SUMMARY
CD4 and CD8 T-cell lineages differentiate through respective thymic selection processes. Here we report cross-differentiation from the CD8 lineage to CD4 T cells, but not vice versa, predominantly in the large-intestine-associated microenvironment. It occurred in the absence or distal presence of cognate antigens. This pathway produced MHC-class-I-restricted CD4+Foxp3+ Treg (CI-Treg) cells. Blocking T-cell-intrinsic TGFβ signaling diminished CI-Treg populations in lamina propria but did not preclude the CD8-to-CD4 conversion. Microbiota were not required for the cross-differentiation but presence of microbiota led to expansion of the converted CD4 T-cell population in the large intestine. CI-Treg cells did not promote tolerance to microbiota per se, but regulated systemic homeostasis of T lymphocytes and protected the large intestine from inflammatory damage. Overall, the clonal conversion from the CD8 lineage to CD4 T-cell subsets occurred regardless of “self” or “nonself”. This lineage plasticity may promote “selfless” tolerance for immune balance.
INTRODUCTION
The development of the immune system has been largely characterized on the basis of discriminating “self”, the host’s own cells, from “nonself”, exemplified by infectious microbes, towards an outcome of either tolerance or immunity (Burnet, 1957). The gut-associated environment (GAE), particularly the large intestine, presents a unique challenge to the immune system with a diversity of food antigens and a superior number of normal floral microorganisms (microbiota) that carry a microbial pattern otherwise typified for initiating immunity (Janeway and Medzhitov, 2002). Directly or indirectly, microbiota affects development of gastrointestinal tract and the host immune system, and performs a number of functions that are beneficial to the host (Hooper and Macpherson, 2010). Thus, a harmonious relationship between the immune system, microbiota and food antigens in the large-intestine-associated microenvironment is vital for the health of a mammalian host.
In vertebrates, the innate immune system discriminates microbial agents by patterns that are distinct from eukaryotic cells, whereas the adaptive immune system is armed with a repertoire of T and B lymphocyte clones with fine specificity to foreign antigens but is tolerant toward the host’s “self” tissue. The “self”-based concept has served as a foundation for modern immunology but its limitations have long been recognized (Matzinger, 1994). How the immune system deals with mutualistic and massive microbiota in the large intestine remains a problem of extensive interest. Extrathymic CD4+Foxp3+ regulatory T (Treg) cells that developed in the periphery through TGFβ signaling were shown to have a critical role in maintaining tolerance at the mucosal surface including in the large intestine (Josefowicz et al., 2012). Indeed, Treg cell clones specific to microbial agents in the large intestine were identified, and the unique repertoire of colonic Treg cells suggested that the differentiation of peripheral Treg cells could occur locally at the intestinal mucosal surface (Lathrop et al., 2011). However, sequencing analyses of the T-cell antigen receptor (TCR) of colonic Treg cells using the TCRmini model, which was constructed to host a diverse but limited repertoire to enable the sequencing studies, suggested that thymus-derived Treg cells may be mainly responsible for tolerance induction to the large intestine microbiota (Cebula et al., 2013).
Nevertheless, one might argue that specific-antigen-based tolerance to microbial organisms must be limited in scope, because a constitutive tolerance toward a broad spectrum of nonpathogenic bacteria can potentially cripple immunity against pathogenic bacteria, which differ minimally from the former in terms of patterns for immune initiation. Indeed, continued presence of microbiota may promote protective immunity overall, as demonstrated in a recent study showing that antibiotic depletion of microbiota impaired antiviral innate and adaptive immunity (Abt et al., 2012).
Therefore, although the CD4 and CD8 lineage specification of αβ T cells occurs in the thymus as a result of a multi-stage, stringent selection process involving recognition of class-I or -II MHC molecules (Doyle and Strominger, 1987; Hedrick, 2012; Norment et al., 1988; Rudd et al., 1988; Veillette et al., 1988), it is possible that in the large-intestine-associated microenvironment, evolution might have shaped unique mechanisms of T-cell plasticity that might not be constrained by “self” versus “nonself” characterization of specific-antigen recognition. We hypothesize that T-cell clones in the large-intestine-associated microenvironment can differentiate at steady-state with lineage plasticity to facilitate immune balance, without regard to “self” or “nonself” denotation of their TCR specificity.
To test this hypothesis, we examined the steady-state T-cell differentiation in the large-intestine-associated microenvironment, tracking the fate of two clones in the CD8 T-cell lineage and two clones in the CD4 T-cell lineage, specific to neither microbiota nor food antigens. Their known specific antigens were either absent in the whole animal or present as a “self” antigen in an organ (the pancreas) distal to GAE. We used the OTI TCR-transgenic model (Hogquist et al., 1994) with Rag1 knockout (Mombaerts et al., 1992) (OT1+Rago), which renders a T-cell repertoire consisting of a single clone of the MHC-class-I restricted CD8 lineage specific to chicken ovalbumin (absent in the mice or their diet). We also used the 8.3 TCR-transgenic model (Verdaguer et al., 1997) on Rag1-deficient (Mombaerts et al., 1992) background (8.3+Rago), which hosts a monoclonal repertoire of the CD8 lineage specific to a “self” antigen that is not in GAE but in the endocrine pancreas. CD4 lineage plasticity in the large-intestine-associated microenvironment was examined with monoclonal TCR-transgenic models in settings analogous to that established for tracking the clonal fate of the CD8 lineage. Although the monoclonal TCR-transgenic model was necessary to track T-cell lineage fate at clonal level in vivo in terms of antigen-specificity, or lack thereof, the exaggerated simplicity of repertoires in this reductionist approach raised question about its physiological relevance. Therefore, the plasticity of the CD8 T-cell lineage was corroborated using T cells with a natural repertoire, in adoptive transfer settings as well as under a condition of immune imbalance simulated by CD4 depletion.
RESULTS
Cross differentiation without regard to selfness
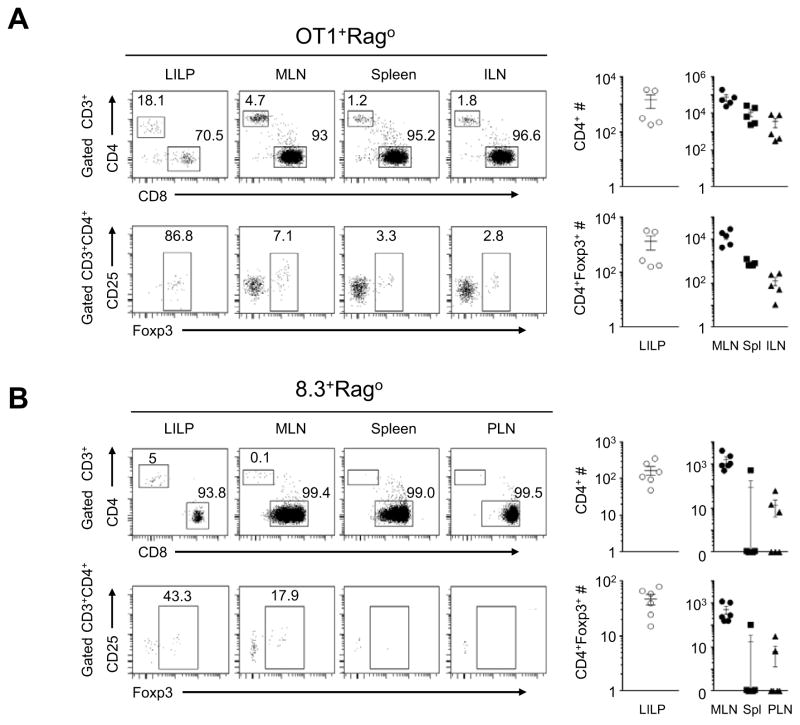
OT1+Rago mice on the C57BL/6 (B6) genetic background, a widely used model in immunological studies, are thought to harbor a monoclone of CD8 T cells with TCR restricted to MHC-class-I recognition. We analyzed adult OT1+Rago B6 mice for T-cell differentiation in GAE. Surprisingly, we found a large population of CD4 T cells (~10–20%) in the large intestine lamina propria (LILP), with a vast majority expressing Foxp3, the transcription factor specifying the CD4+ Treg cell lineage (Fontenot et al., 2003; Hori et al., 2003; Khattri et al., 2003). CD4+Foxp3− and CD4+Foxp3+ T cells also existed as substantial populations in the mesenteric lymph node (MLN) of OT1+Rago mice, but were infrequent in the other organs (Figure 1A). We have analyzed five OT1+Rago mice from 3- to 7 wks of age and did not detect CD4 T-cell populations in intra-epithelial lymphocyte (IEL) preparations of the large or small intestines (not shown). The small intestine lamina propria (SILP) (not shown) in three of the five OT1+Rago mice had CD4 T cells but the percentage of the CD4 T-cell population was 2–10-fold less than that in LILP.
Figure 1. Cross-differentiation of two independent clones in the MHC-class-I-restricted CD8 T-cell lineage to CD4+Foxp3− and CD4+Foxp3+ T cells.
