Abstract
The autosomal dominant Hailey Hailey disease (HHD) is caused by mutations in the ATP2C1 gene encoding for human secretory pathway Ca2+/Mn2+ ATPase protein (hSPCA1) in the Golgi apparatus. Clinically, HHD presents with erosions and hyperkeratosis predominantly in the intertrigines. Here we report an exome next generation sequencing (NGS) based analysis of ATPase genes in a Greek family with 3 HHD patients presenting with clinically atypical lesions mainly localized on the neck and shoulders. By NGS of one HHD-patient and in silico SNP calling and SNP filtering we identified a SNP in the expected ATP2C1 gene and SNPs in further ATPase genes. Verification in all 3 affected family members revealed a heterozygous frameshift deletion at position 2355_2358 in exon 24 of ATP2C1 in all three patients. 7 additional SNPs in 4 ATPase genes (ATP9B, ATP11A, ATP2B3 and ATP13A5) were identified. The SNPs rs138177421 in the ATP9B gene and rs2280268 in the ATP13A5 gene were detected in all 3 affected, but not in 2 non affected family members. The SNPs in the ATP2B3 and ATP11A gene as well as further SNPs in the ATP13A5 gene could not be confirmed in all affected family members. One may speculate that besides the level of functional hSPCA1 protein, levels of other ATPase proteins may influence expressivity of the disease and might also contribute, as in this case, to atypical presentations.
Introduction
The autosomal dominant inherited Hailey Hailey disease (HHD; also benign familiar chronic pemphigus) is caused by mutations (permanent changes of the nucleotide sequence which may affect one or several nucleotides) in the human secretory pathway Ca2+/Mn2+ ATPase protein (hSPCA1) in the Golgi apparatus encoded by the ATP2C1 gene on chromosome 3q21[1,2]. The ATP2C1 gene spans 28 exons and has four splice variants, ATP2C1a–d. A variety of more than eighty mutations lead to dysfunction of the protein [1–3]. In this context, impaired calcium levels in the Golgi and cytoplasm have been described [1].
Clinically, HHD presents with erosions and hyperkeratosis predominantly in the intertrigines. Erosions are caused by intraepidermal acantholysis due to a retraction of keratin filaments from the desmosomal plaque and formation of perinuclear aggregates [4]. Disease onset lies around puberty up to 30–40 years with a complete penetrance but variable expressivity. Heat, sweating, mechanical trauma, infections, and UVB exposure can cause exacerbations. Remissions and relapses characterize the course of HHD[5]. Interestingly, extracutaneous manifestations have not been reported despite the ubiquitous expression of hSPCA1 [5].
Several mechanisms have been suggested to explain how a mutation in the ATP2C1 gene causes blistering in HHD. Most are linked to calcium homeostasis, since it is essential for trafficking of desmosomal proteins to the cell membrane. Calcium and manganese are also required for correct processing, production, and maturation of proteins including glycosylation [6]. Indeed, an overall impaired calcium homeostasis with high resting cytosolic and low Golgi-calcium levels has been shown in keratinocytes of HHD patients [1][7]. In addition, HHD epidermis displayed an altered calcium gradient in vivo [7] which together with abnormal ATP receptor expression may contribute to the blistering [8].
In HHD the majority of patients present with crusted erosions and warty papules on skin folds, mainly the axilla, inguinal folds and groin. Interestingly, a family of Greek origin was presenting with clinically atypical HHD, in which lesions were mainly localized on the non-folded skin of neck and the chest, back and shoulders. All lesions first presented around the 25th year of age in all affected family members and had a tendency to recur during the summer months or after sweating and to heal with postinflammatory pigmentation during the cold months of the year. (Fig 1a). Since ATP2C1 is a member of a large family of genes involved in ATP-dependent ion transport and other members of the ATPase gene family are also involved in calcium homeostasis and desmosomal processing, e.g. ATP2A2, the Darier‘s disease gene [3], all ATPase genes in 5 males (3 affected and 2 non-affected) of this family were investigated for single nucleotide polymorphisms (SNPs) by next generation sequencing (NGS).
