by ELISA. Levels of secreted THBD in cultures of cells exposed to LASV or ML29 were different and correlated with transcriptome data [124]. Exposure to LASV upregulated THBD secretion while exposure to ML29 had the opposite effect. Treating with LPS resulted in lower levels of secreted THBD [124] including LASV-exposed cells. In the presence of thrombin, THBD generates activated protein C (aPC) that inhibits pro-inflammatory and pro-coagulant responses such as factor V activation, fibrinogen cleavage, platelet activation, upregulation of leukocyte adhesion molecules and chemotaxis for neutrophils or monocytes [172–174]. Treating cells with IFN-γ shortens the half-life of THBD mRNA [175]. It is our hypothesis that the higher THBD or aPC and lower IFN responses are responsible for the absence of inflammatory cell infiltrates or hemorrhagic symptoms seen in LF. Our current view is that a massive LASV infection of endothelial or antigen-presenting cells would increase the expression of both membrane and soluble THBD forms, activating aPC and capturing HMGB1 with anti-inflammatory or anticoagulation effects [124]. Given these results, inhibiting THBD could be a therapeutic strategy for experimental LF in primates.
Upregulated coagulation-related genes might also contribute to LF disease. Examples include PLAU that has anticlotting effects, HB-EGF that is involved in blood vessel [176] and kidney pathologies [177,178] and it promotes vascular maturation in conjunction with PD-ECGF (TYMP) [179,180]. The ITGB3 (CD61, platelet marker) was 12-fold upregulated in cells exposed to LASV at 24 h. High-affinity LASV binding to cellular α-dystroglycan receptor perturbs the signaling interaction between α6β1 integrin and dystroglycan, thus inhibiting MEK/ERK signaling [181]. The α-dystroglycan receptor is ubiquitous for extracellular matrix (ECM) proteins that cooperate with β1 integrins to control cell-matrix interactions. Thus, virus-induced perturbation may contribute to functional changes among epithelial and vascular endothelial cells that contribute to shock and death in LF patients [9].
We also know that arenaviruses can bind directly to platelets and inhibits their function [182]. This mechanism would be exacerbated by high viremia and triggered by the upregulation of ITGAM, ITGB3 and PD-ECGF [124]. After arterial injury, leukocytes are recruited and an increase in ITGAM expression allows fibrinogen and platelet binding to the vessel walls [183]. LASV-induction of ITGAM and PD-ECGF/TYMP will increase platelet sequestration in tissues and decreased the number of circulating platelets during hemorrhagic episodes. Differential expression of these genes may contribute to the pathogenic phenotype of LASV and to the attenuated behavior of the vaccine strain ML-29.
In primates infected with LASV, DCs were the initial cellular targets and Kupffer cells, hepatocytes, adrenal cortical cells or endothelial cells were infected at later stages in terminal disease [184]. In PBMC cultures exposed to LASV or ML29, significant transcriptome differences were observed by 24 h after exposure even though most of the mononuclear cells in PBMC cultures are not permissive for LASV, since the virus replicates only in differentiated monocytes or MPs [46]. DCs, representing only approximately 1% of circulating PBMCs, are most permissive for virus replication. Thus, the gene-expression profiles of LASV- or ML29-exposed PBMCs are primarily gene expression of uninfected bystander cells that receive signaling from contact with virus particles, with infected DCs and with DC-produced cytokines.
IFN production in infected cells is inhibited by the interaction of viral NP with IRF-3 [127] or the interaction of Z protein with eIF4E [185]; both complexes affect the NF-κB pathway and favor suppression of chemokines or type I IFNs. Since virulent and nonvirulent viruses have the same capacity to inhibit this pathway, it is primarily virus replication levels that determine which is more effective in vivo. A byproduct of virus replication, noninfectious virus particles, may still trigger uninfected bystander cells to express ISGs. Upregulation of ISGs was observed in PBMC transcriptome studies, and these particles might be driving gene expression during the viremic stage in vivo.
Both LASV and ML29 have identical GP1 envelope proteins capable of binding to α-DG. The ML29 GP2 is different from LASV GP2 due to a K272E substitution located between the fusion domain and the RRLL motif at the cleavage site for cellular subtilase SKI1/S1P [69,120]. Substituting lysine (K) to glutamic acid (E) introduces two negatively charged R groups at positions 272–273 (Asp-Glu) that distinguish ML29 GP2 from any LASV GP2 glycoproteins analyzed so far. This potentially can affect fusion and contribute to postfusion events as well as to ML29 attenuation.
Finally, exposing human PBMCs to viruses, mimics the viremic stage of infection. Exposing PBMCs to LASV or to the ML29 vaccine strain, showed that LASV, but not ML29, affected genes involved in coagulation pathways. For example, LASV exposure strongly upregulated mRNA and protein expression for THBD, suggesting a connection between inflammatory and vascular abnormalities leading to death in late-stage LF patients. Based on our current knowledge about THBD involvement in pro-coagulant and pro-inflammatory pathways, THBD inhibition could be a promising target for therapeutic intervention in LF animal models.
Profiling LASV-infected NHPs
A study of LASV-infected cynomolgus macaques compared surviving and nonsurviving animals [56]. Fatal cases suffered from fever, weight loss, high viremia and acute respiratory distress, whereas survivors had lower viral loads and fewer disease signs. All animals developed strong antibody responses but survivors developed them earlier. Similarly, type I IFNs were detected soon after infection in survivors but only in terminal stages for nonsurvivors. All infected monkeys upregulated chemokine genes CXCL10 (IP-10) and CXCL11 (I-TAC) in peripheral blood mono-nuclear cells and lymph. Unlike people surviving LF [166], macaque survivors had lower levels of IP10. As noted previously in LCMV-WE-infected rhesus macaques [150,186], high levels of plasma IL-6 were detected only in fatalities. In survivors, high counts of activated-monocytes and rising numbers of circulating monocytes contrasted with low circulating monocytes and few activated or proliferating T cells in fatal cases. Early and strong immune responses were needed to control virus replication and were associated with recovery. Fatal infection was characterized by weak immune responses and uncontrolled viral replication. The investigators were surprised that macaques given high doses of 106 pfu LASV were more likely to survive than those given low doses of 103 pfu LASV [56]. This result however, is consistent with our view that a higher dose would increase signaling responses from uninfected bystander cells and poise the animal to resist fatal disease. Bystander cells secrete inflammatory cytokines to attract immune-surveillance at sites of infection. This promotes robust antiviral immunity. Such a phenomenon yields a previremia transcriptome profile similar to what is found in the viremic phase and what is seen in vitro when PBMCs are exposed to LASV. A similar phenomenon is at work when attenuated LASV vaccines are delivered therapeutically after lethal challenge [77,109].
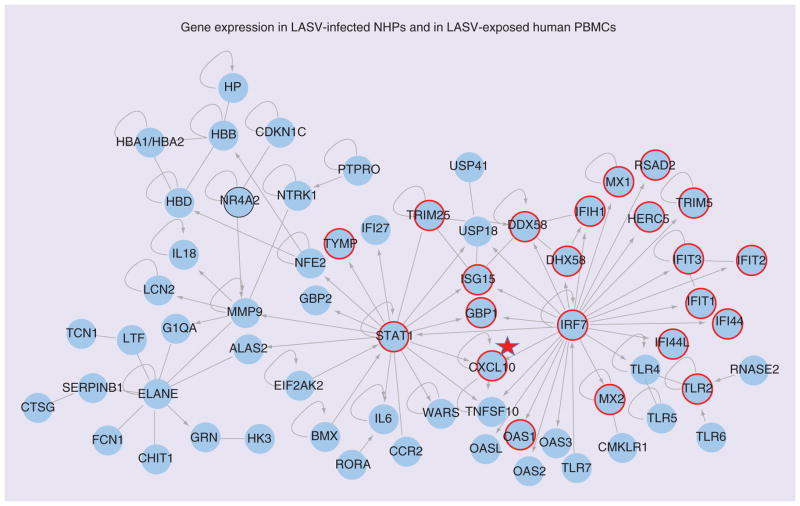
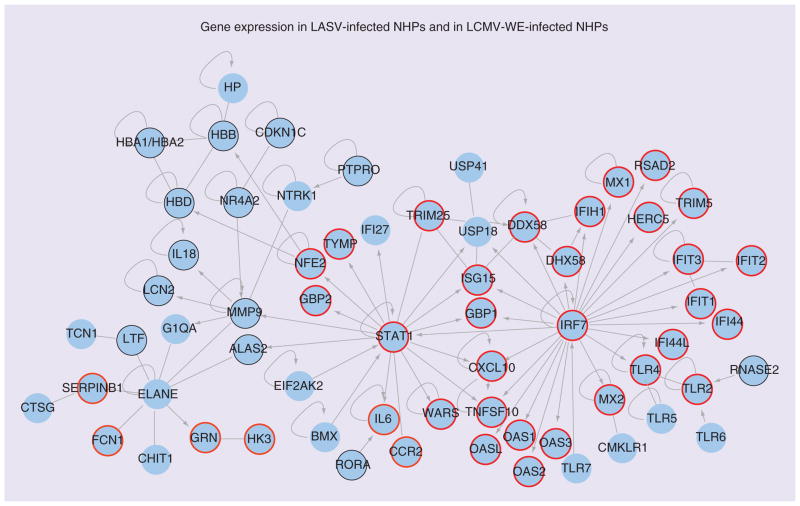
Profiling of LASV-infection in cynomolgus macaques [126] can be compared with our profiles of LASV exposed PBMCs (Figure 2) or profiles of LCMV-infected rhesus macaques (Figure 3). In Figures 2 & 3 there are 65 blue circles representing differentially regulated genes in LASV-infected macaques during the viremic phase. Based on Ingenuity Pathway Analysis, the 65 genes are highly interconnected.
Figure 2. Genes differentially regulated in blood of Lassa virus-infected monkeys and in Lassa virus-exposed periperal blood mononuclear cells.
Ingenuity Pathway Analysis software was used to analyze a list of genes upregulated in PBMCs after Lassa challenge of cynomolgus macaques. Nodes represent gene symbols, the products of which participate in protein–protein interactions, straight lines represent simple interaction, arrows represent control of one gene product by another, and loops represent self-regulation. Those genes encircled in red were also noted to be expressed in LASV-exposed human PBMCs by [124]. Encircled in black are genes downregulated in the Lymphocytic choriomeningitis virus WE strain monkey model. It is notable that the gene products connected to the IRF7 node are similarly upregulated in PBMCs exposed to either virulent LASV or nonvirulent ML29. CXCL10 (IP10) is noted because it is more expressed after exposure to ML29 than after exposure to LASV, and more evident in survivors than in those who succumb to Lassa fever [166].
LASV: Lassa virus; NHP: Nonhuman primate; PBMC: Periperal blood mononuclear cell.
Adapted with permission from Figure 3 of [126].
