Abstract
Clostridium perfringens relies upon plasmid-encoded toxin genes to cause intestinal infections. These toxin genes are associated with insertion sequences that may facilitate their mobilization and transfer, giving rise to new toxin plasmids with common backbones. Most toxin plasmids carry a transfer of clostridial plasmids locus mediating conjugation, which likely explains the presence of similar toxin plasmids in otherwise unrelated C. perfringens strains. The association of many toxin genes with insertion sequences and conjugative plasmids provides virulence flexibility when causing intestinal infections. However, incompatibility issues apparently limit the number of toxin plasmids maintained by a single cell.
Keywords: plasmid-encoded toxin, clostridia, gastrointestinal disease, conjugation, plasmid evolution
1. Introduction to Clostridium perfringens
C. perfringens has a ubiquitous environmental distribution but also ranks amongst the most important pathogens of humans and domestic animals. The virulence of this bacterium is largely attributable to its ~17 toxin arsenal. However, individual strains produce only subsets of this toxin repertoire, which forms the basis for a toxinotyping classification scheme that consigns isolates to five types (A–E), based upon their production of four typing toxins (Table 1). In addition, several toxins not used for toxinotyping are important for pathogenicity, as will be discussed later [1, 2].
Table 1.
Classification of Clostridium perfringens based on the production of the four major typing toxins
| Type | Typing toxin produced: | |||
|---|---|---|---|---|
| Alpha | Beta | Epsilon | Iota | |
| A | + | − | − | − |
| B | + | + | + | − |
| C | + | + | − | − |
| D | + | − | + | − |
| E | + | − | − | + |
C. perfringens causes a panoply of illnesses ranging from histotoxic infections, such as clostridial myonecrosis (gas gangrene), to intestinal infections. The ability of C. perfringens to cause infections originating in the intestines is often dependent upon possession of toxin plasmids, which are the main focus of this review.
2. C. perfringens toxin plasmids and intestinal disease
When producing certain plasmid-encoded toxins, each C. perfringens type (and sometimes even specific subtypes) can cause intestinal infections, as shown in Table 2. These infections include enteritis and enterotoxemias, the latter characterized by toxins produced in the intestines, which then transit into the circulation to affect extra-intestinal organs. The ability of each C. perfringens type/subtype to cause intestinal diseases will now be briefly reviewed, along with a brief description of the plasmids relevant to those illnesses.
Table 2.
C. perfringens toxinotypes, plasmid-encoded toxins, and associated diseases
| Type | Toxin(s) | Human Disease(s) | Animal Disease(s) |
|---|---|---|---|
|
| |||
| A | CPE* | Human food poisoning; non-food- borne GI diseases | Possible enteritis in dogs, pigs, horses, and goats. |
| NetB | Not reported | Necrotizing enteritis in chickens | |
| CPB2 | Not reported | Possible enteritis in pigs; possible enterocolitis in horses | |
| BEC | Possible human food poisoning | Not reported | |
|
| |||
| B | Beta toxin, Epsilon toxin | Not reported | Necrotizing enteritis and enterotoxemia in sheep, cattle, and horses. Rare focal symmetrical encephalomalacia in sheep. |
|
| |||
| C | Beta toxin, CPE | Human enteritis necroticans | Necrotizing enteritis and enterotoxemia in pigs, sheep, cattle, horse, and other spp. (usually neonatal) |
|
| |||
| D | Epsilon toxin | Not reported | Enterotoxemia in sheep and goats; occasionally cattle and other species |
|
| |||
| E | Iota toxin | Not reported | Possible enteritis in rabbits, sheep and cattle |
2.1 Type A C. perfringens
2.1.1 C. perfringens enterotoxin (CPE) plasmids
Type A strains producing CPE are the second most common cause of bacterial food poisoning in the United States, with ~1,000,000 cases/yr at an estimated economic cost of >$300 million USD/yr [3, 4]. Additionally, CPE-producing type A strains are associated with 5–15% of nonfoodborne human intestinal diseases, including antibiotic-associated diarrhea (AAD) and sporadic diarrhea (SD) [5]. The enterotoxin gene (cpe) can be located chromosomally or on plasmids, with ~70% of food poisoning strains harboring a chromosomal copy of cpe, whereas the remaining ~30% of food poisoning strains, and virtually all AAD/SD strains, carry a plasmid-borne cpe gene [6, 7]. All of these strains cause disease when C. perfringens sporulates in the intestine and produces CPE (see below). During this in vivo sporulation, CPE accumulates in the cytoplasm and is finally released into the intestinal lumen when the mother cell lyses [6].
Substantial evidence supports CPE involvement in human intestinal disease. For example: 1) administration of CPE to human volunteers caused the classical diarrhea observed during natural disease [8]; 2) CPE is detectable in the feces of individuals with C. perfringens type A infection [9]; 3) CPE antisera can inhibit intestinal pathology in experimental animal models [10]; and 4) purified CPE damaged human ileal tissue ex vivo [11]. Perhaps the most persuasive evidence for the pathogenic role of CPE was provided by fulfilling molecular Koch’s postulates for strain SM101 (a type A, chromosomal cpe, food poisoning strain) and F4969 (a type A, plasmid cpe, SD strain), which showed that CPE is essential for these two strains to cause histological damage and fluid accumulation in rabbit ileal loops [12].
CPE, an ~35 kDa single polypeptide, consists of a C-terminal binding domain and an N-terminal domain that mediates oligomerization and membrane insertion [6]. CPE action starts when this toxin binds to claudins, including claudin-3, -4, -6, -7, -8, -14, on the apical surface of small intestinal or colonic cells [13–19]. This binding localizes CPE in a small ~90 kDa complex, which then oligomerizes [20] into an ~500 kDa hexameric prepore named CH-1 that forms on the plasma membrane surface [17, 21, 22]. The toxin then uses its amphipathic region named TM1 to insert into membranes and form a pore of 0.5 – 1.0 nm [23]. Both the small complex and CH-1 contain receptor and nonreceptor claudins, as well as CPE [17]. A secondary CPE large complex, named CH-2, can form that contains receptor and nonreceptor claudins, as well as another tight junction protein named occludin [17]. Formation of the CH-1 pore leads to an influx of Ca2+ into the cell and a K+ efflux. The Ca2+ influx activates calpain, which can lead to apoptosis (low toxin dose) or necrosis (high toxin dose) [24, 25]. During in vivo disease, CPE-induced cell death leads to the intestinal lesions that trigger fluid accumulation and diarrhea [10, 18]. Upon prolonged contact with the intestines, CPE can be absorbed into the circulation and cause enterotoxemia, affecting organs such as the liver or kidneys [26]. This enterotoxemia may explain fatalities that occurred during two food poisoning outbreaks in psychiatric hospitals [27, 28]. In mouse models of CPE enterotoxemia, this leads to increased serum K+ and hyperkalemia, which then causes cardiac arrhythmia and death [26].
During type A foodborne illness involving CPE, C. perfringens spores that survive the cooking process germinate in food, multiply and then are ingested [6]. Spore resistance against cooking and other stresses is influenced by which Ssp4 small acid-soluble protein variant is produced by the infecting strain [29, 30]. Foodborne strains carrying a chromosomal cpe gene typically make a Ssp4 variant that binds strongly to spore DNA and thus imparts exceptional heat and chemical resistance properties to C. perfringens spores, while strains carrying a plasmid-borne cpe gene produce a different Ssp4 variant that binds DNA less tightly, resulting in decreased spore resistance properties [29, 30]. These differences in spore resistance properties help to explain why the chromosomal cpe strains are more commonly implicated in food poisoning than the plasmid cpe strains.
