Abstract
Vineyard- and winery-associated lactic acid bacteria (LAB) from two major PDO regions in Greece, Peza and Nemea, were surveyed. LAB were isolated from grapes, fermenting musts, and winery tanks performing spontaneous malolactic fermentations (MLF). Higher population density and species richness were detected in Nemea than in Peza vineyards and on grapes than in fermenting musts. Pediococcus pentosaceus and Lactobacillus graminis were the most abundant LAB on grapes, while Lactobacillus plantarum dominated in fermenting musts from both regions. No particular structure of Lactobacillus plantarum populations according to the region of origin was observed, and strain distribution seems random. LAB species diversity in winery tanks differed significantly from that in vineyard samples, consisting principally of Oenococcus oeni. Different strains were analysed as per their enological characteristics and the ability to produce biogenic amines (BAs). Winery-associated species showed higher resistance to low pH, ethanol, SO2, and CuSO4 than vineyard-associated isolates. The frequency of BA-producing strains was relatively low but not negligible, considering that certain winery-associated Lactobacillus hilgardii strains were able to produce BAs. Present results show the necessity of controlling the MLF by selected starters in order to avoid BA accumulation in wine.
1. Introduction
In winemaking, a secondary fermentation known as malolactic fermentation (MLF) often takes place following the cease of yeast activity. During MLF, L-malate is converted into L-lactate by the lactic acid bacteria (LAB) of wine. This bioconversion is a desirable process in red winemaking and also in the production of certain white wines of high acidity, due to the organoleptic advantages that LAB activity confers. These include a decline in the total acidity and an increase of soft mouth feel, flavour, and microbiological stability of the wine [1]. However, MLF often entails certain risks, that is, the production of off-flavours, reduction in colour, and most importantly the formation of biogenic amines (BAs) [2, 3].
Currently, there is a growing concern regarding the limits of BAs in wines because of their potential health implications [4]. Although not regulated uniformly worldwide, BAs are generally confronted under similar regulations as for allergens. As a matter of fact, wines containing elevated amounts of histamine are rejected from certain markets due to recommended or suggested existing limits [4], while recently the Panel on Biological Hazards of the European Food Safety Authority (EFSA) released a scientific opinion on risk based control of BA formation in fermented foods [5]. Therefore, MLF in wine needs to be regulated to avoid the accumulation of BAs by LAB. This may be accomplished by the use of selected LAB strains tested for low production of BAs [6, 7] or able to degrade BA in wine [8].
Selected strains of Oenococcus oeni, the principal malolactic bacterium, have been launched in the market over the last decades. Nevertheless, wineries often face difficulties when conducting MLF by current commercial starters, as the induction of the process is not always successful [9]. Still several wineries prefer to conduct spontaneous malolactic fermentations by the native microbiota [10]. In these cases, the indigenous bacteria actualize MLF more effectively than commercial O. oeni, since native strains can deal with microbial incompatibilities and are better acclimatized to the local wine and practices [11, 12]. In addition, spontaneous MLF typically involves a composite bacterial community that may confer a more complex flavour to wine [1].
To this end, the wine industry seeks for novel MLF starters bearing positive technological and flavouring attributes [12]. The use of LAB species other than O. oeni is also being considered [13]. Grape resident microbial diversity forms an untapped reservoir of indigenous bacteria strains and may be primarily considered in an MLF starter selection scheme. Here we explored the local vineyard- and winery-associated LAB culturable populations in two key viticultural regions in Greece, Nemea and Peza. By using different molecular techniques various species and strains of enological importance were identified and characterised.
2. Materials and Methods
2.1. Sampling and LAB Isolation
Grape samples belonging to the Greek grapevine (Vitis vinifera) varieties “Vilana” (white), “Mandilaria” (red), and “Kotsifali” (red) were collected from 16 vineyards (1VP–16VP) within the Peza PDO region in Crete. Grapes of the “Agiorgitiko” cultivar (red variety) were collected from 11 vineyards (1VN–11VN) in the Nemea PDO region, Peloponnese. Samples consisting of healthy grape bunches were collected from at least 3 distant sampling points (sites) within each vineyard, placed into sterile plastic bags and transferred at 4°C to the laboratory. Grapes were crushed with a stomacher and let to ferment spontaneously in sterile bottles. Fermentation progress was daily followed by weight determinations. LAB were isolated from grapes or fermenting grape juice at the middle stage (MF) when about 50% of sugars were consumed, the final stage (EF) when sugars were depleted, and after the end of alcoholic fermentation. LAB were also isolated from wine samples collected from 9 tanks (T1–T9) of a winery in Nemea during spontaneous MLF. No spontaneous MLF was conducted in Peza winery. For bacteria enumeration, appropriate dilutions were spread onto MRS agar medium (pH 5.5) supplemented with 100 mg/L cycloheximide and incubated in anaerobic jars at 28°C for 3–8 days. Colonies were randomly selected from plates and examined microscopically. Bacterial colonies were further examined for Gram stain and catalase reaction. Isolates were maintained in liquid cultures in MRS broth with 30% glycerol at −80°C until further analysis.