Figure 3. Genes differentially regulated in periperal blood mononuclear cells of Lassa virus-infected and lymphocytic choriomeningitis virus WE strain-infected monkeys.
Ingenuity Pathway Analysis software was used to analyze a list of genes upregulated in periperal blood mononuclear cells after Lassa challenge of cynomolgus macaques. The blue circles contain symbolic gene names, the products of which participate in protein–protein interactions, straight lines represent simple interaction, arrows represent control of one gene product by another and loops represent self-regulation. Those genes encircled in red were also noted to be expressed in the LCMV-WE-infected nonhuman primate by [80]. It is notable that the gene products connected to the IRF7 node are similarly upregulated in Periperal blood mononuclear cells exposed to either virulent LCMV-WE or nonvirulent LCMV-ARM. In LCMV-WE-infected NHP, no effects were seen on ELANE, strong downregulation (encircled in black) was seen for NR4A2 and mild to insignificant downregulation of hemoglobins (HBA, HBB, HBD), CDKN1C, IL-18, MMP9, LCN2, LTF, ALAS2, RORA and RNASE2 was observed [80].
LASV: Lassa virus; LCMV-ARM: Lymphocytic choriomeningitis virus Armstrong strain; LCMV-WE: Lymphocytic choriomeningitis virus WE strain; NHP: Nonhuman primate.
Adapted with permission from Figure 3 of [126].
For Figure 2, gene symbols corresponding to upregulated genes in the LASV-exposed PBMCs are circled in red while the common gene (NR4A2) that is downregulated is circled in black. Two important nodes, STAT1 and IRF7, represent genes whose products interact with many ISG products and pro-inflammatory molecules. It is notable that these gene products are upregulated equally well by virulent (LASV) and nonvirulent (ML29) in our study with virus-exposed PBMCs. The gene CXCL10 (expressing chemokine IP10) is marked with a star because it is upregulated much more by ML29 than by LASV in our study. IP10 is also outstanding in the study of human LF survivors and in the study of macaque LF survivors, and is important for attracting immunosurveillance to infection sites. The unique feature of this LASV-macaque study [126] is that several neutrophil-specific genes are upregulated, centering on the node ELANE (elastase of neutrophils). Note that PBMCs do not usually contain neutrophils that are removed during purification before RNA extraction. When these cells are included amongst PBMCs, the pathology in LASV-infected cynomolgus macaques is associated with increases in ‘band’ or immature neutrophils [Conner JH, Pers. Comm.].
The downregulated NR4A2 (nuclear receptor 4-A2) is circled in black (Figure 2) and it encodes a transcription factor that induces anti- inflammatory gene products. It is notable that NR4A2 is downregulated in the LASV-exposed PBMCs as well as in the LCMV-WE-infected rhesus macaque, where it is a major biomarker differentiating pathogenic from nonpathogenic infections. It is also downregulated in this group’s LASV-macaque studies according to their latest publication (see Figure 3 in [187]). Although NR4A2 is downregulated early in the previremic stage in LCMV macaques [80], it still needs to be validated as a predictive biomarker of viral hemorrhagic fever (VHF).
Figure 3 compares differentially-regulated genes in LASV-infected macaques [126] with the expression of genes from LCMV-WE-infected rhesus macaques [80]. Once again there is strong agreement between the datasets, except for a few genes in the ‘neutrophil’ cluster. Most of the discordant genes in the LCMV-WE model are either downregulated or barely affected (e.g., hemoglobins: HBA, HBB, HBD; CDKN1C; IL-18; MMP9; LCN2;LTF; ALAS2; RORA; RNASE2). Some of these genes impact the eicosanoid pathways that contribute to edema, myocarditis and a number of other LF disease signs. Although it is tantalizing to speculate about roles for these genes in HF, experimental studies are still needed to document the protein changes and confirm the effects on LASV pathogenesis. These initial studies in NHPs [80,126] should be continued to expand the previremic biomarkers of disease, and to determine which predict the development of pathogenic or nonpathogenic infections.
Conclusion: the importance of early immune response
LHF survivors control viral loads early during the infection. Fatal cases have poor inflammatory responses characterized by lymphopenia of all lymphocyte subsets (CD4, CD8, B and NK cells), along with necrosis of lymphoid organs [17,33,56,184,188]. This immunosuppressive state is established early after virus exposure. In vitro infection of MPs, DCs or endothelial cells downregulates the production of inflammatory mediators [46,47,189] as a key mechanism for immune suppression.
Most RNA viruses activate DCs and MPs by producing dsRNA genomic intermediates that are recognized by pattern recognition molecules. This activation is followed by IFN responses that induce cellular genes involved in the innate or adaptive immune responses and inhibit virus replication [190]. Although sensitive to the antiviral effects of IFN-α and IFN-γ, strains of LASV differ in their control of IFN production in vitro and in vivo [46,47,189,191]. In lethally infected animals, IFN-γ levels were moderately elevated [150] and in human fatal cases IFN-γ levels were increased for some individuals [192]. In monkeys IFN type I was detected early in survivors but only in terminal stages of the fatal disease [56]. Inhibition of IFN by LASV affects pDC maturation and DC production of cytokines or chemokines. Viral nucleoprotein (NP) plays an important role in IFN control by preventing activation plus nuclear translocation of IRF-3, IFN type I activation and downstream induction of the ISGs [127,193–195]. The anti-IFN function is located within the NP C-terminus. Functional and structural studies showed homology of this region with the DEDDH family of 3′-5′ exoribonucleases [196]. The crystal structure and interactions with RIG-I or MDA-5 suggest that NP ribonuclease activity removes viral PAMP RNAs, thereby avoiding recognition by pattern-recognition receptors and inhibiting IFN production [196,197].
In 1999 we reported that LASV infection of monocyte or endothelial cell cultures suppressed the induction of TNF-α and IL-8, an effect that would be expected to inhibit inflammation and neutrophil migration [46]. Subsequently, two more LASV-research groups, one in Lyon [47] and one in Atlanta, GA, USA [189], corroborated the idea that the virulent virus was suppressing expression of chemokines that could stimulate acquired immunity. However, there was a minor disagreement in their results: for one group [189], DCs incubated with both infectious and inactivated LASV produced IL-1β, IL-8, little IL-10 and no TNF-α and for the other group [47], both DCs and MPs incubated with infectious LASV suppressed TNF-α, IL-1β and IL-10 while inactivated virus particles fostered production of those chemokines. The Lyon group concluded that the LASV stock of the Atlanta group ‘probably contained a significant quantity of noninfectious viral products [47],’ implying that replication of infectious particles is needed to suppress innate immunity, a finding later corroborated by others [137]. Two findings in human beings with LF lent weight to the idea that innate immunity is suppressed in severe infection: first, fatal cases of LHF had lower chemokine levels (IL-8 and IP-10) compared with survivors [166] and, second, IL-8 was the only cytokine that peaked in infected contacts of severe LASV cases who did not develop LF disease [71]. In addition, studies of primates and of human PBMCs exposed to virus succeeded in linking virulent infection to undetectable levels of TNF-α, low levels of IL-8 in plasma and inhibition of IL-8 mRNA expression [80,150].
IL-6 appears at high levels in plasma from LHF fatal cases and monkeys infected with LCMV-WE. This high level of IL-6 during the last stages of severe LF could result from hepatic regeneration and may be associated with neutrophilia [28,30,56,150,186]. Another function that appears to be decreased in LASV-infected DCs is the maturation marker CCR7 and the capacity to migrate to secondary lymphoid organs [47].
The mechanism for disease involves LASV targeting of monocytes and DCs, inhibiting their initial immune response and suppressing migration of activated cells to primary sites of infection. The viral effects on host gene expression result in immune suppression, higher viral replication and increased virulence.
Treating humans or animals with immune serum showed various results: some promising, some discouraging [54,86–88]. Neutralizing antibodies might have a role in LASV protection, especially if high titers can be achieved. During natural infection of naive hosts, there is not enough time to mount a protective antibody response. However, more clinical and serological studies are needed to define the role of neutralizing antibodies in LHF recovery or reinfection.
After recovering from acute LF, patients overcome the initial lymphopenia and develop strong CD4+ immune responses against NP or GP2. The NP-CD4+ response is only partially strain-specific and cross-reacts among LASV strains [198]. The LASV-GP2-CD4+ response recognizes a conserved epitope common in the Old World (100% similarity with LCMV) and New World arenaviruses (>90% similarity) [199]. Immunizing monkeys or guinea pigs with LCMV, MOBV, MOPV or ML29 and other nonpathogenic Old World viruses confers some protection against LF disease. In these animals, T-cell responses are more important than B-cell responses. CTL-mediated protection relies more on CD8+ cells than on CD4+ cells. However, the mechanism of protection remains unknown and more studies are needed in human cases of infection to characterize the roles for T cells in LHF protection [17,69,111,114,122,170,200,201].
In LASV-infected monkeys T-cell activation was delayed in fatal cases and in vitro lymphoproliferative responses did not appear [56]. A decrease in CD20+ B cells and downregulation of class II MHC antigens was also seen [55]. In contrast to fatal cases, survivors showed T-cell activation and proliferation after exposure to inactivated LASV particles, and also increases in circulating monocytes [56]. Similarly, ML29 immunization of marmosets increased the CD3+ and CD14+ cell populations after LASV challenge [55].
In conclusion, much evidence suggests that inhibition of pattern-recognition receptors by LASV in early infection and a failure to induce pro-inflammatory cytokines results in immuno-suppression and uncontrolled virus replication. In late VHF disease several different viral infections are characterized by high inflammatory responses (e.g., NLRP3 inflammasomes producing high IL-18) [202]. Thus, an early and strong immune response that controls virus replication is more likely to promote recovery from LHF disease, and NLRP3 inflammasome inhibitors would be reasonable treatments for late disease.
Future perspective
Future efforts in Lassa research depend on improved diagnostics, technology to monitor vaccines, evaluating vaccine candidates and validating therapies based on modulation of host responses. Much of this will be contingent on previremic biomarkers of Lassa infection and how we can incorporate them into diagnostics that both confirm infection status and clearly define the disease stage. Rapid analysis of gene-expression patterns may be used directly or studied in order to identify the best protective biomarkers.
Development and testing of antiviral therapies (such as the drugs ST-193 and T-705) should proceed from animal models to human clinical trials. Transcriptome profiling studies have already pointed to new therapeutic targets (e.g., downregulation of PTGS2 and NR4A2 in the eicosonoid pathways could be counteracted by prostacyclin or bryostatin). Several FDA-approved drugs that are predicted to treat VHF have been suggested [94]. Cell culture studies on the direct effects of infectious and noninfectious virus particles on PBMCs showed that LASV upregulates THBD that interferes with coagulation, so inhibitors of THBD define another class of potential therapies.
