SUMMARY
Human pluripotent stem cell (hPSC) lines exhibit repeated patterns of genetic variation, which can alter in vitro properties as well as suitability for clinical use. We examined associations between copy number variations (CNVs) on chromosome 17 and hPSC mesodiencephalic dopaminergic (mDA) differentiation. Among 24 hPSC lines, two karyotypically-normal lines, BG03 and CT3, and BG01V2, with trisomy 17, exhibited amplification of the WNT3/WNT9B region and rapid mDA differentiation. In hPSC lines with amplified WNT3/WNT9B, bFGF signaling through MAPK/ERK amplifies canonical WNT signaling by phosphorylating LRP6, resulting in enhanced undifferentiated proliferation. When bFGF is absent, non-canonical WNT signaling becomes dominant due to up-regulation of SIAH2, enhancing JNK signaling and promoting loss of pluripotency. When bFGF is present during mDA differentiation, stabilization of canonical WNT signaling causes up-regulation of LMX1A and mDA induction. Therefore CNVs in 17q21.31, a “hot spot” for genetic variation, have multiple and complex effects on hPSC cellular phenotype.
INTRODUCTION
While CNVs make major contributions to human genetic variation (Zhang et al., 2009), there have been few opportunities to examine their roles in altered cellular function and development. We have taken advantage of variations in differentiation capacity between a series of hPSC lines, including BG01V2 with trisomy 17, to explore the role of chromosome 17 CNVs in mDA neuronal differentiation.
The utility of both human embryonic stem cells (hESCs) and human induced pluripotent stem cells (hiPSCs) depends on differentiation to specific somatic cell types. Clinical applications include transplantation to replace or restore the mDA neurons lost in Parkinson’s disease. hPSC lines differ in features that include growth in the undifferentiated state (Werbowetski-Ogilvie et al., 2009), potential for tumor formation (Ben-David and Benvenisty, 2011), and differentiation to selected phenotypes (Kim et al., 2011). These factors impact the utility of hPSCs both for transplantation and as models of human cell types and organ systems for in vitro studies of development and drug discovery.
Parallels between hPSC cytogenetic changes and neoplastic progression (Werbowetski-Ogilvie et al., 2009) emphasize the need for genetic analyses of hPSCs. The genomes of many individual humans contain CNVs which span between a few hundred to more than a million bases (McCarroll et al., 2008). CNVs are found frequently in hESCs as well as hiPSCs (Närvä et al., 2010) and have the potential to impact hPSC function.
The present study focused on functional characterization of CNVs on chromosome 17. Chromosome 17 has been implicated in pluripotency of hESCs (Ben-Yehudah et al., 2010). Gain of chromosome 17, especially 17q, has been repeatedly observed in hESC lines (Baker et al, 2007). Previous studies have correlated genetic alterations on 17q21.31 with neurobehavioral and neurodegenerative disorders, including mDA related disorders such as Parkinson’s disease (Grisart et al., 2009, Spencer et al., 2011). We have therefore sought evidence for involvement of chromosome 17 in variations between hPSC lines. We report that variations in the 17q21.31-17q21.32 region which include the WNT3-WNT9B gene cluster alter hPSC proliferation and mDA differentiation phenotypes.
RESULTS
BG01V2 and BG03 Display Enhanced Undifferentiated Proliferation in the Presence of bFGF and Increased Loss of Pluripotency Following Growth Factor Withdrawal
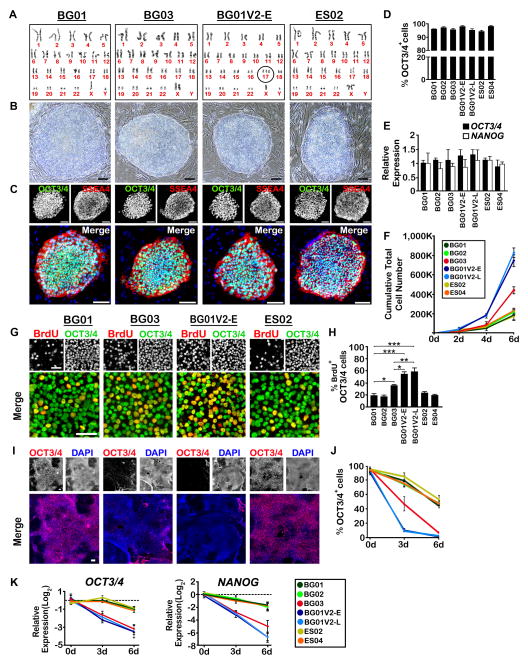
We characterized five karyotypically-normal hESC lines: BG01, BG02, BG03, ES02, and ES04, as well as BG01V2, a line with a trisomy 17 (Vazin et al., 2008) (Figures 1A and S1A). BG01V2 was studied at two passage levels (> 24 passages apart) to provide a test for genomic stability. Undifferentiated colonies of all hESC lines displayed well-defined boundaries when cultured in a feeder-dependent or -independent manner (Figures 1B, S1B and S1D), a feature normally absent in transformed hESCs (Werbowetski-Ogilvie et al., 2009). All undifferentiated lines expressed the pluripotency markers OCT3/4, SSEA4, and NANOG (Figures 1C, S1C and S1E; NANOG not shown). More than 90% of cells expressed OCT3/4 (Figures 1D and S1F). There were no significant differences in OCT3/4 or NANOG expression (Figures 1E and S1G).
Figure 1. BG01V2 and BG03 are Phenotypically Distinct from Other Karyotypically Normal hESC Lines.
(A) Representative G-banding karyotype analysis of hESC lines. Black circle indicates gain of chromosome 17 in BG01V2-E. BG01 (P165), BG03 (P52), BG01V2-E (P37), and ES02 (P46). (B) Phase-contrast images of hESC lines in feeder-dependent cultures showing well-defined colony boundaries. Scale bar, 100 μm. (C) Expression of OCT3/4 and SSEA4 by immunocytochemistry in each hESC line. Scale bar, 100 μm. (D, E) Percentage of OCT3/4+ cells (D) and relative expression of OCT3/4 and NANOG (E) in feeder-dependent culture for each hESC line. n = 3. All values in (E) were expressed as fold-changes with respect to BG01. (F) Cumulative growth of hESC lines over 6 d in feeder-free culture. n = 3. (G, H) Expression of BrdU and OCT3/4 by immunocytochemistry under feeder-free conditions for each hESC line after 6 d. Percentage of OCT3/4+ cells incorporating BrdU are shown in (H). Scale bar, 50 μm. n = 3. *P < 0.05, **P < 0.01, ***P < 0.001. (I, J) Expression of OCT3/4 by immunocytochemistry (I) under feeder-free conditions for each hESC line after 6 d of bFGF withdrawal. Percentage of cells remaining OCT3/4+ after 6 d are shown in (J). Scale bar, 100 μm. n = 3. (K) Relative expression in log2 form of OCT3/4 and NANOG following bFGF withdrawal over 6 d under feeder-free conditions. n = 3. Error bars, SEM.
Cumulative growth curves over 6 d showed a ~4-fold increase in BG01V2 cell number at both early and late passages (BG01V2-E and –L, respectively) and a ~2-fold increase in BG03, as compared to the other lines (Figure 1F). These data are consistent with increased percentages of OCT3/4+ cells labeled by bromodeoxyuridine (BrdU, 10 μM, 1 h exposure at the end of 6 d culture) in BG01V2 (-E and -L) and BG03 (Figures 1G, 1H and S1H). Expression of the apoptotic marker Annexin V in OCT3/4+ cells indicated that preferential cell death was not responsible for these differences in cell accretion (Figures S1J and S1K). Thus, BG01V2 and BG03 proliferate faster than the remaining lines.
Following withdrawal of basic fibroblast growth factor (bFGF), essential for maintaining pluripotency, BG01V2 (-E and –L) and to a lesser degree BG03 lost pluripotency more rapidly than the other cell lines (Figures 1I, 1J and S1I). OCT3/4 and NANOG expression also decreased more rapidly over 6 d in BG01V2 (-E and –L) and in BG03 (Figure 1K). These data suggest that an enhanced propensity for departure from pluripotency distinguishes BG01V2 (-E and -L) and BG03 from the characteristic in vitro behavior of transformed or tumorigenic hESC variants (Werbowetski-Ogilvie et al., 2009), despite their increased rates of proliferation.
BG01V2 and BG03 Exhibit Enhanced mDA Differentiation
We compared mDA differentiation using a well-established protocol (Yan et al., 2005) that employs midbrain trophic factors to generate mDA neurons via distinct stages of differentiation in parallel to in utero mDA development. Undifferentiated hESC colonies were converted to embryoid bodies (EBs), then differentiated for 10 d in the presence of bFGF then FGF8 to yield neuroepithelial (NE) rosettes with radially-organized columnar cells. Rosettes were isolated and cultured in the presence of FGF8 and SHH to yield mDA progenitors (mDP) by d31. These mDP were further differentiated to reach mature mDA neuronal identity by d45 (Figure 2A).
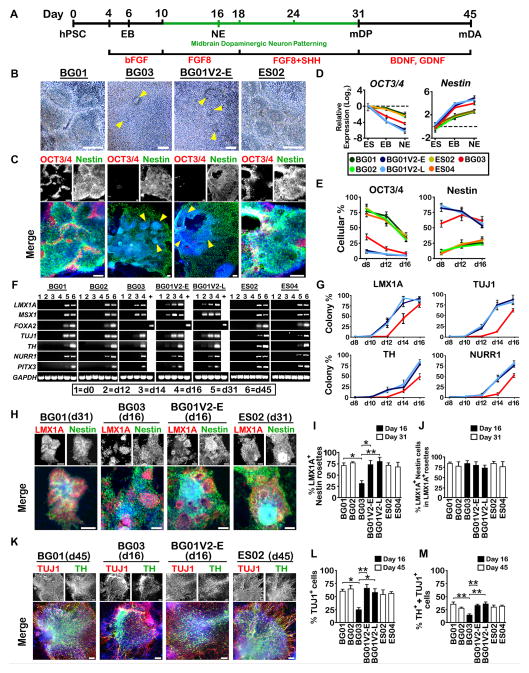
Figure 2. BG01V2 and BG03 Exhibit Highly Efficient mDA Differentiation.