2.2. Species Identification
DNA was extracted as previously described [14]. The 16S rDNA region of bacteria isolates was PCR-amplified using primers pA and pH [15]. For restriction analysis of the amplified 16S rDNA region (16S-ARDRA), approximately 500 ng of PCR product was digested with the restriction endonuclease MseI [15] and fragments were analyzed by agarose gel electrophoresis. For the differentiation of Lactobacillus plantarum, Lactobacillus pentosus, and Lactobacillus paraplantarum, a multiplex PCR assay was performed with the recA gene-based primers paraF, pentF, planF, and pREV, according to Torriani et al. [16]. For sequence analysis, the V1–V3 region of 16S rDNA was amplified using the primers P1V1 and P4V3 as previously described [17]. PCR products of representative isolates per distinct PCR-ARDRA pattern were sequenced (Macrogen; http://www.macrogen.com/). BLAST searches of sequences were performed at the NCBI/GenBank database.
2.3. Strain Typing and Genetic Analysis
Repetitive element sequence-based PCR (rep-PCR) using the single primer (GTG)5 or the primer pair REP1R-Dt and REP2R-Dt [18, 19] and RAPD analysis using the single primer RAPD1 or RAPD2 [20], 5′-ACGCGCCCT-3′ [21], and 1283 [22] were initially evaluated. The banding patterns corresponding to isolates from the same vineyard were considered as a vineyard population. UPGMA clustering of vineyard populations was conducted by using the PopGene 1.32 software [23].
2.4. Detection of BA-Producing Genes
For simultaneous detection of four genes involved in the production of major BAs in wine by LAB, that is, histamine (hdc), tyramine (tyrdc), and putrescine (odc and agdi), a multiplex PCR assay was applied as described elsewhere [24]. Briefly, the hdc and tyrdc genes were targeted with the primer pairs HDC3/HDC4 and TD2/TD5, respectively, while the primers ODC1/ODC2 and AGD1/AGD2 were used for the detection of agdi and odc genes, respectively. The 16S rRNA gene was concomitantly targeted with the universal primers BSF8/BSR1541 [25].
2.5. Technological Characterization of LAB
Tests were performed on MRS agar (pH 4) containing 7% ethanol unless otherwise stated. Ethanol tolerance was determined at ethanol contents of 10, 12, or 14%. SO2 resistance was evaluated at 5, 15, or 30 mg/L. Tolerance to low pH was determined at pH values of 3.0, 3.5, 4.0, or 5.5 adjusted by the addition of HCl. CuSO4 tolerance was evaluated at concentrations of 5 or 20 mg/L. Biogenic amines formation was determined on modified decarboxylating agar (MDA) plates (per litre: 5.0 g tryptone, 8.0 g meat extract, 4.0 g yeast extract, 0.5 g Tween 80, 0.2 g MgSO4, 0.05 g MnSO4, 0.04 g FeSO4, 0.1 g CaCO3, 0.06 g bromocresol purple, and 20.0 g agar) supplemented with 2% of either tyrosine, histidine, or arginine. The formation of biogenic amines was indicated by a purple halo around the bacterial colony as a result of amino acid decarboxylation [26]. Isolates were spot inoculated (ca. 106 cells/mL) on the surface of agar medium. Growth was evaluated after anaerobic incubation for up to 8 days at 28°C.
3. Results and Discussion
3.1. Bacterial Abundance
Grapes and wine fermentations constitute complex microbial ecosystems consisting of highly dynamic yeast and bacteria communities. Despite the importance of LAB populations in shaping the wine quality, our current knowledge on the spatiotemporal distribution of LAB populations in grapes and musts during the alcoholic or malolactic fermentation is still limited. Here we analyzed the LAB culturable communities in two distant viticultural zones in Greece, Peza in Crete and Nemea in Peloponnese. Samples included grapes and the respective fermenting musts. Sampling was also conducted after the end of the alcoholic fermentation (AF) and in situ in winery tanks during spontaneous MLF.
LAB were detected at relatively low frequencies on grapes. About 28% of grape samples from the Nemea region harbored bacteria at populations ranging from 1.4 to 3.8 log CFU/mL. In grapes from Peza, the bacterial populations were below the detection limit. The low incidence of LAB populations on wine grapes, as detected here, is in accordance with previous studies that suggest limited LAB population density (<3 log CFU/g) in vineyards, due to their nutritional requirements [10, 27–31].
Musts from grape samples were allowed to ferment spontaneously and at the middle stage of the AF (MF stage) bacteria could be recovered from 16% of the samples from either region. In the case of Nemea, population densities were relatively low (1.4–3.7 log CFU/mL), except for a single population that reached 8.7 log CFU/mL. Similarly, in Peza samples, populations at stage MF ranged from 0.9 to 3.3 log CFU/mL, except for one sample (ca. 7.2 log CFU/mL). At the end of the AF (EF stage), the number of Nemea samples with detectable populations decreased to 9%, while counts ranged from 1.3 to 7.0 log CFU/mL. As opposed, the respective percentage of Peza samples increased (24%), with populations ranging from 1.9 to 4.3 log CFU/mL. No bacterial populations were detected in samples from Nemea or Peza regions after the completion of AF. Present results show that, with a few exceptions, the bacterial growth is limited during the AF. Similarly low bacterial densities during the AF, ranging from 2 to 4 log CFU/mL, have been recorded previously [32]. These populations may further decline at the end of AF, with the exception of O. oeni [28, 32–36]. It is most likely that bacterial growth is prevented by the accumulating ethanol, the lack of nutrients, or the competition with indigenous yeast biota [28, 36]. Contracting this general observation, tumultuous bacterial growth during AF, as reported here, has been occasionally associated with musts infected with certain Lactobacillus spp. [28]. As opposed to vineyard-associated samples, relatively high bacterial densities (ca. 7 log CFU/mL) were recovered from winery tanks T1–T6. Populations of ca. 4 log CFU/mL were detected in tanks T7 and T8. Bacteria were below the detection limit in tank T9.