Besides developing sensitive early diagnostics, prevention through public health education and vaccine research are high priorities. Biomarkers can be validated for use in vaccine studies, as we did in our study of the Lassa vaccine candidate ML29 in a rhesus macaque model of AIDS [122]. In that study we characterized gene-expression changes usually associated with benign infection indicating that the ML29 vaccine was attenuated even in an immunosuppressed animal. There are many benefits of attenuated LASV vaccine candidates that have received insufficient attention so far and have not advanced to clinical trials.
Forty years after LHF disease was described, molecular biology of LASV is well known but we lack tools needed to achieve serious reductions in the prevalence and severity of Lassa HF. Rapid diagnostics, effective vaccines and a clear understanding of LASV pathogenesis are needed to advance the development of therapeutics. Molecular diagnostics led by gene-expression profiling, offer a solid contribution to improving our understanding of LF, and will contribute to prevention and treatment of disease.
EXECUTIVE SUMMARY.
Background
-
History:
Lassa emerged in West Africa in 1969;
Endemic areas of West Africa have 50% seroprevalence and 3–5000 fatalities.
-
Clinical manifestations:
Severe disease presents with myocarditis, edema, respiratory distress, elevated AST/ALT, viremia, high plasma IL-6, encephalitis, vascular leakage and hearing loss in a third of hospitalized cases.
-
Lassa virus infection of animal models:
Disease signs most resemble human Lassa fever (LF) in rhesus and cynomolgus macaques, less so in marmosets and guinea pigs;
Disease in mice is dissimilar to human LF, requires high dose and is strongly mmunopathogenic.
-
Diagnosis:
No diagnostics are designed to detect host responses before viremia;
Current diagnostics detect viral antigens and antiviral immune responses;
Virus isolation is the gold standard for diagnosis.
-
Treatment:
Ribavirin is limited to early treatment;
Combinations of ribavirin + immune serum or ribavirin + IFN-α showed promise;
New drugs ST-193, T-705 show promise.
-
Vaccine approaches:
Obstacles to vaccine studies include the need for BSL-4, difficult animal models and the broad genetic diversity of Lassa virus (LASV);
Successful vaccines elicit cell-mediated immunity as well as humoral responses;
Primate-tested vaccines include Mopeia virus, ML29, a vaccinia vector expressing LASV-GP (VV-LASGP) and a vesiculostomatitis virus vector expressing VSV-LASGP. ML29 shows the broadest protection.
Profiling disease progression
-
Profiling of lymphocytic choriomeningitis virus-infected nonhuman primate, a model for LASV infection:
Infection of primates has a previremic stage, a viremic stage and a final stage of recovery or morbidity;
The blood transcriptome revealed at least 90 previremic genes discriminating virulent and benign infections;
Changes in lymphocytic choriomeningitis virus WE strain (LCMV-WE) blood transcriptome showed major disruptions in eicosnoid (COX-2), immune response and signaling pathways;
Disease signs of fatigue, hemorrhage, coagulation defects and myocarditis could all be related to changes seen in blood and liver transcriptome during previremic stage;
Biomarkers of disease enable predictions of possible treatments;
Meta-analysis and guinea pig results revealed similar transcription factors controlling viral hemorrhagic fever in the different animal models;
Liver transcriptome in LCMV-WE monkeys revealed impacts on fatty acid and glucose metabolism, steroid metabolism, complement and coagulation cascades, MAPK signaling pathway and cell adhesion;
Human and cynomolgus macaque survivors of LF have higher CXC chemokines (IL-8 and IP-10).
-
Profiling of LASV-exposed human cells:
Transcriptome profile of human periperal blood mononuclear cells exposed to LASV is a model for the viremic stage of in vivo infection;
Profile is dominated by IFN-sensitive genes as a common antiviral pattern, even in response to inactivated virus particles and in noninfected cells;
Direct effects of viral genes and virus particles have been demonstrated in cell culture, but most of the transcriptome profile in vivo is from indirect effects of virus on uninfected ‘bystander’ cells;
Exposure of periperal blood mononuclear cells to LASV or to the ML29 vaccine strain, showed that LASV, but not ML29, affected genes involved in coagulation pathways.
-
Profiling of LASV-infected nonhuman primates:
A study in Lyon showed cynomolgus macaques infected with high doses of LASV survived more frequently than those given low doses. It is proposed that the high doses insured early effective innate immunity;
The Ft. Detrick study did not compare the LASV infection to infection with a nonpathogenic virus so they did not see that most of their upregulated genes are upregulated in response to most viral infections;
The Ft. Detrick study was distinct from the LCMV-WE model for LASV infection by having more neutrophil markers upregulated;
Nonhuman primate studies should be continued in order to validate more previremic markers and to test therapies.
Conclusions: the importance of early immune response
There is an immunosuppressive state established early after LASV exposure characterized by poor inflammatory responses that leads to uncontrolled virus replication.
Surviving individuals showed early increased IFN levels while lethal cases only showed IFN in terminal stages.
In lethally-infected animals, IFN- levels were moderately elevated and in human fatal cases IFN- levels were increased for some individuals.
High serum IL-6 documented in Lassa cases and in LCMV-WE monkeys is not explained by LASV infection of hepatocytes or monocyte-derived macrophages (where LASV suppresses IL-6 production). Therefore, there must be a ‘bystander’ source.
Future perspective
Gene-expression profiling can expand our knowledge of Lassa hemorrhagic fever disease as well as improve its diagnosis and treatment options.
Footnotes
For reprint orders, please contact: reprints@futuremedicine.com
Financial & competing interests disclosure
Neither MS Salvato nor JC Zapata have financial interests in the production of vaccines or therapeutics for viral hemorrhagic fevers. NIH grants to MS Salvato (AI053619, AI053620 and a subcontract from the Mid-Atlantic Regional Centers of Excellence and Emerging Infectious Disease Research [MARCE; U54 AI057168 to M Levine] for diagnostics for flu-like diseases). The bioinformatics data analysis at VBI was funded by Department of Defense grant DA AD 13-02-C-0018 to B Sobral. Profiling of LASV-exposed cells occurred at Biosafety Level-4 facilities managed by Jean Patterson and Ricardo Carrion with financial support through NIH to the Western Regional Center of Excellence (U54 AI057156, subcontract to JL Patterson and R Carrion). The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as:
• of interest;
•• of considerable interest
- 1.CDC. Imported Lassa Fever-New Jersey. MMWR Morb Mortal Wky Rep. 2004;53:894–897. [PubMed] [Google Scholar]
- 2.Speir RW, Wood O, Liebhaber H, Buckley SM. Lassa fever, a new virus disease of man from West Africa. IV Electron microscopy of Vero cell cultures infected with Lassa virus. Am J Trop Med Hyg. 1970;19(4):692–694. doi: 10.4269/ajtmh.1970.19.692. [DOI] [PubMed] [Google Scholar]
- 3.Frame JD, Baldwin JM, Jr, Gocke DJ, Troup JM. Lassa fever, a new virus disease of man from west Africa. I Clinical description and pathological findings. Am J Trop Med Hyg. 1970;19(4):670–676. doi: 10.4269/ajtmh.1970.19.670. [DOI] [PubMed] [Google Scholar]
- 4.Troup JM, White HA, Fom AL, Carey DE. An outbreak of Lassa fever on the Jos plateau, Nigeria, in January-February 1970. A preliminary report. Am J Trop Med Hyg. 1970;19(4):695–696. doi: 10.4269/ajtmh.1970.19.695. [DOI] [PubMed] [Google Scholar]
- 5.Khan SH, Goba A, Chu M, et al. New opportunities for field research on the pathogenesis and treatment of Lassa fever. Antiviral Res. 2008;78(1):103–115. doi: 10.1016/j.antiviral.2007.11.003. [DOI] [PubMed] [Google Scholar]
- 6.Monath TP, Mertens PE, Patton R, et al. A hospital epidemic of Lassa fever in Zorzor, Liberia, March–April 1972. Am J Trop Med Hyg. 1973;22(6):773–779. doi: 10.4269/ajtmh.1973.22.773. [DOI] [PubMed] [Google Scholar]
- 7.Monath TP, Maher M, Casals J, Kissling RE, Cacciapuoti A. Lassa fever in the Eastern Province of Sierra Leone, 1970–1972. II Clinical observations and virological studies on selected hospital cases. Am J Trop Med Hyg. 1974;23(6):1140–1149. doi: 10.4269/ajtmh.1974.23.1140. [DOI] [PubMed] [Google Scholar]
- 8.Fraser DW, Campbell CC, Monath TP, Goff PA, Gregg MB. Lassa fever in the Eastern Province of Sierra Leone, 1970–1972. I Epidemiologic studies. Am J Trop Med Hyg. 1974;23(6):1131–1139. doi: 10.4269/ajtmh.1974.23.1131. [DOI] [PubMed] [Google Scholar]
- 9.Fisher-Hoch SP, McCormick JB. Patho-physiology and treatment of Lassa fever. Curr Top Microbiol Immunol. 1987;134:231–239. doi: 10.1007/978-3-642-71726-0_10. [DOI] [PubMed] [Google Scholar]
- 10.Lukashevich IS, Clegg JC, Sidibe K. Lassa virus activity in Guinea: distribution of human antiviral antibody defined using enzyme-linked immunosorbent assay with recombinant antigen. J Med Virol. 1993;40(3):210–217. doi: 10.1002/jmv.1890400308. [DOI] [PubMed] [Google Scholar]
- 11.Bausch DG, Demby AH, Coulibaly M, et al. Lassa fever in Guinea. I Epidemiology of human disease and clinical observations. Vector Borne Zoonotic Dis. 2001;1(4):269–281. doi: 10.1089/15303660160025903. [DOI] [PubMed] [Google Scholar]
- 12.Henderson BE, Gary GW, Jr, Kissling RE, Frame JD, Carey DE. Lassa fever. Virological and serological studies. Trans R Soc Trop Med Hyg. 1972;66(3):409–416. doi: 10.1016/0035-9203(72)90272-6. [DOI] [PubMed] [Google Scholar]
- 13.Monath TP. Lassa fever: review of epidemiology and epizootiology. Bull World Health Organ. 1975;52(4–6):577–592. [PMC free article] [PubMed] [Google Scholar]