(A) Representative flow cytometry of the large intestinal lamina propria (LILP), mesenteric lymph nodes (MLN), spleen and inguinal lymph nodes (ILN) in 5-week old OT1+Rago mice, followed by total cell counts for CD4+ and CD4+ Foxp3+ T cells (n=5, mean±SEM). (B) Representative flow cytometry of LILP, MLN, spleen and pancreatic lymph nodes (PLN) of 7-wk-old NOD/B6.8.3+Rago mice, followed by CD4+ and CD4+Foxp3+ cell numbers in different organs (n=6, mean±SEM). The numbers in the flow cytometry plots represent percentages of gated populations. Each data point represents one animal.
We next examined the clonal fate of 8.3+Rago T cells in the large-intestine-associated microenvironment. We detected low frequencies of CD4+, but not CD4+Foxp3+, T cells in 8.3+Rago mice on the NOD genetic background (Figure S1A–B). However, in 8.3+Rago mice on the NOD/B6 mixed genetic background (NOD/B6.8.3+Rago), we found substantial populations of CD4+Foxp3− and CD4+Foxp3+ T cells. Interestingly, CD4+Foxp3− and CD4+Foxp3+ T cells were detected in the LILP and MLN but not in the pancreatic lymph node (PLN), despite the presence of natural self-antigens specific for the 8.3 clone in the pancreas (Verdaguer et al., 1997) (Figure 1B; Figure S1A–B). Homozygosity of the NOD MHC locus on the NOD/B6 mixed genetic background did not preclude the cross-differentiation, and thus was unlikely responsible for the lack of the cross-differentiated CI-Treg cells in the NOD genetic background (Figure S1C), indicating a non-MHC genetic element(s) that influences CD8-to-CD4 cross-differentiation. These observations, together with the findings in the OT1+Rago model, suggest that cross-differentiation from the CD8 T-cell lineage to CD4+Foxp3− and CD4+Foxp3+ T cells occurs in the large-intestine-associated microenvironment regardless of the lack of, or distance to, cognate antigens.
Conversion from the CD8 lineage to CD4 T cells with MHC-class-I-restricted TCR clonotype
The OT1 clonotypic TCR is restricted to MHC-class-I recognition. Using Rag-deficient OT1 transgenic mice, previous studies established that MHC-class-I-based positive selection is essential for maturation of OT1 T-cell clone to the CD8 lineage in the thymus (Kirchner and Bevan, 1999). To examine the TCR clonotype of the CD4+ and CD4+Foxp3+ cells in the OT1+Rago mice, we did flow cytometry analyses of the TCR α and β chain. Indeed, CD4+ and CD4+Foxp3+ cells in the OT1+Rago mice expressed the OT1 clonotypic TCR (Figure S2A). In addition, the CD4 T cells produced in OT1+Rago mice responded to stimulation with the MHC-class-I-restricted peptide (SIINFEKL) of the chicken ovalbumin and proliferated like the CD8 OT1 cells (Figure S2B). In vivo, purified CD4 OT1 cells adoptively transferred into B6 hosts responded to immunization with SIINFEKL peptide (Figure S2C). Therefore, the CD4+Foxp3− and CD4+Foxp3+ subsets cross-differentiated from the CD8 lineage differed from the MHC-class-II-restricted CD4 T-cell subsets associated with the gut environment in previous studies (Esplugues et al., 2011; Ivanov et al., 2009; Wu et al., 2010), in their MHC-class-I restricted responses. The development of MHC-class-I-restricted CD4+Foxp3+ Treg (CI-Treg) cells represents a novel pathway of CD4+ Treg cell generation in the periphery.
With regard to their differentiation of effector function, the cross-differentiated CD4 T cells is also distinct from typical gut-associated, MHC-Class-II-restricted CD4 T cells (Esplugues et al., 2011; Ivanov et al., 2009; Wu et al., 2010): they were predominantly Th1-like, rather than Th17-like (Figure S2D–E). We also analyzed suppressive cytokine expression in purified OT1 CD4+Foxp3+ cells and OT1 CD4+Foxp3− cells, in comparison to that in CD4+Foxp3+ cells from B6 mice. By quantitative RT-PCR, we did not detect IL10 mRNA in OT1 CD4+Foxp3+ cells, whereas IL10 mRNA levels in OT1 CD4+Foxp3− cells were comparable to that in B6 Treg cells. TGFβ mRNA levels were similar in OT1 CD4+Foxp3+, OT1 CD4+Foxp3− and B6 Treg cells (1.44±0.02, 1.38±0.02 and 1.78±0.5 relative units, respectively).
Lack of lineage plasticity in the MHC-class-II-restricted CD4 T cell clones in the large intestine microenvironment at steady state
To examine whether T-cell clones in the MHC-class-II-restricted CD4 T-cell lineage also have steady-state plasticity in the large-intestine-associated microenvironment, we used models analogous to OT1+Rago and 8.3+Rago mice, respectively: OTII TCR-transgenic model (Barnden et al., 1998) deficient in Rag1 (OTII+Rago), with a monoclonal repertoire of MHC-class-II restricted CD4 T-cell lineage specific to the chicken ovalbumin; BDC2.5 TCR-transgenic model (Katz et al., 1993) deficient in Rag1 (BDC2.5+Rago), with a monoclonal repertoire of MHC-class-II-restricted CD4 lineage reactive to a natural self-antigen in the endocrine pancreas. We did not detect conversion of the CD4 T-cell clones to CD8+ or Foxp3+ cells in LILP or MLN (Figure S3).
Two recent studies demonstrated the reprogramming of CD4 T cells to CD8 T cells in the small intestine, triggered by microbial antigens (Mucida et al., 2013; Reis et al., 2013). Those studies suggest novel molecular pathways operating in the small-intestine-associated microenvironment for the generation of CD4+CD8α+ cytotoxic T lymphocytes (CTLs) restricted to MHC-class-II recognition (Anderson, 2013). Although our studies of T-cell clonal fate with known antigen-specificities were limited to the two clones in the CD4 T-cell lineage, the results suggest that the CD4 T-cell clones may lack lineage plasticity in steady-state of the large-intestine-associated microenvironment, unlike their CD8 counterparts, the OT1 and 8.3 clones, in the same setting. On the other hand, our observations of CI-Treg-cell development in the large-intestine-associated microenvironment provide a parallel to the findings of MHC-class-II-restricted generation of CTLs in the small-intestine-associated microenvironment (Mucida et al., 2013; Reis et al., 2013).
MHC-class-II-dependency of cross-differentiation from the CD8 lineage to CD4 T cells
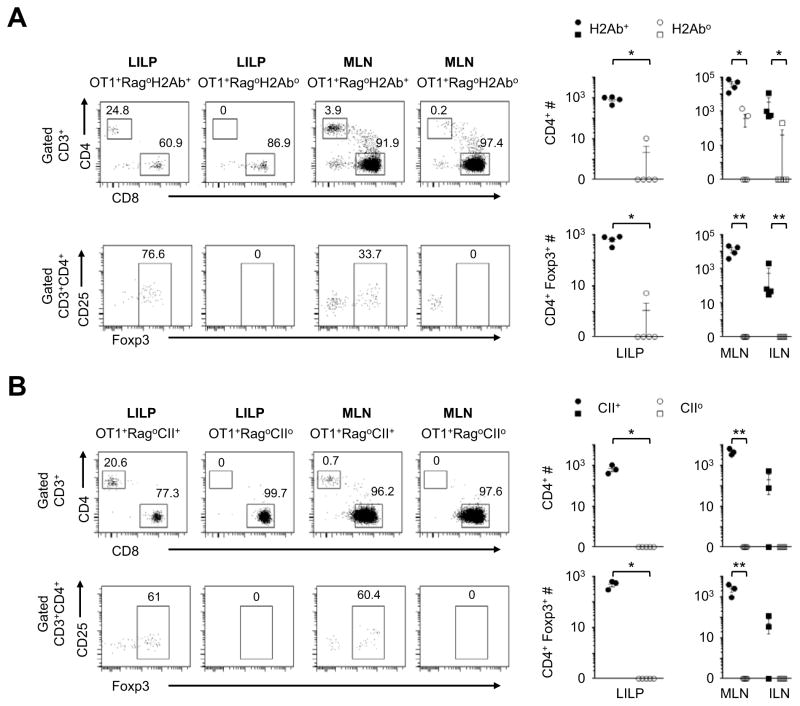
While the cross-differentiation generated MHC-class-I-restricted CD4 T cells, the cross-differentiation process may require CD4 interaction with MHC-class II (MHCII) molecules. To examine this possibility, we used two independent models (Grusby et al., 1991; Madsen et al., 1999) of MHCII-deficiencies. Indeed, MHCII-deficiencies caused by either H-2Ab knockout or complete knockout of the whole MHCII locus (Grusby et al., 1991; Madsen et al., 1999) abrogated the CD8-to-CD4 cross-differentiation (Figure 2). Possibly, during the cross-differentiation, CD8 silencing and CD4 induction may lead to engagement of both class I and class II of MHC on the same antigen-presenting cell, by the MHC-class-I-restricted TCR and CD4, respectively.