(A) Schematic representation of the mDA neuronal differentiation procedure. (B, C) Phase contrast (B) and expression of OCT3/4 and nestin by immunocytochemistry (C) on d16 for each hESC line. For BG01V2-E and BG03, rosettes are interspersed among a layer of cells, and are indicated by yellow arrowheads. Scale bar, 50 μm (B) and 100 μm (C). (D, E) Relative expression in log2 form (D) and percentages of cells (E) positive for OCT3/4 and nestin at various stages of differentiation. n = 3. (F) RT-PCR analysis showing expression of genes related to mDA development at various stages of differentiation, +, positive control representing mDA stage of the BG01 line at d45. (G) Percentages of colonies positive for various mDA-related markers from d8 to d16 of differentiation in BG01V2 (-E and -L) and BG03, as denoted by color legend in (D) and (E). n = 3. (H) Expression of LMX1A and nestin by immunocytochemistry at d16 for BG01V2-E and BG03, and at d31 for the remaining lines. Scale bar, 100 μm. (I) Percentage of LMX1A+ rosettes among total nestin+ rosettes per colony. n = 3. (J) Percentage of LMX1A+ cells among total nestin+ cells in LMX1A+ rosettes. n = 3. (K) Expression of TH and TUJ1 by immunocytochemistry at d16 for BG01V2-E and BG03, and at d45 for the remaining lines. Scale bar, 100 μm. (L, M) Percentages of TUJ1+ cells (L) and cells positive for both TH and TUJ1 (M) among total cells. n = 3. Error bars, SEM. *P < 0.05, **P < 0.01. Abbreviations: hPSC, human pluripotent stem cell; EB, embryoid body; NE, neuroepithelial; mDP, midbrain dopaminergic progenitor, mDA, midbrain dopaminergic neuron.
At d16, whole BG01V2 (-E and -L) and BG03 colonies displayed a layer of loosely-arranged nestin+ cells interspersed with nestin+ rosettes, retaining only trace areas of OCT3/4+ cells (Figures 2B, 2C, S2A and S2B). In other hESC lines, nestin+ rosettes were restricted to the colony centers, and were surrounded by abundant OCT3/4+ cells (Figures 2B, 2C, S2A and S2B). Temporal gene expression analysis revealed that OCT3/4 expression declined with concomitant increased nestin expression from d0 to d16 in BG01V2 (-E and -L) and BG03 (Figure 2D). The rapid loss of pluripotency found in BG01V2 and BG03 was thus associated with accelerated differentiation toward a neuroectodermal lineage.
We further quantified OCT3/4+ and nestin+ cells during the NE stage. As early as day 8, there were significant disparities between numbers of OCT3/4+ and nestin+ cells in BG01V2 (-E and -L), BG03, and the remaining lines (Figure 2E). Surprisingly, nestin+ cellular percentages in BG01V2 (-E and -L) declined from d8-16 (cellular percentage in BG03 started declining at d12), diverging from ordinary patterns observed in Figure 2D. Percentages of TUNEL-labeled nestin+ progenitors did not significantly increase in BG01V2 (-E and -L) or BG03 from d8-16 (Figures S2C and S2D). Thus accelerated differentiation of nestin+ cells, rather than apoptosis, seems to have been responsible this decline in nestin+ cells.
We next examined mDA differentiation. The transcription factor LMX1A and its downstream target, MSX1, are both expressed in mDPs and required for mDA phenotype specification (Cai et al., 2009). Both were present at d12 in BG01V2 (-E and -L) and at d14 in BG03, but appeared much later (d31) in the remaining lines (Figures 2F and 2G). BG01V2 (-E and -L) displayed greater numbers of LMX1A+ rosettes per colony than BG03 on d16 while still containing similar LMX1A+ cellular percentages in those rosettes (Figures 2H–2J and S2E). In non-rosette areas, ~50% of nestin+ cells in BG01V2 (-E and -L), and ~20% in BG03, differentiated to LMX1A+ cells (Figures S2F and S2G). Thus, BG01V2 and BG03 displayed more efficient conversion to mDPs than the remaining lines, with BG03 showing a less efficient conversion than BG01V2.
Moreover, the neuronal marker TUJ1, the dopaminergic marker TH, and the midbrain-specific transcription factors NURR1 and PITX3 appeared earlier in BG01V2 (-E and -L) than BG03, while appearing much later in the remaining lines (Figures 2F and 2G). BG01V2 (-E and -L), and to a lesser degree BG03, showed comparable mDA differentiation at d16 relative to other lines at d45 (Figures 2K–2M and S2H). Nevertheless, all lines ultimately displayed a similar mature mDA phenotype, manifested by high percentages of TH+ cells that expressed NURR1 (Figures S2I and S2J). Therefore, mDA conversion was accelerated in BG01V2 and BG03.
In contrast to LMX1A and MSX1, FOXA2, another mDP transcription factor expressed in the midbrain floor plate and a downstream target of the SHH signaling pathway (Ferri et al., 2007), was absent from BG01V2 (-E and -L) and BG03, yet was present in the remaining hESC lines starting on d31 (Figures 2F and S2K). These data indicate that LMX1A-MSX1, but not the SHH-FOXA2 pathway, is responsible for the accelerated mDA specification in BG01V2 and BG03. Complete mDA specification, with FOXA2 expression, in BG01V2 and BG03 could be achieved by adding SHH from d10-16 (Figure S2L).
Identification of the WNT3/WNT9B Gene Cluster at 17q21.31 in Relationship to mDA Differentiation
Next, we attempted to determine the genetic contributors to enhanced mDA differentiation in BG01V2 and BG03. We developed an a priori list of 103 genes, including the entire WNT, SHH, and FGF8 signaling families, that were potentially related to mDA differentiation based on literature review (Table S1). We then focused on the seven genes located on chromosome 17 (Figure 3A) due to the enhanced mDA differentiation potential of BG01V2, which contains a trisomy 17.
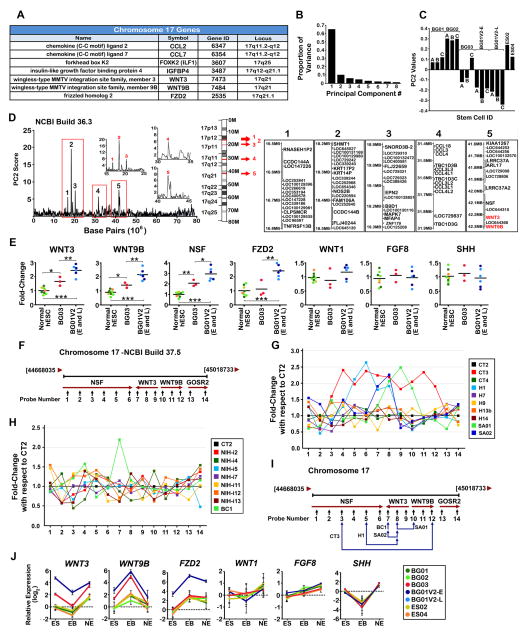
Figure 3. The WNT3/WNT9B Gene Cluster is Amplified in BG01V2 and BG03.
(A) A priori list of genes potentially related to mDA neuronal differentiation on chromosome 17. (B) PCA analysis of CNV and SNP probes located on chromosome 17, indicating overall contribution of each principal component to the overall variance in the data set obtained from 3 distinct passage numbers for BG01, 02, 03, and 01V2 (-E and –L), and one passage number for ES02 and 04. (C) PC2 values derived from the hybridization intensities of DNA samples. A, B, and C represent different passage numbers, see Figure S3C. (D) The PC2 scores highlight five regions on chromosome 17 that may contribute to the phenotypic differences. Each of the five regions is enlarged and represented by a red number on the ideogram (middle diagram). Genes located in each amplified region were obtained using NCBI Build 36.3 and are listed in the tables. The genes of interest, WNT3 and WNT9B, are located between 41.6 MB and 42.3 MB, and are highlighted in red. (E) qPCR analysis of seven genes in different hESC lines; BG01, BG02, BG03, BG01V2 (-E and –L) (n=3), ES02 and ES04 (n=1). Values are expressed as fold-changes with respect to BG01. Horizontal lines represent the mean value for each data set. P* < 0.05, P** < 0.01, ***P < 0.001. (F) Schematic representation of probe numbers and locations for the 17q21.31 target region. (G, H) qPCR analysis of copy number variations in this region in additional hESC (G) and hiPSC (H) lines using designed probes. Values are expressed as fold-changes with respect to CT2 (black line). (I) Schematic showing extent of the amplified regions (above 1.5 fold-change) for the indicated hESC and hiPSC lines. NCBI Build 37.5 used for (F-I). (J) Relative expression of genes at different stages of mDA differentiation. Values are expressed as fold-changes, in log2 form, with respect to BG01. n=3. Error bars, SEM.