Both chromosomal and plasmid-borne cpe genes are only expressed when C. perfringens sporulates; during disease, this sporulation occurs in the intestines. CPE production during sporulation is dependent upon three sporulation-specific sigma factors named SigF, SigE, and SigK. SigK and SigE bind to promoters upstream of cpe genes and positively-regulate toxin expression, while SigF indirectly controls CPE expression by controlling the production of SigE and SigK [31, 32]. CPE production and sporulation are also positively regulated by the Agr-like quorum sensing (QS) system [33] and the CcpA protein [34]. In contrast, the virX small RNA negatively regulates cpe expression during early sporulation [35].
Two cpe-carrying plasmids of type A strains were the first fully-sequenced C. perfringens toxin plasmids (Fig. 1B) [36]. pCPF4969, a plasmid of 70.5 kb from type A strain F4969, carries a functional copy of the cpe gene but lacks genes encoding any other toxins or putative toxins. In contrast, the cpe-encoding toxin plasmid pCPF5603 in type A SD strain F5603 is ~75.3 kb and also carries the cpb2 gene encoding the C. perfringens beta2-toxin (CPB2) toxin (Table 3). The plasmid-borne cpe gene in both strains is flanked by a 5′ IS1469, though the 3′ end of cpe can be flanked by either IS1151 (for pCPF5603) or IS1470 (for pCPF4969) [36].
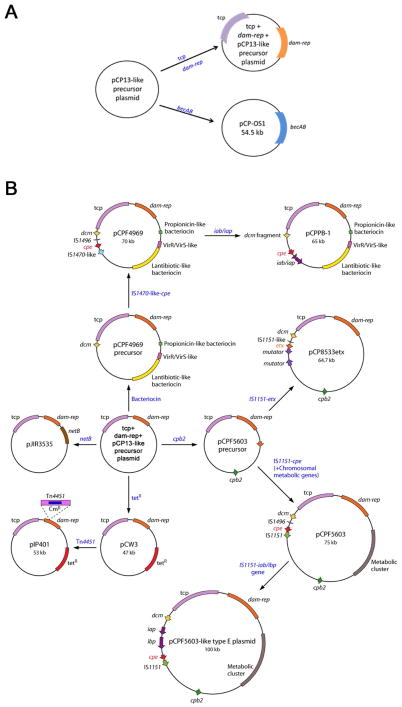
Figure 1. Comparison of C. perfringens plasmid diversity and organization.
The sequenced plasmids of C. perfringens are represented graphically. Panel A shows an aligned comparison of tcp-negative plasmids pCP13 (AP003515.1 [72]) and BEC-encoding plasmid pCP-OS1 (AP013033 [71]) demonstrating significant homology between these two plasmids. Panel B shows plasmids with a pCP13-like backbone that harbors the tcp locus. Depicted are plasmids: pCW3 (DQ366035 [129]); pJIR3844 (JN689217 [61]); pJIR3535 (JN689219 [61]); pCP8533etx (AB444205 [70]); pCPF5603 (AB236337 [36)]; pCPPB-1 (AB604032 [44]); pCPF4969 (AB236337 [36]). The conserved region of these C. perfringens plasmids is shown at the top of Panel B, with variable regions displayed graphically below. Arrows represent ORFs, and are colored as follows in Panel B: red arrows – tcp conjugation locus; dark blue arrows – conserved ORFs; yellow arrows – plasmid replication region; light blue arrows – ORFs unique to each plasmid; fuchsia arrows – tetracycline resistance genes; green arrows – cpb2; purple arrow – netB; pink arrow – etx; gray arrows – cpe; dark gray arrows – iap/ibp. Asterisks designate toxin genes. Modified with permission from [122].
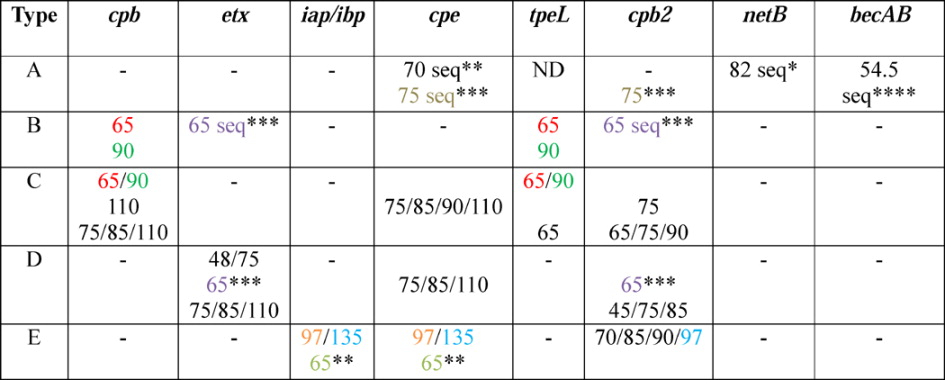
Table 3.
Size and diversity of C. perfringens toxin-encoding plasmids
|
Shared colors other than black indicate a similar/identical plasmid. Modified with permission from[122].
“Seq” indicates a sequenced plasmid; numbers are size in kb
indicates plasmid from pCW3-like family
indicates plasmid from pCPF4969 family
indicates plasmid from pCPF5603 family
indicates plasmid from pCP13-like family
Both pCP5603 and pCP4969 share nearly 50% homology. Of note, both plasmids carry a complete tcp (transfer clostridial plasmids) gene locus, which is known (as discussed later) to mediate conjugative plasmid transfer of other C. perfringens plasmids [36]. The presence of this tcp locus likely explains the demonstrated ability of pCPF4969 to conjugatively transfer and predicts that pCP5603 should also be conjugative [37].
Variable regions of these two prototype cpe+ type A plasmids include genes exclusively encoded on pCPF4969, i.e., genes encoding a putative bacteriocin, a biosynthetic operon for the production and secretion of peptide-based lantibiotics, and two component regulatory systems, including one resembling the VirS/VirR system. Conversely, pCPF5603 differs from pCPF4969 not only by encoding CPB2 but also carrying several genes encoding proteins involved in carbohydrate and lipid metabolism [36] (Fig. 1).
Additional studies [36] revealed that pCPF5603 and pCPF4969 represent the vast majority of cpe-encoding plasmids found in type A strains, particularly AAD and SD strains. However, some different cpe-encoding plasmids have been identified in type A soil strains, although they have not yet been sequenced [38]. CPE can also be encoded for by plasmids of types C, D, and E (to be discussed in later sections) [39–44].
2.1.2 NetB plasmids
Certain type A strains cause necrotic enteritis (NE), a debilitating intestinal disease that affects several poultry species. This disease has been demonstrated to involve the Necrotic Enteritis Beta-like toxin (NetB) (see below) [45, 46]. Clinically, birds with the acute or peracute form of the disease present with diarrhea, ruffled feathers, anorexia and depression; sudden death without clinical signs being observed can occasionally occur. Most peracute cases result in death within hours of the onset of symptoms, with flock mortality levels reaching 50%. Birds may develop subclinical disease which is characterized clinically by a drop in production with little or no diarrhea. Although mortality rates for this form of the disease are typically very low, feed conversion is negatively affected by the disease, resulting in significantly longer than normal grow-out periods. Pathologically, all three forms of NE are characterized by multifocal to coalescent intestinal necrosis, and are frequently covered by a pseudomembrane [47–49].
NetB has been implicated as the major toxin involved in avian NE through several lines of evidence. First, C. perfringens alpha toxin null mutants constructed in NE strains retained full virulence during experimental challenge [50]. Second, a netB null mutant failed to produce lesions during experimental challenge in poultry, whereas the wild-type parent strain produced disease in 45% of the challenged animals [45]. This effect was reversed by complementation of the netB gene. Finally, numerous studies have assayed strains from NE outbreaks, identifying 60–90% of outbreak strains as NetB-positive [51–54].
NetB is expressed as a 323 amino acid protein that is processed prior to secretion to remove a 30 amino acid signal sequence generating a mature 33 kDa protein [45]. The crystalline structure of NetB has been solved, identifying this protein as a member of the β-pore-forming toxin (PFT) family [55, 56]. Although complete structure-function analysis of NetB has not been performed, several site directed mutants generated in the predicted rim domain of the protein showed a reduced binding phenotype using chicken hepatocellular carcinoma cells [55].