Figure 2. Cross-differentiation from the CD8 lineage to CD4 T cells with a requirement of MHC Class II.
(A) OT1+Rago B6 mice were crossed with the H-2Ab knockout line to generate H-2Ab-deficient OT1+Rago mice (n=5) and controls (n=4). On the B6 genetic background, H-2Ab knockout effectively renders deficiencies in MHC class II. The animals were analyzed at 6 to 10 wks of age. Representative flow cytometry plots are followed with cell counts of CD4+ and CD4+Foxp3+ T cells. Closed and open symbols denote H2-Ab+ controls and H2-Ab− mice, respectively. Each symbol represents one animal (mean±SEM). (B) OT1+Rago mice were crossed with the knockout line of the whole MHC class II locus to generate class-II-deficient OT1+Rago mice (n=5) and controls (n=3). The animals were analyzed at 4 to 21 wks of age. Representative flow cytometry plots are followed with cell counts of CD4+ and CD4+Foxp3+ T cells. Closed and open symbols denote MHCII+ and MHCII− mice, respectively. Each data point represents one animal (mean±SEM). *, p<0.05; **, p<0.01.
Essential role of MLN but not the thymus nor the immunocompromised status
To identify the organs wherein the cross-differentiation first occurred, we analyzed cohorts of OT1+Rago mice starting when they were young. We did not detect CD4+Foxp3+ populations in the thymus. CD4+Foxp3− and CD4+Foxp3+ T cells first appeared in the MLN and then in LILP around two weeks of age (Figure S4A). This predominant occurrence in the large-intestine-associated microenvironment was also similar to what was found with the NOD/B6.8.3+Rago model (Figure 1B).
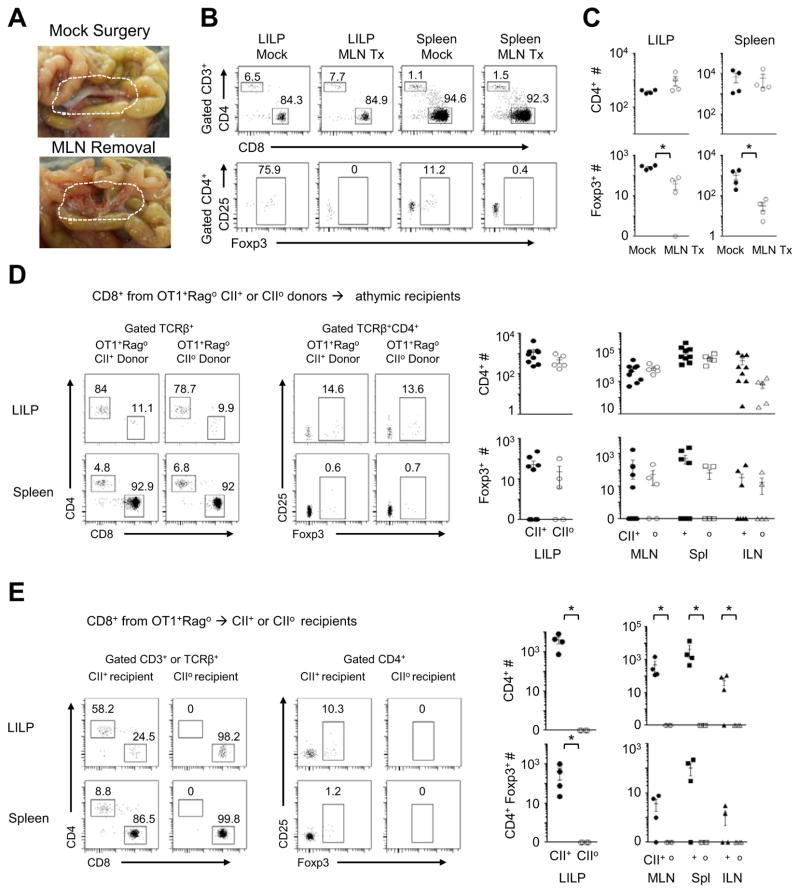
The kinetic evidence from both OT1+Rago and 8.3+Rago models suggests a role for MLN in cross-differentiation from the CD8 lineage to CD4 T cells. Previous studies showed that in the MLN, both dendritic cells and stromal cells presented antigens to stimulate OT1 cell responses (Lee et al., 2007). We assessed the role of MLN at a whole organ level by surgical experiments. We surgically removed the MLN in 2-wk-old OT1+Rago mice and analyzed the animals 2 wks later. Removing MLN diminished the CD4+Foxp3+ population. However, it did not reduce total CD4 T cell counts in OT1+Rago mice (Figure 3A–C).
Figure 3. A role of MLN but not the thymus in cross-differentiation from the CD8 T-cell lineage to CD4+Foxp3+ CI-Treg cells.
(A–C) Analyses of OT1+Rago mice 2 wks post MLN removal: (A) success of MLN removal; (B) representative flow cytometry plots (the numbers in the flow cytometry plots represent percentages of gated populations); (C) Summary of CD4+ and CD4+Foxp3+ cell counts (n=4, mean±SEM). (D) Conversion of purified peripheral OT1+CD8+CD4−CD25−Foxp3− cells after transfer into athymic hosts. MHCII+ or MHCIIo OT1+Rago donors were used to purify the peripheral CD8 T cells to examine whether presence of MHCII during maturation of the CD8 T cells enables a lasting potential for their conversion to CD4+ cells in a new host. (E) MHCII+ or MHCIIo Rago recipients were used to examine the requirements of MHCII during conversion of adoptively transferred OT1+Rago CD8 T cells. Representative flow cytometry plots are followed by a summary of CD4+ and CD4+Foxp3+ cell counts. Each data point represents one animal (n=5–7, mean±SEM).*, p<0.05.
To determine whether mature CD8 T cells can indeed convert to CD4 T cells without a thymic input, we purified CD8+CD4−CD25−Foxp3− T cells from the peripheral lymphoid organs (the spleen and lymph nodes) of adult OT1+Rago mice and injected the cells into Foxn1-deficient “nude” mice that lack a functional thymus (Nehls et al., 1994). In the athymic recipients, purified CD8+CD4−CD25−Foxp3− OT1 T cells efficiently converted to CD4 T cells, such that in LILP, CD4 T cells, rather than CD8 T cells, predominated (>50%-80%) (Figure 3D). Still, there was a possibility that the purified peripheral CD8 T-cell population contained a fraction of cells “mis-selected” or “imprinted” by MHCII in thymic selection but exported to the peripheral CD8 T-cell repertoire in a metastable state. To test this possibility, we purified CD8 T cells from OT1+RagoMHCIIo or OT1+RagoMHCII+ donors and transferred them into nude mice. As shown in Figure 3D, CD8 T cells from MHC+ or MHCIIo donors both converted to CD4 T cells efficiently. In this setting of MHCIIo, the donor CD8 T cells had never been exposed to MHCII prior to transfer. The recipient mice were athymic. Therefore, the CD8-to-CD4 cross-differentiation reflects peripheral conversion rather than an undetected effect of “mis-selection” or “imprinting” by thymic MHCII.
Conversely, to test whether prior exposure to MHCII potentiates CD8-to-CD4 conversion in a new MHCIIo host, we compared conversion of CD8 T cells from OT1+RagoMHC+ donors in RagoMHCIIo versus RagoMHCII+ hosts. The absence of MHCII in the new host abrogated CD8-to-CD4 conversion, despite possible interaction of the CD8 T cells with MHCII in the thymus prior to adoptive transfer (Figure 3E).
In addition, we examined whether the immunocompromised status of the monoclonal TCR-transgenic model was required for the cross-differentiation. We reconstituted OT1+Rago neonates with splenocytes from normal B6 mice. The reconstitution with a large number of polyclonal immune cells did not preclude cross-differentiation of the OT1 clone to CD4 T cells, although it reduced the population size of the converted cells, possibly due to limitation of their expansion as a result of pre-occupancy of “niche” by the large population of cells transferred into the neonate recipients (Figure S4B–C).