Principal component analysis (PCA) of hybridization intensity data from chromosome 17 CNV and SNP probes on Affymetrix 6.0 SNP arrays were used to search for evidence of chromosome 17 CNVs among the hESC lines. PCA of the hybridization of material prepared from cell line DNA to CNV and SNP probes on these arrays identified five principal components (PC1 – PC5) that contributed substantially to the overall variance among these cell lines (Figure 3B). PC2 was strongly related to mDA differentiation (Figures 3C and S3A). PC2 scores were significantly lower for BG01V2 (-E and –L) and BG03 than for the remaining lines. Mapping PC2 scores onto chromosome 17 identified loci at 16.5–16.8 MB, 18.2–18.5 MB, 18.9–19.3 MB, 31.4–31.9 MB, and 41.6–42.3 MB (NCBI Build 36.3) (Figure 3D), that contributed substantially to the overall PC2 scores. The 17q21.31, 41.6–42.3 MB locus contains WNT3 and WNT9B, two of the seven a priori chromosome 17 candidates for involvement with mDA differentiation (Figure 3A). SNP genotyping confirmed that BG01V2 and BG03 were not genetically related by descent (Figure S3B).
We then examined whether the DNA levels of WNT3, WNT9B and NSF, which is located within the same peak as the WNT3-WNT9B cluster (Figure 3D), varied between hESC lines. There were increased levels of DNA for each of these genes in both BG01V2 and BG03 (Figure 3E). By contrast, levels of DNA encoding the WNT-receptor FZD2, also found on chromosome 17 but not located within the PC2-derived peaks, were increased only in the trisomic BG01V2 line (Figures 3A and 3E). Neither line displayed increased DNA levels for WNT1, FGF8, or SHH (Figure 3E) which all play important roles in mDA differentiation (Burbach et al., 2006), but are located on chromosomes 12, 10, and 7, respectively.
It is generally acknowledged that there are laboratory- and cell line-specific variations in hESCs and hiPSCs (Guenther et al., 2010, Tomoda et al., 2012). We thus screened ten additional hESC and eight hiPSC lines from various laboratories for extra copies of WNT3/WNT9B, by using fourteen probes across NSF, WNT3, WNT9B, and GOSR2 from chromosome 17 44668035-45018733 (NCBI Build 37.5) (Figure 3F). Two hESC lines, CT3 and SA01, showed increased levels of both WNT3 and WNT9B DNA, while hESC lines H1 and SA02 and hiPSC line BC1 showed increased WNT3 DNA only (Figures 3G–3I). These data support the likelihood that genomic amplifications in the WNT3-WNT9B region are not unusual. These genomic differences were accompanied by phenotypic differences. CT3, with complete amplification of the WNT3-WNT9B region, exhibited enhanced proliferation in the undifferentiated state in the presence of bFGF, accelerated loss of pluripotency after removal of bFGF, and highly efficient mDA induction (18.2% of cells positive for both TUJ1 and TH in CT3 at d16), similar to BG01V2 and BG03 (Figures S3D–S3F).
We next examined expression of mRNAs encoded by the amplified WNT genes during mDA differentiation. Because BG01V2-E and BG01V2-L did not differ markedly, we used BG01V2-E for further experiments. We observed greater WNT3 and WNT9B mRNA levels in BG01V2, BG03, and CT3 during mDA differentiation, paralleling expression of these genes in other hESC lines (Figures 3J and S3G, and fold-changes are shown in Table S2). Changes in mRNA were as large as 10- to 56-fold, greatly exceeding the corresponding changes in DNA. WNT3 expression was predominant at ES and NE stages of differentiation, while WNT9B expression was predominant during the EB stage. FZD2 expression was increased in BG01V2 only, at all stages of differentiation. No differences in expression of WNT1, FGF8 or SHH were observed (Figures 3J and S3G). Taken together, these findings suggest that amplification of the WNT3/WNT9B gene cluster contributes to the acceleration of mDA differentiation seen in BG01V2, BG03, and CT3. Expression patterns suggest distinct roles of this increased expression at different stages of mDA differentiation.
Canonical WNT3 and Non-canonical WNT9B Signaling Pathways are Activated at Different Stages of mDA Differentiation
WNT3 and WNT9B act mainly via the β-catenin-dependent (canonical) and the Rho/JNK (non-canonical) signaling pathways, respectively (Lie et al., 2005, Karner et al., 2009). FZD2 is involved in both canonical and non-canonical pathways (Sato et al., 2010). The active (dephosphorylated) form of β-catenin was elevated during ES and NE stages for BG01V2, BG03, and CT3 as compared to the other hESC lines, and to a smaller extent at the EB stage for BG01V2 (Figures 4A, 4B, and S4A). In contrast, active forms of Rho and JNK (GTP-bound Rho and phosphorylated JNK) were increased only at the EB stage (Figures 4C, 4D, and S4C). These data are consistent with the expression patterns of WNT3 and WNT9B (Figures 3J and S3G). Together the results suggest that WNT3/β-catenin signaling is activated predominantly at the ES and NE stages, while WNT9B/JNK signaling is activated primarily at the EB stage.
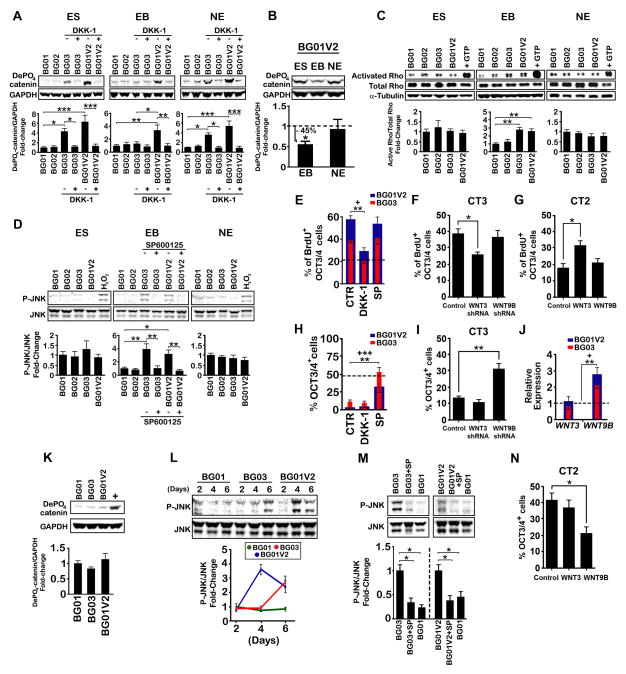
Figure 4. WNT3/β-catenin and WNT9B/JNK Signaling are Differentially Activated at Various Stages of mDA Differentiation and Play a Role in Undifferentiated Proliferation and Pluripotency.
(A) Immunoblot of dephosphorylated (active) β-catenin levels at various stages of mDA differentiation in the presence or absence of 500 ng/ml DKK-1. ES represents 6 d of feeder-free culture supplemented with bFGF, EB represents d0-6 of differentiation, and NE represents d6-d16 of differentiation. ES (n = 4), EB (n = 3), NE (n = 4). (B) Immunoblot showing dephosphorylated (active) β-catenin levels in BG01V2 at various stages of mDA differentiation. Values were expressed as a ratio to BG01V2 at the ES stage (dashed line). ES (n = 4), EB (n = 3), NE (n = 4). (C) Immunoblot of active Rho levels in hESC lines at various stages of mDA differentiation. Addition of GTP to activate Rho in BG01 was used as a positive control; ES, EB, NE (n = 3). (D) Immunoblot showing phosphorylated JNK (P-JNK) levels in hESC lines at various stages of mDA differentiation. H202 (10−3M, 1 hr) in BG01 was used as a positive control for the ES and NE stages. For the EB stage BG01V2 and BG03 were treated with 10 μM SP600125; ES, EB, NE (n = 4). (E–G) Percentage of OCT3/4+ cells incorporating BrdU for BG01V2 and BG03 treated with 500 ng/ml DKK-1 or 10 μM SP600125 (E), for lentivirus-mediated WNT3 or WNT9B knockdown in CT3 (F), or for lentivirus-mediated WNT3 or WNT9B overexpression in CT2 (G) over 6 d in feeder-free culture with bFGF. BG01 (dashed line) was used for comparison in E. n = 3. (H, I) Quantification of percentages of cells remaining OCT3/4+ after 6 d of bFGF withdrawal for BG01V2 and BG03 treated with 500 ng/ml DKK-1 or 10 μM SP600125 (H) or for lentivirus-mediated WNT3 or WNT9B knockdown in CT3 (I), under feeder-free conditions. BG01 (dashed line) was used for comparison in (H). n = 3. (J) Relative expression of WNT3 and WNT9B in BG01V2 and BG03 under feeder-free conditions after 6 d of bFGF withdrawal. All values are expressed as fold-changes with respect to BG01 (dashed line) at d6. n = 4. (K) Immunoblot of dephosphorylated (active) β-catenin levels on d6 under feeder-free conditions after 6 d of bFGF withdrawal. BG01V2 at the ES stage in the presence of bFGF was used as a positive control. n = 4. (L, M) Immunoblot of P-JNK in BG01V2 and BG03 under feeder-free conditions at various stages over a 6 d period following bFGF withdrawal (L) and in the presence or absence of 10 μM SP600125 on d6 (M). BG01 was used for comparison. n = 3. (N) Quantification of percentages of cells remaining OCT3/4+ after 6 d of bFGF withdrawal for lentivirus-mediated WNT3 or WNT9B overexpression in CT2 under feeder-free conditions. n = 3. All immunoblot values were normalized to internal control (GAPDH [A, B, K], total Rho [C], or JNK [D, L, M]) and expressed as ratios to BG01 (A, C, D, J), to BG01V2 at the ES stage (B, dashed line), to BG01 at d2 (L), or to control (M). SP = SP600125. Error bars, SEM. P-values for BG03 and BG01V2 in E, H, and J are represented by + and *, respectively. +P and *P < 0.05; **P< 0.01; +++P and ***P < 0.001.