As with all β-PFT’s, NetB-induced cell death is induced through the formation of unregulated ion channels in the membrane of susceptible cells; NetB pores have a pore diameter of ~1.4–1.6 nm. Formation of the pore begins with binding of the monomeric toxin to a currently unknown receptor. Binding is followed by oligomerization of the toxin into a prepore, a process linked to direct interactions of the toxin with cholesterol [55]. The prepore then inserts into the host cell membrane, likely utilizing an amphipathic domain identified in crystallization studies [55, 56]. The specific ions traversing the pore are not currently known, but initial experiments and model predictions suggest that the ion channel may be cation-selective [55, 56]. LD50 levels for NetB have not been determined; however, reports in the literature indicate doses as low as 2.5 μg/ml of NetB are capable of causing LMH cell rounding and lysis in vitro.
Studies have determined that netB expression is under the control of the VirS/VirR two component system. In this work, two VirR-binding boxes were identified directly upstream of the netB promoter. Testing of a virR null mutant demonstrated reduced NetB production as compared to wild type, which was restored by complementation [57]. Genes under control of VirS/VirR system are often also under control of the Agr-like QS system in C. perfringens [33, 58, 59]; however, it remains to be proven if the Agr system also regulates NetB production.
Type A necrotic enteritis strains typically carry 2 to 5 highly conserved, low copy number plasmids ranging in size from 50 to 100 kb in size [60]. The netB gene maps to a 42 kb pathogenicity locus called NEloc1 that is present on a plasmid of 80–85 kb in size, with the sequenced plasmid, pJIR3535, being 82 kb [60–62]. This plasmid is distinct from the plasmid carrying CPB2 in CPB2-positive type A avian necrotic enteritis strains, although there is a large common region. A 5.6 kb putative pathogenicity locus, named NEloc3, was also shown to be plasmid-borne, mapping to a second, 70 kb plasmid (Table 3). All of the sequenced large plasmids from type A NE isolates have been shown to carry the C. perfringens tcp locus (Fig. 1). Conjugative conversion of strains has been demonstrated experimentally using type A strain EHE-NE18 [61]. In this work, transfer of all three large plasmids present in EHE-NE18, including the plasmids harboring netB and tet(P) genes, to a recipient strain was demonstrated.
2.1.3 CPB2 plasmids
Many type A strains encode another toxin named beta-2 toxin (CPB2). CPB2 has also been found in types B, C, D and E C. perfringens isolates. CPB2 is expressed as a 31 kDa prototoxin that is subsequently cleaved during secretion into the mature 28 kDa toxin [63]. This toxin is active in vitro, causing cell rounding and death of both I407 and CHO cell lines at CPB2 concentrations >20 μg/ml [63]. It is not currently known how CPB2 causes cell death, but disruption of the cellular membrane or pore formation has been proposed as a possible explanation [64].
CPB2 has an unclear etiological role in disease since, to date, molecular Koch’s postulates have not been reported in the literature for this toxin. Indirect evidence supporting CPB2 having a role in disease comes mainly from the isolation of cpb2-positive strains from diseased animals. However, many normal flora isolates from healthy animals also carry this gene, making it challenging to draw conclusions about CPB2 contributions to disease. Gilbert et al.[63] did report that 3 μg of purified CPB2 delivered intravenously to mice was lethal. Additionally, one study found more pronounced disease from a cpa+/cpb2+ strain as compared to cpa+/cpe+ or cpa+ strains in a bovine ligated intestinal loop model, possibly suggesting CPB2 and CPA have synergistic effects in vivo [65, 66].
The regulation of CPB2 expression has been examined in C. perfringens strain 13. In this type A stain, CPB2 expression is under the control of the VirS/VirR two-component system. Furthermore, VirS/VirR regulation appears to be indirect, involving a sRNA named VR-RNA [67]. The Agr QS system was also shown to have regulatory effects on CPB2 expression in a type A strain, although not in two type B strains [33, 68].
In C. perfringens strains, CPB2 is encoded on plasmids ranging in size from 45–97 kb [40, 41, 43, 63, 69]. CPB2 maps to the same plasmid harboring epsilon toxin (ETX) in type B and D strains, and the same plasmid as CPE in some type A strains, but has not been found on the same plasmid as CPB in type C strains [40, 69, 70] (Table 3). Furthermore, the cpb2 gene has been mapped to plasmids independent of the iota toxin plasmids in type E strains [43]. Only some of the cpb2 plasmids in type E strains carry a tcp conjugation locus. In type A chicken necrotic enteritis strains, the cpb2 gene is located on a conjugative plasmid that is distinct from the plasmid that carries the netB gene [61].
2.1.4 BEC plasmids
Non-CPE-producing strains of type A C. perfringens were recently implicated in two food-borne gastroenteritis outbreaks in Japan [71]. Culture supernatants from those isolates were able to cause fluid accumulation in rabbit ileal loops. An enterotoxic protein was purified and its N-terminal region was sequenced. Genome sequencing revealed the presence of a binary toxin that matched the N-terminal sequence of the purified enterotoxic protein; this toxin was found to share approximately 43% identity to the binding and enzymatic components of iota toxin (discussed in 2.5.1). This novel toxin, named binary enterotoxin of C. perfringens (BEC), was found to ADP-ribosylate actin (similarly as other binary toxins of clostridial species) via the BECa subunit and also caused fluid accumulation in a suckling mouse model. Lastly, an isogenic becB (the gene encoding the BEC binding subunit) null mutant lost fluid-accumulating activity in the suckling mouse model, suggesting this binary toxin is the major mediator of gastrointestinal disease for the newly identified C. perfringens isolates [71].
BEC was found to be encoded on large plasmids of ~54 kb (Table 3). Sequencing showed these plasmids encode 39–55 potential ORFs, 16 of which have an assigned function. The BEC-encoding plasmids, pCP-OS1 and pCP-TS1, share significant homology with pCP13 over a span of ~38 kb and contain a number of partitioning and replication-associated genes common to pCP13 [72]. Like pCP13, these plasmids lack a tcp locus. Interestingly, and similarly as other plasmids of C. perfringens, these two plasmids encoded a putative transposon resolvase, which may play a role in transfer of the becAB genes to other plasmids and/or strains (Fig. 1) [71].
2.2 Toxin plasmids of type B C. perfringens
While also associated with hemorrhagic enteritis in goats, calves, and foals, type B strains are primarily known as the etiological agent of lamb dysentery [73]. Infection begins with transfer of the organism either directly from the dam or the environment to the lamb, usually within the first few days of life. Once in the intestine, the type B strain rapidly divides and produces toxins, resulting in enteritis with extensive necrosis and hemorrhage of the small intestine and enterotoxemia. Occasionally, focal symmetrical encephalomalacia is also present. In acute cases, clinical signs consist mainly of hemorrhagic diarrhea, abdominal pain, neurological signs, and as disease progresses, recumbency and death occur within 24 h of clinical signs onset. In peracute cases, no signs are observed and sudden death is the only indicator of disease. During type B outbreaks, infection rates can exceed 30%, with lethality rates approaching 100%. However, outbreaks of type B disease are rare and restricted to the United Kingdom, South Africa and the Middle East [73–76].
Both beta toxin (CPB) and epsilon toxin (ETX) appear to be important for the lethal enterotoxemias caused by type B strains [77]. This conclusion is based upon results where supernatants from 19 type B isolates were treated with or without trypsin (note that trypsin is necessary for ETX toxin activation) and in the presence or absence of toxin neutralizing antibodies, before those supernatants were injected intravenously into mice in a lethality model of type B enterotoxaemia. In this work, a positive correlation was noted between LD50 and CPB levels present in type B culture supernatants. Furthermore, a neutralizing MAb against CPB (but not ETX or CPA) reduced the lethality of non-trypsin treated culture supernatants. In contrast, antibodies against both CPB and ETX were necessary to prevent the lethality of trypsin-treated culture supernatants, despite the trypsin-labile nature of CPB [78].