Critical but non-essential role for microbiota in CD8-to-CD4 cross-differentiation
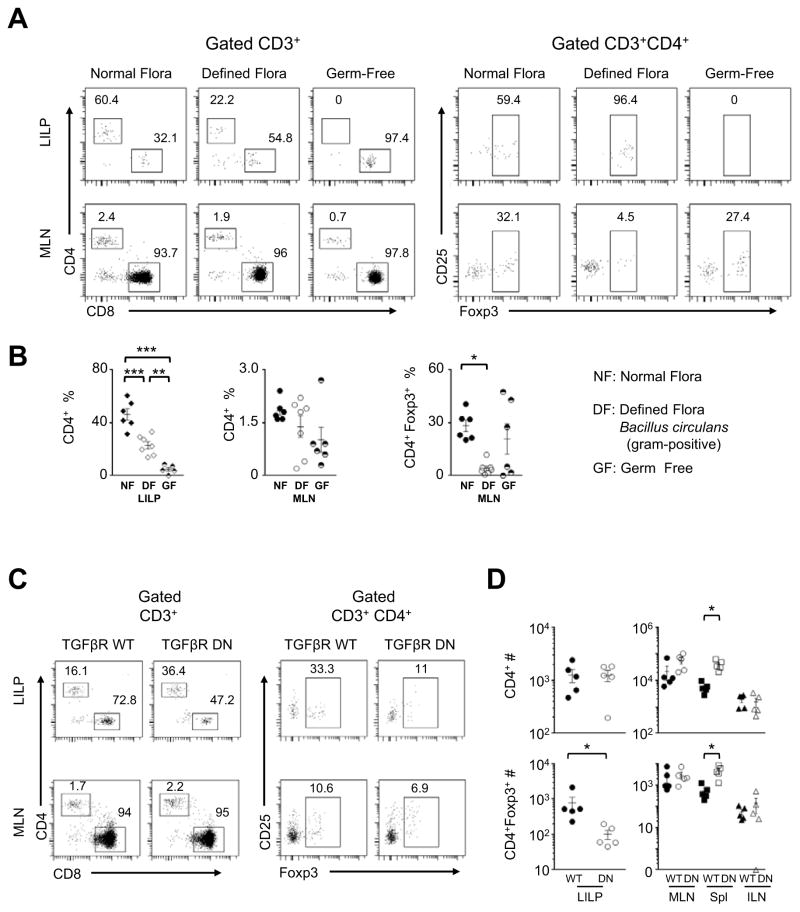
To determine whether normal microbial flora triggered the CD8 T-cell lineage plasticity in the large-intestine-associated microenvironment, we re-derived OT1+Rago mice under germ-free conditions, and then analyzed germ-free OT1+Rago mice. As shown in Figure 4A–B, CD4+Foxp3− or CD4+Foxp3+ populations were diminished in LILP of the germ-free animals. However, germ-free conditions did not abrogate the cross-differentiated CD4+Foxp3− or CD4+Foxp3+ populations in MLN of the OT1+Rago mice. Thus, the cross-differentiation did not require microbiota, but microbiota possibly promote homing to and / or expansion in LILP for the converted CD4+Foxp3− or CD4+Foxp3+ T cells.
Figure 4. Roles of microbiota and TGFβ in the CD8-to-CD4 cross-differentiation.
(A) Representative flow cytometry of OT1+Rago mice from germ-free (GF) facility, compared to animals in specific pathogen-free facility with normal flora (NF), or with a defined flora (DF) of the gram-positive Bacillus circulans. (B) Summary of CD4+ and CD4+Foxp3+ percentages in the OT1+Rago mice under GF versus NF and DF conditions (n=5–7, mean±SEM). (C–D) OT1+Rago mice were crossed with the CD4-dnTGFBRII transgenic line (TGFβR DN), to examine the effect of T-cell intrinsic TGFβ signaling in the cross-differentiation. Representative flow cytometry of OT1+Rago TGFβR DN mice versus OT1+Rago controls (C), followed by summary of cell counts (n=5, mean±SEM; closed and open symbols, control and TGFβR DN mice, respectively) (D). The numbers in the flow cytometry plots represent percentages of gated populations. Each data point represents one animal. *, p<0.05; **, p<0.01; ***, p<0.001.
Furthermore, a defined microbial flora consisting of the gram-positive Bacillus circulans was sufficient to restore the CD4+Foxp3− and CD4+Foxp3+ populations in LILP of OT1+Rago mice (Figure 4A–B). In addition, we tested whether the CD8-to-CD4 cross-differentiation depended on a particular facility or diet. The animals were raised in different buildings in combination with different diets. The cross differentiation of the OT1 T-cell clone to CD4+Foxp3− and CD4+Foxp3+ T cells occurred efficiently in the large-intestine-associated microenvironment in all variations of animal housing and diet we have examined (Figure S5).
Minimal role for T-cell-intrinsic TGFβ signaling in CD8-to-CD4 cross-differentiation
One of the major factors that influence T-cell differentiation in GAE is TGFβ (Li and Flavell, 2008). We tested whether TGFβ-mediated intrinsic signaling in T cells impacts the cross-differentiation from the CD8 lineage to CD4 T cells. We crossed OT1+Rago mice with a transgenic line carrying a dominant-negative mutant of TGFβ receptors, CD4-dnTGFBRII (Gorelik and Flavell, 2000). Blocking TGFβ signaling in T cells did not suppress cross-differentiation of the OT1 clone to CD4 T cells. It substantially reduced CD4+Foxp3+ population in LILP (Figure 4C), but not in lymphoid organs of OT1+Rago mice. Notably, it increased CD4+ and CD4+Foxp3+ populations in the spleen (Figure 4D), an effect that remains to be explained.
Conversion from lineage-marked mature CD8 T cells with a physiological repertoire
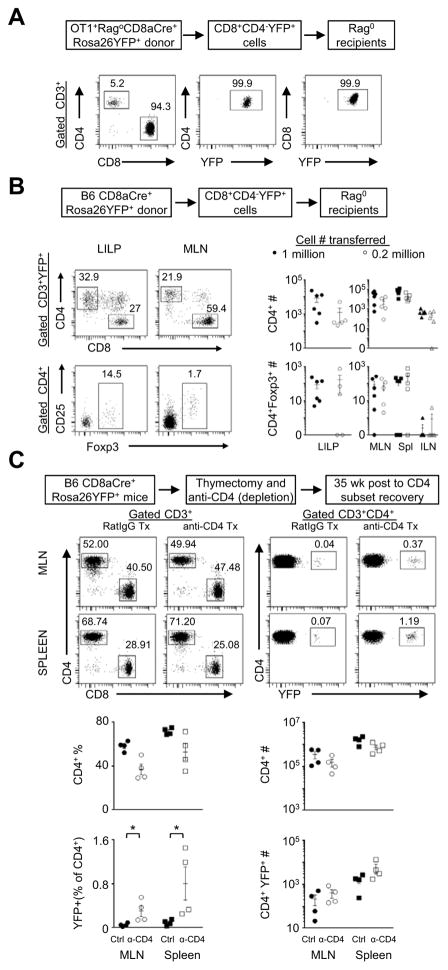
To study whether mature T cells in the CD8 lineage can convert to CD4 T cells, we used the CD8aCre (Maekawa et al., 2008) × Rosa26YFP (Srinivas et al., 2001) model, in which the Cre-transgene expression driven by an engineered CD8 promoter was demonstrated to mark mature CD8 T cells in the periphery (Maekawa et al., 2008). In the OT1+RagoCD8aCre+Rosa26YFP+ mice, CD8aCre+Rosa26YFP+ only marked a minority of CD8 T cells in the periphery (Figure S6A). CD8+YFP+ OT1 cells were purified from the peripheral lymphoid organs of OT1+RagoCD8aCre+Rosa26YFP+ mice and transferred into Rago recipients. Indeed, the transferred CD8+YFP+ OT1 cells converted to CD4 T cells (Figure 5A), consistent with the results from adoptive transfer experiments using CD8+ OT1 cells from OT1+Rago donors, as described before (Figure 3D).
Figure 5. Conversion of lineage-marked CD8 T cells to CD4+ and CD4+Foxp3+ T cells.
(A) Conversion of lineage-marked OT1 CD8 T cells. CD8+YFP+CD4−CD25− cells purified from OT1+RagoCD8aCre+Rosa26YFP+ mice were transferred into Rago recipients and analyzed ~6wks later (n=4). Representative flow cytometry plots of MLN. The numbers in the plots indicate the percentages of gated populations. (B) Conversion of lineage-marked polyclonal CD8 T cells. CD8+YFP+CD4−CD25−Foxp3− cells were purified from CD8aCre+Rosa26YFP+ B6 mice and adoptively transferred into Rago hosts, each receiving 1.0×106 cells (closed symbols) or 2×105 cells (open symbols). The animals were analyzed ~6 wks later. Representative flow plots of T cells in LILP and MLN were followed by summary of CD4+ and CD4+Foxp3+ cell counts. Each data point represents one animal (n=5–6, mean±SEM). (C) Influence of CD4-depletion. Adult B6 CD8aCre+Rosa26YFP+ mice were thymectomized (to eliminate thymic development). Two weeks later, mice were treated with two doses of Rat IgG controls or anti-CD4 depleting antibodies. Mice were analyzed by flow cytometry at 35–40 wks post treatment (after the CD4 T-cell population recovered from depletion). Representative flow cytometry plots are followed by summary of percentages and cell counts. Each data point represents one animal (mean±SEM, n=4–5). *, p<0.05.