WNT3/β-catenin Contributes to Enhanced Proliferation while WNT9B/JNK Contributes to Loss of Pluripotency
To further elucidate the effects of WNT3 and WNT9B in hPSC lines with trisomy/gains in chromosome 17, we employed (i) DKK-1 and SP600125, inhibitors of canonical β-catenin signaling and non-canonical JNK signaling, respectively, (ii) lentiviral vectors to overexpress WNTs, or (iii) to silence WNT3 and WNT9B expression through delivery of shRNAs at various stages of mDA differentiation (Figures 4A, 4D, S4B, S4D, and S7A, Table S6).
First, we explored the roles of WNTs in undifferentiated proliferation and pluripotency in BG01V2, BG03, and CT3. At the undifferentiated ES stage DKK-1 administration or WNT3-knockdown, but neither SP600125 nor WNT9B-knockdown, reversed the enhanced proliferation of these lines, indicated by fewer BrdU-labeled OCT3/4+ cells (Figures 4E and 4F). DKK-1 also reversed the increased expression of cyclin D1 (Figure S4E), a downstream target of β-catenin that regulates cell cycle progression, to levels comparable to controls. These data suggest that WNT3/β-catenin signaling is involved in the enhanced proliferation of BG01V2, BG03, and CT3. In addition, overexpression of WNT3, but not WNT9B, in CT2 increased the percentage of BrdU-labeled OCT3/4+ cells (Figure 4G), further evidence that WNT3 amplification can enhance proliferation of hPSCs.
Conversely, accelerated loss of pluripotency in BG01V2, BG03, and CT3 following bFGF withdrawal was reversed by SP600125 treatment or WNT9B-knockdown, but not by DKK-1 administration or WNT3-knockdown, as indicated by increased percentages of OCT3/4+ cells (Figures 4H and 4I). SP600125 also increased expression of OCT3/4 and NANOG (Figure S4F), after bFGF withdrawal, to levels similar to controls. These results are concordant with the increased expression of WNT9B, but not WNT3, in BG01V2 and BG03 6 d after withdrawal of bFGF (Figure 4J). The results are also consistent with the activation of JNK, but not β-catenin, seen on d6 (Figures 4K and 4L). JNK activation peaked earlier in BG01V2 than in BG03 (Figure 4L) and was lowered by SP600125 to control levels (Figure 4M). Together, these results indicate that WNT9B/JNK signaling accelerates the loss of pluripotency in BG01V2, BG03, and CT3 following bFGF withdrawal. Overexpression of WNT9B, but not WNT3, in CT2 decreased the percentage of OCT3/4+ cells after bFGF withdrawal, suggesting that WNT9B amplification can accelerate ES differentiation (Figure 4N).
WNT9B/JNK Promotes mDA Differentiation through Enhanced Loss of Pluripotency
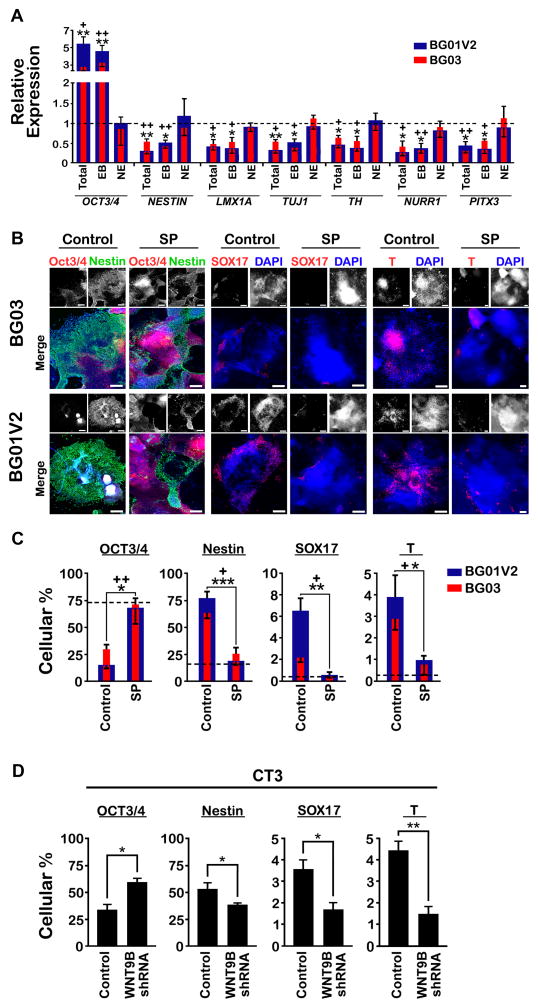
To investigate the roles of WNTs in mDA differentiation in hPSC lines with chromosome 17 abnormalities, the overall 16-d mDA differentiation process was subdivided into total (d0-16), EB (d0-6), and NE (d6-16) epochs. Interestingly, SP600125 treatment during either the total or EB epoch, but not NE, increased expression of OCT3/4 and decreased expression of nestin and other mDA neuronal markers at d16 (Figure 5A). These results implicate WNT9B/JNK in early mDA differentiation, and are consistent with the enhanced WNT9B expression and activation of WNT9B/JNK signaling primarily at the EB stage (Figures 3J, 4D, S3G, and S4C).
Figure 5. WNT9B/JNK Signaling Regulates mDA Differentiation during Early NE.
(A) Relative expression of genes involved in mDA differentiation following treatment with 10 μM SP600125 every other day during three distinct periods: total (d0-16), EB (d0-6), and NE (d6-16). All BG01V2 and BG03 values are expressed as fold-changes with respect to the control condition, i.e., without SP600125 (dashed line). n = 3. (B) Expression of germ layer markers (nestin, SOX17, and T) by immunocytochemistry at d8 for BG01V2 and BG03 in the presence or absence of 10 μM SP600125. Scale bar, 100 μm. (C, D) Percentages of cells positive for OCT3/4, nestin, SOX17, and T at d8 for BG01V2 and BG03 in the presence or absence of 10 μM SP600125 or for CT3 after control or WNT9B shRNA lentivirus-mediated infection. n = 3. The dashed line represents BG01 in C. Error bars, SEM. P-values for BG03 and BG01V2, with respect to the control condition, are represented by + and *, respectively. +P and *P < 0.05; ++P and **P < 0.01; ***P < 0.001.
The disruption of mDA differentiation by administration of SP600125 during the EB epoch could be due either to enhanced pluripotency or to non-ectodermal germ layer differentiation. At d8, after EBs have adhered and flattened, SP600125 administration or WNT9B-knockdown produced an increase in OCT3/4+ cells and a decrease in markers for all three germ layer lineages, including nestin, SOX17, and Brachyury (T) (Figures 5B, 5C, and 5D). These data indicate that blockage of WNT9B/JNK during the EB stage interrupts hPSC departure from pluripotency, consistent with previous pluripotency assays (Figures 4H, 4I, and S4F). Therefore, in chromosome 17-abnormal hPSC lines, WNT9B/JNK enhances departure of hPSCs from pluripotency, leading to an increase in neuroectodermal precursors that will later adopt an mDA fate at the NE stage.
WNT3/β-catenin Promotes Expansion of NE cells, Neuronal Induction and mDA Differentiation
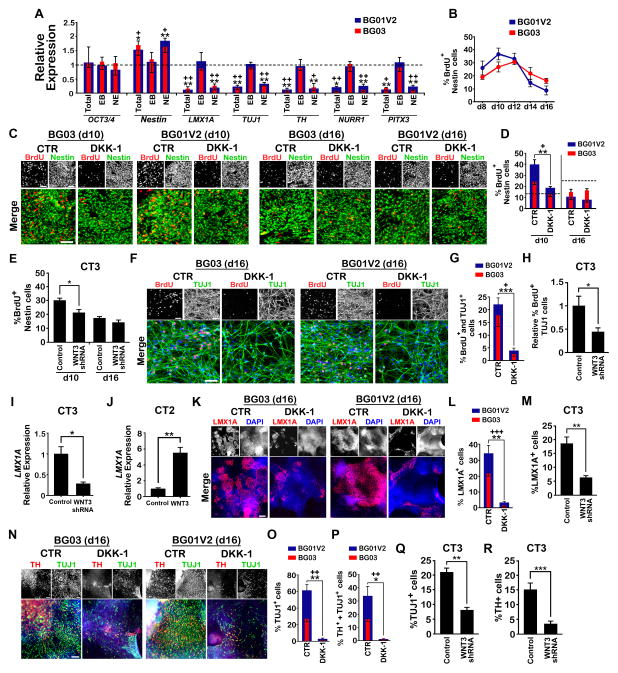
We next studied the role of WNT3/β-catenin during mDA differentiation in hPSC lines with amplified WNTs. In contrast to SP600125, DKK-1 treatment did not significantly alter OCT3/4 expression (Figure 6A), suggesting that WNT3/β-catenin is not involved in the regulation of pluripotency. However, DKK-1 treatment during either total or NE epochs, but not during the EB epoch, decreased the expression of all mDA markers (Figure 6A). Thus WNT3/β-catenin is involved in late mDA differentiation. This finding is consistent with the enhanced WNT3 expression and activation of WNT3/β-catenin signaling primarily at the NE stage in BG01V2, BG03, and CT3 (Figures 3J, 4A, S3G, and S4A).
Figure 6. WNT3/β-catenin Regulates mDA Differentiation during Late NE.