2.2.1 CPB plasmids
CPB is the second most potent of the C. perfringens toxins, with an LD50 of 400 ng/kg body weight in mice [79]. This toxin is expressed as a 336 amino acid polypeptide containing a 27 amino acid leader sequence that is cleaved during secretion to generate a mature protein of ~35 kDa. The CPB structure has not been resolved, but this toxin is predicted to be a β-PFT based on sequence homology to other toxins with related amino acid sequences and known structures. Cell death caused by CPB occurs through the creation of unregulated ion channels in host cell plasma membranes. Pore formation begins with monomeric CPB binding to susceptible cells via an unknown receptor. During acute infection, CPB can bind to vascular endothelial cells in the intestine and it has been speculated, although not yet proven, that this causes thrombosis which might be responsible for the intestinal necrosis characteristic of the disease [80]; whether direct CPB damage to enterocytes is involved in disease is less clear. Once bound, CPB oligomerizes on the host cell surface into heptameric or hexameric prepores [81]. These prepores then rapidly insert into the plasma membrane using their amphipathic transmembrane domain, resulting in pore formation. The CPB channels have a pore size of approximately 12Å and allow the rapid efflux of K+ and influx of Na+, Ca2+ and Cl−, resulting in cellular swelling and lysis [81, 82]. A recent study suggested that CPB-induced cell death involves programmed necrosis [83].
Studies of CPB production regulation by type B strains are limited. Using AgrB mutants and complementing strains, the Agr QS system was shown to regulate CPB production by type B strains CN1793 and CN1795. The defect in CPB production by the AgrB mutants of type B strains involved a decrease in cpb transcription [68].
C. perfringens type B strains carry their CPB- and ETX-encoding genes on separate plasmids [69]. The cpb gene in type B strains is often carried on a plasmid of 90kDa, although a few isolates possess a 65 kDa cpb plasmid that is distinct from the 65 kDa plasmid carrying the etx gene in some type B strains (see below) (Table 3). Furthermore, the cpb plasmid can carry additional toxin genes encoding CPB2, the large clostridial cytotoxin (TpeL) or both of those toxins. The tcp locus is present on most or all cpb plasmids, indicating that these plasmids are likely to be conjugative. On type B plasmids, IS1551-like sequences are present upstream of the cpb gene; in addition, cpb-carrying circular DNA forms have been identified, indicating that the cpb gene can be excised from plasmids and suggesting that it may be carried on a transposable genetic element [69].
2.2.2 ETX plasmids
ETX, the most potent C. perfringens toxin, is also produced by type B (as well as type D) strains. ETX is secreted as a nearly-inactive, ~33 kDa prototoxin [84–86]. In the intestines, proteases such as trypsin and chymotrypsin remove the 13 N-terminal amino acids (trypsin) [84] and, as required for activation and cytotoxic activity, the 23-(trypsin) or 29-(chymotrypsin) C-terminal amino acid residues [85, 86]. This activation results in a nearly 1000-fold decrease in mouse LD50 [87] and increased cytotoxicity in cultured Madine-Darby Canine Kidney (MDCK) cells [88, 89]. ETX action is similar to that of other pore-forming toxins [90] in that the activated ETX binds to an unknown receptor on the surface of cells present in anatomical niches such as the intestines of goats, or the kidney and other organs of other mammals [1, 75, 76, 91]. Once bound, ETX oligomerizes into a surface prepore [89]. After insertion of transmembrane loops to form a pore, ion dysregulation causes eventual host cell death [92].
In contrast to the regulation of CPB production in type B strains, isogenic AgrB null mutants of two type B strains still produced wild-type levels of ETX, indicating the the Agr-like QS system does not regulate expression of all plasmid-encoded toxins in all strains of C. perfringens [68]. However, the Agr-like QS system does regulate ETX production in type D strains (to be discussed later) [93].
One etx-carrying plasmid, i.e., pCP8533etx from the type B strain CN8533, has been completely sequenced [70]. This etx plasmid is ~65 kb and also carries the cpb2 gene, like pCPF5603 (Table 3). In fact, pCP8533etx and pCPF5603 share approximately 80% of the same ORFs, with pCP8533etx lacking the ORFs on pCPF5603 discussed in 2.1.1 that encode for proteins involved in carbohydrate and lipid metabolism [36, 70] (Fig. 1).
Diversity amongst etx plasmids in other type B strains has been addressed by overlapping PCR and by pulsed-field gel electrophoresis (PFGE) with Southern blotting using an etx-specific probe. Those studies detected the presence of a similar, if not identical, ~65 kb etx plasmid in all surveyed type B isolates (Table 3) [69]. The plasmid pCP8533etx also carries the tcp gene locus required for conjugative transfer of C. perfringens plasmids, suggesting that type B etx-carrying plasmids, can undergo conjugative transfer between isolates, as proven for the etx plasmids of type D strains CN3718 and CN1020 [69, 70, 94]. Finally, an IS1151-like insertion sequence and a transposase gene are located adjacent to the etx gene, providing evidence that mobile DNA intermediates may have played a role in the evolution of etx plasmids of type B C. perfringens strains [69, 70].
2.3 Type C C. perfringens
2.3.1 CPB plasmids
C. perfringens type C infections, which occur in both humans and several animal species (horses, sheep, cattle, and pigs amongst others), manifest as necrotic enteritis that may be accompanied by enterotoxemia. The majority of type C disease is observed in neonatal animals, presenting in an acute or peracute form and characterized by severe abdominal pain, bloody diarrhea and depression. Occasionally, neurological signs are also observed. The rapidity of disease is likely due to the lack of competing flora and the trypsin-inhibiting effects of colostrum, which create an ideal environment for disease by protecting trypsin-labile CPB. Affected animals are colonized within a few hours of birth, likely from contact with contaminated fecal material shed by the dam. Outbreaks of the disease occur in unvaccinated herds, with rapid onset of signs and lethality rates in excess of 50%. In addition to affecting neonates, chronic disease is also seen in unvaccinated mature sheep and horses, where it presents as a chronic blood-free diarrhea that leads to dehydration [95].
In humans, type C disease presents as enteritis necroticans (EN), also referred to as Darmbrand or Pigbel, which is a severe intestinal infection marked by the presence of abdominal pain, bloody stool, vomiting and, in severe cases, a rapid toxemia causing death within 48 hours [96, 97]. The majority of EN cases happen in developing countries, where limited diets and consumption of staple foods rich in trypsin inhibitors, such as sweet potato, provide an ideal environment for epidemic disease, and support the importance of trypsin as a natural host defense against EN. EN occurs occasionally in developed countries, where it is generally restricted to people with pancreatic dysfunction, such as diabetics [98, 99].
Strong evidence supports the role of CPB as the major toxin for type C infections. Early studies demonstrated that neutralization of CPB in type C culture supernatants was necessary and sufficient to protect mice from lethal intravenous challenge with those samples [100]. Later studies utilizing highly purified CPB mixed with trypsin inhibitor reproduced the intestinal lesions typical of type C disease seen in rabbit intestinal loops. Further, pre-incubation of purified CPB with monoclonal antibodies specific for CPB blocked the pathological effects of the purified toxin [101, 102]. The most persuasive evidence supporting CPB’s role as the primary toxin in type C disease comes from fulfilling molecular Koch’s postulates. Isogenic toxin knockout mutants of strain CN3685, which produces chromosomally-encoded alpha toxin (CPA) and perfringolysin O (PFO) as well as CPB, demonstrated full pathogenicity of the cpa and pfo mutants, whereas the cpb KO mutant was completely attenuated unless the mutation was reversed [101]).