- 14.McCormick JB, Webb PA, Krebs JW, Johnson KM, Smith ES. A prospective study of the epidemiology and ecology of Lassa fever. J Infect Dis. 1987;155(3):437–444. doi: 10.1093/infdis/155.3.437. [DOI] [PubMed] [Google Scholar]
- 15.Tomori O, Fabiyi A, Sorungbe A, Smith A, McCormick JB. Viral hemorrhagic fever antibodies in Nigerian populations. Am J Trop Med Hyg. 1988;38(2):407–410. doi: 10.4269/ajtmh.1988.38.407. [DOI] [PubMed] [Google Scholar]
- 16.McCormick JB. Lassa fever: epidemiology, therapy and vaccine development. Kansenshogaku Zasshi. 1988;62(Suppl):353–366. [PubMed] [Google Scholar]
- 17•.McCormick JB, Fisher-Hoch SP. Lassa fever. Curr Top Microbiol Immunol. 2002;262:75–109. doi: 10.1007/978-3-642-56029-3_4. Excellent updated review of Lassa fever pathogenesis in humans and in animal models. [DOI] [PubMed] [Google Scholar]
- 18.Richmond JK, Baglole DJ. Lassa fever: epidemiology, clinical features, and social consequences. BMJ. 2003;327(7426):1271–1275. doi: 10.1136/bmj.327.7426.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shaffer JG, Grant DS, Schieffelin JS, et al. Lassa fever in post-conflict sierra leone. PLoS Negl Trop Dis. 2014;8(3):e2748. doi: 10.1371/journal.pntd.0002748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bowen MD, Rollin PE, Ksiazek TG, et al. Genetic diversity among Lassa virus strains. J Virol. 2000;74(15):6992–7004. doi: 10.1128/jvi.74.15.6992-7004.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21•.Peters CJ, Jahrling PB, Liu CT, Kenyon RH, Mckee KT, Jr, Barrera Oro JG. Experimental studies of arenaviral hemorrhagic fevers. Curr Top Microbiol Immunol. 1987;134:5–68. doi: 10.1007/978-3-642-71726-0_2. Comprehensive description of Lassa virus (LASV) infection in different animal models. [DOI] [PubMed] [Google Scholar]
- 22.Andersen KG, Shylakhter I, Tabrizi S, Grossman SR, Happi CT, Sabeti PC. Genome-wide scans provide evidence for positive selection of genes implicated in Lassa fever. Philos Trans R Soc Lond B Biol Sci. 2012;367(1590):868–877. doi: 10.1098/rstb.2011.0299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sabeti PC, Varilly P, Fry B, et al. Genome-wide detection and characterization of positive selection in human populations. Nature. 2007;449(7164):913–918. doi: 10.1038/nature06250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stephenson EH, Larson EW, Dominik JW. Effect of environmental factors on aerosol-induced Lassa virus infection. J Med Virol. 1984;14(4):295–303. doi: 10.1002/jmv.1890140402. [DOI] [PubMed] [Google Scholar]
- 25.Rotz LD, Khan AS, Lillibridge SR, Ostroff SM, Hughes JM. Public health assessment of potential biological terrorism agents. Emerg Infect Dis. 2002;8(2):225–230. doi: 10.3201/eid0802.010164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26•.McCormick JB. Epidemiology and control of Lassa fever. Curr Top Microbiol Immunol. 1987;134:69–78. doi: 10.1007/978-3-642-71726-0_3. Excellent overiew of early attempts to manage Lassa outbreaks. [DOI] [PubMed] [Google Scholar]
- 27.Johnson KM, McCormick JB, Webb PA, Smith ES, Elliott LH, King IJ. Clinical virology of Lassa fever in hospitalized patients. J Infect Dis. 1987;155(3):456–464. doi: 10.1093/infdis/155.3.456. [DOI] [PubMed] [Google Scholar]
- 28.Schmitz H, Kohler B, Laue T, et al. Monitoring of clinical and laboratory data in two cases of imported Lassa fever. Microbes Infect. 2002;4(1):43–50. doi: 10.1016/s1286-4579(01)01508-8. [DOI] [PubMed] [Google Scholar]
- 29.Oldstone MB, Campbell KP. Decoding arenavirus pathogenesis: essential roles for alpha-dystroglycan-virus interactions and the immune response. Virology. 2011;411(2):170–179. doi: 10.1016/j.virol.2010.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McCormick JB, Walker DH, King IJ, et al. Lassa virus hepatitis. a study of fatal Lassa fever in humans. Am J Trop Med Hyg. 1986;35(2):401–407. doi: 10.4269/ajtmh.1986.35.401. [DOI] [PubMed] [Google Scholar]
- 31.Winn WC, Jr, Monath TP, Murphy FA, Whitfield SG. Lassa virus hepatitis. Observations on a fatal case from the 1972 Sierra Leone epidemic. Arch Pathol. 1975;99(11):599–604. [PubMed] [Google Scholar]
- 32.Fisher-Hoch SP, Mitchell SW, Sasso DR, Lange JV, Ramsey R, McCormick JB. Physiological and immunologic disturbances associated with shock in a primate model of Lassa fever. J Infect Dis. 1987;155(3):465–474. doi: 10.1093/infdis/155.3.465. [DOI] [PubMed] [Google Scholar]
- 33.Fisher-Hoch S, McCormick JB, Sasso D, Craven RB. Hematologic dysfunction in Lassa fever. J Med Virol. 1988;26(2):127–135. doi: 10.1002/jmv.1890260204. [DOI] [PubMed] [Google Scholar]
- 34.Knobloch J, McCormick JB, Webb PA, Dietrich M, Schumacher HH, Dennis E. Clinical observations in 42 patients with Lassa fever. Tropenmed Parasitol. 1980;31(4):389–398. [PubMed] [Google Scholar]
- 35.Lange JV, Mitchell SW, McCormick JB, Walker DH, Evatt BL, Ramsey RR. Kinetic study of platelets and fibrinogen in Lassa virus-infected monkeys and early pathologic events in Mopeia virus-infected monkeys. Am J Trop Med Hyg. 1985;34(5):999–1007. doi: 10.4269/ajtmh.1985.34.999. [DOI] [PubMed] [Google Scholar]
- 36.Cummins D, Fisher-Hoch SP, Walshe KJ, et al. A plasma inhibitor of platelet aggregation in patients with Lassa fever. Br J Haematol. 1989;72(4):543–548. doi: 10.1111/j.1365-2141.1989.tb04321.x. [DOI] [PubMed] [Google Scholar]
- 37•.Peters CJ, Liu CT, Anderson GW, Jr, Morrill JC, Jahrling PB. Pathogenesis of viral hemorrhagic fevers: Rift Valley fever and Lassa fever contrasted. Rev Infect Dis. 1989;11(Suppl 4):S743–S749. doi: 10.1093/clinids/11.supplement_4.s743. Describes platelet defect responsible in part for causing vascular leakage in Lassa fever. [DOI] [PubMed] [Google Scholar]
- 38.Cummins D, McCormick JB, Bennett D, et al. Acute sensorineural deafness in Lassa fever. JAMA. 1990;264(16):2093–2096. [PubMed] [Google Scholar]
- 39.Solbrig MV. Headache syndromes in Sierra Leone, west Africa. Headache. 1991;31(6):419. [PubMed] [Google Scholar]
- 40.Price ME, Fisher-Hoch SP, Craven RB, McCormick JB. A prospective study of maternal and fetal outcome in acute Lassa fever infection during pregnancy. BMJ. 1988;297(6648):584–587. doi: 10.1136/bmj.297.6648.584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cao W, Henry MD, Borrow P, et al. Identification of alpha-dystroglycan as a receptor for lymphocytic choriomeningitis virus and Lassa fever virus. Science. 1998;282(5396):2079–2081. doi: 10.1126/science.282.5396.2079. [DOI] [PubMed] [Google Scholar]
- 42.Kunz S, Rojek JM, Perez M, Spiropoulou CF, Oldstone MB. Characterization of the interaction of Lassa fever virus with its cellular receptor alpha-dystroglycan. J Virol. 2005;79(10):5979–5987. doi: 10.1128/JVI.79.10.5979-5987.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shimojima M, Stroher U, Ebihara H, Feldmann H, Kawaoka Y. Identification of cell surface molecules involved in dystroglycan-independent Lassa virus cell entry. J Virol. 2012;86(4):2067–2078. doi: 10.1128/JVI.06451-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Callis RT, Jahrling PB, Depaoli A. Pathology of Lassa virus infection in the rhesus monkey. Am J Trop Med Hyg. 1982;31(5):1038–1045. doi: 10.4269/ajtmh.1982.31.1038. [DOI] [PubMed] [Google Scholar]
- 45.Walker DH, McCormick JB, Johnson KM, et al. Pathologic and virologic study of fatal Lassa fever in man. Am J Pathol. 1982;107(3):349–356. [PMC free article] [PubMed] [Google Scholar]
- 46••.Lukashevich IS, Maryankova R, Vladyko AS, et al. Lassa and Mopeia virus replication in human monocytes/macrophages and in endothelial cells. different effects on IL-8 and TNF-alpha gene expression. J Med Virol. 1999;59(4):552–560. First paper to contradict the ‘cytokine storm’ view of Lassa fever pathogenesis showing that in cell culture LASV can suppress TNF and IL-8 expression. [PMC free article] [PubMed] [Google Scholar]
- 47.Baize S, Kaplon J, Faure C, Pannetier D, Georges-Courbot MC, Deubel V. Lassa virus infection of human dendritic cells and macrophages is productive but fails to activate cells. J Immunol. 2004;172(5):2861–2869. doi: 10.4049/jimmunol.172.5.2861. [DOI] [PubMed] [Google Scholar]
- 48.Pannetier D, Reynard S, Russier M, et al. Human dendritic cells infected with the nonpathogenic Mopeia virus induce stronger T-cell responses than those infected with Lassa virus. J Virol. 2011;85(16):8293–8306. doi: 10.1128/JVI.02120-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Geisbert W, Jahrling P. Exotic emerging viral diseases: progress and challenges. Nat Med. 2004;10(12 Suppl):S110–S121. doi: 10.1038/nm1142. [DOI] [PubMed] [Google Scholar]
- 50.Roberts PJ, Cummins D, Bainton AL, et al. Plasma from patients with severe Lassa fever profoundly modulates f-met-leu-phe induced superoxide generation in neutrophils. Br J Haematol. 1989;73(2):152–157. doi: 10.1111/j.1365-2141.1989.tb00245.x. [DOI] [PubMed] [Google Scholar]
- 51.Gilden DH, Cole GA, Monjan AA, Nathanson N. Immunopathogenesis of acute central nervous system disease produced by lymphocytic choriomeningitis virus. I Cyclophosphamide-mediated induction by the virus-carrier state in adult mice. J Exp Med. 1972;135(4):860–873. doi: 10.1084/jem.135.4.860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McCormick JB. In: Emergence and Control of Rodent-Borne Viral Diseases. Saluzzo JF, Dodet B, editors. Elsevier; Paris, France: 1999. pp. 177–175. [Google Scholar]
- 53.Colonna M. Toll-like receptors and IFN-alpha: partners in autoimmunity. J Clin Invest. 2006;116(9):2319–2322. doi: 10.1172/JCI29879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McCormick JB, King IJ, Webb PA, et al. Lassa fever. Effective therapy with ribavirin. N Engl J Med. 1986;314(1):20–26. doi: 10.1056/NEJM198601023140104. [DOI] [PubMed] [Google Scholar]