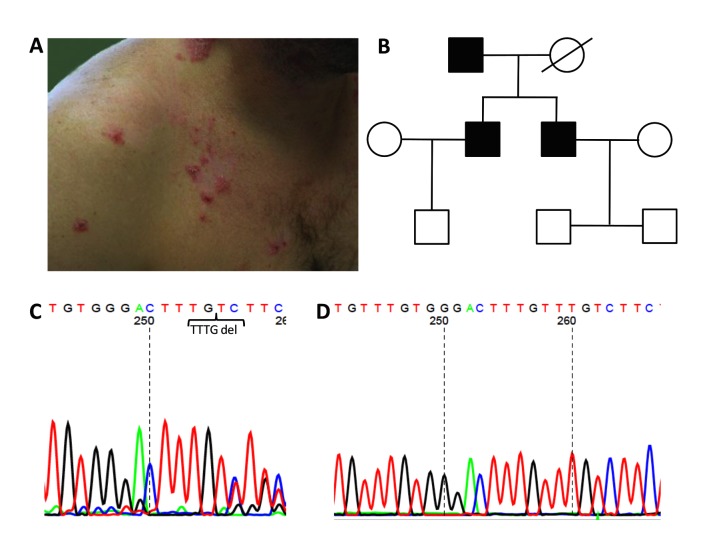
Fig 1. Clinical picture, pedigree, and electropherogram of the Hailey-Hailey disease (HHD) family.
Erythematous slightly scaling vesicles, papules, and plaques on the right breast, shoulder, and neck of HHD-affected 1 (a). Pedigree of the HHD family. Black squares represent affected males (aged 70, 39 and 35 years), white squares unaffected males (aged 4, 9 and 9 years), white circles represent unaffected women, icons with a crossing line represent family members that had passed away (b). ATP2C1 mutation of HHD-affected 2 shown by electropherogram at position 2355_2358 in exon 24 resulting in subsequent frameshift displayed by heterozygous mismatch (c) Electropherogram of the unaffected HHD- healthy 1displaying the reference allele (d).
Materials and Methods
Blood was drawn from all 5 family members and non-lesional skin of one affected male was obtained after informed consent following the declarations of Helsinki. The participants provided their verbal and written consent and our group has obtained an approval from the ethics committee of the University of Lübeck to conduct genome wide association studies and genomic sequencing of pemphigus patients (approval number 08–156).
Sequence data of one HHD patient gained by NGS Exome Sequencing with Nimblegen Exon array, depth 50x, paired end 100bp reads (BGI, Shenzhen, China) were analyzed for quality, assembled and aligned to the human genome reference build HG19. In silico SNP calling and SNP filtering by I) per base quality score II) Phred score, III) 1,000 genomes project frequency (version April 2012_ALL), IV) AV-SIFT Score <0.05 and V) non-synonymous predicted effects identified 66 SNPs, including a deletion in the expected ATP2C1 gene. Verification by conventional Sanger- resequencing in this patient and two further affected and two unaffected family members was carried out after DNA extraction with Qiagen DNA mini Kit (Qiagen, Hilden, Germany) and PCR using Phusion High- fidelity PCR Kit (New England Biolabs, Germany) following the manufactures instructions (S1 Table). Sanger sequencing was performed at Beckman Coulter Genomics (Essex, United Kingdom). SNP confirmation analysis was performed with Chromas Lite and GENtle.
Results and Discussion
The SNP verification revealed a heterozygous frameshift deletion at position 2355_2358 in exon 24 of ATP2C1 in all three patients. This mutation resembles a mutation reported earlier [1,2] and could not be detected in 2 unaffected family members. Interestingly, 7 additional SNPs in 4 ATPase genes, ATP9B, ATP11A, ATP2B3 and ATP13A5, were identified and confirmed in various family members (Table 1).