Several type C human EN strains produce CPE in addition to CPB [39, 103]. Recent work using sporulating culture lysates (SCLs) demonstrated that inactivating either CPB or CPE production by directed mutation rendered the SCLs of EN strain CN3758 unable to cause intestinal lesions in rabbit small intestinal loops, whereas SCLs of wild-type CN3758 produced fulminant disease. Quantification of toxin in wild-type CN3758 SCLs detected a relatively low presence of CPB and CPE, explaining why SCLs from mutants producing either CPE or CPB alone failed to cause disease. Consistent with that conclusion, challenge of rabbit loops with low doses of purified CPB or CPE mirrored the mutant SCL results, further demonstrating synergistic effects for these two toxins when present together in the small intestine at low concentrations [104].
The VirS/VirR system, which regulates the expression of CPA and PFO [105, 106], also modulates CPB expression in type C strains. Initial in vitro experiments demonstrated a rapid increase in CPB production in the presence of cultured Caco-2 cells [107]. Involvement of the VirS/VirR system was shown when an isogenic virR null mutant failed to increase CPB production in the presence of Caco-2 cells; complementation of virR rescued the mutant. Later work showed that, in contrast to the virulence of wild-type CN3685, an isogenic virR null mutant was attenuated in rabbit intestinal loops. Furthermore, Western blot analysis of intestinal fluid detected in vivo CPB production by the parent but not the mutant. Complementation completely restored the virR mutant’s virulence and wild-type CPB production levels, indicating that VirS/VirR is necessary for virulence because it controls in vivo CPB production. These strains were also tested for lethality in a mouse ID challenge model. Again, a significant reduction in mortality was observed in mice treated with the virR mutant compared to challenge with the wild-type strain [108]. In addition to the VirS/VirR system, type C strains also utilize the Agr-like QS system for virulence. Western blotting of intestinal fluid samples collected from rabbit small intestinal loops challenged with CN3685 or a agrB null mutant demonstrated that the Agr-like QS system is required for in vivo CPB expression by type C strains [59].
The cpb gene in type C strains is encoded on large plasmids ranging from 65–110 kb in size. Other toxins may be encoded on the same plasmid as CPB, such as TpeL or CPE, but these three toxin genes have not yet been found together on the same plasmid [39, 40]. To date, no plasmid in type C strains have been shown to encode both CPB and CPB2.
Interestingly, CPB and CPE are located on the same ~85 kb plasmid in some type C strains, but on different plasmids (110 kb and 65–75 kb for CPE and CPB respectively) in other strains [39] (Table 3). The presence of IS sequences flanking both the CPB and CPE toxin genes provides a potential mechanism for the generation of single plasmids harboring both toxins, where one toxin gene has been mobilized and inserted onto a plasmid already harboring the other toxin gene.
Like most other toxin plasmids, CPB plasmids of type C strains typically possess a tcp region, indicating their potential for conjugative transfer. Additionally, IS1151 sequences are associated with the cpb gene, suggesting that type C strains arise when the cpb gene inserts into a plasmid in a type A strain, converting it to type C [40]. In support of this theory, circular cpb containing transposon intermediates have been identified in CPB positive strains, demonstrating the potential mobility of the cpb gene [39].
2.4 Type D C. perfringens
2.4.1 ETX plasmids
Type D infections occur mostly in sheep and goats, with occasional cases observed in cattle and other animal species. The acute, sub-acute and chronic cases are clinically characterized by neurological and respiratory alterations, although hemorrhagic diarrhea can be observed in goats. Peracute cases can present with sudden death without clinical signs being observed in all species. Pathologically, the disease in all species is mainly characterized by pulmonary and cerebral edema, the latter being observed mostly in a perivascular location which is considered to be a rather specific diagnostic feature. In goats, necrotizing and hemorrhagic enterocolitis is characteristic of the sub-acute and chronic form of the disease [75, 76, 90].
ETX, described in 2.2.2, is the major toxin required for the virulence of type D strains of C. perfringens when mice, goats, or sheep are challenged intraduodenally with washed type D cells [91]. It has been proposed that the Agr system signals through the VirS/VirR two-component regulatory system. However, while ETX expression was found to be positively regulated by the Agr-like QS system in type D strain CN3718, inactivating VirS/VirR in this strain had no effect on ETX production levels, indicating that the Agr-like QS system can sometimes regulate gene expression independently of VirS/VirR [93]. In addition to Agr, the regulator CodY also positively regulates ETX production by binding to sequences directly upstream of the etx gene in strain CN3718 [109].
In contrast to the single etx plasmid found in type B strains, the etx plasmids of type D strains show considerable variability, ranging in size from ~45 kb to ~110 kb, based upon PFGE Southern blotting with an etx-specific probe [41]. The diversity of etx plasmid size in type D strains correlates with carriage of cpe and/or cpb2 genes, i.e., etx is typically carried on plasmids of ~48 kb (though rarely 75 kb) in cpe−/cpb2− strains, but etx is carried on larger plasmids of either 75 or 110 kb in cpe+/cpb2+ strains (Table 3). Most etx plasmids of type D strains carry a functional tcp conjugative transfer locus [41, 94], and conjugative transfer of etx plasmids has been demonstrated for type D strains CN1020 and CN3718 [94]. Interestingly, a few type D strains carry the same ~65 kb etx and cpb2 plasmid that is also found in most or all type B strains of C. perfringens [41, 69, 70] (Table 3).
2.5 Type E C. perfringens
2.5.1 Iota toxin plasmids
Iota toxin (ITX) is a typing toxin produced only by type E strains [1, 110], which have been implicated in enteritis in rabbits, lambs, and cattle. However, the role of ITX in these type E-associated diseases has not yet been carefully examined [1]. Nor has the regulation of ITX production been explored [43, 44].
ITX is a member of the binary toxin family that also includes BEC of C. perfringens, CDT of C. difficile, C. botulinum C2 toxin, and C. spiroforme toxin CST [110, 111]. ITX is comprised of an enzymatic subunit (Ia) and a binding component (Ib) [110, 111]. The itx genes are transcribed in an operon. The regulation of ITX production remains unexplored [43, 44]. ITX has cytotoxic activity in numerous cell culture models [110, 112, 113]. The action of this toxin begins when Ia and Ib propeptides are activated by proteases such as alpha-chymotrypsin, pepsin, proteinase K, subtilisin, and thermolysin [114]. The activated Ib toxin subunit then binds to a surface-localized receptor named lipolysis-stimulated lipoprotein receptor [115] and, possibly, to the mammalian protein CD44 [116]. Once bound to its receptor, ITX-Ib oligomerizes to form a heptamer, which then binds the Ia enzymatic subunit [117–119]. The holotoxin is endocytosed [119, 120] and, after escaping into the cytoplasm, Ia then ADP-ribosylates actin, leading to a disassembly of the cellular cytoskeleton and cell death [112, 113].
The ITX plasmids of type E strains have been well characterized, with two major kinds of ITX toxin plasmids identified in these strains. These include plasmids of either 97 or 135 kb that encode ITX (iap and ibp), as well as urease and lambda-protease. This 97/135 kb ITX plasmid family also encodes silent cpe sequences (Table 3) [43, 44]. The backbone of these plasmids can resemble pCPF5603, where an IS1151-like insertion sequence is located immediately adjacent to the ITX encoding genes and the silent cpe sequences [43, 44, 121]. This may explain the evolution of these ITX plasmids, as discussed later. The ITX plasmids present in these type E plasmids carry a tcp locus, which suggests they can transfer horizontally [43, 44].