3.2. Species Identification
16S-ARDRA grouped 626 isolates according to their banding profiles (profiles I to IX) (Table 1; Figure 1). Phylogenetic analysis of the V1–V3 region of 16S rDNA of representative isolates from each group assigned them to the species Lactobacillus graminis, Lactobacillus hilgardii, Lactobacillus pentosus/plantarum, Lactococcus lactis, Oenococcus oeni, Pediococcus parvulus, P. pentosaceus, Staphylococcus epidermidis, and Weissella sp. According to the above analysis, isolates within group III showed 100% sequence similarity to both Lactobacillus pentosus JCM 1558T (D79211) and Lactobacillus plantarum NRRL B-14768T (AJ965482) followed by 99.8% to Lactobacillus paraplantarum DSM 10667T. Since 16S rDNA sequence is identical or highly similar among these species, a multiplex PCR assay with recA gene-based primers was applied for the identification of isolates within group III, as previously suggested [16], revealing that all isolates belong to the species Lactobacillus plantarum.
Table 1.
Species identification of bacteria isolates based on 16S-ARDRA profiles and sequence analysis.
| Profile | Approximate Sizes of Restriction Fragments (bp) | Species |
|---|---|---|
| I | 610 + 280 + 260 + 190 + 130 + 90 | Lactobacillus graminis |
| II | 420 + 270 + 200 + 130 + 110 + 90 | Lactobacillus hilgardii |
| III | 480 + 290 + 270 + 160 + 140 + 110 + 90 | Lactobacillus plantarum* |
| IV | 400 + 380 + 270 + 180 + 160 + 140 | Lactococcus lactis |
| V | 610 + 250 + 200 + 130 | Oenococcus oeni |
| VI | 400 + 270 + 230 + 150 + 130 + 80 | Pediococcus parvulus |
| VII | 290 + 260 + 250 + 130 + 120 + 110 + 90 | Pediococcus pentosaceus |
| VIII | 610 + 410 + 290 + 140 + 80 | Staphylococcus epidermidis |
| IX | 400 + 270 + 240 + 200 + 140 + 80 | Weissella sp. |
* Lactobacillus plantarum was differentiated from L. pentosus and L. paraplantarum with a multiplex PCR assay using recA gene-derived primers.
Figure 1.
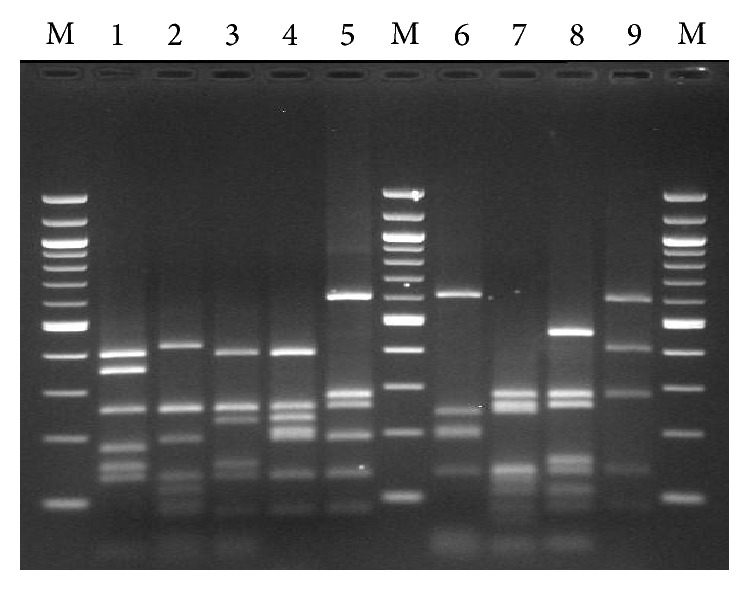
16S-ARDRA patterns obtained after digestion with MseI. Lanes: 1, Lactococcus lactis; 2, Lactobacillus hilgardii; 3, Pediococcus parvulus; 4, Weissella sp.; 5, Lactobacillus graminis; 6, Oenococcus oeni; 7, Pediococcus pentosaceus; 8, Lactobacillus plantarum; 9, Staphylococcus epidermidis; M, 100 bp molecular marker.
3.3. LAB Species Diversity and Succession
Pediococcus pentosaceus and Lactobacillus graminis were the most abundant LAB species in grape samples from Nemea (12.5 and 9.4%, resp.), followed by Weissella sp. and Lactococcus lactis at percentages lower than 7%. Typically, LAB species diversity associated with grape surfaces is rather limited mainly due to their nutritional requirements [28]. Species that have been reported to occur on grapes belong to the genera Lactobacillus (Lactobacillus casei, Lactobacillus hilgardii, Lactobacillus kunkeei, Lactobacillus lindneri, Lactobacillus mali, and Lactobacillus plantarum), Pediococcus, and Leuconostoc [29, 37, 38]. By applying a culture independent approach Renouf et al. [39] revealed a broader LAB diversity than previously described, including species within the genera Enterococcus and Weissella. Here we also detected Lactococcus lactis, a species that is quite scarce on grapes and a potentially novel Weissella species.