(A) Relative expression of genes involved in mDA differentiation following treatment with 500 ng/ml DKK-1 every other day during the total (d0-16), EB (d0-6), and NE (d6-16) differentiation epochs. BG01V2 and BG03 values are expressed as fold-changes with respect to the control condition, i.e., in the absence of DKK-1 (dashed line). n = 3. (B) Percentage of BrdU-labeled nestin+ cells from d8 to d16. n = 3. (C, D) Expression of BrdU and nestin (C) by immunocytochemistry for BG01V2 and BG03 at d10 and d16 in presence or absence of DKK-1. Cells were treated with DKK-1 (500 ng/ml) for 2 d prior to the end of culture. Corresponding BrdU-labeled nestin+ cellular percentages are shown in (D). Scale bar, 50 μm. n = 3. (E) BrdU-labeled nestin+ cellular percentages for lentivirus-mediated WNT3 knockdown (4d prior to end of culture) in CT3 at d10 and d16. n = 3. (F, G) Expression of BrdU and TUJ1 (F) by immunocytochemistry for BG01V2 and BG03 at d16 in presence or absence of DKK-1. Percentages of BrdU+ and TUJ1+ cells are shown in (G). Scale bar, 50 μm, n = 3. (H) Relative percentage of BrdU-labeled TUJ1 cells for lentivirus-mediated WNT3 knockdown (d12-d16) in CT3. n=6. (I, J) Relative expression of LMX1A for lentivirus-mediated WNT3 knockdown in CT3 (I) and WNT3 overexpression in CT2 (J) from d12-16. n = 3. (K, L) Expression of LMX1A (K) by immunocytochemistry for BG01V2 and BG03 at d16 in the presence or absence of DKK-1. Percentages of LMX1A+ cells are shown in (L). Scale bar, 100 μm. n = 3. (M) Percentages of LMX1A+ cells for lentivirus-mediated WNT3 knockdown (d12-d16) in CT3. n = 3. (N-P) Expression of TUJ1 and TH (N) by immunocytochemistry for BG01V2 and BG03 at d16 in the presence or absence of DKK-1. Percentages of TUJ1+ (O) and TH+ and TUJ1+ (P) cells are shown. Scale bar, 100 μm. n = 3. (Q, R) Percentages of TUJ1+ (Q) and TH+ (R) cells for lentivirus-mediated WNT3 knockdown (d12-d16) in CT3. n = 3 (Q), n = 6 (R). For F, G, K, L, and N-P cells were treated with DKK-1 for 4 d prior to the end of culture. Error bars, SEM. P-values for BG03 are represented by + and for BG01V2 and CT3 are represented by *. +P and *P < 0.05; ++P and **P < 0.01; +++P and ***P < 0.001.
During the NE stage, DKK-1 treatment increased nestin expression (Figure 6A), either due to increased NE cell survival or proliferation, or to reduced NE cell differentiation. We ruled out an effect on cell survival by quantifying percentages of nestin+ cells labeled by TUNEL (Figure S5A). NE cell proliferation was further examined by studying BrdU-labeled nestin+ cells (10 μM, 1 h exposure at the end of culture) at d10 and d16, when NE proliferation was either predominant (d10) or greatly decreasing (d16) (Figure 6B). At d10, but not d16, DKK-1 administration or WNT3-knockdown decreased cell proliferation, and DKK1 decreased cyclin D1 expression (Figures 6C–6E and S5B). Thus, the DKK-1-mediated increase in nestin expression at d16 was not linked to enhanced proliferation of NE cells.
We then determined whether DKK-1 administration or WNT3-knockdown influences neuronal fate. As shown in Figures 2F and 2G, neuronal differentiation began around d12-14 in BG01V2 and BG03. We therefore examined the effect of DKK-1 treatment or WNT3-knockdown on neurogenesis during late NE (d12-16) in BG01V2, BG03, and CT3. Percentages of BrdU-labeled (1 μM, 24h exposure from d14-15) Tuj1+ cells were decreased by DKK-1 administration or knockdown of WNT3 (Figures 6F–6H), signifying that WNT3/β-catenin is involved in neurogenesis during the later NE period. Taken together, these data explain the seemingly paradoxical observation that blockade of the WNT3/β-catenin pathway in early NE (at d10) reduced nestin+ cell populations by decreasing NE cell proliferation, while blocking the pathway during a later NE period interrupted neurogenesis, thereby causing an accumulation of NE cells and an increase in nestin expression (Figure 6A).
LMX1A is essential for mDA differentiation of hESCs (Cai et al., 2009). In mouse ESCs (mESCs), Lmx1A expression is up-regulated by WNT/β-catenin signaling and coordinates several downstream transcription factors including Msx1, Nurr1 and Pitx3 to induce the mDA phenotype (Chung et al., 2009). To study whether LMX1A was also up-regulated by WNT3 through β-catenin in chromosome 17-abnormal lines, we examined LMX1A expression following DKK-1 treatment during late NE (d12-16) and observed a dose-dependent decrease in LMX1A expression in BGO1V2 and BGO3 (Figure S5C). This finding was confirmed using another WNT signaling inhibitor, sFRP2 (Figure S5C), which binds to WNT directly as opposed to DKK-1 which binds to the WNT receptor complex (Kawano and Kypta, 2003), and by WNT3-knockdown in CT3 during late NE (Figure 6I). In addition, overexpression of WNT3 in CT2 during late NE increased LMX1A expression (Figure 6J). These data suggest that WNT3/β-catenin up-regulates LMX1A expression during mDA differentiation in these abnormal hPSC lines. As expected, blockade of WNT3/β-catenin signaling in those lines by DKK-1 administration or WNT3-knockdown during late NE reduced the production of LMX1A+ mDPs (Figures 6K–6M), and interrupted mDA development as indicated by reductions in TUJ1+ neurons and TH+ mDA neurons (Figures 6N–6R). Therefore, up-regulation of LMX1A through the WNT3/β-catenin pathway during late NE promotes specification of the mDA fate in BG01V2, BG03, and CT3.
Other Possible Factors Contributing to Rapid mDA Differentiation in hPSC Lines with Chromosome 17 Abnormalities
To assess the possible involvement of other pathways in mDA differentiation of chromosome 17 abnormal hPSC lines, we first examined the roles of FGF8 and SHH in their rapid mDA differentiation. The mDA differentiation protocol (Yan et al., 2005) includes addition of FGF8 from d10- d16 to promote ventral midbrain patterning, yet FGF8 withdrawal or treatment with a SHH inhibitor (cyclopamine) from d0-16 did not alter expression of OCT3/4, nestin, or mDA neuronal markers (Figure S5D). Moreover, expression of FGF8 and SHH at different mDA stages did not significantly differ between control and abnormal lines (Figures 3J and S3G), suggesting that neither FGF8 nor SHH signaling was involved in the rapid mDA differentiation of hPSCs with chromosome 17 abnormalities.
To exclude the possibility that WNT3 and WNT9B involvement in mDA differentiation was selective for the particular differentiation protocol used for this study, we used a directed, non-EB-based, neural differentiation protocol based on dual SMAD inhibition (Chambers et al., 2009). This procedure induces neural progenitor cells from ES cells in 7–10d, as well as inducing an mDA fate by d19 with the addition of SHH at d5 and FGF8 at d9 through d12. To examine mDA induction in the abnormal lines, we adopted a 10d (NE) differentiation period (Figure S6A), excluding SHH and FGF8, based on the results of our original protocol which showed that mDA induction could be obtained in the abnormal lines by the NE stage.
Gene expression analysis at d5 indicated that CT3 rapidly lost OCT3/4, with a simultaneous increase in nestin expression (Figure S6B). Therefore, CT3 shows more efficient neural progenitor conversion from the ES stage. WNT9B-knockdown, but not WNT3-knockdown, blocked OCT3/4 down-regulation and nestin up-regulation, indicating that WNT9B promotes NE induction through enhanced loss of pluripotency in CT3 (Figure S6C). No change in the expression of SOX17 or T was caused by WNT9B-knockdown (Figure S6C), possibly due to dual SMAD inhibitor-mediated suppression of SOX17 and T (Chambers et al., 2009). However, CT3 failed to show efficient mDA induction on d10 (Figure S6D), and measurement of WNT activity during 10d differentiation indicated that only WNT9B/JNK signaling was activated in CT3, as compared to CT2 (Figures S6F and S6G). Interestingly, adding bFGF to CT3 cultures through d4-d10, as per our original protocol, resulted in a switch from non-canonical to canonical WNT signaling (Figures S6F and S6G), and resulted in substantial mDA induction on d10 (Figures S6D and S6E).
Knockdown of WNT3 but not WNT9B during d4-10 effectively inhibited bFGF-induced mDA induction in CT3 (Figures S6D and S6E), suggesting a role of WNT3 in mDA induction. This was confirmed by inducing mDA differentiation in CT2 by overexpression of WNT3 from d4-d10, with bFGF included (Figure S6H). Complete mDA differentiation with FOXA2 expression could be achieved at d10 in CT3 with addition of both bFGF and SHH through d4-10 (Figure S6I). These results are consistent with our previous findings suggesting that WNT9B/JNK and WNT3/β-catenin perform independent, distinct roles at each stage of differentiation to achieve potent mDA neuronal conversion in hPSC lines with amplified WNT3/WNT9B.
bFGF Determines whether WNT3/β-catenin or WNT9B/JNK Acts to Regulate Distinct Processes of mDA Differentiation of hPSCs
The expression patterns of WNT3 and WNT9B during mDA differentiation (Figures 3J and S3G) may contribute to mechanisms whereby WNT3/WNT9B amplification in hPSCs can influence both undifferentiated proliferation and differentiation. Due to the correlation between WNT3/β-catenin activation and the presence of bFGF, WNT9B/JNK activation when bFGF is absent (Figures S6F and S6G), and because it has been shown that WNT and FGF pathways cooperate in various cellular processes, including pluripotency (Ding et al., 2010), we examined the signaling crosstalk between WNTs and bFGF in mDA differentiation in hPSCs with amplified WNT3/WNT9B.