A more recent study identified type E strains carrying an ~65 kb plasmid (named pCPPB-1) that carries a variant cpe locus and a variant iap/ibp operon. This ITX plasmid also possesses a tcp locus and is related to the pCFP4969 cpe plasmid (Table 3). For example, pCPPB-1 shares several features of pCPF4969 including encoding a VirR/VirS-like two-component regulatory system, a bacteriocin, and enzymes involved in the synthesis and secretion of lantibiotics (Fig. 1). Several isolates carrying pCPPB-1 were found in the feces of healthy individuals or the environment, though a clear role for the variant CPE or ITX in the pathogenesis of type E strains has not been identified [36, 43, 44, 121].
Type E strains carrying silent cpe sequences also commonly possess a second plasmid of 75–97 kb that carries the cpb2 gene. This plasmid also carries IS1151 sequences, but does not always carry the tcp locus needed for conjugative transfer [43].
3. C. perfringens plasmid biology
3.1 Conjugation of C. perfringens toxin plasmids
Conjugative transfer has been demonstrated for several C. perfringens toxin and antibiotic resistance plasmids [37, 61, 94]. All known conjugative plasmids of C. perfringens have a novel conjugation region called the tcp locus [122]. Using the paradigm conjugative C. perfringens plasmid pCW3, a 47 kb tetracycline resistance plasmid [61], mutagenesis studies demonstrated that the tcp locus is essential for conjugative transfer [61, 123–125]. A model for the C. perfringens conjugation system has been proposed based on functional studies of several Tcp proteins [122, 126]. The presence of the tcp genes on most studied C. perfringens toxin plasmids suggests that this model represents a conserved mechanism of transfer that contributes to the spread of toxin genes and resistance determinants in C. perfringens [122].
Conjugative plasmid transfer involves a type IV secretion system (T4SS) that has recently been structurally resolved for Gram-negative systems [127]. Recent studies have classified the T4SS, or mating pair formation complex, encoded by the tcp locus of pCW3 as belonging to the MPFFA class, which includes Tn916 and ICEBs1 from Bacillus subtilis [128]. This classification is consistent with the original finding of similarity between Tcp proteins and products of the conjugation region of Tn916 [129]. The Gram-positive T4SS are predicted to be minimized systems and, unsurprisingly, lack homologs for proteins that form the outer membrane core complex in Gram-negatives [130, 131]. Conserved protein families have been identified in both the Gram-negative and Gram-positive systems, including the pCW3 system (Fig. 2) [128, 130, 131].
Figure 2. The genetic organization of the pCW3 tcp locus.
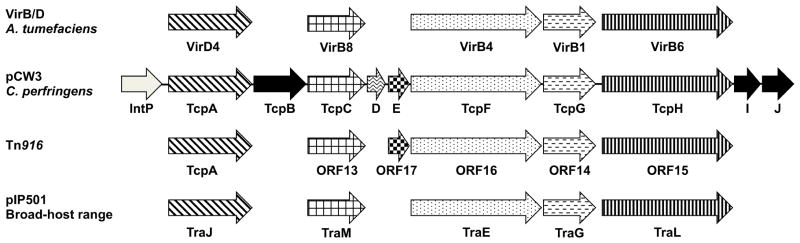
Proteins encoded by the various genes are stated below the arrows. The patterned arrows indicate Tcp proteins involved in pCW3 conjugative transfer, black arrows depict non-essential Tcp proteins and the grey arrow the IntP protein, which is currently uncharacterized. Proteins with sequence, functional or structural similarity from the paradigm VirB/D system from the Ti plasmid from the Gram-negative Agrobacterium tumefaciciens, the conjugation region from Tn916 and the broad-host range plasmid pIP501 from the Gram-positive Streptococcus agalactiae are represented by arrows with similar patterns. Each of these conjugation regions has homologs of the putative coupling proteins, VirD4 and TcpA, VirB8-like proteins, VirB4-like ATPases, VirB1-like lytic transglycolases and VirB6-like proteins. Based on data from [128, 129, 135].
Domains from proteins that form the inner membrane complex in Gram-negative T4SS, i.e., VirB6 and VirB8, were identified in TcpH and TcpC, respectively, suggesting that they form the core of the T4SS in C. perfringens [125, 129, 132]. Essential for pCW3 transfer, TcpH was identified as an integral membrane protein with eight putative transmembrane domains (TMDs), which localizes TcpH to the cell envelope at the poles of C. perfringens donor cells [129]. Based on its similarity to VirB6 proteins, TcpH is postulated to play a similar core role in T4SS assembly and stabilization [132]. The VirB6 domain, the N-terminal domain and the 242VQQPW246 conserved motif were shown to be essential for TcpH function [132]. The N-terminal domain mediates TcpH interactions with itself, TcpA and TcpC, while the VirB6 domain is crucial for interaction with the other postulated core component, TcpC [126, 132].
TcpC was identified as a 359 amino acid biotopic membrane protein that is required for efficient transfer of pCW3 and which is localized to the cell envelope by two essential N-terminal TMDs [125]. Structural resolution of the stable TcpC99–359 derivative lacking these TMDs identified two linked structural domains that each had an unexpectedly similar fold to biotopic VirB8-like proteins. TcpC is the first VirB8-like protein to have two domains with a VirB8 fold and represents a novel class of this family of proteins [133]. TcpC was shown to interact with itself, TcpA, TcpG and TcpH, which is consistent with its postulated role as an assembly and scaffolding protein, similar to other VirB8-like proteins [125, 126]. The TMDs were shown to be essential for TcpC interactions, probably due to their role in localization and oligomerization [125]. The central domain, which is buried within the trimeric TcpC structure, was involved in TcpC self-interaction and interactions with TcpG. Deletion of the C-terminal domain completely abolished interactions with TcpA, TcpG and TcpH, a result consistent with its localization on the external surface of the TcpC trimer.
TcpF has a putative VirB4-like ATPase domain and therefore is related to a protein family that is a signature of all T4SS [129]. TcpF was shown to be essential for conjugative transfer of pCW3 and is predicted to energize the C. perfringens conjugation system. Immunofluorescence studies showed that TcpF co-localizes with TcpH at the poles of C. perfringens donor cells, suggesting that it forms part of the pCW3 T4SS, although no protein-protein interactions have been identified between TcpF and the other Tcp proteins [132].
Two gene products encoded by the tcp locus, TcpG and TcpI, were identified as putative peptidoglycan hydrolases [129], which are postulated to be important for the assembly of the T4SS in the Gram-positive cell envelope [134]. TcpG is required for efficient conjugative transfer of pCW3, but TcpI is not required [123]. TcpG was shown to have peptidoglycan hydrolyzing activity on purified peptidoglycan from C. perfringens and has two functional catalytic domains that are required for activity. Interactions between TcpG, TcpC and TcpA are postulated to direct the localized assembly of the transfer apparatus in C. perfringens donor cells [123, 126].
Three hypothetical proteins are encoded on the tcp locus: TcpD, TcpE and TcpJ [129]. The only homology identified was for TcpE, which has 27% sequence identity to ORF17-like proteins of unknown function that are only present in the MPFFA class of T4SS [128, 129]. TcpD and TcpE are essential for pCW3 transfer, whereas TcpJ is not required (J.A. Wisniewski, W.L. Teng, T.L. Bannam and J.I. Rood, unpublished). The functional role of these novel proteins in C. perfringens conjugative transfer remains to be determined.
A family of single-stranded DNA translocases known as type IV coupling proteins (T4CP) are associated with T4SS systems that have the ability to transfer DNA [135]. The T4CPs are DNA-dependent ATPases that link the T4SS system with its nucleoprotein substrate [136]. TcpA was postulated to be the DNA translocase of the pCW3 conjugation system based on the presence of an FtsK-like domain similar to that present in the FtsK/SpoIIIE family of double-stranded DNA translocases [124]. Homologs of TcpA have been identified in other systems from the MPFFA class, which all lack the classic VirD4-like T4CP, supporting the hypothesis that these systems have acquired a dsDNA translocase to drive DNA transfer [135]. TcpA is essential for conjugative transfer of pCW3, with the ATP-binding motifs in the FtsK-like domain essential for TcpA function, indicative of potential ATPase activity [124]. The FtsK domain was also important for TcpA self-interaction, as well as interactions with components of the T4SS, specifically TcpC, TcpG and TcpH [126]. Two N-terminal TMDs were necessary for wild-type TcpA function since their deletion resulted in a reduced transfer frequency that may be explained by a loss of TcpA oligomerisation and an inability to interact with TcpC and TcpG [124, 126].