Table 1. ATPase genes in 3 HHD patients and 2 unaffected family members where SNPs could be verified to a variable extend.
| Gene | ref | Observed by NGS | dbSNP 135 | Verification by Sanger sequencing | |||||
|---|---|---|---|---|---|---|---|---|---|
| affected 1 | affected 2 | affected 3 | healthy 1 | healthy 2 | affected 2/ skin | ||||
| ATP2C1 | TTTG | heterozygous frameshift deletion TTTG | n.a. | TTTG/0 | TTTG/0 | TTTG/0 | TTTG/ TTTG | TTTG/ TTTG | TTTG/0 |
| ATP11A | G | heterozygous A/G | rs61746637 | G/G | A/G | A/G | A/G | A/G | A/G |
| ATP9B | A | heterozygous A/T | rs138177421 | A/T | A/T | A/T | A/A | A/A | A/T |
| ATP2B3 | C | homozygous T | n.a. | C/C | T/T | C/C | C/C | C/C | T/T |
| ATP13A5Snp1 | A | heterozygous G/A | rs2271791 | G/A | G/A | G/A | G/A | G/A | G/A |
| ATP13A5Snp2 | C | heterozygous C/T | rs2280268 | C/T | C/T | C/T | T/ T | T/T | C/T |
| ATP13A5Snp3 | C | heterozygous C/G | rs6797429 | C/C | C/G | C/G | C/C | C/C | C/G |
| ATP13A5Snp4 | G | heterozygous G/T | rs12637558 | T/T | G/T | G/T | G/T | G/T | G/T |
ref = reference allele, NGS = exome next generation sequencing performed on affected 2, dbSNP135 = dbSNP database release 135, skin = non-lesional skin of affected 2, n.a. = not available
Remarkably, in the ATP9B gene, a heterozygous SNP rs138177421 was detected in all 3 affected, but not in the 2 non affected family members. This SNP is highly conserved and infrequent in populations e.g. 1000genome project frequency 0.0009 (see Table 2). Although little is known about ATP9B, it has recently been identified as a class 2 P4-ATPase, a putative phospholipid flippase translocating phospholipids to the cytoplasm, that localizes to the trans- Golgi network [9]. This type of ATPase is involved in transport vesicle formation and cell polarity by pumping an unknown ion [9]. These alterations in cell polarity might add on to the compromised hSPCA1 function in HHD during Golgi stress. Golgi- and ER- stress have been discussed as being involved in desmosomal processing [8]. Indeed, desmosomal proteins are in particular susceptible to cleavage during apoptosis. Thus, altered cell polarity due to a mutation in ATP9B may further challenge the Golgi stress response towards apoptosis resulting in acantholysis.
Table 2. Conservation scores and frequencies of SNPs in ATPase genes found in a Greek Family with atypical Hailey Hailey disease.