- 55•.Carrion R, Jr, Brasky K, Mansfield K, et al. Lassa virus infection in experimentally infected marmosets. liver pathology and immunophenotypic alterations in target tissues. J Virol. 2007;81(12):6482–6490. doi: 10.1128/JVI.02876-06. First description of the LASV-infected marmoset model for human Lassa fever. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Baize S, Marianneau P, Loth P, et al. Early and strong immune responses are associated with control of viral replication and recovery in Lassa virus-infected cynomolgus monkeys. J Virol. 2009;83(11):5890–5903. doi: 10.1128/JVI.01948-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McCormick JB, King IJ, Webb PA, et al. A case–control study of the clinical diagnosis and course of Lassa fever. J Infect Dis. 1987;155(3):445–455. doi: 10.1093/infdis/155.3.445. [DOI] [PubMed] [Google Scholar]
- 58.Bausch DG, Rollin PE, Demby AH, et al. Diagnosis and clinical virology of Lassa fever as evaluated by enzyme-linked immunosorbent assay, indirect fluorescent-antibody test, and virus isolation. J Clin Microbiol. 2000;38(7):2670–2677. doi: 10.1128/jcm.38.7.2670-2677.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bloch A. A serological survey of Lassa fever in Liberia. Bull World Health Organ. 1978;56(5):811–813. [PMC free article] [PubMed] [Google Scholar]
- 60.Arnold RB, Gary GW. A neutralization test survey for Lassa Fever activity in Lassa, Nigeria. Trans R Soc Trop Med Hyg. 1977;71(2):152–154. doi: 10.1016/0035-9203(77)90085-2. [DOI] [PubMed] [Google Scholar]
- 61.Goldwasser RA, Elliott LH, Johnson KM. Preparation and use of erythrocyte-globulin conjugates to Lassa virus in reversed passive hemagglutination and inhibition. J Clin Microbiol. 1980;11(6):593–599. doi: 10.1128/jcm.11.6.593-599.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tomori O, Johnson KM, Kiley MP, Elliott LH. Standardization of a plaque assay for Lassa virus. J Med Virol. 1987;22(1):77–89. doi: 10.1002/jmv.1890220110. [DOI] [PubMed] [Google Scholar]
- 63.Wulff H, Lange JV. Indirect immuno-fluorescence for the diagnosis of Lassa fever infection. Bull World Health Organ. 1975;52(4–6):429–436. [PMC free article] [PubMed] [Google Scholar]
- 64.Van Der Waals FW, Pomeroy KL, Goudsmit J, Asher DM, Gajdusek DC. Hemorrhagic fever virus infections in an isolated rainforest area of central Liberia. Limitations of the indirect immunofluorescence slide test for antibody screening in Africa. Trop Geogr Med. 1986;38(3):209–214. [PubMed] [Google Scholar]
- 65.Niklasson BS, Jahrling PB, Peters CJ. Detection of Lassa virus antigens and Lassa virus-specific immunoglobulins G and M by enzyme-linked immunosorbent assay. J Clin Microbiol. 1984;20(2):239–244. doi: 10.1128/jcm.20.2.239-244.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jahrling PB, Niklasson BS, McCormick JB. Early diagnosis of human Lassa fever by ELISA detection of antigen and antibody. Lancet. 1985;1(8423):250–252. doi: 10.1016/s0140-6736(85)91029-3. [DOI] [PubMed] [Google Scholar]
- 67.Ivanov AP, Tkachenko EA, Van Der Groen G, Butenko AM, Konstantinov OK. Indirect immunoenzyme method for the laboratory diagnosis of Lassa and Ebola hemorrhagic fevers. Vopr Virusol. 1986;31(2):186–190. [PubMed] [Google Scholar]
- 68.Barber GN, Clegg JC, Lloyd G. Expression of the Lassa virus nucleocapsid protein in insect cells infected with a recombinant baculovirus: application to diagnostic assays for Lassa virus infection. J Gen Virol. 1990;71(Pt 1):19–28. doi: 10.1099/0022-1317-71-1-19. [DOI] [PubMed] [Google Scholar]
- 69••.Lukashevich IS, Patterson J, Carrion R, et al. A live attenuated vaccine for Lassa fever made by reassortment of Lassa and Mopeia viruses. J Virol. 2005;79(22):13934–13942. doi: 10.1128/JVI.79.22.13934-13942.2005. Landmark description of the reassortant Lassa vaccine candidate, ML29 in mice, guinea pigs and macaques. This is the first Lassa vaccine candidate to give robust cell-mediated immune responses prior to lethal challenge. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Emmerich P, Thome-Bolduan C, Drosten C, et al. Reverse ELISA for IgG and IgM antibodies to detect Lassa virus infections in Africa. J Clin Virol. 2006;37(4):277–281. doi: 10.1016/j.jcv.2006.08.015. [DOI] [PubMed] [Google Scholar]
- 71.Grove JN, Branco LM, Boisen ML, et al. Capacity building permitting comprehensive monitoring of a severe case of Lassa hemorrhagic fever in Sierra Leone with a positive outcome., case report. Virol J. 2011;8:314. doi: 10.1186/1743-422X-8-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ter Meulen J, Koulemou K, Wittekindt T, et al. Detection of Lassa virus anti-nucleoprotein immunoglobulin G (IgG) and IgM antibodies by a simple recombinant immunoblot assay for field use. J Clin Microbiol. 1998;36(11):3143–3148. doi: 10.1128/jcm.36.11.3143-3148.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lunkenheimer K, Hufert FT, Schmitz H. Detection of Lassa virus RNA in specimens from patients with Lassa fever by using the polymerase chain reaction. J Clin Microbiol. 1990;28(12):2689–2692. doi: 10.1128/jcm.28.12.2689-2692.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Trappier SG, Conaty AL, Farrar BB, Auperin DD, McCormick JB, Fisher-Hoch SP. Evaluation of the polymerase chain reaction for diagnosis of Lassa virus infection. Am J Trop Med Hyg. 1993;49(2):214–221. doi: 10.4269/ajtmh.1993.49.214. [DOI] [PubMed] [Google Scholar]
- 75.Demby AH, Chamberlain J, Brown DW, Clegg CS. Early diagnosis of Lassa fever by reverse transcription-PCR. J Clin Microbiol. 1994;32(12):2898–2903. doi: 10.1128/jcm.32.12.2898-2903.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Drosten C, Gottig S, Schilling S, et al. Rapid detection and quantification of RNA of Ebola and Marburg viruses, Lassa virus, Crimean-Congo hemorrhagic fever virus, Rift Valley fever virus, dengue virus, and yellow fever virus by real-time reverse transcription-PCR. J Clin Microbiol. 2002;40(7):2323–2330. doi: 10.1128/JCM.40.7.2323-2330.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77•.Lukashevich IS, Carrion R, Jr, Salvato MS, et al. Safety, immunogenicity, and efficacy of the ML29 reassortant vaccine for Lassa fever in small non-human primates. Vaccine. 2008;26(41):5246–5254. doi: 10.1016/j.vaccine.2008.07.057. Safety studies showing that ML29 elicits sterilizing immunity to LASV at low doses (103 pfu); at high doses (106 pfu), ML20 can cause pathology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Vieth S, Drosten C, Lenz O, et al. RT-PCR assay for detection of Lassa virus and related Old World arenaviruses targeting the L gene. Trans R Soc Trop Med Hyg. 2007;101(12):1253–1264. doi: 10.1016/j.trstmh.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 79.Panning M, Emmerich P, Olschlager S, et al. Laboratory diagnosis of Lassa fever, liberia. Emerg Infect Dis. 2010;16(6):1041–1043. doi: 10.3201/eid1606.100040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80••.Djavani MM, Crasta OR, Zapata JC, et al. Early blood profiles of virus infection in a monkey model for Lassa fever. J Virol. 2007;81(15):7960–7973. doi: 10.1128/JVI.00536-07. Transcriptome analysis of blood from rhesus macaques comparing pathogenic and nonpathogenic lymphocytic choriomeningitis virus (LCMV) infections. Previremic, viremic and terminal stages of disease are defined by distinct gene–expression responses. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zaas AK, Chen M, Varkey J, et al. Gene expression signatures diagnose influenza and other symptomatic respiratory viral infections in humans. Cell Host Microbe. 2009;6(3):207–217. doi: 10.1016/j.chom.2009.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Woods CW, Mcclain MT, Chen M, et al. A host transcriptional signature for presymptomatic detection of infection in humans exposed to influenza H1N1 or H3N2. PLoS ONE. 2013;8(1):e52198. doi: 10.1371/journal.pone.0052198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Enria DA, Briggiler AM, Levis S, Vallejos D, Maiztegui JI, Canonico PG. Tolerance and antiviral effect of ribavirin in patients with Argentine hemorrhagic fever. Antiviral Res. 1987;7(6):353–359. doi: 10.1016/0166-3542(87)90017-9. [DOI] [PubMed] [Google Scholar]
- 84.Kilgore PE, Ksiazek TG, Rollin PE, et al. Treatment of Bolivian hemorrhagic fever with intravenous ribavirin. Clin Infect Dis. 1997;24(4):718–722. doi: 10.1093/clind/24.4.718. [DOI] [PubMed] [Google Scholar]
- 85.Bausch DG, Hadi CM, Khan SH, Lertora JJ. Review of the literature and proposed guidelines for the use of oral ribavirin as postexposure prophylaxis for Lassa fever. Clin Infect Dis. 2010;51(12):1435–1441. doi: 10.1086/657315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Fisher-Hoch SP, McCormick JB. Towards a human Lassa fever vaccine. Rev Med Virol. 2001;11(5):331–341. doi: 10.1002/rmv.329. [DOI] [PubMed] [Google Scholar]
- 87.Leifer E, Gocke DJ, Bourne H. Lassa fever, a new virus disease of man from West Africa. II Report of a laboratory-acquired infection treated with plasma from a person recently recovered from the disease. Am J Trop Med Hyg. 1970;19(4):677–679. doi: 10.4269/ajtmh.1970.19.677. [DOI] [PubMed] [Google Scholar]
- 88.Jahrling PB, Frame JD, Rhoderick JB, Monson MH. Endemic Lassa fever in Liberia. IV Selection of optimally effective plasma for treatment by passive immunization. Trans R Soc Trop Med Hyg. 1985;79(3):380–384. doi: 10.1016/0035-9203(85)90388-8. [DOI] [PubMed] [Google Scholar]
- 89.Jahrling PB, Peters CJ, Stephen EL. Enhanced treatment of Lassa fever by immune plasma combined with ribavirin in cynomolgus monkeys. J Infect Dis. 1984;149(3):420–427. doi: 10.1093/infdis/149.3.420. [DOI] [PubMed] [Google Scholar]
- 90.Gowen BB, Smee DF, Wong MH, et al. Combinatorial ribavirin and interferon alfacon-1 therapy of acute arenaviral disease in hamsters. Antivir Chem Chemother. 2006;17(4):175–183. doi: 10.1177/095632020601700402. [DOI] [PubMed] [Google Scholar]