Conjugative plasmids are transferred as a nucleoprotein complex from the donor cell to a recipient cell [137]. Prior to transfer, a strand of the plasmid at the oriT site is cleaved and subsequently bound by a relaxase protein, a family of site- and strand-specific transferases that possess at least one nucleophilic tyrosine residue [138]. No relaxase-encoding gene or oriT site has been identified on pCW3 or any of the other conjugative toxin plasmids [129, 138]. However, the first gene in the tcp operon is a potential tyrosine recombinase, IntP, which is postulated to act as an atypical relaxase in the pCW3 conjugation system [122].
3.2 Plasmid compatibility in C. perfringens
It is well established that many C. perfringens isolates carry more than one toxin plasmid and that these plasmids are very closely related (see earlier), sharing up to 40 kb of almost identical sequences [122]. These shared regions include the tcp conjugation locus and genes involved in plasmid replication and maintenance. For example, two separate studies have shown that individual NetB toxin-producing C. perfringens type A strains can carry at least three separate, closely related conjugative plasmids, with one plasmid encoding NetB toxin, another encoding CPB2-toxin and the third a tetracycline resistance determinant [61, 62]. Other studies have shown that C. perfringens type B, C and D isolates can also carry multiple toxin plasmids that are closely related as already discussed [40, 41, 69].
How are such closely related plasmids stably maintained in the same cell? Examination of the plasmid replication and maintenance region of the tetracycline resistance plasmid pCW3 [129] revealed the presence of a parMRC locus that appears to encode a classical type II actin-like plasmid partitioning system [139, 140]. Subsequent studies have shown that the sequence of this locus in different C. perfringens plasmids varies subtly, with individual plasmids that are in the same isolate having slightly different parMRC sequences [61, 62]. Based on these observations and bioinformatic analysis, it was further proposed that these differences could account for the coexistence of these plasmids in the same cell and it was postulated that the toxin and resistance plasmids could be divided into four separate parMRC incompatibility groups (now designated as ParMRCA to D; J. Rood, V. Adams & J. Prescott, unpublished), with no more than one member of an incompatibility group being found in any one strain [62, 122, 141]. Subsequent studies have revealed the presence of a fifth C. perfringens incompatibility group, ParMRCE [141]. Further experimental studies are required to prove that the observed variation at this locus is responsible for the coexistence of more than one toxin or resistance plasmid in the same C. perfringens strain.
3.3 Plasmid replication of C. perfringens plasmids
Plasmid replication generally involves Rep proteins that bind to specific plasmid-specific DNA sequences and assist in initiation of plasmid replication [142]. However, initial bioinformatic analyses of C. perfringens plasmid-encoded proteins, including their amino acid identity and predicted domain architecture, failed to identify a Rep protein homologue [129]. In order to identify a Rep protein for these plasmids, portions of pCW3 were subcloned and assessed for their ability to replicate independently. This analysis revealed an ~4 kb fragment that afforded a plasmid the ability to independently replicate in C. perfringens. Transposon mutagenesis of this region was later performed, revealing that insertions mapping to a specific gene abrogated the ability of that plasmid to replicate in C. perfringens [129]. This 831 bp ORF (now called rep) encodes a Rep protein that is present on 95–100% of all characterized C. perfringens toxin and resistance plasmids, indicating that the mechanism of replication of these C. perfringens plasmids is likely to be identical [122, 129]. This Rep protein has a predicted pI of 10, consistent with this being a DNA-binding protein [122, 129, 142]. The Rep protein of C. perfringens plasmids does not share similarity, motifs or domains with Rep proteins of other species, which probably explains why the C. perfringens plasmids have not been observed in other clostridial species or bacterial genera [143, 144].
3.4 Evolution and diversity of C. perfringens toxin plasmids
As mentioned above, all C. perfringens toxin plasmids share sequences with pCP13 [72]. Most of these plasmids also contain the tcp locus that mediates conjugative transfer between two isolates, as well as a common dam-rep region that is required for plasmid replication [36, 41, 43, 44, 69, 70, 121]. Given the nature of plasmid carriage between C. perfringens types and strains, a number of events apparently led to the evolution and diversity of the toxin plasmids characterized to this date.
Of likely significance for C. perfringens toxin plasmid evolution is the close association between most toxin genes and insertion sequences, as described in Section 2 of this Review. While insertion sequence-directed movement of toxin genes between plasmids has not yet been formally demonstrated, this possibility is supported by the detection, using PCR-based approaches, of several excised circular intermediates carrying toxin genes [39, 41, 43, 69, 122]. As mentioned earlier, circular intermediates have been detected that carry the cpe gene in type A strains [36, 121], cpb-tpeL genes in type B strains [69], cpb and cpe genes in type C strains [39], cpe and etx genes in type D strains [41], and the iap/ibp genes in type E strains [43]. These circular intermediates may represent transposon intermediates capable of integration into C. perfringens DNA, particularly into plasmid backbones. Many plasmid-borne toxin genes are present adjacent to the dcm gene, which may represent a preferential location on plasmids for insertion of mobile genetic elements carrying toxin genes [36, 41, 43, 44, 69, 70, 121]. If so, this would help to explain why the plasmid-encoded toxin genes are not commonly found on the chromosome.
Based upon the characterization and sequencing of several C. perfringens plasmids, a model for evolution of these plasmids can be proposed (Fig. 3). This model entails a common precursor plasmid resembling pCP13 that gave rise to a variety of C. perfringens plasmids, including both the characterized toxin and antibiotic resistance plasmids. By homologous recombination or another mechanism, pCP13-like precursor plasmids acquired either the becAB locus or the tcp locus (Fig. 3A) [122]. At least once, the tcp-carrying plasmid then acquired the netB gene adjacent to the dam-rep region, producing plasmid pJIR3535 [61]. A similar type of event may have occurred to create pCW3, which is related to pJIR3535, but instead of netB possesses a tet(P) operon that encodes tetracycline resistance (Figs. 1 and 3B) [129].
Figure 3. Model for evolution of the C. perfringens toxin plasmids.
A model of evolution for sequenced and characterized C. perfringens toxin plasmids is shown. A) A pCP13-like plasmid acquires the becAB locus (pCP-OS1 and pCP-TS1 [71]) or the tcp locus (pCP13 [72]). B) The further evolution of pCW3 [129], pJIR3535 [61], pCP8533etx [70], pCPF5603 [36], pCPPB-1 [44], and pCPF4969 [36] are diagrammed. See section 3.3 for a discussion of evolution of these plasmids. Note that important plasmid regions are color coded. Modified with permission from [122].
To form the pCPF5603 family of toxin plasmids, the tcp-carrying, pCP13-related precursor plasmid likely obtained the gene containing cpb2 gene and a metabolic gene cluster via homologous recombination or another mechanism. Those acquisitions formed a pCPF5603 precursor that, via a transposition intermediate, then gained the cpe gene to create pCPF5603 [36, 69]. A pCPF5603 plasmid may later have obtained the iap/ibp genes via transposition to form the pCPF5603-like plasmid of type E strains [36, 43, 44]. This genetic element apparently inserted into the promoter region of the cpe gene, silencing that gene [43]. Alternatively, another pCPF5603 precursor plasmid appears to have obtained etx via a transposition intermediate containing an IS1511-like insertion sequence to form pCP8533etx (Fig. 3B) [43, 69].