| Gene | ExonicFunc | Aminoacid Change | ESP5400_ALL | 1000genomes_ALL | dbSNP135 | AVSIFT | LJB_PhyloP | LJB_SIFT | LJB_PolyPhen2 |
|---|---|---|---|---|---|---|---|---|---|
| ATP2C1 | frameshift deletion | NM_001199182:c.2355_2358del:p.785_786del | NA | NA | NA | NA | NA | NA | NA |
| ATP13A5 | nonsynonymous SNV | NM_198505:c.T3392C:p.V1131A | 0.368935 | 0.36 | rs2271791 | 0.16 | 0.998904 | 0.99 | 35 |
| ATP13A5 | nonsynonymous SNV | NM_198505:c.G2215A:p.G739S | 0.538855 | 0.55 | rs2280268 | 0.53 | 0.857449 | 0.56 | 0.0030 |
| ATP13A5 | nonsynonymous SNV | NM_198505:c.G397C:p.E133Q | 0.549080 | 0.53 | rs6797429 | 0.96 | 0.99508 | 0.4 | 0.0 |
| ATP13A5 | nonsynonymous SNV | NM_198505:c.C287A:p.S96Y | 0.371537 | 0.41 | rs12637558 | 0.02 | 0.992706 | 0.98 | 994 |
| ATP11A | nonsynonymous SNV | NM_015205:c.G2743A:p.A915T | 0.020450 | 0.02 | rs61746637 | 0.14 | 0.999472 | 0.83 | 0.682206 |
| ATP9B | nonsynonymous SNV | NM_198531:c.A617T:p.Q206L | 0.001952 | 0.0009 | rs138177421 | 0.23 | 0.980726 | 0.63 | 0.0010 |
| ATP2B3 | nonsynonymous SNV | NM_001001344:c.C422T:p.S141L | NA | NA | NA | 0.12 | 0.980682 | 0.8 | 0.0 |
Gene = Gene name for variant, ExonicFunc = synonymous, non-synonymous, indel, etc., SNV = single nucleotide variant, Aminoacid Change = variant change in nucleotide and protein format, ESP5400_ALL = MAF in Exome Sequencing Project dataset (5,400 exomes) for all populations, 1000genomes_ALL = MAF in 1000Genomes February 2012 release, dbSNP135 = RS# from the dbSNP database, AVSIFT = SIFT Pathogenicity score: closer to 0 is more damaging, LJB_PhyloP = Pathogenicity score from dbNSFP: conserved > 0.95, not conserved < 0.95, LJB_SIFT = Pathogenicity score from dbNSFP: tolerated < 0.95, deleterious > 0.95, LJB_PolyPhen2 = Pathogenicity score from dbNSFP: probably damaging > 0.85, possibly damaging 0.85–0.15, benign < 0.15, NA = not available.
Furthermore, we identified a second SNP in ATP13A5 being unique for the affected family members. ATP13A5 SNP 2 rs2280268 showed a heterozygous state in all affected individuals studied. In contrast, the 2 non-affected family members displayed a homozygous mutation C→T distinct from the reference genome. Further SNPs in the ATP13A5 gene were present in the unaffected family members as well (ATP13A5 SNP 1 and 4) or not present in all of the affected family members (ATP13A5 SNP 3) (Table 1). Since these SNPs are common and conserved in the general population (Table 2), only an accumulation of SNPs in this particular gene may contribute to disease susceptibility in HHD. ATP13A5 belongs to the group of P5-ATPases of which the substrate specificity is unknown. However, recent work suggests that these enzymes affect the intracellular level of different cations, as they localize to vacuolar/lysosomal or apical membranes [10]. In ATP11A, a SNP was confirmed by Sanger sequencing in all samples tested except for one affected family member analyzed (Table1) and a homozygous SNP of the ATP2B3 gene occurred in one affected family member and his skin. These results fit the understanding of HHD as a disease in which the loss of function mutation in ATP2C1 on its own is not sufficient to cause the HHD symptoms[11]. The phenotype is strongly influenced by genetic modifiers and environmental triggers[11]. With the identification of ATP9B and ATP13A5 SNP2 we provide two possible novel genetic modifiers in the orchestra of ionic balance in the Golgi apparatus and hSPCA1.
Conclusion
Taken together although the level of functional hSPCA1 protein in epidermal cells seems critical [6], levels of other ATPase proteins may influence expressivity of Hailey-Hailey disease. Additionally affected ATPase genes might also contribute, as possibly in this case, to the atypical presentation of the disease. Effects of these mutations could further add on Golgi stress and ionic imbalance, thus might be leaving the skin in affected individuals more vulnerable to environmental triggers than in those patients only carrying an ATP2C1 mutation. Elucidating ATPase SNP frequencies in diseased patients as well as their role in the pathogenesis of HHD and their functional effects requires further population-based studies and functional investigations in the future.