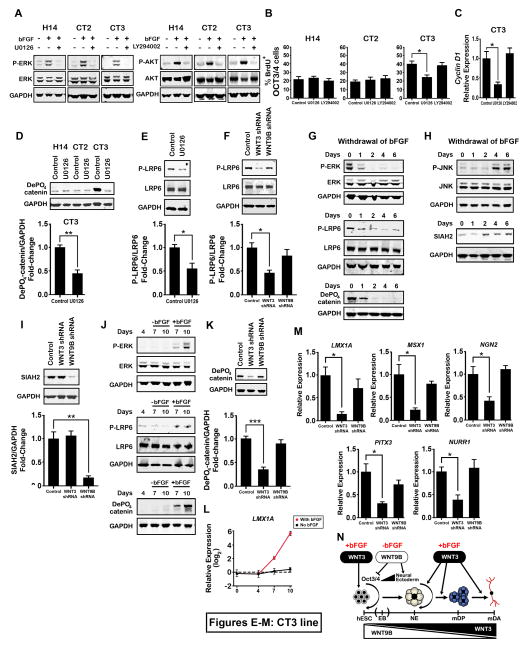
We examined whether PI3K/AKT and MAPK/ERK pathways, which are activated by bFGF in hPSCs (Ding et al., 2010), can be activated by bFGF in hPSCs with WNT3/WNT9B amplification. In CT3 starved for 24 h, by omission of KSR and bFGF, both pathways were activated by bFGF (Figure 7A), suggesting that both PI3K/AKT and MAPK/ERK are downstream targets of bFGF signaling in abnormal hPSCs. Next, we determined if these two pathways are involved in enhanced undifferentiated proliferation. Using U0126 or LY294002 to block ERK or AKT activities respectively (Figure 7A), we found that inhibition of ERK, but not AKT, significantly reduced proliferation rate and cyclin D1 expression in CT3; though neither of these pathways seemed to be involved in normal hPSC line proliferation (Figures 7B and 7C). These data indicate that the bFGF/ERK pathway, but not bFGF/AKT, is involved in the enhanced proliferation of undifferentiated hPSCs caused by amplification of WNT3/WNT9B.
Figure 7. Molecular Mechanisms Involved in WNT3/WNT9B-regulated Proliferation in the Undifferentiated State and mDA Differentiation.
(A) Immunoblot showing phosphorylated ERK (P-ERK) and phosphorylated AKT (P-AKT) levels in hESC lines, with KSR and bFGF withdrawn overnight, treated with the ERK inhibitor U0126 or the AKT inhibitor LY294002 1 hr prior to adding 20 ng/ml bFGF for 30 min. (B) Percentage of OCT3/4+ cells incorporating BrdU under feeder-free conditions for each hESC line with bFGF and in presence or absence of U0126 or LY294002 over 6 d. n = 3. (C) Relative expression of cyclin D1 in CT3 in the presence or absence of U0126 or LY294002 cultured in feeder-free conditions with bFGF for 6 d. All values are expressed as fold-changes with respect to the control. n = 3. (D) Immunoblot showing dephosphorylated (active) β-catenin levels in hESC lines in the presence or absence of U0126 over 6 d in feeder-free culture with bFGF. n = 3. (E, F) Immunoblot showing phosphorylated LRP6 (P-LRP6) levels in CT3 in the presence or absence of U0126 (E), or for control, WNT3, or WNT9B shRNA lentivirus-mediated infection (F), for 6 d of feeder-free culture with bFGF. n = 3. (G, H) Immunoblot of P-ERK, P-LRP6, and dephosphorylated (active) β-catenin (G) and P-JNK, SIAH2 (H) levels for CT3 under feeder-free conditions during 6 d of bFGF withdrawal. (I) Immunoblot showing SIAH2 levels for lentivirus-mediated WNT3 or WNT9B knockdown in CT3 under feeder-free conditions after 6 d of bFGF withdrawal. n = 3. (J) Immunoblot showing P-ERK, P-LRP6, and dephosphorylated (active) β-catenin levels in CT3 during dual-SMAD inhibition in the presence or absence of bFGF. (K) Immunoblot showing dephosphorylated (active) β-catenin levels for lentivirus-mediated WNT3 or WNT9B knockdown (d4-d10) in CT3 for dual-SMAD inhibition in the presence of bFGF. n = 3. (L) Relative expression of LMX1A in CT3 during dual-SMAD inhibition in the presence or absence of bFGF. Values are expressed as fold-changes, in log2 form, with respect to CT3 at d0. n=3. (M) Relative expression of LMX1A, MSX1, NGN2, PITX3, and NURR1 for lentivirus-mediated WNT3 or WNT9B knockdown (d4-d10) in CT3, using dual-SMAD inhibition, in the presence of bFGF. n=3. (N) Schematic illustrating the roles of WNT3 and WNT9B in mDA differentiation in human pluripotent stem cells with WNT3/WNT9B amplification. Error bars, SEM. *P < 0.05; **P < 0.01; ***P < 0.01.
We next asked how bFGF/ERK cooperates with WNTs to enhance proliferation in the abnormal lines. Activity of β-catenin was significantly decreased after U0126 treatment in CT3 but not in normal hPSCs, suggesting that MAPK/ERK is involved in enhanced β-catenin activity in hPSC lines with amplified WNTs (Figure 7D). It has been suggested that FGF signaling through ERK phosphorylates LRP6, a WNT co-receptor for the canonical signaling pathway, at PPPS/TP motifs (Krejci et al., 2012), creating additional high affinity binding sites for ASIN1/GSK3, causing ASIN1/GSK3 to be sequestered from the β-catenin destruction complex, thereby increasing β-catenin stability and amplifying canonical WNT signaling. Therefore we examined whether MAPK/ERK signaling is implicated in the phosphorylation of LRP6 at Ser1490, in a PPPS/TP motif. U0126 significantly decreased phosphorylation of LRP6 in CT3 cells (Figure 7E). In addition, WNT3-knockdown, but not WNT9B-knockdown, decreased LRP6 phosphorylation (Figure 7F). These data suggest that bFGF signaling through MAPK/ERK amplifies the canonical WNT signaling pathway by phosphorylating LRP6, resulting in enhanced undifferentiated proliferation of hPSCs with amplified WNTs.
Conversely, when bFGF was withdrawn, phosphorylation of ERK and LPR6, and activity of β-catenin gradually decreased, followed by an increase in activity of JNK in CT3 (Figures 7G and 7H). It has been suggested that non-canonical WNT antagonizes the canonical WNT pathway through up-regulation of SIAH2, which promotes β-catenin degradation (Topol et al., 2003). We found that SIAH2 was up-regulated following bFGF withdrawal, and that this up-regulation was reversed by knockdown of WNT9B but not WNT3 in CT3 (Figures 7H and 7I). These data suggest that in hPSCs with WNT3/WNT9B amplification, without bFGF/ERK/LPR6 to stabilize and amplify the WNT/β-catenin pathway, non-canonical WNT becomes dominant due to up-regulation of SIAH2 and contributes to enhanced loss of pluripotency through JNK signaling.
Finally, we studied the crosstalk between WNTs and bFGF in mDA differentiation using the dual SMAD inhibition protocol to avoid the complexities of EB formation. As discussed above, treatment with bFGF through d4-d10 switched non-canonical to canonical WNT signaling, and induced mDA differentiation (Figures S6D, S6F, and S6G). We also found that phosphorylation of ERK and LRP6, and activity of β-catenin, were gradually increased during d4-d10 in the presence of bFGF in CT3 (Figure 7J), and knockdown of WNT3 but not WNT9B inhibited β-catenin activation (Figure 7K). These data suggest that bFGF signaling through ERK/LPR6 enables canonical WNT signaling to become dominant by stabilizing and amplifying canonical WNT signaling. In addition, temporal gene expression analysis revealed that LMX1A, a downstream target of canonical WNT signaling, was increased by bFGF treatment from d4-d10 in CT3 (Figure 7L), and knockdown of WNT3 but not WNT9B induced a substantial loss of LMX1A and expression of its downstream mDA-specific transcription factors (Figure 7M). These data indicate that bFGF stabilizes canonical WNT signaling, contributing to efficient mDA induction through up-regulation of LMX1A in chromosome 17-abnormal lines, and reveals the role of bFGF in the interplay of WNT3 and WNT9B signaling at each stage of differentiation (Figure 7N).
DISCUSSION
Our study has identified amplification of the WNT3/WNT9B cluster located at the border between 17q21.31 and 17q21.32 as a major contributor to several important phenotypic properties of hPSCs: enhanced proliferation in the undifferentiated stage, accelerated loss of pluripotency after withdrawal of bFGF, and rapid mDA differentiation. Trisomy 17 has been repeatedly observed in hESC cultures (Baker et al, 2007), possibly due to selective pressure exerted by culture conditions. Even in the absence of trisomy 17, however, gains of small regions on chromosome 17, including 17q21.31, seem to be fairly common in hESCs (Närvä et al., 2010).
While CNVs at 17q21.31 occur in phenotypically normal humans (Table S3), genetic anomalies at 17q21.31 have been linked to Parkinson’s disease, disorders of neural development (Spencer et al., 2011, Grisart et al., 2009), and autism (Girirajan et al., 2011). Understanding the functions of the WNT3-WNT9B cluster at 17q21.31 may shed light on the molecular mechanisms of these disorders.