- 91.Cashman KA, Smith MA, Twenhafel NA, et al. Evaluation of Lassa antiviral compound ST-193 in a guinea pig model. Antiviral Res. 2011;90(1):70–79. doi: 10.1016/j.antiviral.2011.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mendenhall M, Russell A, Smee DF, et al. Effective oral favipiravir (T-705) therapy initiated after the onset of clinical disease in a model of arenavirus hemorrhagic fever. PLoS Negl Trop Dis. 2011;5(10):e1342. doi: 10.1371/journal.pntd.0001342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Garcia CC, Djavani M, Topisirovic I, Borden KL, Salvato MS, Damonte EB. Arenavirus Z protein as an antiviral target: virus inactivation and protein oligomerization by zinc finger-reactive compounds. J Gen Virol. 2006;87(Pt 5):1217–1228. doi: 10.1099/vir.0.81667-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94••.Zapata JC, Pauza CD, Djavani MM, et al. Lymphocytic choriomeningitis virus (LCMV) infection of macaques. a model for Lassa fever. Antiviral Res. 2011;92(2):125–138. doi: 10.1016/j.antiviral.2011.07.015. Review describing animal models used to understand arenaviral disease. It describes current antivirals and future possible use of US FDA-approved drugs that modulate specific host responses. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Linero FN, Sepulveda CS, Giovannoni F, et al. Host cell factors as antiviral targets in arenavirus infection. Viruses. 2012;4(9):1569–1591. doi: 10.3390/v4091569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Grant-Klein RJ, Altamura LA, Schmaljohn CS. Progress in recombinant DNA-derived vaccines for Lassa virus and filoviruses. Virus Res. 2011;162(1–2):148–161. doi: 10.1016/j.virusres.2011.09.005. [DOI] [PubMed] [Google Scholar]
- 97.McCormick JB, Mitchell SW, Kiley MP, Ruo S, Fisher-Hoch SP. Inactivated Lassa virus elicits a non protective immune response in rhesus monkeys. J Med Virol. 1992;37(1):1–7. doi: 10.1002/jmv.1890370102. [DOI] [PubMed] [Google Scholar]
- 98.Krasnianskii VP, Potryvaeva NV, Borisevich IV, Gradoboev VN, Pashanina TP, Pshenichnov VA. A trial to produce an inactivated Lassa fever vaccine. Vopr Virusol. 1993;38(6):276–279. [PubMed] [Google Scholar]
- 99.Salvato MS, Lukashevich IS. Vaccines against Lassa Fever’ in New Generation Vaccines. Marcel Dekker, Inc; NY, USA: 2009. pp. 895–904. [Google Scholar]
- 100.Branco LM, Grove JN, Geske FJ, et al. Lassa virus-like particles displaying all major immunological determinants as a vaccine candidate for Lassa hemorrhagic fever. Virol J. 2010;7:279. doi: 10.1186/1743-422X-7-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ignat’ev GM. Immunogenic and protective characteristics of recombinant Lassa virus NP protein. Vopr Virusol. 2002;47(2):28–31. [PubMed] [Google Scholar]
- 102.Liu F, Feuer R, Hassett DE, Whitton JL. Peptide vaccination of mice immune to LCMV or vaccinia virus causes serious CD8 T cell-mediated, TNF-dependent immunopathology. J Clin Invest. 2006;116(2):465–475. doi: 10.1172/JCI25608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Rodriguez-Carreno MP, Nelson MS, Botten J, Smith-Nixon K, Buchmeier MJ, Whitton JL. Evaluating the immunogenicity and protective efficacy of a DNA vaccine encoding Lassa virus nucleoprotein. Virology. 2005;335(1):87–98. doi: 10.1016/j.virol.2005.01.019. [DOI] [PubMed] [Google Scholar]
- 104.Djavani M, Yin C, Xia L, Lukashevich IS, Pauza CD, Salvato MS. Murine immune responses to mucosally delivered Salmonella expressing Lassa fever virus nucleoprotein. Vaccine. 2000;18(15):1543–1554. doi: 10.1016/s0264-410x(99)00439-9. [DOI] [PubMed] [Google Scholar]
- 105.Clegg JC, Lloyd G. Vaccinia recombinant expressing Lassa-virus internal nucleocapsid protein protects guineapigs against Lassa fever. Lancet. 1987;2(8552):186–188. doi: 10.1016/s0140-6736(87)90767-7. [DOI] [PubMed] [Google Scholar]
- 106.Auperin DD, Esposito JJ, Lange JV, et al. Construction of a recombinant vaccinia virus expressing the Lassa virus glycoprotein gene and protection of guinea pigs from a lethal Lassa virus infection. Virus Res. 1988;9(2–3):233–248. doi: 10.1016/0168-1702(88)90033-0. [DOI] [PubMed] [Google Scholar]
- 107.Morrison HG, Bauer SP, Lange JV, Esposito JJ, McCormick JB, Auperin DD. Protection of guinea pigs from Lassa fever by vaccinia virus recombinants expressing the nucleoprotein or the envelope glycoproteins of Lassa virus. Virology. 1989;171(1):179–188. doi: 10.1016/0042-6822(89)90525-4. [DOI] [PubMed] [Google Scholar]
- 108•.Fisher-Hoch SP, Hutwagner L, Brown B, McCormick JB. Effective vaccine for Lassa fever. J Virol. 2000;74(15):6777–6783. doi: 10.1128/jvi.74.15.6777-6783.2000. Review on Lassa vaccine candidates detailing extensive monkey studies on LASV-constructs expressed from vaccinia vectors; VV-GpC were best. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Garbutt M, Liebscher R, Wahl-Jensen V, et al. Properties of replication-competent vesicular stomatitis virus vectors expressing glyco-proteins of filoviruses and arenaviruses. J Virol. 2004;78(10):5458–5465. doi: 10.1128/JVI.78.10.5458-5465.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Geisbert TW, Jones S, Fritz EA, et al. Development of a new vaccine for the prevention of Lassa fever. PLoS Med. 2005;2(6):e183. doi: 10.1371/journal.pmed.0020183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Pushko P, Geisbert J, Parker M, Jahrling P, Smith J. Individual and bivalent vaccines based on alphavirus replicons protect guinea pigs against infection with Lassa and Ebola viruses. J Virol. 2001;75(23):11677–11685. doi: 10.1128/JVI.75.23.11677-11685.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Jiang X, Dalebout TJ, Bredenbeek PJ, et al. Yellow fever 17D-vectored vaccines expressing Lassa virus GP1 and GP2 glycoproteins provide protection against fatal disease in guinea pigs. Vaccine. 2011;29(6):1248–1257. doi: 10.1016/j.vaccine.2010.11.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Stephen EL, Jahrling PB. Experimental Lassa fever virus infection successfully treated with ribavirin. Lancet. 1979;1(8110):268–269. doi: 10.1016/s0140-6736(79)90790-6. [DOI] [PubMed] [Google Scholar]
- 114.Kiley MP, Lange JV, Johnson KM. Protection of rhesus monkeys from Lassa virus by immunisation with closely related Arenavirus. Lancet. 1979;2(8145):738. doi: 10.1016/s0140-6736(79)90659-7. [DOI] [PubMed] [Google Scholar]
- 115.Jahrling PB, Hesse RA, Eddy GA, Johnson KM, Callis RT, Stephen EL. Lassa virus infection of rhesus monkeys: pathogenesis and treatment with ribavirin. J Infect Dis. 1980;141(5):580–589. doi: 10.1093/infdis/141.5.580. [DOI] [PubMed] [Google Scholar]
- 116.Walker DH, Johnson KM, Lange JV, Gardner JJ, Kiley MP, McCormick JB. Experimental infection of rhesus monkeys with Lassa virus and a closely related arenavirus, Mozambique virus. J Infect Dis. 1982;146(3):360–368. doi: 10.1093/infdis/146.3.360. [DOI] [PubMed] [Google Scholar]
- 117.Walker DH, McCormick JB, Johnson KM, et al. Pathologic and virologic study of fatal Lassa fever in man. Am J Pathol. 1982;107(3):349–356. [PMC free article] [PubMed] [Google Scholar]
- 118.Lukashevich IS. Generation of reassortants between African arenaviruses. Virology. 1992;188(2):600–605. doi: 10.1016/0042-6822(92)90514-p. [DOI] [PubMed] [Google Scholar]
- 119•.Moshkoff DA, Salvato MS, Lukashevich IS. Molecular characterization of a reassortant virus derived from Lassa and Mopeia viruses. Virus Genes. 2007;34(2):169–176. doi: 10.1007/s11262-006-0050-3. Sequence comparison of the ML29 reassortant with its parental viruses (Mopeia virus contributing the L RNA segment and LASV contributing the S segment) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lukashevich IS, Salvato MS. Lassa virus genome. Curr Genomics. 2006;7(6):351–379. [Google Scholar]
- 121•.Carrion R, Jr, Patterson JL, Johnson C, et al. A ML29 reassortant virus protects guinea pigs against a distantly related Nigerian strain of Lassa virus and can provide sterilizing immunity. Vaccine. 2007;25(20):4093–4102. doi: 10.1016/j.vaccine.2007.02.038. Describes the broad protective capabilities of the ML29 Lassa vaccine candidate. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zapata JC, Poonia B, Bryant J, et al. An attenuated Lassa vaccine in SIV-infected rhesus macaques does not persist or cause arenavirus disease but does elicit Lassa virus-specific immunity. Virol J. 2013;10:52. doi: 10.1186/1743-422X-10-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Querec TD, Akondy RS, Lee EK, et al. Systems biology approach predicts immunogenicity of the yellow fever vaccine in humans. Nat Immunol. 2009;10(1):116–125. doi: 10.1038/ni.1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124••.Zapata JC, Carrion R, Jr, Patterson JL, et al. Transcriptome analysis of human peripheral blood mononuclear cells exposed to Lassa virus and to the attenuated Mopeia/Lassa reassortant 29 (ML29), a vaccine candidate. PLoS Negl Trop Dis. 2013;7(9):e2406. doi: 10.1371/journal.pntd.0002406. Comparison of the transcriptome profiles of periperal blood mononuclear cells exposed to LASV or to ML29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Muller S, Geffers R, Gunther S. Analysis of gene expression in Lassa virus-infected HuH-7 cells. J Gen Virol. 2007;88(Pt 5):1568–1575. doi: 10.1099/vir.0.82529-0. [DOI] [PubMed] [Google Scholar]