We further examined cross-differentiation in a non-TCR transgenic setting. To unequivocally demonstrate that mature T cells in the CD8 lineage with a natural repertoire can indeed convert to CD4 T cells, we purified peripheral CD8+YFP+CD4−Foxp3−CD25− cells from CD8aCre+Rosa26YFP+ mice on the B6 genetic background, and adoptively transferred the cells into Rago/B6 recipients. The mature CD8aCre+Rosa26YFP+ cells converted to CD4+Foxp3− and CD4+Foxp3+ cells in the new hosts, with high frequencies of CD4 T cells in LILP (average ~20%) followed by in MLN (Figure 5B). In unperturbed CD8aCre+Rosa26YFP+ mice, CD4+YFP+ cells existed rarely (Figure S6B). Depletion of the CD4 T-cell population enhanced cross-differentiation from the CD8 lineage to CD4 T cells (Figure 5C). Although these experiments had the caveats of the artificial promoter constructed to mark mature CD8 T cells in the periphery (Maekawa et al., 2008), and unknown antigen-specificity of transferred polyclonal CD8 T cells, the results indicate that naturally matured T cells in the CD8 lineage can indeed convert to CD4 T cells in the large-intestine-associated microenvironment.
A physiological role of CI-Treg cells in selfless tolerance induction
Foxp3 expression in the OT1+Rago mice, identified with the Foxp3-IRES-RFP reporter (Wan and Flavell, 2005) or anti-Foxp3 antibodies, was only detected in the CD4 T-cell population but not in the CD8 T cells (Figure S7). To examine the physiological role of the CD4+Foxp3+ CI-Treg cells in immune tolerance and homeostasis, we generated Foxp3-deficient OT1+Rago mice, by crossing OT1+Rago mice with the Foxp3sf line with a null mutation of the Foxp3 gene (Wildin et al., 2001). The Foxp3sf line is a commonly used model for studying the role of Foxp3+ Treg cells in their natural settings.
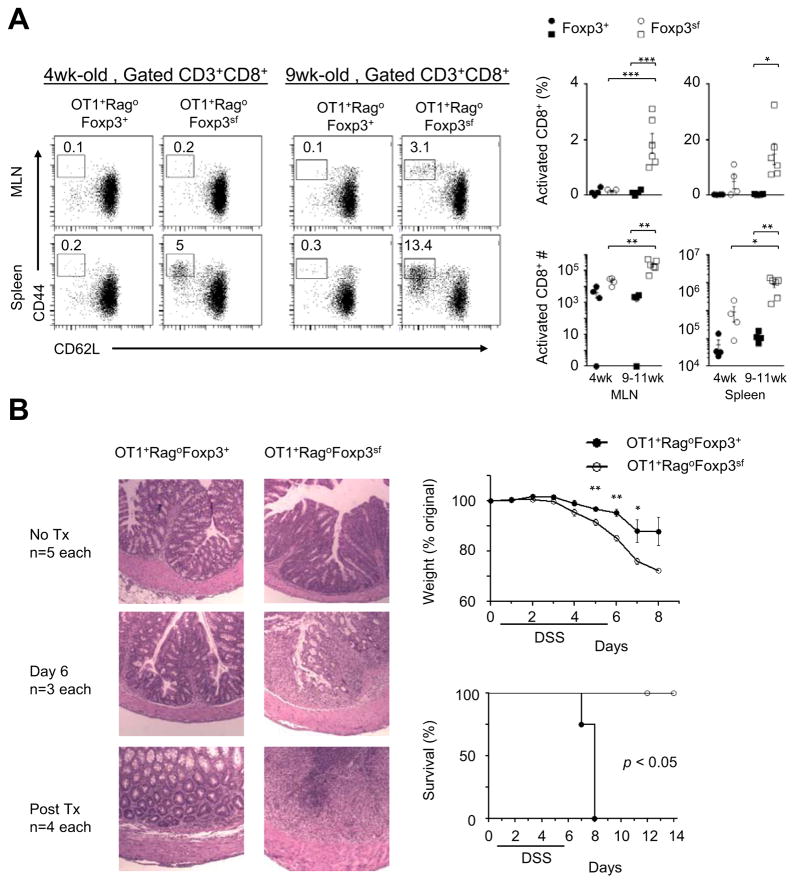
The OT1+RagoFoxp3sf mice developed healthily like OT1+RagoFoxp3+ controls. Importantly, a functional Foxp3 gene was not required for the cross-differentiation from the CD8 lineage to CD4 T cells (Figure S7C). However, CD8 T cells in the OT1+RagoFoxp3sf mice, when compared to the controls, exhibited an age-dependent increase in total counts and activation status in lymphoid organs (Figure 6A). These results indicate that the CD4+Foxp3+ T cells converted from the CD8 lineage have a role as Treg cells in their natural settings to regulate homeostasis of the conventional T-cell compartments.
Figure 6. Function of CD4+Foxp3+ CI-Treg cells in their natural settings in regulating immune homeostasis and protecting the large intestine from inflammatory damage.
(A) The effect of Foxp3-deficiencies in OT1+Rago mice on homeostasis and activation of CD8 T cells. Representative flow cytometry followed by cell counts (closed symbols, OT1+RagoFoxp3+ mice; open symbols, OT1+RagoFoxpsf mice). The numbers in the flow cytometry plots represent percentages of gated populations. Each data point represents one animal (n=4–6; mean±SEM). (B) Colitis development in OT1+RagoFoxp3+ versus OT1+RagoFoxp3sf mice treated with DSS drinking water. Left, histopathology of colon tissue sections (original magnification × 5). Right, weight loss and survival curves (n=4–7 pooled from two experiments, mean±SEM). *, p<0.05; **, p<0.01; ***, p<0.001.
Despite the accumulation of activated CD8 T cells in the OT1+RagoFoxp3sf mice, the animals exhibited growth and weight gains comparable to that of OT1+RagoFoxp3+ controls. In consistency, pathology examination did not reveal substantial damage of the intestine tissues at the interface of the large intestine and microbiota in the OT1+RagoFoxp3sf mice (Figure 6B, left, top panels). Therefore, the CD4+Foxp3+ CI-Treg cells did not appear to have a role in tolerance induction to the constituents in the large intestine including microbiota, in contrast to other types of thymic or induced colonic Treg cells (Cebula et al., 2013; Josefowicz et al., 2012; Lathrop et al., 2011).
We then examined if CD4+Foxp3+ CI-Treg cells protect the large intestine from inflammatory assault. We used a mouse model of colitis that was induced by oral administration of dextran sodium sulfate (DSS) (Okayasu et al., 1990). This is a commonly used model, in which the colitis was not caused by specific immune targeting, but due to chemically (DSS) induced inflammatory damage. DSS treatment induced colitis in both OT1+RagoFoxp3sf mice and OT1+RagoFoxp3+ controls. After termination of the DSS treatment, OT1+RagoFoxp3+ controls quickly recovered and there was no mortality in this group. However, OT1+RagoFoxp3sf mice could not heal from the colitis after discontinuing the DSS treatment and all animals in this group progressed to moribund conditions (Figure 6B). Thus, the CD4+Foxp3+ CI-Treg cells in their natural settings, specific to neither “self” nor microbial agents, not only regulated systemic immune homeostasis but also facilitated inflammation control in the large intestine.
A protective role of adoptively transferred CI-Treg cells in T-cell-mediated immune damage
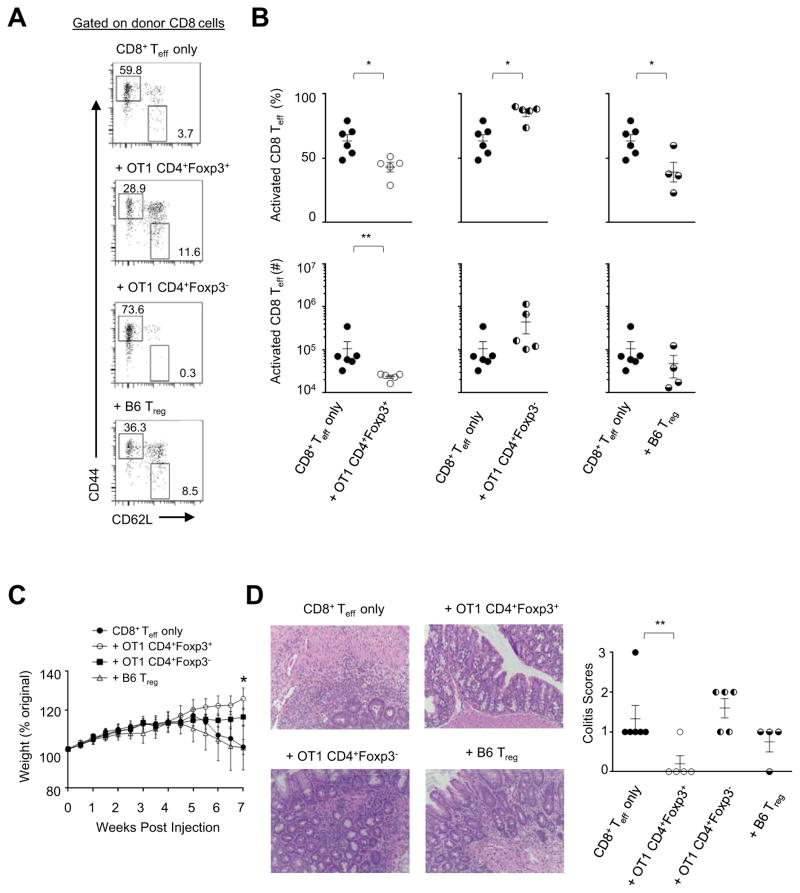
We then examined the regulatory potential of CI-Treg cells in an adoptive transfer setting. Led by the findings shown in Figure 6 that CI-Treg cells reduced activation of CD8 T cells, we used a model of colitis induced by transfer of naïve CD8 effector T (Teff) cells into Rago recipients (Tajima et al., 2008). OT1 CD4+Foxp3+ CI-Treg cells co-transferred at 1:3 ratio with the CD8 Teff cells indeed suppressed the activation of the CD8 Teff cells (Figure 7A–B), improved weight maintenance (Figure 7C) and suppressed colitis development (Figure 7D). Co-transfer of OT1 CD4+Foxp3− cells did not suppress but slightly increased the activation of CD8 Teff cells. It did not inhibit colitis development. Of note, in the OT1 CD4+Foxp3− co-transfer group, ~30% of OT1 CD4 cells became Foxp3+ in the new host (not shown). The CI-Treg cells appeared to be more consistent than standard CD4+Foxp3+ Treg cells from B6 mice in suppressing immune damage in this model (Figure 7C–D). Further experimentations at a larger scale and with various cell doses are needed to test if CI-Treg cells might be more effective than standard CD4+Foxp3+ Treg cells in this and other conditions of CD8 T-cell-mediated damage. Future studies are also needed to examine whether CI-Treg cells function in other settings of inflammatory damage, including immune damage mediated by CD4 Teff cells. Taken together, the results from Figure 6 and Figure 7 suggest that CI-Treg cells can indeed be effective in suppressing inflammatory damage to intestinal tissue.