At the MF stage in Nemea samples, Pediococcus pentosaceus showed a higher level of persistence compared to the other species encountered on grapes. All other grape-associated populations were undetectable except for Lactobacillus graminis, which replaced S. epidermidis in one case. Lactobacillus plantarum emerged for the first time in two out of five samples, in which initial LAB populations on grapes were below the detection limit. At the EF stage, LAB were detected in three samples and all isolates were identified as Lactobacillus plantarum. Although in Peza grape samples bacteria were below the detection limit, LAB populations then emerged during the AF. At stage MF, Lactobacillus plantarum was the only species detected in all samples. At the EF stage, all samples were exclusively dominated by Lactobacillus plantarum, except for one sample in which P. pentosaceus thrived.
Previous studies have also shown that Lactobacillus plantarum is scarce on grapes [29, 30], but frequent in fermenting musts [10]. Oenococcus oeni, the principal malolactic bacterium often isolated from wines, was not detected on grapes or fermenting musts, collaborating previous suggestions about the absence or low population of this species in Greek vineyards [10].
The dominant population in winery-associated samples was O. oeni that could be recovered from all tanks performing spontaneous MLF. In 75% of the samples, Pediococcus parvulus was also isolated, albeit at significant lower populations than O. oeni. In one case, Lactobacillus hilgardii was also isolated along with P. parvulus, again at much lower population density than O. oeni (ca. 3 versus 7 log CFU/mL, resp.). The high occurrence of P. parvulus in the present samples needs further consideration since it is often associated with ropiness and oiliness of wine [40]. Furthermore, P. parvulus and Lactobacillus hilgardii were identified as the main spoilage, high histamine producing bacteria [41]; therefore their presence during MLF needs to be controlled.
3.4. Genotypic Diversity
For the discrimination of different LAB genotypes, various PCR-based fingerprinting methods were initially evaluated, including rep-PCR using the primer (GTG)5 or the primer set REP1R-Dt/REP2R-Dt and RAPD analysis with various primers. Among them, PCR using the primer RAPD2 (RAPD2-PCR) generated clear and reproducible banding patterns and also showed the highest discriminatory capacity in our tests (data not shown). Therefore, it was retained as the fingerprinting method of choice in the present genotyping analysis. The primer RAPD2 has been successfully applied previously in RAPD-PCR assays to differentiate strains within various LAB species [20, 42].
In the case of Lactobacillus plantarum isolates, RAPD2-PCR generated a total of 45 polymorphic bands and 14 distinct banding patterns (hereafter referred to as genotypes) were identified (Table 2). The number of different genotypes detected within a vineyard (all sampling points included) ranged from 1 to 5 (Figure 2). Recent metagenomic studies by using next generation sequencing technology suggest that different wine-growing regions may maintain different microbial communities [43, 44]. As far as regional variation in wine characteristics may be influenced by the local grape microflora, the so-called microbial “terroir” concept, it is very important to examine in more detail the spatiotemporal distribution of various strains. In this study, population genetic analysis was conducted in isolates of different vineyards (populations) and the existence of genetic structure between populations of the two geographical zones of origin (groups of Peza and Nemea) was evaluated. Results from UPGMA cluster analysis showed that the spatial distribution of genotypes within a vineyard is rather random (data not shown). Measures of genetic identity (Nei's coefficient) showed that most vineyard populations shared a relatively high degree of genetic similarity (>0.7). The UPGMA tree of vineyard populations showed no clustering according to the zone of origin (Figure 3).
Table 2.
Distinct genotypes according to RAPD2-PCR patterns of vineyard-associated LAB populations.
| LAB species | Region of origin | No. of isolates | No. of distinct patterns | Percentage of biodiversity* | Common patterns among vineyards | Common patterns between regions |
|---|---|---|---|---|---|---|
| Lactobacillus plantarum | Nemea Peza |
64 319 |
3 13 |
4.7 4.1 |
1 3 |
2 |
|
| ||||||
| Pediococcus pentosaceus | Nemea Peza |
61 16 |
5 1 |
8.2 6.3 |
4 — |
1 |
|
| ||||||
| Lactobacillus graminis | Nemea Peza |
37 nd** |
5 — |
13.5 — |
— — |
— |
|
| ||||||
| Lactococcus lactis | Nemea Peza |
21 nd |
3 — |
14.3 — |
— — |
— |
|
| ||||||
| Weissella sp. | Nemea Peza |
11 nd |
2 — |
18.2 — |
— — |
— |
*Ratio between the number of patterns and the number of isolates [45].
**Not detected.
Figure 2.
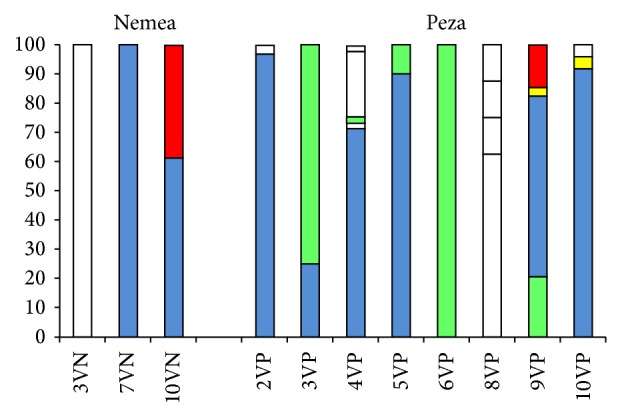
Distribution of Lactobacillus plantarum genotypes (%) in different vineyards of Nemea and Peza regions. Common genotypes are represented with the same colour. Unique genotypes are shown in white colour.
Figure 3.