- 126••.Malhotra S, Yen JY, Honko AN, et al. Transcriptional profiling of the circulating immune response to Lassa virus in an aerosol model of exposure. PLoS Negl Trop Dis. 2013;7(4):e2171. doi: 10.1371/journal.pntd.0002171. Transcriptome profile of cynomolgus macaques challenged with LASV. Many IFN-stimulated genes are upregulated. Marked distinction of results from the lymphocytic choriomeningitis virus WE strain model. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Martinez-Sobrido L, Zuniga EI, Rosario D, Garcia-Sastre A, De La Torre JC. Inhibition of the type I interferon response by the nucleoprotein of the prototypic arenavirus lymphocytic choriomeningitis virus. J Virol. 2006;80(18):9192–9199. doi: 10.1128/JVI.00555-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Gaucher D, Therrien R, Kettaf N, et al. Yellow fever vaccine induces integrated multilineage and polyfunctional immune responses. J Exp Med. 2008;205(13):3119–3131. doi: 10.1084/jem.20082292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Scherer CA, Magness CL, Steiger KV, et al. Distinct gene expression profiles in peripheral blood mononuclear cells from patients infected with vaccinia virus, yellow fever 17D virus, or upper respiratory infections. Vaccine. 2007;25(35):6458–6473. doi: 10.1016/j.vaccine.2007.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Loke P, Hammond SN, Leung JM, et al. Gene expression patterns of dengue virus-infected children from nicaragua reveal a distinct signature of increased metabolism. PLoS Negl Trop Dis. 2010;4(6):e710. doi: 10.1371/journal.pntd.0000710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Nascimento EJ, Braga-Neto U, Calzavara-Silva CE, et al. Gene expression profiling during early acute febrile stage of dengue infection can predict the disease outcome. PLoS ONE. 2009;4(11):e7892. doi: 10.1371/journal.pone.0007892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Geimonen E, Neff S, Raymond T, Kocer SS, Gavrilovskaya IN, Mackow ER. Pathogenic and nonpathogenic hantaviruses differentially regulate endothelial cell responses. Proc Natl Acad Sci USA. 2002;99(21):13837–13842. doi: 10.1073/pnas.192298899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Rubins KH, Hensley LE, Wahl-Jensen V, et al. The temporal program of peripheral blood gene expression in the response of nonhuman primates to Ebola hemorrhagic fever. Genome Biol. 2007;8(8):R174. doi: 10.1186/gb-2007-8-8-r174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Wahl-Jensen V, Kurz S, Feldmann F, et al. Ebola virion attachment and entry into human macrophages profoundly effects early cellular gene expression. PLoS Negl Trop Dis. 2011;5(10):e1359. doi: 10.1371/journal.pntd.0001359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Bowick GC, Fennewald SM, Scott EP, et al. Identification of differentially activated cell-signaling networks associated with pichinde virus pathogenesis by using systems kinomics. J Virol. 2007;81(4):1923–1933. doi: 10.1128/JVI.02199-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Fennewald SM, Scott EP, Zhang L, et al. Thioaptamer decoy targeting of AP-1 proteins influences cytokine expression and the outcome of arenavirus infections. J Gen Virol. 2007;88(Pt 3):981–990. doi: 10.1099/vir.0.82499-0. [DOI] [PubMed] [Google Scholar]
- 137.Hayes M, Salvato M. Arenavirus evasion of host anti-viral responses. Viruses. 2012;4(10):2182–2196. doi: 10.3390/v4102182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Bowick GC, Mcauley AJ. Meta-analysis of high-throughput datasets reveals cellular responses following hemorrhagic fever virus infection. Viruses. 2011;3(5):613–619. doi: 10.3390/v3050613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Djavani M, Crasta OR, Zhang Y, et al. Gene expression in primate liver during viral hemorrhagic fever. Virol J. 2009;6:20. doi: 10.1186/1743-422X-6-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Smith MW, Yue ZN, Korth MJ, et al. Hepatitis C virus and liver disease. global transcriptional profiling and identification of potential markers. Hepatology. 2003;38(6):1458–1467. doi: 10.1016/j.hep.2003.09.024. [DOI] [PubMed] [Google Scholar]
- 141.Pythoud C, Rodrigo WW, Pasqual G, et al. Arenavirus nucleoprotein targets interferon regulatory factor-activating kinase IKKepsilon. J Virol. 2012;86(15):7728–7738. doi: 10.1128/JVI.00187-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142•.Fowler BJ, Gelfand BD, Kim Y, et al. Nucleoside reverse transcriptase inhibitors possess intrinsic anti-inflammatory activity. Science. 2014;346(6212):1000–1003. doi: 10.1126/science.1261754. Nucleoside reverse transcriptase inhibitors are cheap FDA-approved compounds that could treat inflammasome-based pathologies such as those associated with viral hemorrhagic fever. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Tarumi T, Sawada K, Sato N, et al. Interferon-alpha-induced apoptosis in human erythroid progenitors. Exp Hematol. 1995;23(12):1310–1318. [PubMed] [Google Scholar]
- 144.Clore JN, Glickman PS, Helm ST, Nestler JE, Blackard WG. Evidence for dual control mechanism regulating hepatic glucose output in nondiabetic men. Diabetes. 1991;40(8):1033–1040. doi: 10.2337/diab.40.8.1033. [DOI] [PubMed] [Google Scholar]
- 145.Shlomai A, Paran N, Shaul Y. PGC-1alpha controls hepatitis B virus through nutritional signals. Proc Natl Acad Sci USA. 2006;103(43):16003–16008. doi: 10.1073/pnas.0607837103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Ogino T, Iwama M, Kinouchi J, Shibagaki Y, Tsukamoto T, Mizumoto K. Involvement of a cellular glycolytic enzyme, phosphoglycerate kinase, in Sendai virus transcription. J Biol Chem. 1999;274(50):35999–36008. doi: 10.1074/jbc.274.50.35999. [DOI] [PubMed] [Google Scholar]
- 147.El-Bacha T, Menezes MMT, Azevedo E, Silva MC, Sola-Penna M, Da Poian AT. Mayaro virus infection alters glucose metabolism in cultured cells through activation of the enzyme 6-phosphofructo 1-kinase. Mol Cell Biochem. 2004;266(1–2):191–198. doi: 10.1023/b:mcbi.0000049154.17866.00. [DOI] [PubMed] [Google Scholar]
- 148.De BP, Gupta S, Zhao H, Drazba JA, Banerjee AK. Specific interaction in vitro and in vivo of glyceraldehyde-3-phosphate dehydrogenase and LA protein with cis-acting RNAs of human parainfluenza virus type 3. J Biol Chem. 1996;271(40):24728–24735. doi: 10.1074/jbc.271.40.24728. [DOI] [PubMed] [Google Scholar]
- 149.Kawaguchi T, Yoshida T, Harada M, et al. Hepatitis C virus down-regulates insulin receptor substrates 1 and 2 through up-regulation of suppressor of cytokine signaling 3. Am J Pathol. 2004;165(5):1499–1508. doi: 10.1016/S0002-9440(10)63408-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150•.Lukashevich IS, Tikhonov I, Rodas JD, et al. Arenavirus-mediated liver pathology. acute lymphocytic choriomeningitis virus infection of rhesus macaques is characterized by high-level interleukin-6 expression and hepatocyte proliferation. J Virol. 2003;77(3):1727–1737. doi: 10.1128/JVI.77.3.1727-1737.2003. Compares pathogenic and nonpathogenic LCMV in macaques to show high IL-6 and liver pathology in severe disease. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Delano MJ, Moldawer LL. The origins of cachexia in acute and chronic inflammatory diseases. Nutr Clin Pract. 2006;21(1):68–81. doi: 10.1177/011542650602100168. [DOI] [PubMed] [Google Scholar]
- 152.Hildeman D, Muller D. Immunopathologic weight loss in intracranial LCMV infection initiated by the anorexigenic effects of IL-1beta. Viral Immunol. 2000;13(3):273–285. doi: 10.1089/08828240050144617. [DOI] [PubMed] [Google Scholar]
- 153.Nonogaki K, Fuller GM, Fuentes NL, et al. Interleukin-6 stimulates hepatic triglyceride secretion in rats. Endocrinology. 1995;136(5):2143–2149. doi: 10.1210/endo.136.5.7720663. [DOI] [PubMed] [Google Scholar]
- 154.Cahill GF., Jr Starvation in man. N Engl J Med. 1970;282(12):668–675. doi: 10.1056/NEJM197003192821209. [DOI] [PubMed] [Google Scholar]
- 155.Kash JC, Basler CF, Garcia-Sastre A, et al. Global host immune response. pathogenesis and transcriptional profiling of type A influenza viruses expressing the hema-gglutinin and neuraminidase genes from the 1918 pandemic virus. J Virol. 2004;78(17):9499–9511. doi: 10.1128/JVI.78.17.9499-9511.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Pannetier D, Faure C, Georges-Courbot MC, Deubel V, Baize S. Human macrophages, but not dendritic cells, are activated and produce alpha/beta interferons in response to Mopeia virus infection. J Virol. 2004;78(19):10516–10524. doi: 10.1128/JVI.78.19.10516-10524.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Brazda V, Coufal J, Liao JC, Arrowsmith CH. Preferential binding of IFI16 protein to cruciform structure and superhelical DNA. Biochem Biophys Res Commun. 2012;422(4):716–720. doi: 10.1016/j.bbrc.2012.05.065. [DOI] [PubMed] [Google Scholar]
- 158.Brass AL, Huang IC, Benita Y, et al. The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell. 2009;139(7):1243–1254. doi: 10.1016/j.cell.2009.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Feeley EM, Sims JS, John SP, et al. IFITM3 inhibits influenza A virus infection by preventing cytosolic entry. PLoS Pathog. 2011;7(10):e1002337. doi: 10.1371/journal.ppat.1002337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Huang IC, Bailey CC, Weyer JL, et al. Distinct patterns of IFITM-mediated restriction of filoviruses, SARS coronavirus, and influenza A virus. PLoS Pathog. 2011;7(1):e1001258. doi: 10.1371/journal.ppat.1001258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Toniato E, Chen XP, Losman J, Flati V, Donahue L, Rothman P. TRIM8/GERP RING finger protein interacts with SOCS-1. J Biol Chem. 2002;277(40):37315–37322. doi: 10.1074/jbc.M205900200. [DOI] [PubMed] [Google Scholar]
- 162.Stremlau M, Owens CM, Perron MJ, Kiessling M, Autissier P, Sodroski J. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature. 2004;427(6977):848–853. doi: 10.1038/nature02343. [DOI] [PubMed] [Google Scholar]
- 163.Chee AV, Lopez P, Pandolfi PP, Roizman B. Promyelocytic leukemia protein mediates interferon-based anti-herpes simplex virus 1 effects. J Virol. 2003;77(12):7101–7105. doi: 10.1128/JVI.77.12.7101-7105.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Rumbaugh J, Turchan-Cholewo J, Galey D, et al. Interaction of HIV Tat and matrix metalloproteinase in HIV neuropathogenesis. a new host defense mechanism. FASEB J. 2006;20(10):1736–1738. doi: 10.1096/fj.05-5619fje. [DOI] [PubMed] [Google Scholar]