Figure 7. Control of T-cell-mediated damage by CD4+Foxp3+ CI-Treg cells in adoptive transferring settings.
Purified naïve CD8 Teff cells from wildtype B6 mice were adoptively transferred into Rago recipients. The CD8 Teff cells (2×105) were injected alone, or co-transferred with 7×104 CD4+Foxp3+ or CD4+Foxp3− cells purified from OT1+Rago mice, or CD4+Foxp3+ cells from B6 mice. Mice were monitored for weight. (A) Representative flow cytometry. The numbers in the flow-cytometry plots represent percentages of gated populations. (B) Summary of percentages and cell counts for the activated CD8 T cells. Each data point represents one animal (n=4–6; mean±SEM). (C) Weight of the animals after T cell transfer. (D) Pathology assessment of colitis development. Left, H–E staining of colon tissue sections (original magnification × 12.5). Right, histopathology score (mean±SEM; n=4–6 pooled from 4 experiments). *, p<0.05; **, p<0.01.
DISCUSSION
Originally formulated to explain the biological relationship of a vertebrate host organism and microbial pathogens, the concept of selfhood-centered tolerance versus immunity has led to much of immunological understandings. Its limitations (Matzinger, 1994) have been highlighted with the recent debates of immune tolerance induction to microbiota in the large intestine (Cebula et al., 2013; Josefowicz et al., 2012; Lathrop et al., 2011). A healthy mammal typically hosts a chimeric composition of cells from diverse genetic origins. The steady-state symbiosis with a superior number of normal flora microbial agents in the large intestine compels a reconsideration of the concept of “self” and “nonself” discrimination, as the mutualistic microbial organisms in a mammalian host do not fit into the traditional definition of “self” or “nonself”.
This study demonstrates conversion of T-cell clones in the CD8 lineage to CD4+Foxp3− and CD4+ Foxp3+ Treg cells in the large-intestine-associated microenvironment, in the absence of its nominal antigen, or despite the remote presence of a “self” antigen in a distal organ. This observation suggests that, in a steady state without apparent “danger” (Matzinger, 1994), T cells do not necessarily delineate a dichotomized discrimination of “self” from “nonself”, but can differentiate with “selfless”, intrinsic plasticity to remedy an imbalanced constitution. Conversion of the CD8 T-cell lineage in the large-intestine-associated microenvironment could be driven by environmental cues without regard to the “self” or “nonself” origin of specific antigenic signals. In this regard, although microbial-specific tolerance could indeed be detected in the large intestine (Cebula et al., 2013; Lathrop et al., 2011), the effect of gut flora on T-cell differentiation may not always be limited to the gut. For example, CD4 Th17 subset differentiation induced by gut-microbial flora could trigger autoimmune damage in the joint, a site distal from the gut (Wu et al., 2010).
The two independent T-cell clones in the MHC-class-I-restricted CD8 lineage showed lineage plasticity in the large-intestine-associated microenvironment. This plasticity may be T-cell intrinsic, since it was not apparently induced by antigenic stimulation from microbiota or the body’s self. To an extent, the observation offers some reconciliation between “selfhood”-centered immunological discrimination and the chimeric composition of a healthy mammal. In this regard, we suggest distinction between central and peripheral T-cell development. Robust evidence exists for T-cell clonal selection based on “self / nonself” discrimination in a central lymphoid organ, the thymus (Anderson et al., 2002; Hsieh et al., 2004; Kisielow et al., 1988). However, in the periphery, in addition to the various mechanisms of tolerance induction to self-antigens, T-cell differentiation at a heterogeneous interface such as GAE could occur independently of “self”-denotation (Matzinger, 1994), in a “selfless” mode of clonal plasticity. Such kind of plasticity may facilitate tolerance and integrity with respect to a wholesome biological entity of a mammalian organism. In other words, there is an additional, peripheral mechanism (s) of T-cell immunity versus tolerance induction that may occur regardless of “self” or “nonself”, but is attuned to environmental cues and evolved for a systemic balance, with an intrinsic cellular plasticity equipped in the CD8 T-cell lineage.
Technically, it is a challenge to study T-cell clonal fate in a natural history of peripheral development without resorting to a reductionist approach, particularly in complex settings such as the large-intestine-associated microenvironment. On the other hand, how microbiota affects the clonal fate of T lymphocytes remains a question of extensive interest. This study used monoclonal TCR transgenic models that made it possible to analyze the T-cell clonal fate in terms of known antigen-specificities. The nature of such in vivo studies limited the practicality to only a few clones. Of note, the OT1+Rago mouse is a standard model generally used for studying CD8 T cells. Still, the limitation of such an approach is apparent: how physiologically relevant is such an exaggerated simplicity of T-cell repertoire in a context of relative lymphopenia?
A previous study showed that a well-established T-cell lineage such as the CD4 Treg cell was stable in lineage specification even during lymphopenia-driven proliferation (Rubtsov et al., 2010). In our studies, lymphopenia was unlikely the original trigger for the CD8-to-CD4 lineage plasticity either. Rectifying the monoclonal and lymphopenic condition by neonatal reconstitution with a natural repertoire of immune cells did not preclude the cross-differentiation, but reduced the size of converted populations, suggesting that the CD8-to-CD4 cross-differentiation may occur at a basal level, and its expansion depends on available “niche”. Consistent with this possibility, the CD8-to-CD4 conversion of polyclonal T cells in CD8aCre+Rosa26YFP+ mice was enhanced in CD4-depleted animals. Of note, the CD8aCre transgene was constructed with an unnatural promoter (Maekawa et al., 2008). Although it labeled most of the peripheral CD8 T cells in B6 mice, it only marked a minority of OT1 CD8 T cells in peripheral lymphoid organs. Therefore, an accurate assessment of the extent of CD8-to-CD4 lineage cross-differentiation in a natural repertoire awaits an alternative tool for efficient lineage tracing unaffected by clonal variations. Nevertheless, CD8-to-CD4 lineage conversion revealed in this study could be relevant in adoptive immune therapies using CD8 T cells, or in clinical settings of immune imbalance due to CD4 T-cell deficit, such as AIDS or immunoablation conditioning.
It remains to be determined whether the CD4+Foxp3− and CD4+Foxp3+ cells derived from cross-differentiation of the CD8 lineage have a stable lineage specification or merely reflect a metastable differentiation state. Lineage stability in various settings has been demonstrated for MHC-class-II-restricted CD4+Foxp3+ Treg cells (Rubtsov et al., 2010). In our study, the functional capacity of CD4+Foxp3+ CI-Treg cells was indeed evident in vivo, but their lineage stability needs to be characterized by genetic tracing. With regard to their therapeutic potentials, unlike the known subsets of thymic and peripheral CD4+Foxp3+ Treg cells which recognize MHC-class-II molecules, expression of which is limited to antigen-presenting cells, the CD4+Foxp3+ CI-Treg cells are distinct in recognizing constitutively and ubiquitously expressed MHC-class-I molecules. Therefore, the “selfless” plasticity of the CD8 lineage cross-differentiating to CD4+Foxp3+ CI-Treg cells suggests novel possibilities for immune tolerance induction.