Supporting Information
PCR conditions: 10μl HF buffer, 1μl dNTPs, 0.5μl phusion polymerase; all Phusion High fidelity Kit; 2.5μl forward and reverse primer (BiomersGmbH, Ulm, Germany), 100ng sample DNA and nuclease free water adding up to a final volume of 50μl; 98°C 1 min, 32 cycles of 98°C 5 sec/ 68°C 20 sec / 72°C 25 sec, 72°C 10 min. Nonsyn. SNV = nonsynonymous single nucleotide variant.
(DOCX)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
These authors have no support or funding to report.
References
- 1. Hu Z, Bonifas JM, Beech J, Bench G, Shigihara T, Ogawa H, et al. Mutations in ATP2C1, encoding a calcium pump, cause Hailey-Hailey disease. Nat Genet. 2000;24: 61–65. [DOI] [PubMed] [Google Scholar]
- 2. Sudbrak R, Brown J, Dobson-Stone C, Carter S, Ramser J, White J, et al. Hailey–Hailey disease is caused by mutations in ATP2C1 encoding a novel Ca2+ pump. Human Molecular Genetics. 2000;9: 1131–1140. [DOI] [PubMed] [Google Scholar]
- 3. Szigeti R, Kellermayer R. Autosomal-Dominant Calcium ATPase Disorders. J Invest Dermatol. 2006;126: 2370–2376. [DOI] [PubMed] [Google Scholar]
- 4. Hashimoto K, Fujiwara K, Tada J, Harada M, Setoyama M, Eto H. Desmosomal dissolution in Grover’s disease, Hailey-Hailey’s disease and Darier’s disease. J Cutan Pathol. 1995;22: 488–501. [DOI] [PubMed] [Google Scholar]
- 5. Burge SM. Hailey-Hailey disease: the clinical features, response to treatment and prognosis. Br J Dermatol. 1992;126: 275–282. [DOI] [PubMed] [Google Scholar]
- 6. Foggia L, Hovnanian A. Calcium pump disorders of the skin. Am J Med Genet C Semin Med Genet. 2004;131C: 20–31. [DOI] [PubMed] [Google Scholar]
- 7. Behne MJ, Tu C-L, Aronchik I, Epstein E, Bench G, Bikle DD, et al. Human Keratinocyte ATP2C1 Localizes to the Golgi and Controls Golgi Ca2+ Stores. Journal of Investigative Dermatology. 2003;121: 688–694. [DOI] [PubMed] [Google Scholar]
- 8. Shull GE, Miller ML, Prasad V. Secretory Pathway Stress Responses as Possible Mechanisms of Disease Involving Golgi Ca2+ Pump Dysfunction. Biofactors. 2011;37: 150–158. 10.1002/biof.141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Takatsu H, Baba K, Shima T, Umino H, Kato U, Umeda M, et al. ATP9B, a P4-ATPase (a Putative Aminophospholipid Translocase), Localizes to the trans-Golgi Network in a CDC50 Protein-independent Manner. J Biol Chem. 2011;286: 38159–38167. 10.1074/jbc.M111.281006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. De La Hera DP, Corradi GR, Adamo HP, De Tezanos Pinto F. Parkinson’s disease-associated human P5B-ATPase ATP13A2 increases spermidine uptake. Biochem J. 2013;450: 47–53. 10.1042/BJ20120739 [DOI] [PubMed] [Google Scholar]
- 11. Kellermayer R. Hailey-Hailey disease from a clinical perspective. Cell Calcium. 2008;43: 105–106. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PCR conditions: 10μl HF buffer, 1μl dNTPs, 0.5μl phusion polymerase; all Phusion High fidelity Kit; 2.5μl forward and reverse primer (BiomersGmbH, Ulm, Germany), 100ng sample DNA and nuclease free water adding up to a final volume of 50μl; 98°C 1 min, 32 cycles of 98°C 5 sec/ 68°C 20 sec / 72°C 25 sec, 72°C 10 min. Nonsyn. SNV = nonsynonymous single nucleotide variant.
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.