- 165.Myskiw C, Arsenio J, Van Bruggen R, Deschambault Y, Cao J. Vaccinia virus E3 suppresses expression of diverse cytokines through inhibition of the PKR, NF-kappaB, and IRF3 pathways. J Virol. 2009;83(13):6757–6768. doi: 10.1128/JVI.02570-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166•.Mahanty S, Bausch DG, Thomas RL, et al. Low levels of interleukin-8 and interferon-inducible protein-10 in serum are associated with fatal infections in acute Lassa fever. J Infect Dis. 2001;183(12):1713–1721. doi: 10.1086/320722. Landmark study of humans with LASV infection comparing survivors and nonsurvivors. [DOI] [PubMed] [Google Scholar]
- 167.Scott EP, Aronson JF. Cytokine patterns in a comparative model of arenavirus haemorrhagic fever in guinea pigs. J Gen Virol. 2008;89(Pt 10):2569–2579. doi: 10.1099/vir.0.2008/002048-0. [DOI] [PubMed] [Google Scholar]
- 168.Kunz S. The role of the vascular endothelium in arenavirus haemorrhagic fevers. Thromb Haemost. 2009;102(6):1024–1029. doi: 10.1160/TH09-06-0357. [DOI] [PubMed] [Google Scholar]
- 169.Moraz ML, Kunz S. Pathogenesis of arenavirus hemorrhagic fevers. Expert Rev Anti Infect Ther. 2011;9(1):49–59. doi: 10.1586/eri.10.142. [DOI] [PubMed] [Google Scholar]
- 170.Fisher-Hoch SP, McCormick JB. Lassa fever vaccine. Expert Rev Vaccines. 2004;3(2):189–197. doi: 10.1586/14760584.3.2.189. [DOI] [PubMed] [Google Scholar]
- 171.Esmon CT. The roles of protein C and thrombomodulin in the regulation of blood coagulation. J Biol Chem. 1989;264(9):4743–4746. [PubMed] [Google Scholar]
- 172.Waugh JM, Yuksel E, Li J, et al. Local overexpression of thrombomodulin for in vivo prevention of arterial thrombosis in a rabbit model. Circ Res. 1999;84(1):84–92. doi: 10.1161/01.res.84.1.84. [DOI] [PubMed] [Google Scholar]
- 173.Van De Wouwer M, Collen D, Conway EM. Thrombomodulin-protein C-EPCR system: integrated to regulate coagulation and inflammation. Arterioscler Thromb Vas Biol. 2004;24(8):1374–1383. doi: 10.1161/01.ATV.0000134298.25489.92. [DOI] [PubMed] [Google Scholar]
- 174.Cui W, Wilson JT, Wen J, et al. Thrombomodulin improves early outcomes after intraportal islet transplantation. Am J Transplant. 2009;9(6):1308–1316. doi: 10.1111/j.1600-6143.2009.02652.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Navarro A, Frevel M, Gamero AM, Williams BR, Feldman G, Larner AC. Thrombo-modulin RNA is destabilized through its 3′-untranslated element in cells exposed to IFN-gamma. J Interferon Cytokine Res. 2003;23(12):723–728. doi: 10.1089/107999003772084833. [DOI] [PubMed] [Google Scholar]
- 176.Chansel D, Ciroldi M, Vandermeersch S, et al. Heparin binding EGF is necessary for vasospastic response to endothelin. FASEB J. 2006;20(11):1936–1938. doi: 10.1096/fj.05-5328fje. [DOI] [PubMed] [Google Scholar]
- 177.Feng L, Garcia GE, Yang Y, et al. Heparin-binding EGF-like growth factor contributes to reduced glomerular filtration rate during glomerulonephritis in rats. J Clin Invest. 2000;105(3):341–350. doi: 10.1172/JCI2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Kirkland G, Paizis K, Wu LL, Katerelos M, Power DA. Heparin-binding EGF-like growth factor mRNA is upregulated in the peri-infarct region of the remnant kidney model: in vitro evidence suggests a regulatory role in myofibroblast transformation. J Am Soc Nephrol. 1998;9(8):1464–1473. doi: 10.1681/ASN.V981464. [DOI] [PubMed] [Google Scholar]
- 179.Nanba D, Higashiyama S. Dual intracellular signaling by proteolytic cleavage of membrane-anchored heparin-binding EGF-like growth factor. Cytokine Growth Factor Rev. 2004;15(1):13–19. doi: 10.1016/j.cytogfr.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 180.Stratman AN, Schwindt AE, Malotte KM, Davis GE. Endothelial-derived PDGF-BB and HB-EGF coordinately regulate pericyte recruitment during vasculogenic tube assembly and stabilization. Blood. 2010;116(22):4720–4730. doi: 10.1182/blood-2010-05-286872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Rojek JM, Moraz ML, Pythoud C, et al. Binding of Lassa virus perturbs extracellular matrix-induced signal transduction via dystroglycan. Cell Microbiol. 2012;14(7):1122–1134. doi: 10.1111/j.1462-5822.2012.01784.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Zapata JC, Cox D, Salvato MS. The role of platelets in the pathogenesis of viral hemorrhagic fevers. PLoS Negl Trop Dis. 2014;8(6):e2858. doi: 10.1371/journal.pntd.0002858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Simon DI, Dhen Z, Seifert P, Edelman ER, Ballantyne CM, Rogers C. Decreased neointimal formation in Mac-1−/− mice reveals a role for inflammation in vascular repair after angioplasty. J Clin Invest. 2000;105(3):293–300. doi: 10.1172/JCI7811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184•.Hensley LE, Smith MA, Geisbert JB, et al. Pathogenesis of Lassa fever in cynomolgus macaques. Virol J. 2011;8:205. doi: 10.1186/1743-422X-8-205. First paper to transcriptome profile LASV in nonhuman primate. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Volpon L, Osborne MJ, Capul AA, De La Torre JC, Borden KL. Structural characterization of the Z RING-eIF4E complex reveals a distinct mode of control for eIF4E. Proc Natl Acad Sci USA. 2010;107(12):5441–5446. doi: 10.1073/pnas.0909877107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186•.Lukashevich IS, Rodas JD, Tikhonov Ii, et al. LCMV-mediated hepatitis in rhesus macaques. WE but not ARM strain activates hepatocytes and induces liver regeneration. Arch Virol. 2004;149(12):2319–2336. doi: 10.1007/s00705-004-0385-9. Compares pathogenic and nonpathogenic LCMV in macaques with focus on liver pathology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Caballero IS, Yen JY, Hensley LE, Honko AN, Goff AJ, Connor JH. Lassa and Marburg viruses elicit distinct host transcriptional responses early after infection. BMC Genomics. 2014;15:960. doi: 10.1186/1471-2164-15-960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Edington GM, White HA. The pathology of Lassa fever. Trans R Soc Trop Med Hyg. 1972;66(3):381–389. doi: 10.1016/0035-9203(72)90268-4. [DOI] [PubMed] [Google Scholar]
- 189.Mahanty S, Hutchinson K, Agarwal S, Mcrae M, Rollin PE, Pulendran B. Cutting edge: impairment of dendritic cells and adaptive immunity by Ebola and Lassa viruses. J Immunol. 2003;170(6):2797–2801. doi: 10.4049/jimmunol.170.6.2797. [DOI] [PubMed] [Google Scholar]
- 190.Jacobs BL, Langland JO. When two strands are better than one: the mediators and modulators of the cellular responses to double-stranded RNA. Virology. 1996;219(2):339–349. doi: 10.1006/viro.1996.0259. [DOI] [PubMed] [Google Scholar]
- 191.Asper M, Sternsdorf T, Hass M, et al. Inhibition of different Lassa virus strains by alpha and gamma interferons and comparison with a less pathogenic arenavirus. J Virol. 2004;78(6):3162–3169. doi: 10.1128/JVI.78.6.3162-3169.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Branco LM, Grove JN, Boisen ML, et al. Emerging trends in Lassa fever. redefining the role of immunoglobulin M and inflammation in diagnosing acute infection. Virol J. 2011;8:478. doi: 10.1186/1743-422X-8-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Martinez-Sobrido L, Giannakas P, Cubitt B, Garcia-Sastre A, De La Torre JC. Differential inhibition of type I interferon induction by arenavirus nucleoproteins. J Virol. 2007;81(22):12696–12703. doi: 10.1128/JVI.00882-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Martinez-Sobrido L, Emonet S, Giannakas P, Cubitt B, Garcia-Sastre A, De La Torre JC. Identification of amino acid residues critical for the anti-interferon activity of the nucleoprotein of the prototypic arenavirus lymphocytic choriomeningitis virus. J Virol. 2009;83(21):11330–11340. doi: 10.1128/JVI.00763-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Hastie KM, Kimberlin CR, Zandonatti MA, Macrae IJ, Saphire EO. Structure of the Lassa virus nucleoprotein reveals a dsRNA-specific 3′ to 5′ exonuclease activity essential for immune suppression. Proc Natl Acad Sci USA. 2011;108(6):2396–2401. doi: 10.1073/pnas.1016404108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Qi X, Lan S, Wang W, et al. Cap binding and immune evasion revealed by Lassa nucleoprotein structure. Nature. 2010;468(7325):779–783. doi: 10.1038/nature09605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Zhou S, Cerny AM, Zacharia A, Fitzgerald KA, Kurt-Jones EA, Finberg RW. Induction and inhibition of type I interferon responses by distinct components of lymphocytic choriomeningitis virus. J Virol. 2010;84(18):9452–9462. doi: 10.1128/JVI.00155-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Ter Meulen J, Badusche M, Kuhnt K, et al. Characterization of human CD4(+) T-cell clones recognizing conserved and variable epitopes of the Lassa virus nucleoprotein. J Virol. 2000;74(5):2186–2192. doi: 10.1128/jvi.74.5.2186-2192.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Meulen J, Badusche M, Satoguina J, et al. Old and New World arenaviruses share a highly conserved epitope in the fusion domain of the glycoprotein 2, which is recognized by Lassa virus-specific human CD4+ T-cell clones. Virology. 2004;321(1):134–143. doi: 10.1016/j.virol.2003.12.013. [DOI] [PubMed] [Google Scholar]
- 200.Jahrling PB, Peters CJ. Serology and virulence diversity among Old-World arenaviruses, and the relevance to vaccine development. Med Microbiol Immunol. 1986;175(2–3):165–167. doi: 10.1007/BF02122441. [DOI] [PubMed] [Google Scholar]
- 201.Goicochea MA, Zapata JC, Bryant J, Davis H, Salvato MS, Lukashevich IS. Evaluation of Lassa virus vaccine immunogenicity in a CBA/J-ML29 mouse model. Vaccine. 2012;30(8):1445–1452. doi: 10.1016/j.vaccine.2011.12.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Hensley LE, Young HA, Jahrling PB, Geisbert TW. Proinflammatory response during Ebola virus infection of primate models: possible involvement of the tumor necrosis factor receptor superfamily. Immunol Lett. 2002;80(3):169–179. doi: 10.1016/s0165-2478(01)00327-3. [DOI] [PubMed] [Google Scholar]