EXPERIMENTAL PROCEDURES
Mice
The transgenic and knockout mouse models were described previously: OT1 (Hogquist et al., 1994), 8.3 (Verdaguer et al., 1997), OTII (Barnden et al., 1998), BDC2.5 (Katz et al., 1993), Rag1o (Mombaerts et al., 1992), Foxp3sf (Wildin et al., 2001), MHC class II H-2Ab knockout (Grusby et al., 1991), MHC-class-II KO (Madsen et al., 1999), Foxp3FIR (Foxp3-IRIS-RFP “knockin”) mice (Wan and Flavell, 2005), CD8aCre (Maekawa et al., 2008), Rosa-26-YFP (Srinivas et al., 2001), TGFβ1 Receptor dominant negative (DN) transgenic mice (Gorelik and Flavell, 2000). These lines were crossed in various combinations to generate the model for this study. Germ-free (GF) mice were re-derived by embryo transfer at Taconic (Hudson, NY). During germ-free re-derivation of OT1+RagoFoxp3FIR mice, one of the isolators was found to be specific-pathogen-free but an aerobic normal microbial flora was detected during monitoring. The flora was identified as gram-positive Bacillus circulans. The animals from this isolator, although of an unsuccessful attempt in germ-free re-derivation, were deemed with a “defined flora” (DF), and analyzed in comparison to animals that are maintained in standard specific-pathogen-free facility (SPF) with normal flora (NF) or germ-free (GF) conditions. Other animals were maintained in a specific-pathogen-free (SPF) barrier facility, with diet Teklad #2018 or #2019 (Harlan Laboratories, Madison, WI), or LabDiet diet #5021(LabDiet, St. Louis, MO). The studies are approved by the Institutional Animal Care and Use Committee at the University of Miami.
Surgical removal of mesenteric lymph nodes and thymectomy
Mock surgery or surgical removal of MLN were conducted on 13- to 14-day old OT1+Rago mice with a procedure similar to previously reported (Griffin et al., 2011). Thymectomy was conducted according to a standard procedure. Both are described in detail in the SUPPLEMENTAL EXPERIMENTAL PROCEDURES.
Antibody depletion of CD4 T cells
For CD4 T cell depletion, mice were injected i.p. with anti-CD4 antibody (GK1.5) or Rat IgG controls, 12 μg/g weight, at day 0 and 5. Efficacy of anti-CD4 T cell depletion was verified by tail vein blood sampling after completion of treatment. The mice were then monitored for CD4 T-cell population recovery. The end point experiments were conducted 35–40 weeks after completion of anti-CD4 depletion.
Cell preparation from intestinal tissue
Intestines were minced and treated with EDTA to isolate IELs. Collagenase was used to dissociate intestinal tissues for lymphocyte isolation from lamina propria. Detailed methods were described in the SUPPLEMENTAL EXPERIMENTAL PROCEDURES.
Flow cytometry, cell sorting and adoptive transfer experiments
Flow cytometry analyses, including intracellular cytokine staining, were conducted with a standard procedure (Devarajan et al., 2014; Miska et al., 2014; Miska et al., 2012), described in the Supplemental Procedures. In adoptive transfer experiments, CD8 T cells were sorted as CD8+CD4−CD11b−CD11c−Ter119−B220−NK1.1−CD25−Foxp3− from the spleen and lymph nodes of OT1+RagoFoxp3FIR+ mice or as CD8+YFP+CD4−CD11b−CD11c−Ter119−B220−NK1.1−CD25−Foxp3− from CD8aCre+ Rosa26YFP+ mice. The purified CD8 T cells (purity > 99.7%), with or without the YFP marker, were injected into recipients. For neonate reconstitution of OT1+RagoFoxp3FIR+ mice, whole splenocyte preparation from B6 mice were injected, 2 × 107 cells per recipient, by intraperitoneal injection.
Colitis studies
Colitis induction by DSS was done following previously described methods (Okayasu et al., 1990), which are described in details in the Supplemental Procedure. Colitis induction by transfer of purified naïve CD8 effector T (Teff) cells into Rago recipients and pathology scoring were done as described before (Tajima et al., 2008) except a lower dose of CD8 T cells were used.
Statistics
Student’s t tests or Mann-Whitney U tests were used when appropriate for single comparisons. For multiple groups, ANOVA analyses followed up with Tukey’s post-hoc test or Mann-Whitney U tests followed by Bonferroni’s correction were used. For survival analysis, log-rank test was conducted. P values equal to or less than 0.05 were considered significant. *, P<0.05; **, P<0.01; ***, P<0.001.
Supplementary Material
Acknowledgments
This work was supported in part by a grant from the National Institute of Health (DP3DK085696 to Z.C.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding agency. We thank Mr. K. Johnson, Mr. J. Enten, and Ms. P. Guevara and Drs. O. Umland, S. Opiela, and J. Miska for their expert assistance and advice.
Footnotes
Supplemental information includes 7 figures and Supplemental Procedures.
DISCLOSURE
The authors declare no competing financial interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abt MC, Osborne LC, Monticelli LA, Doering TA, Alenghat T, Sonnenberg GF, Paley MA, Antenus M, Williams KL, Erikson J, et al. Commensal bacteria calibrate the activation threshold of innate antiviral immunity. Immunity. 2012;37:158–170. doi: 10.1016/j.immuni.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson MK. Changing course by lymphocyte lineage redirection. Nat Immunol. 2013;14:199–201. doi: 10.1038/ni.2544. [DOI] [PubMed] [Google Scholar]
- Anderson MS, Venanzi ES, Klein L, Chen Z, Berzins SP, Turley SJ, von Boehmer H, Bronson R, Dierich A, Benoist C, et al. Projection of an immunological self shadow within the thymus by the aire protein. Science. 2002;298:1395–1401. doi: 10.1126/science.1075958. [DOI] [PubMed] [Google Scholar]
- Barnden MJ, Allison J, Heath WR, Carbone FR. Defective TCR expression in transgenic mice constructed using cDNA-based alpha- and beta-chain genes under the control of heterologous regulatory elements. Immunol Cell Biol. 1998;76:34–40. doi: 10.1046/j.1440-1711.1998.00709.x. [DOI] [PubMed] [Google Scholar]
- Burnet FM. A modification of Jerne’s theory of antibody production using the concept of clonal selection. Aust J Sci. 1957;20:67–69. doi: 10.3322/canjclin.26.2.119. [DOI] [PubMed] [Google Scholar]
- Cebula A, Seweryn M, Rempala GA, Pabla SS, McIndoe RA, Denning TL, Bry L, Kraj P, Kisielow P, Ignatowicz L. Thymus-derived regulatory T cells contribute to tolerance to commensal microbiota. Nature. 2013;497:258–262. doi: 10.1038/nature12079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devarajan P, Miska J, Lui JB, Swieboda D, Chen Z. Opposing Effects of CTLA4 Insufficiency on Regulatory versus Conventional T Cells in Autoimmunity Converge on Effector Memory in Target Tissue. J Immunol. 2014;193:4368–4380. doi: 10.4049/jimmunol.1400876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle C, Strominger JL. Interaction between CD4 and class II MHC molecules mediates cell adhesion. Nature. 1987;330:256–259. doi: 10.1038/330256a0. [DOI] [PubMed] [Google Scholar]
- Esplugues E, Huber S, Gagliani N, Hauser AE, Town T, Wan YY, O’Connor W, Jr, Rongvaux A, Van Rooijen N, Haberman AM, et al. Control of TH17 cells occurs in the small intestine. Nature. 2011;475:514–518. doi: 10.1038/nature10228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontenot JD, Gavin MA, Rudensky AY. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat Immunol. 2003;4:330–336. doi: 10.1038/ni904. [DOI] [PubMed] [Google Scholar]
- Gorelik L, Flavell RA. Abrogation of TGFbeta signaling in T cells leads to spontaneous T cell differentiation and autoimmune disease. Immunity. 2000;12:171–181. doi: 10.1016/s1074-7613(00)80170-3. [DOI] [PubMed] [Google Scholar]
- Griffin AJ, Li LX, Voedisch S, Pabst O, McSorley SJ. Dissemination of persistent intestinal bacteria via the mesenteric lymph nodes causes typhoid relapse. Infect Immun. 2011;79:1479–1488. doi: 10.1128/IAI.01033-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grusby MJ, Johnson RS, Papaioannou VE, Glimcher LH. Depletion of CD4+ T cells in major histocompatibility complex class II-deficient mice. Science. 1991;253:1417–1420. doi: 10.1126/science.1910207. [DOI] [PubMed] [Google Scholar]