We performed preliminary experiments to examine the neuronal differentiation ability for different hPSC lines using the protocol in Figure 2A but withdrawing all of the trophic factors, such as bFGF and FGF8. At d16, BG01, BG02, ES02, and ES04 exhibited a dorsal forebrain neural progenitor cell fate, expressing the anterior telencephalic transcription factors BF1 and OTX2 and the dorsal forebrain marker PAX6. For the abnormal lines including BG01V2 and BG03, however, most colonies exhibited abundant projections and a neuronal phenotype. Small percentages of the neural progenitors in BG01V2 and BG03 expressed PAX6, but most expressed LMX1A. In addition, these progenitors did not express the hindbrain/spinal cord marker HOXB4 or the ventral telencephalic marker NKX2.1. We further examined the specific types of neurons generated from BG01V2 and BG03 differentiation and found a significant induction of TH+ neurons, with only a small percentage of glutamate neurons and no GABA or motor neuron marker-HB9-expressing neurons. These data indicate that normal hPSC lines have a natural tendency to differentiate to a dorsal telencephalon phenotype, but the abnormal lines examined here have a tendency to become mDA neurons. Due to these preliminary data we focused on mDA differentiation for hPSCs with amplification of 17q21.31/WNT3-WNT9B. Figure S7B provides additional data, with statistics, regarding the mDA differentiation of eleven hPSC lines; seven normal hPSC lines: BG01, BG02, ES02, ES04, H7, H14, and CT2; and four abnormal hPSC lines: BG03, BG01V2-E, CT3, and H1 (with similar genotype as shown in Figure 3I) using the protocol shown in Figure 2A in the absence of trophic factors. These data show that at d16 the abnormal lines produced significantly more DA neurons than the normal lines.
It is remarkable that BG01V2, BG03, and CT3 display more rapid departure from pluripotency and differentiation despite exhibiting enhanced proliferation in the undifferentiated state. The increase in DNA encoding WNT3 and WNT9B was modest, yet consistent with the presence of additional copies of these genes. Increases in mRNA were much greater, supporting altered regulation as well as altered copy number (Figures 3D, 3E, 3J, and Table S2). The greater enhancement of WNT3 and WNT9B expression than DNA might be caused by genetic alterations resulting in greater enhancer or promoter efficiency, epigenetic modifications such as differential methylation of the additional DNA copies, or other unknown regulatory mechanisms. Our findings emphasize the importance of elucidating cellular functional changes in hPSCs caused by even modest-sized genomic changes.
Epigenetic factors are important regulators in neural differentiation from hPSCs. Kim and coworkers (2011) showed a neural differentiation bias based on expression of miR-371-3. By using Agilent-016436 Human miRNA Microarray 1.0 G4472A (miRNA ID version), Figure S7C show the differential expression of miR-371 among eight hPSC lines including six normal hPSC lines: BG01, BG02, ES02, ES04, H7, and H14, and two abnormal lines: BG03 and H1. Although there are differences in miR-371 between the BG lines and ES lines as compared to the H lines, these are not correlated with the overall functional variance among our hPSC lines. We also did not find other significant correlations for epigenetic factors, miRNA or methylation, and the functional variance among these hPSC lines based on the NIH Human Pluripotent Stem Cell Database, compiled by the NIH stem cell unit (Mallon et al., 2013, Mallon et al., 2014) with whom we collaborated. While it is certainly true that multiple factors, such as CNVs, SNPs, miRNA, and methylation can influence the differentiation of hPSCs, here we have focused on just one such factor, 17q21.31/WNT3-WNT9B amplification; however, we do not mean to imply that this is the only reason that hPSC lines differ from one another.
A number of previous studies have identified roles of WNT1, WNT3A, and WNT5A in mDA differentiation (Castelo-Branco et al., 2003, Chung et al., 2009, Kriks et al., 2011). We found that DKK-1 treatment or WNT3-knockdown during NE down-regulates expression of LMX1A, MSX1, NGN2, NURR1, and PITX3 (Figures 6A and S5E). In hPSCs with amplified WNTs, enhanced WNT3/β-catenin signaling accelerates mDA specification via up-regulation of LMX1A, but without FOXA2 expression. FOXA2 induction could, however, be achieved by addition of SHH, consistent with reports that both WNT-LMX1A and SHH-FOXA2 signaling pathways contribute to mDA neuronal differentiation (Chung et al., 2009, Kriks et al., 2011).
Furthermore, it has been suggested that WNT5A acts on non-canonical pathways to increase rodent mDA neuronal differentiation (Castelo-Branco et al., 2003). Since WNT9B/JNK blockade during NE did not affect mDA differentiation in our model, WNT5A and WNT9B may act via different non-canonical pathways. In this study we identify a novel role for non-canonical WNT signaling during early mDA differentiation, suggesting that various WNT-regulated pathways can work cooperatively in promoting mDA differentiation.
Due to activity of the β-catenin destruction complex, cytoplasmic β-catenin levels are normally low. WNT-mediated phosphorylation of mature (transmembrane) LRP6 occurs when the signalosome complex is formed by assembly of WNT and LRP6. This is in contrast to ERK, which is able to phosphorylate both mature and immature (intracellular) LRP6. ERK-phosphorylated LRP6 is able to divert a minor amount of ASIN1/GSK3 from the β-catenin destruction complex; however, this is not sufficient to stabilize β-catenin on its own. When WNT is present, ERK-phosphorylated LRP6 is able to dramatically contribute to stabilizing β-catenin and amplifying canonical WNT signaling (Krejci et al., 2012). Our findings also indicate that, without bFGF, abnormal hPSCs with amplified WNTs exhibited only limited activity of β-catenin, no different from normal hPSCs. However, activity of β-catenin is enhanced by bFGF, through phosphorylation of LRP6 at PPPS/TP motifs. Once bFGF is withdrawn, without the subsequent ERK/LPR6 pathway to stabilize β-catenin, non-canonical WNT becomes the dominant mode of WNT signaling.
Our results demonstrate that bFGF regulates whether WNT3/β-catenin or WNT9B/JNK is dominant in regulating distinct processes of mDA differentiation. WNT9B, acting through a non-canonical pathway, guides hPSCs away from pluripotency and towards neuroectodermal differentiation. WNT3 accelerates undifferentiated hPSC proliferation, furthers expansion of neuroectodermal cells, and directs them to an mDA fate (Figure 7N). Although WNT3 and WNT9B are not known to be involved in normal human midbrain development, their role in orchestrating mDA differentiation of hPSCs under conditions of 17q21.31-17q21.32 amplification suggests that CNVs in this region have the capacity to cause profound alterations in neural differentiation and development.
EXPERIMENTAL PROCEDURES
hESC culture
The hESC lines BG01 (P163-187), BG02 (P39-69), and BG03 (P47-64), provided by BresaGen, Inc., BG01V2-E (P32-54) and BG01V2-L (P78-96) derived from BG01 (Vazin et al., 2008), ES02 (P45-58) and ES04 (P65-74), provided by ES Cell International (Singapore), H1 (P40-44), H7 (P43-45), H14 (P40-58), provided by WiCell Research Institute, and CT2 (P90-109) and CT3 (P27-55), provided by University of Connecticut Stem Cell Core were cultured as previously described (Vazin et al., 2008). For more details, see Supplemental Information.
SNP Array 6.0 and Analytical Methods
DNA was extracted from undifferentiated hESC lines, quantitated and genotyped using Affymetrix 6.0 microarrays to the manufacturer’s instructions. Hybridization intensities for all chromosome 17 SNP and CNV probes were subjected to principal components analyses, described in Supplemental Experimental Procedures. Quantification of cell numbers, proteins, RNA and DNA was performed using immunocytochemistry, Western blotting, RT-PCR, RT-qPCR, and qPCR using standard methods as described in supplementary information and primers as listed in Tables S4 and S5.
Statistical Analysis
Data are shown as means ± SEM. Means were compared by two-tailed t-tests, or by one-way ANOVA followed by Tukey’s compromise post-hoc test for multiple comparisons, using GraphPad InStat Version 3 software. The criterion for statistical significance was P < 0.05.
Supplementary Material
Acknowledgments
Research supported by the IRP of NIDA, NIH. We thank Teruo Hayashi, Donna Walther and Aaron Russell for assistance.
Footnotes
ACCESSION NUMBERS
The GEO accession number for the Affymetrix 6.0 microarray data is GSE64241.