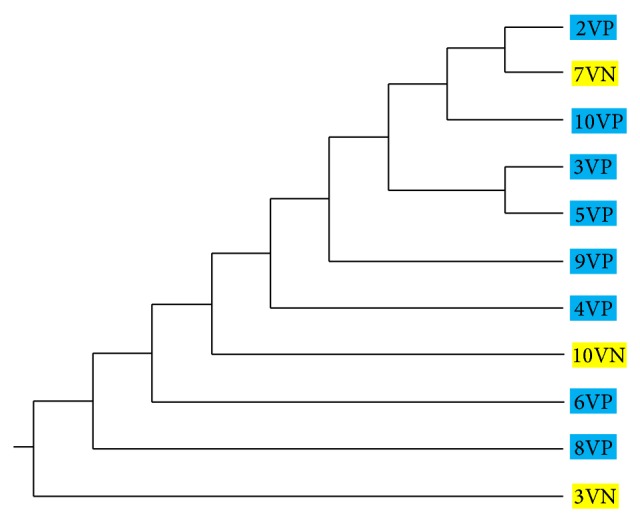
UPGMA dendrogram based on Nei's genetic distances among Lactobacillus plantarum vineyard populations. Populations from Nemea and Peza are yellow- and blue-highlighted, respectively.
The isolates from four more vineyard-associated LAB populations belonging to the species Lactobacillus graminis, Lactococcus lactis, P. pentosaceus, and Weissella sp. were analysed by RAPD2-PCR. Five distinct genotypes of P. pentosaceus were identified in samples originating from the Nemea region. Peza samples harbored a single P. pentosaceus genotype, which was also found in Nemea suggesting that it may be a cosmopolitan genotype. The species Lactobacillus graminis, Lactococcus lactis, and Weissella sp. were only detected in the Nemea region. The number of isolates analysed, the distinct banding patterns per population, and the percentage of biodiversity are summarized in Table 2.
Three different bacterial populations were associated with spontaneously fermenting wines in winery tanks. These included 12, 23, and 3 distinct genotypes for O. oeni, P. parvulus, and Lactobacillus hilgardii, respectively. The number of genotypes identified in different tanks is presented in Table 3. One up to five O. oeni distinct genotypes were isolated from the same tank. The respective range for P. parvulus was 2 to 7. In the case of Lactobacillus hilgardii all different genotypes were isolated from the same tank. Present results suggest that the genetic biodiversity of LAB species within a winery may be quite high (Table 3). Most importantly, different strains of the same LAB species may coexist in the same tank during MLF.
Table 3.
Distinct genotypes according to RAPD2-PCR patterns of winery-associated LAB populations.
| LAB species | Tank (T1–T9) | No. of isolates | No. of distinct patterns | Percentage of biodiversity* |
|---|---|---|---|---|
| Lactobacillus hilgardii | T6 | 4 | 3 | 75.0 |
| Oenococcus oeni | T1–T8 | 46 | 12 | 26.1 |
| Pediococcus parvulus | T1–T6 | 38 | 23 | 60.5 |
*Ratio between the number of patterns and the number of isolates [45].
3.5. Technological Characterization
Distinct genotypes within each species were evaluated as per their technological and enological characteristics (Table 4). Among LAB species, only O. oeni and P. parvulus isolates were able to grow at low pH, that is, at 3 or 3.5 in the presence of 7% ethanol. Growth at pH 4 was supported by all other species, albeit at different percentages. Winery-associated species showed higher resistance to SO2 than vineyard-associated isolates. Among the latter, several isolates of Lactobacillus plantarum, Lactococcus lactis, and Pediococcus pentosaceus tolerated up to 30 mg/L SO2. Lactobacillus graminis exhibited a moderate resistance, while Weissella sp. could grow only up to 30 mg/L SO2. Differences between winery- and vineyard-associated species were more profoundly reflected in ethanol tolerance. All vineyard-associated isolates could grow only up to 10% ethanol, except Weissella sp. A percentage of 43% of Lactobacillus plantarum strains could withstand 12% ethanol. Yet, winery-associated isolates could be considered as highly ethanol tolerant, resisting up to 14% ethanol. Again, the winery-associated isolates showed higher resistance to CuSO4 than vineyard-associated isolates did. P. pentosaceus was the most sensitive species to CuSO4, as none of the strains could tolerate a concentration of 20 mg/L.
Table 4.
Technological characteristics and biogenic amines production of vineyard- and winery-associated LAB species. The total number of strains analysed per species and the number of strains that produced positive reactions are indicated.
| LAB species | No of strains | Biogenic amines | pH | SO2 (mg/L) | Ethanol (%) | CuSO4 (mg/L) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Putrescine | Tyramine | Histamine | 3.0 | 3.5 | 4 | 5 | 15 | 30 | 10 | 12 | 14 | 5 | 20 | ||
| Lactobacillus graminis | 5 | 1 | 0 | 1 | 0 | 0 | 2 | 2 | 2 | 2 | 1 | 0 | 0 | 2 | 1 |
| Lactobacillus hilgardii | 3 | 1 | 3 | 0 | 0 | 0 | 3 | 3 | 3 | 3 | 3 | 3 | 2 | 3 | 3 |
| Lactobacillus plantarum | 14 | 0 | 0 | 0 | 0 | 0 | 11 | 11 | 11 | 8 | 8 | 6 | 0 | 11 | 4 |
| Lactococcus lactis | 3 | 1 | 0 | 0 | 0 | 0 | 3 | 3 | 3 | 3 | 1 | 0 | 0 | 3 | 3 |
| Pediococcus parvulus | 23 | 0 | 0 | 0 | 17 | 21 | 22 | 21 | 21 | 21 | 21 | 21 | 21 | 21 | 21 |
| Pediococcus pentosaceus | 5 | 5 | 0 | 0 | 0 | 0 | 5 | 5 | 5 | 2 | 4 | 0 | 0 | 5 | 0 |
| Oenococcus oeni | 12 | 0 | 0 | 0 | 3 | 9 | 11 | 10 | 10 | 6 | 12 | 10 | 10 | 10 | 9 |
| Weissella sp. | 2 | 0 | 0 | 0 | 0 | 0 | 1 | 2 | 1 | 0 | 0 | 0 | 0 | 1 | 1 |
3.6. BA-Producing LAB
LAB are the main producers of biogenic amines (BAs) in wine. Therefore, LAB should be evaluated for their ability to produce BAs, before being used as malolactic starters. By using appropriate culture media [26], we analysed the different strains identified in this study for their ability to produce the three major BAs in wine, that is, putrescine, tyramine, and histamine. As it is shown in Table 4, except for Lactobacillus plantarum, P. parvulus, and Weissella sp., certain strains from the other species were able to produce putrescine. The percentage of putrescine-producing strains was rather low, except for Pediococcus pentosaceus. Tyramine was found to be produced only by Lactobacillus hilgardii strains.