- Hedrick SM. Positive selection in the thymus: an enigma wrapped in a mystery. J Immunol. 2012;188:2043–2045. doi: 10.4049/jimmunol.1200077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogquist KA, Jameson SC, Heath WR, Howard JL, Bevan MJ, Carbone FR. T cell receptor antagonist peptides induce positive selection. Cell. 1994;76:17–27. doi: 10.1016/0092-8674(94)90169-4. [DOI] [PubMed] [Google Scholar]
- Hooper LV, Macpherson AJ. Immune adaptations that maintain homeostasis with the intestinal microbiota. Nat Rev Immunol. 2010;10:159–169. doi: 10.1038/nri2710. [DOI] [PubMed] [Google Scholar]
- Hori S, Nomura T, Sakaguchi S. Control of regulatory T cell development by the transcription factor Foxp3. Science. 2003;299:1057–1061. doi: 10.1126/science.1079490. [DOI] [PubMed] [Google Scholar]
- Hsieh CS, Liang Y, Tyznik AJ, Self SG, Liggitt D, Rudensky AY. Recognition of the peripheral self by naturally arising CD25+ CD4+ T cell receptors. Immunity. 2004;21:267–277. doi: 10.1016/j.immuni.2004.07.009. [DOI] [PubMed] [Google Scholar]
- Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janeway CA, Jr, Medzhitov R. Innate immune recognition. Annu Rev Immunol. 2002;20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]
- Josefowicz SZ, Niec RE, Kim HY, Treuting P, Chinen T, Zheng Y, Umetsu DT, Rudensky AY. Extrathymically generated regulatory T cells control mucosal TH2 inflammation. Nature. 2012;482:395–399. doi: 10.1038/nature10772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz JD, Wang B, Haskins K, Benoist C, Mathis D. Following a diabetogenic T cell from genesis through pathogenesis. Cell. 1993;74:1089–1100. doi: 10.1016/0092-8674(93)90730-e. [DOI] [PubMed] [Google Scholar]
- Khattri R, Cox T, Yasayko SA, Ramsdell F. An essential role for Scurfin in CD4+CD25+ T regulatory cells. Nat Immunol. 2003;4:337–342. doi: 10.1038/ni909. [DOI] [PubMed] [Google Scholar]
- Kirchner J, Bevan MJ. ITM2A is induced during thymocyte selection and T cell activation and causes downregulation of CD8 when overexpressed in CD4(+)CD8(+) double positive thymocytes. J Exp Med. 1999;190:217–228. doi: 10.1084/jem.190.2.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisielow P, Bluthmann H, Staerz UD, Steinmetz M, von Boehmer H. Tolerance in T-cell-receptor transgenic mice involves deletion of nonmature CD4+8+ thymocytes. Nature. 1988;333:742–746. doi: 10.1038/333742a0. [DOI] [PubMed] [Google Scholar]
- Lathrop SK, Bloom SM, Rao SM, Nutsch K, Lio CW, Santacruz N, Peterson DA, Stappenbeck TS, Hsieh CS. Peripheral education of the immune system by colonic commensal microbiota. Nature. 2011;478:250–254. doi: 10.1038/nature10434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JW, Epardaud M, Sun J, Becker JE, Cheng AC, Yonekura AR, Heath JK, Turley SJ. Peripheral antigen display by lymph node stroma promotes T cell tolerance to intestinal self. Nat Immunol. 2007;8:181–190. doi: 10.1038/ni1427. [DOI] [PubMed] [Google Scholar]
- Li MO, Flavell RA. TGF-beta: a master of all T cell trades. Cell. 2008;134:392–404. doi: 10.1016/j.cell.2008.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen L, Labrecque N, Engberg J, Dierich A, Svejgaard A, Benoist C, Mathis D, Fugger L. Mice lacking all conventional MHC class II genes. Proc Natl Acad Sci U S A. 1999;96:10338–10343. doi: 10.1073/pnas.96.18.10338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa Y, Minato Y, Ishifune C, Kurihara T, Kitamura A, Kojima H, Yagita H, Sakata-Yanagimoto M, Saito T, Taniuchi I, et al. Notch2 integrates signaling by the transcription factors RBP-J and CREB1 to promote T cell cytotoxicity. Nat Immunol. 2008;9:1140–1147. doi: 10.1038/ni.1649. [DOI] [PubMed] [Google Scholar]
- Matzinger P. Tolerance, danger, and the extended family. Annu Rev Immunol. 1994;12:991–1045. doi: 10.1146/annurev.iy.12.040194.005015. [DOI] [PubMed] [Google Scholar]
- Miska J, Abdulreda MH, Devarajan P, Lui JB, Suzuki J, Pileggi A, Berggren PO, Chen Z. Real-time immune cell interactions in target tissue during autoimmune-induced damage and graft tolerance. J Exp Med. 2014;211:441–456. doi: 10.1084/jem.20130785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miska J, Bas E, Devarajan P, Chen Z. Autoimmunity-mediated antitumor immunity: tumor as an immunoprivileged self. Eur J Immunol. 2012;42:2584–2596. doi: 10.1002/eji.201242590. [DOI] [PubMed] [Google Scholar]
- Mombaerts P, Iacomini J, Johnson RS, Herrup K, Tonegawa S, Papaioannou VE. RAG-1-deficient mice have no mature B and T lymphocytes. Cell. 1992;68:869–877. doi: 10.1016/0092-8674(92)90030-g. [DOI] [PubMed] [Google Scholar]
- Mucida D, Husain MM, Muroi S, van Wijk F, Shinnakasu R, Naoe Y, Reis BS, Huang Y, Lambolez F, Docherty M, et al. Transcriptional reprogramming of mature CD4(+) helper T cells generates distinct MHC class II-restricted cytotoxic T lymphocytes. Nat Immunol. 2013;14:281–289. doi: 10.1038/ni.2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nehls M, Pfeifer D, Schorpp M, Hedrich H, Boehm T. New member of the winged-helix protein family disrupted in mouse and rat nude mutations. Nature. 1994;372:103–107. doi: 10.1038/372103a0. [DOI] [PubMed] [Google Scholar]
- Norment AM, Salter RD, Parham P, Engelhard VH, Littman DR. Cell-cell adhesion mediated by CD8 and MHC class I molecules. Nature. 1988;336:79–81. doi: 10.1038/336079a0. [DOI] [PubMed] [Google Scholar]
- Okayasu I, Hatakeyama S, Yamada M, Ohkusa T, Inagaki Y, Nakaya R. A novel method in the induction of reliable experimental acute and chronic ulcerative colitis in mice. Gastroenterology. 1990;98:694–702. doi: 10.1016/0016-5085(90)90290-h. [DOI] [PubMed] [Google Scholar]
- Reis BS, Rogoz A, Costa-Pinto FA, Taniuchi I, Mucida D. Mutual expression of the transcription factors Runx3 and ThPOK regulates intestinal CD4(+) T cell immunity. Nat Immunol. 2013;14:271–280. doi: 10.1038/ni.2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubtsov YP, Niec RE, Josefowicz S, Li L, Darce J, Mathis D, Benoist C, Rudensky AY. Stability of the regulatory T cell lineage in vivo. Science. 2010;329:1667–1671. doi: 10.1126/science.1191996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudd CE, Trevillyan JM, Dasgupta JD, Wong LL, Schlossman SF. The CD4 receptor is complexed in detergent lysates to a protein-tyrosine kinase (pp58) from human T lymphocytes. Proc Natl Acad Sci U S A. 1988;85:5190–5194. doi: 10.1073/pnas.85.14.5190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, Costantini F. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima M, Wakita D, Noguchi D, Chamoto K, Yue Z, Fugo K, Ishigame H, Iwakura Y, Kitamura H, Nishimura T. IL-6-dependent spontaneous proliferation is required for the induction of colitogenic IL-17-producing CD8+ T cells. J Exp Med. 2008;205:1019–1027. doi: 10.1084/jem.20071133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veillette A, Bookman MA, Horak EM, Bolen JB. The CD4 and CD8 T cell surface antigens are associated with the internal membrane tyrosine-protein kinase p56lck. Cell. 1988;55:301–308. doi: 10.1016/0092-8674(88)90053-0. [DOI] [PubMed] [Google Scholar]
- Verdaguer J, Schmidt D, Amrani A, Anderson B, Averill N, Santamaria P. Spontaneous autoimmune diabetes in monoclonal T cell nonobese diabetic mice. J Exp Med. 1997;186:1663–1676. doi: 10.1084/jem.186.10.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan YY, Flavell RA. Identifying Foxp3-expressing suppressor T cells with a bicistronic reporter. Proc Natl Acad Sci U S A. 2005;102:5126–5131. doi: 10.1073/pnas.0501701102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildin RS, Ramsdell F, Peake J, Faravelli F, Casanova JL, Buist N, Levy-Lahad E, Mazzella M, Goulet O, Perroni L, et al. X-linked neonatal diabetes mellitus, enteropathy and endocrinopathy syndrome is the human equivalent of mouse scurfy. Nat Genet. 2001;27:18–20. doi: 10.1038/83707. [DOI] [PubMed] [Google Scholar]
- Wu HJ, Ivanov II, Darce J, Hattori K, Shima T, Umesaki Y, Littman DR, Benoist C, Mathis D. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity. 2010;32:815–827. doi: 10.1016/j.immuni.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.