AUTHOR CONTRIBUTION
C-TL, GRU, and WJF conceived and designed the study. C-TL, RMB, AAK, LTW, MPW, BSM, BKH, CTR, RSH, JC, SLE, and S-YT performed the experiments involving stem cell culture, mDA differentiation, PCR, Western blotting, and Lentiviral Transduction. TD performed SNP array and analysis. C-TL, RMB, GRU, and WJF analyzed and interpreted data. C-TL, RMB, AAK, GRU, and WJF drafted the manuscript. C-TL, RMB, AAK, LTW, MPW, TD, BSM, BKH, CTR, RSH, JC, SLE, S-YT, GRU, and WJF performed critical revision of the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baker DE, Harrison NJ, Maltby E, Smith K, Moore HD, Shaw PJ, Heath PR, Holden H, Andrews PW. Adaptation to culture of human embryonic stem cells and oncogenesis in vivo. Nat Biotechnol. 2007;25:207–215. doi: 10.1038/nbt1285. [DOI] [PubMed] [Google Scholar]
- Ben-David U, Benvenisty N. The tumorigenicity of human embryonic and induced pluripotent stem cells. Nat Rev Cancer. 2011;11:268–277. doi: 10.1038/nrc3034. [DOI] [PubMed] [Google Scholar]
- Ben-Yehudah A, Navara CS, Redinger CJ, Mich-Basso JD, Castro CA, Oliver S, Chensny LJ, Richards TJ, Kaminski N, Schatten G. Pluripotency genes overexpressed in primate embryonic stem cells are localized on homologues of human chromosomes 16, 17, 19, and X. Stem Cell Res. 2010;4:25–37. doi: 10.1016/j.scr.2009.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burbach JP, Smidt MP. Molecular programming of stem cells into mesodiencephalic dopaminergic neurons. Trends Neurosci. 2006;29:601–603. doi: 10.1016/j.tins.2006.09.003. [DOI] [PubMed] [Google Scholar]
- Cai J, Donaldson A, Yang M, German MS, Enikolopov G, Iacovitti L. The role of Lmx1a in the differentiation of human embryonic stem cells into midbrain dopamine neurons in culture and after transplantation into a Parkinson’s disease model. Stem Cells. 2009;1:220–229. doi: 10.1634/stemcells.2008-0734. [DOI] [PubMed] [Google Scholar]
- Castelo-Branco G, Wagner J, Rodriguez FJ, Kele J, Sousa K, Rawal N, Pasolli HA, Fuchs E, Kitajewski J, Arenas E. Differential regulation of midbrain dopaminergic neuron development by Wnt-1, Wnt-3a, and Wnt-5a. Proc Natl Acad Sci USA. 2003;100:12747–12752. doi: 10.1073/pnas.1534900100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers SM, Fasano CA, Papapetrou EP, Tomishima M, Sadelain M, Studer L. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat Biotechnol. 2009;27:275–80. doi: 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury R, Bois PR, Feingold E, Sherman SL, Cheung VG. Genetic analysis of variation in human meiotic recombination. PLoS Genet. 2009;5:e1000648. doi: 10.1371/journal.pgen.1000648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung S, Leung A, Han BS, Chang MY, Moon JI, Kim CH, Hong S, Pruszak J, Isacson O, Kim KS. Wnt1-lmx1a forms a novel autoregulatory loop and controls midbrain dopaminergic differentiation synergistically with the SHH-FoxA2 pathway. Cell Stem Cell. 2009;5:646–658. doi: 10.1016/j.stem.2009.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding VM, Ling L, Natarajan S, Yap MG, Cool SM, Choo AB. FGF-2 modulates Wnt signaling in undifferentiated hESC and iPS cells through activated PI3-K/GSK3beta signaling. J Cell Physiol. 2010;225:417–428. doi: 10.1002/jcp.22214. [DOI] [PubMed] [Google Scholar]
- Ferri AL, Lin W, Mavromatakis YE, Wang JC, Sasaki H, Whitsett JA, Ang SL. Foxa1 and Foxa2 regulate multiple phases of midbrain dopaminergic neuron development in a dosage-dependent manner. Development. 2007;15:2761–2769. doi: 10.1242/dev.000141. [DOI] [PubMed] [Google Scholar]
- Girirajan S, Brkanac Z, Coe BP, Baker C, Vives L, Vu TH, Shafer N, Bernier R, Ferrero GB, et al. Relative burden of large CNVs on a range of neurodevelopmental phenotypes. PLoS Genet. 2011;7:e1002334. doi: 10.1371/journal.pgen.1002334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grisart B, Willatt L, Destrée A, Fryns JP, Rack K, de Ravel T, Rosenfeld J, Vermeesch JR, Verellen-Dumoulin C, Sandford R. 17q21.31 microduplication patients are characterised by behavioural problems and poor social interaction. J Med Genet. 2009;46:524–530. doi: 10.1136/jmg.2008.065367. [DOI] [PubMed] [Google Scholar]
- Guenther MG, Frampton GM, Soldner F, Hockemeyer D, Mitalipova M, Jaenisch R, Young RA. Chromatin structure and gene expression programs of human embryonic and induced pluripotent stem cells. Cell Stem Cell. 2010;7:249–257. doi: 10.1016/j.stem.2010.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karner CM, Chirumamilla R, Aoki S, Igarashi P, Wallingford JB, Carroll TJ. Wnt9b signaling regulates planar cell polarity and kidney tubule morphogenesis. Nat Genet. 2009;7:793–799. doi: 10.1038/ng.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawano Y, Kypta R. Secreted antagonists of the Wnt signalling pathway. J Cell Sci. 2003;116:2627–2634. doi: 10.1242/jcs.00623. [DOI] [PubMed] [Google Scholar]
- Kim H, Lee G, Ganat Y, Papapetrou EP, Lipchina I, Socci ND, Sadelain M, Studer L. miR-371-3 expression predicts neural differentiation propensity in human pluripotent stem cells. Cell Stem Cell. 2011;8:695–706. doi: 10.1016/j.stem.2011.04.002. [DOI] [PubMed] [Google Scholar]
- Krejci P, Aklian A, Kaucka M, Sevcikova E, Prochazkova J, Masek JK, Mikolka P, Pospisilova T, Spoustova T, Weis M, et al. Receptor tyrosine kinases activate canonical WNT/β-catenin signaling via MAP kinase/LRP6 pathway and direct β-catenin phosphorylation. PLoS One. 2012;7:e35826. doi: 10.1371/journal.pone.0035826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriks S, Shim JW, Piao J, Ganat YM, Wakeman DR, Xie Z, Carrillo-Reid L, Auyeung G, Antonacci C, Buch A, et al. Dopamine neurons derived from human ES cells efficiently engraft in animal models of Parkinson’s disease. Nature. 2011;480:547–551. doi: 10.1038/nature10648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lie DC, Colamarino SA, Song HJ, Désiré L, Mira H, Consiglio A, Lein ES, Jessberger S, Lansford H, Dearie AR, et al. Wnt signaling regulates adult hippocampal neurogenesis. Nature. 2005;437:1370–1375. doi: 10.1038/nature04108. [DOI] [PubMed] [Google Scholar]
- Mallon BS, Chenoweth JG, Johnson KR, Hamilton RS, Tesar PJ, Yavatkar AS, Tyson LJ, Park K, Chen KG, Fann YC, McKay RD. StemCellDB: the human pluripotent stem cell database at the National Institutes of Health. Stem Cell Res. 2013;10:57–66. doi: 10.1016/j.scr.2012.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallon BS, Hamilton RS, Kozhich OA, Johnson KR, Fann YC, Rao MS, Robey PG. Comparison of the molecular profiles of human embryonic and induced pluripotent stem cells of isogenic origin. Stem Cell Res. 2014;12:376–86. doi: 10.1016/j.scr.2013.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarroll SA, Kuruvilla FG, Korn JM, Cawley S, Nemesh J, Wysoker A, Shapero MH, de Bakker PI, Maller JB, Kirby A, et al. Integrated detection and population-genetic analysis of SNPs and copy number variation. Nat Genet. 2008;40:1166–1174. doi: 10.1038/ng.238. [DOI] [PubMed] [Google Scholar]
- Närvä E, Autio R, Rahkonen N, Kong L, Harrison N, Kitsberg D, Borghese L, Itskovitz-Eldor J, Rasool O, Dvorak P, et al. High-resolution DNA analysis of human embryonic stem cell lines reveals culture-induced copy number changes and loss of heterozygosity. Nat Biotechnol. 2010;28:371–377. doi: 10.1038/nbt.1615. [DOI] [PubMed] [Google Scholar]
- Rocchi A, Pelliccia F. Synergistic effect of DAPI and thymidylate stress conditions on the induction of common fragile sites. Cytogenet Cell Genet. 1988;48:51–54. doi: 10.1159/000132585. [DOI] [PubMed] [Google Scholar]
- Sato A, Yamamoto H, Sakane H, Koyama H, Kikuchi A. Wnt5a regulates distinct signalling pathways by binding to Frizzled2. EMBO J. 2010;29:41–54. doi: 10.1038/emboj.2009.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer CC, Plagnol V, Strange A, Gardner M, Paisan-Ruiz C, Band G, Barker RA, Bellenguez C, Bhatia K, Blackburn H, et al. Dissection of the genetics of Parkinson’s disease identifies an additional association 59 of SNCA and multiple associated haplotypes at 17q21. Hum Mol Genet. 2011;20:345–353. doi: 10.1093/hmg/ddq469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomoda K, Takahashi K, Leung K, Okada A, Narita M, Yamada NA, Eilertson KE, Tsang P, Baba S, White MP, et al. Derivation conditions impact X-inactivation status in female human induced pluripotent stem cells. Cell Stem Cell. 2012;11:91–99. doi: 10.1016/j.stem.2012.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topol L, Jiang X, Choi H, Garrett-Beal L, Carolan PJ, Yang Y. Wnt-5a inhibits the canonical Wnt pathway by promoting GSK-3-independent beta-catenin degradation. J Cell Biol. 2003;162:899–908. doi: 10.1083/jcb.200303158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazin T, Chen J, Spivak CE, Amable R, Gabitzsch E, Lee CT, Lupica CR, Freed WJ. Dopaminergic neurons derived from BG01V2, a variant of human embryonic stem cell line BG01. Restor Neurol Neurosci. 2008;26:447–458. [PMC free article] [PubMed] [Google Scholar]
- Werbowetski-Ogilvie TE, Bossé M, Stewart M, Schnerch A, Ramos-Mejia V, Rouleau A, Wynder T, Smith MJ, Dingwall S, Carter T, et al. Characterization of human embryonic stem cells with features of neoplastic progression. Nat Biotechnol. 2009;27:91–97. doi: 10.1038/nbt.1516. [DOI] [PubMed] [Google Scholar]
- Yan Y, Yang D, Zarnowska ED, Du Z, Werbel B, Valliere C, Pearce RA, Thomson JA, Zhang SC. Directed differentiation of dopaminergic neuronal subtypes from human embryonic stem cells. Stem Cells. 2005;23:781–790. doi: 10.1634/stemcells.2004-0365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Gu W, Hurles ME, Lupski JR. Copy number variation in human health, disease, and evolution. Annu Rev Genomics Hum Genet. 2009;10:451–481. doi: 10.1146/annurev.genom.9.081307.164217. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.