Recently, a PCR method was developed for the simultaneous detection of four genes involved in the production of the above BAs [24]. We applied this multiplex PCR to screen the above LAB strains. The PCR results were in good agreement with those obtained by the culture method. There was only one mismatch regarding a Lactobacillus hilgardii strain that produced tyramine but the corresponding gene (tyrdc) was not amplified. Thus the percentage of mismatching was rather low (1.5% of the strains), being slightly lower than the one detected by Coton et al. [24] (2.5%). It is likely that this discrepancy may be attributed to the existence of novel BA-producing genes not amplifiable by the present degenerate primers [24]. The relatively low frequency of BA-producing strains identified in this study is in accordance to previous results for wine-associated LAB, particularly as regards the low percentage of histamine-producing strains [24].
All three Lactobacillus hilgardii strains isolated from one winery tank performing spontaneous MLF produced tyramine and/or putrescine. Present results show the necessity of controlling the MLF by selected starters in order to avoid BA accumulation in the final product, since spontaneous fermentation may allow the occurrence of BA-producing strains.
4. Conclusions
The present study shows that the LAB species richness and population densities on grapes may differ considerably between regions or vineyards. Yet, Lactobacillus plantarum was the most abundant species in both regions and dominated the alcoholic fermentations. However, there was not any genetic structure in the Lactobacillus plantarum populations examined. As expected, O. oeni was quantitatively the principal LAB in the winery tanks during the MLF. Present results point to relatively high genotypic and phenotypic diversity within most LAB species identified, including O. oeni. Most importantly, various strains of the same species may coexist in the same tank during the MLF. Winery-associated species showed higher resistance to low pH, ethanol, SO2, and CuSO4 than vineyard-associated isolates. Most LAB strains did not produce BAs in our tests. Further PCR analysis targeting BA-producing genes verified that the frequency of BA-producing LAB was low. However, a few LAB strains isolated from a winery tank conducting MLF did produce major BAs, strengthening the need for novel superior LAB starters to control the MLF.
Acknowledgment
This work is funded by EU FP7 under Grant Agreement 315065-WILDWINE (http://www.wildwine.eu/).
Conflict of Interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
References
- 1.Lonvaud-Funel A. Lactic acid bacteria in the quality improvement and depreciation of wine. Antonie van Leeuwenhoek. 1999;76(1–4):317–331. doi: 10.1023/A:1002088931106. [DOI] [PubMed] [Google Scholar]
- 2.Henick-Kling T. Malolactic fermentation. In: Fleet G. H., editor. Wine Microbiology and Biotechnology. Chur, Switzerland: Harwood Academic Publisher; 1993. pp. 289–326. [Google Scholar]
- 3.Liu S.-Q. Malolactic fermentation in wine—beyond deacidification. Journal of Applied Microbiology. 2002;92(4):589–601. doi: 10.1046/j.1365-2672.2002.01589.x. [DOI] [PubMed] [Google Scholar]
- 4.Smit A. Y., du Toit W. J., du Toit M. Biogenic amines in wine: understanding the headache. South African Journal of Enology & Viticulture. 2008;29(2):109–127. [Google Scholar]
- 5.EFSA Panel on Biological Hazards (BIOHAZ) Scientific opinion on risk based control of biogenic amine formation in fermented foods. EFSA Journal. 2011;9(10, article 2393) [Google Scholar]
- 6.Martín-Álvarez P. J., Marcobal Á., Polo C., Moreno-Arribas M. V. Influence of technological practices on biogenic amine contents in red wines. European Food Research and Technology. 2006;222(3-4):420–424. doi: 10.1007/s00217-005-0142-7. [DOI] [Google Scholar]
- 7.Spano G., Russo P., Lonvaud-Funel A., et al. Biogenic amines in fermented foods. European Journal of Clinical Nutrition. 2010;64:95–100. doi: 10.1038/ejcn.2010.218. [DOI] [PubMed] [Google Scholar]
- 8.García-Ruiz A., González-Rompinelli E. M., Bartolomé B., Moreno-Arribas M. V. Potential of wine-associated lactic acid bacteria to degrade biogenic amines. International Journal of Food Microbiology. 2011;148(2):115–120. doi: 10.1016/j.ijfoodmicro.2011.05.009. [DOI] [PubMed] [Google Scholar]
- 9.López R., López-Alfaro I., Gutiérrez A. R., et al. Malolactic fermentation of Tempranillo wine: contribution of the lactic acid bacteria inoculation to sensory quality and chemical composition. International Journal of Food Science and Technology. 2011;46(11):2373–2381. doi: 10.1111/j.1365-2621.2011.02759.x. [DOI] [Google Scholar]
- 10.Nisiotou A. A., Rantsiou K., Iliopoulos V., Cocolin L., Nychas G.-J. E. Bacterial species associated with sound and Botrytis-infected grapes from a Greek vineyard. International Journal of Food Microbiology. 2011;145(2-3):432–436. doi: 10.1016/j.ijfoodmicro.2011.01.017. [DOI] [PubMed] [Google Scholar]
- 11.Maicas S., Pardo I., Ferrer S. The effects of freezing and freeze-drying of Oenococcus oeni upon induction of malolactic fermentation in red wine. International Journal of Food Science and Technology. 2000;35(1):75–79. doi: 10.1046/j.1365-2621.2000.00359.x. [DOI] [Google Scholar]
- 12.Solieri L., Genova F., De Paola M., Giudici P. Characterization and technological properties of Oenococcus oeni strains from wine spontaneous malolactic fermentations: a framework for selection of new starter cultures. Journal of Applied Microbiology. 2010;108(1):285–298. doi: 10.1111/j.1365-2672.2009.04428.x. [DOI] [PubMed] [Google Scholar]
- 13.Capozzi V., Russo P., Ladero V., et al. Biogenic amines degradation by Lactobacillus plantarum: toward a potential application in wine. Frontiers in Microbiology. 2012;3, article 122 doi: 10.3389/fmicb.2012.00122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Manzano M., Cocolin L., Cantoni C., Comi G. Bacillus cereus, Bacillus thuringiensis and Bacillus mycoides differentiation using a PCR-RE technique. International Journal of Food Microbiology. 2003;81(3):249–254. doi: 10.1016/S0168-1605(02)00222-2. [DOI] [PubMed] [Google Scholar]
- 15.Rodas A. M., Ferrer S., Pardo I. 16S-ARDRA, a tool for identification of lactic acid bacteria isolated from grape must and wine. Systematic and Applied Microbiology. 2003;26(3):412–422. doi: 10.1078/072320203322497446. [DOI] [PubMed] [Google Scholar]
- 16.Torriani S., Felis G. E., Dellaglio F. Differentiation of Lactobacillus plantarum, L. pentosus, and L. paraplantarum by recA gene sequence analysis and multiplex PCR assay with recA gene-derived primers. Applied and Environmental Microbiology. 2001;67(8):3450–3454. doi: 10.1128/AEM.67.8.3450-3454.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klijn N., Weerkamp A. H., de Vos W. M. Identification of mesophilic lactic acid bacteria by using polymerase chain reaction-amplified variable regions of 16S rRNA and specific DNA probes. Applied and Environmental Microbiology. 1991;57(11):3390–3393. doi: 10.1128/aem.57.11.3390-3393.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Versalovic J., Schneider M., De Bruijn F. J., Lupski J. R. Genomic fingerprinting of bacteria using repetitive sequence-based polymerase chain reaction. Methods in Molecular and Cellular Biology. 1994;5(1):25–40. [Google Scholar]
- 19.Gevers D., Huys G., Swings J. Applicability of rep-PCR fingerprinting for identification of Lactobacillus species. FEMS Microbiology Letters. 2001;205(1):31–36. doi: 10.1016/S0378-1097(01)00439-6. [DOI] [PubMed] [Google Scholar]
- 20.Fontana C., Cocconcelli P. S., Vignolo G. Monitoring the bacterial population dynamics during fermentation of artisanal Argentinean sausages. International Journal of Food Microbiology. 2005;103(2):131–142. doi: 10.1016/j.ijfoodmicro.2004.11.046. [DOI] [PubMed] [Google Scholar]
- 21.Johansson M.-L., Quednau M., Molin G., Ahrne S. Randomly amplified polymorphic DNA (RAPD) for rapid typing of Lactobacillus plantarum strains. Letters in Applied Microbiology. 1995;21(3):155–159. doi: 10.1111/j.1472-765X.1995.tb01030.x. [DOI] [PubMed] [Google Scholar]
- 22.Spano G., Beneduce L., Tarantino D., Zapparoli G., Massa S. Characterization of Lactobacillus plantarum from wine must by PCR species-specific and RAPD-PCR. Letters in Applied Microbiology. 2002;35(5):370–374. doi: 10.1046/j.1472-765X.2002.01200.x. [DOI] [PubMed] [Google Scholar]
- 23.Yeh F. C., Boyle T. J. B. Population genetic analysis of co-dominant and dominant markers and quantitative traits. Belgian Journal of Botany. 1997;129:p. 157. [Google Scholar]
- 24.Coton M., Romano A., Spano G., et al. Occurrence of biogenic amine-forming lactic acid bacteria in wine and cider. Food Microbiology. 2010;27(8):1078–1085. doi: 10.1016/j.fm.2010.07.012. [DOI] [PubMed] [Google Scholar]
- 25.Edwars U., Rogall T., Blocker H., Emde M., Bottger E. C. Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Research. 1989;17(19):7843–7853. doi: 10.1093/nar/17.19.7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maijala R. L. Formation of histamine and tyramine by some lactic acid bacteria in MRS-broth and modified decarboxylation agar. Letters in Applied Microbiology. 1993;17(1):40–43. doi: 10.1111/j.1472-765X.1993.tb01431.x. [DOI] [Google Scholar]
- 27.Lafon Lafourcade S., Carre E., Ribéreau-Gayon P. Occurrence of lactic acid bacteria during the different stages of vinification and conservation of wines. Applied and Environmental Microbiology. 1983;46(4):874–880. doi: 10.1128/aem.46.4.874-880.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fugelsang K. C., Edwards C. G. Microbial ecology during vinification. In: Fugelsang K. C., Edwards C. G., editors. Wine Microbiology: Practical Applications and Procedures. 2nd. New York, NY, USA: Springer; 2007. [Google Scholar]
- 29.Bae S., Fleet G. H., Heard G. M. Lactic acid bacteria associated with wine grapes from several Australian vineyards. Journal of Applied Microbiology. 2006;100(4):712–727. doi: 10.1111/j.1365-2672.2006.02890.x. [DOI] [PubMed] [Google Scholar]
- 30.Francesca N., Settanni L., Sannino C., Aponte M., Moschetti G. Ecology and technological capability of lactic acid bacteria isolated during Grillo grape vinification in the Marsala production area. Annals of Microbiology. 2011;61(1):79–84. doi: 10.1007/s13213-010-0109-1. [DOI] [Google Scholar]
- 31.Barata A., Malfeito-Ferreira M., Loureiro V. Changes in sour rotten grape berry microbiota during ripening and wine fermentation. International Journal of Food Microbiology. 2012;154(3):152–161. doi: 10.1016/j.ijfoodmicro.2011.12.029. [DOI] [PubMed] [Google Scholar]
- 32.Ribéreau-Gayon P., Dubourdieu D., Donèche B., Lonvaud A. The microbiology of wine and vinifications. In: Ribéreau-Gayon P., editor. Handbook of Enology. Vol. 1. Chichester, UK: John Wiley & Sons; 2006. [Google Scholar]
- 33.Wibowo D., Eschenbruch R., Davis D. R., Fleet G. H., Lee T. H. Occurrence and growth of lactic acid bacteria in wine: a review. The American Journal of Enology and Viticulture. 1985;36(4):302–313. [Google Scholar]
- 34.Lonvaud-Funel A., Joyeux A., Ledoux O. Specific enumeration of lactic acid bacteria in fermenting grape must and wine by colony hybridization with non-isotopic DNA probes. The Journal of Applied Bacteriology. 1991;71(6):501–508. doi: 10.1111/j.1365-2672.1991.tb03824.x. [DOI] [Google Scholar]
- 35.van Vuuren H. J. J., Dicks L. M. T. Leuconostoc oenos: a review. American Journal of Enology and Viticulture. 1993;44(1):99–112. [Google Scholar]
- 36.Volschenk H., van Vuuren H. J. J., Viljoen-Bloom M. Malic acid in wine: origin, function and metabolism during vinification. South African Journal of Enology and Viticulture. 2006;27(2):123–136. [Google Scholar]
- 37.Jackson R. S. Origin and growth of lactic acid bacteria. In: Jackson R. S., editor. Wine Science: Principles and Applications. San Diego, Calif, USA: Academic Press; 2008. pp. 394–402. [Google Scholar]
- 38.du Toit M., Engelbrecht L., Lerm E., Krieger-Weber S. Lactobacillus: the next generation of malolactic fermentation starter cultures—an overview. Food and Bioprocess Technology. 2011;4(6):876–906. doi: 10.1007/s11947-010-0448-8. [DOI] [Google Scholar]
- 39.Renouf V., Claisse O., Lonvaud-Funel A. Inventory and monitoring of wine microbial consortia. Applied Microbiology and Biotechnology. 2007;75(1):149–164. doi: 10.1007/s00253-006-0798-3. [DOI] [PubMed] [Google Scholar]
- 40.Dols-Lafargue M., Lee H. Y., le Marrec C., Heyraud A., Chambat G., Lonvaud-Funel A. Characterization of gtf, a glucosyltransferase gene in the genomes of Pediococcus parvulus and Oenococcus oeni, two bacterial species commonly found in wine. Applied and Environmental Microbiology. 2008;74(13):4079–4090. doi: 10.1128/AEM.00673-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Landete J. M., Ferrer S., Pardo I. Which lactic acid bacteria are responsible for histamine production in wine? Journal of Applied Microbiology. 2005;99(3):580–586. doi: 10.1111/j.1365-2672.2005.02633.x. [DOI] [PubMed] [Google Scholar]
- 42.Belfiore C., Raya R. R., Vignolo G. M. Identification, technological and safety characterization of Lactobacillus sakei and Lactobacillus curvatus isolated from Argentinean anchovies (Engraulis anchoita) SpringerPlus. 2013;2(1, article 257) doi: 10.1186/2193-1801-2-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bokulich N. A., Thorngate J. H., Richardson P. M., Mills D. A. Microbial biogeography of wine grapes is conditioned by cultivar, vintage, and climate. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(1):E139–E148. doi: 10.1073/pnas.1317377110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gilbert J. A., van der Lelie D., Zarraonaindia I. Microbial terroir for wine grapes. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(1):5–6. doi: 10.1073/pnas.1320471110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schuller D., Alves H., Dequin S., Casal M. Ecological survey of Saccharomyces cerevisiae strains from vineyards in the Vinho Verde Region of Portugal. FEMS Microbiology Ecology. 2005;51(2):167–177. doi: 10.1016/j.femsec.2004.08.003. [DOI] [PubMed] [Google Scholar]