To create the pCPF4969 plasmid family, the tcp-carrying, pCP13-related precursor plasmid may have initially gained loci encoding a peptide bacteriocin or a lantibiotic-like bacteriocin gene cluster, as well as a VirS/VirR-like two-component regulatory system. Via a IS1470-like transposition intermediate, this pCPF4969 precursor then acquired a cpe gene to form pCPF4969 [36]. At least once, this pCPF4969 plasmid picked-up a functional iap/ibp gene locus, to form pCPPB-1, which possesses both functional cpe and iap/ibp genes (Fig. 3B) [36, 44].
Summarizing, many toxin plasmids of C. perfringens, including those belonging to the pCPF4969, pCPF5603, or pCW3 plasmid families, are hypothesized to have evolved from a common pCP13-like precursor plasmid [36, 41, 43, 44, 69, 70, 121, 129]. This model offers potential understanding of the origin and evolution of these mobile genetic elements carrying the toxins that impart virulence plasticity to C. perfringens types and strains.
4. Conclusions
Toxin-encoding plasmids often play an essential virulence role when C. perfringens causes the intestinal infections that are major problems in humans and livestock. Toxin plasmids characterized to date fall within four families, i.e., the non tcp-carrying pCP13-like BEC plasmid, pCW3-like plasmids, pCPF4969-like plasmids and pCPF5603-like plasmids. Except for the BEC-encoding plasmid, all of the toxin plasmids share a conserved region carrying, in part, the tcp locus that mediates conjugative transfer. Conjugative transfer likely explains the presence of toxin genes on different plasmids amongst genetically-variable strains of this bacterium. It also favors accumulation of toxin plasmids, as evident from identification of C. perfringens strains carrying three distinct toxin plasmids [122].
The association of toxin genes with conjugative plasmids may also directly contribute to disease. C. perfringens is a common component of the normal microbiota in the intestines. Those normal flora strains are generally type A strains that do not produce toxins with a proven involvement in intestinal disease, but are presumably proficient at intestinal colonization. Therefore, when strains carrying toxin genes important for intestinal disease are introduced into the intestines, the presence of these toxin genes on conjugative plasmids capable of high frequency transfer could result in conversion of the colonization-proficient normal microbiota strains into virulent strains capable of causing gastrointestinal disease.
Plasmid-borne toxin genes are also often closely associated with insertion sequences, which can excise these toxin genes from plasmid backbones. This close association between many toxin genes and insertion sequences likely contributes to the virulence flexibility of C. perfringens. However, incompatibility issues place some limitation on the total number of toxin plasmids that can be accumulated by one strain. Again, the common association of toxin plasmids with insertion sequences may provide a potential mechanism to overcome this barrier as it could explain why single plasmids carrying four different toxin genes have been observed.
Further study of C. perfringens toxin plasmids is essential. For example, additional toxin plasmids exist that do not fall into the four known toxin plasmid families and those plasmids should be characterized. A greater understanding of plasmid incompatibility mechanisms is needed. Lastly, attempts should be made to demonstrate the insertion into plasmids of mobile genetic elements carrying insertion sequences and toxin genes.
Acknowledgments
This work was supported in part by National Institutes of Health (NIH) grants: R37AI19844-30 to B.A.M; R01AI056177-09 to B.A.M, J.I.R., and F.A.U.; a project in Middle Atlantic Regional Centers of Excellence-2 grant 2U54AI57168-09 (M. Levine, overall principal investigator) awarded to B.A.M.; and T32 AI060525 (J. Flynn, PI) to J.C.F. and J.R.T. Work at Monash University (J.I.R.) was supported by the Australian Research Council (ARC) through funding of the ARC Centre of Excellence in Structural and Functional Microbial Genomics and by a grant from the Australian National Health and Medical Research Council. J.A.W. was supported by the provision of an Australian Postgraduate Award.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Uzal FA, Vidal JE, McClane BA, Gurjar AA. Toxins involved in mammalian veterinary diseases. Open Toxinology J. 2010;2:24–42. [PMC free article] [PubMed] [Google Scholar]
- 2.Uzal FA, Freedman JC, Shrestha A, Theoret JR, Garcia J, Awad MM, et al. Towards an understanding of the role of Clostridium perfringens toxins in human and animal disease. Future Microbiol. 2014;9:361–77. doi: 10.2217/fmb.13.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson M, Roy S, et al. Foodborne illness acquired in the United States-major pathogens. Emerg Infect Dis. 2011;17:7–15. doi: 10.3201/eid1701.P11101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Batz MB, Hoffmann S, Morris JG., Jr Ranking the disease burden of 14 pathogens in food sources in the United States using attribution data from outbreak investigations and expert elicitation. J Food Prot. 2012;75:1278–91. doi: 10.4315/0362-028X.JFP-11-418. [DOI] [PubMed] [Google Scholar]
- 5.Carman RJ. Clostridium perfringens in spontaneous and antibiotic-associated diarrhoea of man and other animals. Rev Med Microbiol. 1997;8(supplement 1):S43–S45. [Google Scholar]
- 6.McClane BA, Robertson SL, Li J. Clostridium perfringens. In: Doyle MP, Buchanan RL, editors. Food Microbiology: Fundamentals and Frontiers. ASM press; Washington D.C: 2013. pp. 465–89. [Google Scholar]
- 7.Grant K, Kenyon S, Nwafor I, Plowman J, Ohai C, Halford-Maw R, et al. The identification and characterization of Clostridium perfringens by real-time PCR, location of enterotoxin gene, and heat resistance. Foodborne Pathog Dis. 2008;5:629–39. doi: 10.1089/fpd.2007.0066. [DOI] [PubMed] [Google Scholar]
- 8.Skjelkvale R, Uemura T. Experimental diarrhea in human volunteers following oral administration of Clostridium perfringens enterotoxin. J Appl Bacteriol. 1977;46:281–86. doi: 10.1111/j.1365-2672.1977.tb00752.x. [DOI] [PubMed] [Google Scholar]
- 9.Birkhead G, Vogt RL, Heun EM, Snyder JT, McClane BA. Characterization of an outbreak of Clostridium perfringens food poisoning by quantitative fecal culture and fecal enterotoxin measurement. J Clin Microbiol. 1988;26:471–74. doi: 10.1128/jcm.26.3.471-474.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garcia JP, Li J, Shrestha A, Freedman JC, Beingesser J, McClane BA, et al. Clostridium perfringens type A enterotoxin damages the rabbit colon. Infect Immun. 2014;82:2211–8. doi: 10.1128/IAI.01659-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fernandez Miyakawa ME, Pistone Creydt V, Uzal FA, McClane BA, Ibarra C. Clostridium perfringens enterotoxin damages the human intestine in vitro. Infect Immun. 2005;73:8407–10. doi: 10.1128/IAI.73.12.8407-8410.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sarker MR, Carman RJ, McClane BA. Inactivation of the gene (cpe) encoding Clostridium perfringens enterotoxin eliminates the ability of two cpe-positive C. perfringens type A human gastrointestinal disease isolates to affect rabbit ileal loops. Mol Microbiol. 1999;33:946–58. doi: 10.1046/j.1365-2958.1999.01534.x. [DOI] [PubMed] [Google Scholar]
- 13.Fujita K, Katahira J, Horiguchi Y, Sonoda N, Furuse M, Tskuita S. Clostridium perfringens enterotoxin binds to the second extracellular loop of claudin-3, a tight junction membrane protein. FEBS Lett. 2000;476:258–61. doi: 10.1016/s0014-5793(00)01744-0. [DOI] [PubMed] [Google Scholar]
- 14.Katahira J, Inoue N, Horiguchi Y, Matsuda M, Sugimoto N. Molecular cloning and functional characterization of the receptor for Clostridium perfringens enterotoxin. J Cell Biol. 1997;136:1239–47. doi: 10.1083/jcb.136.6.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Katahira J, Sugiyama H, Inoue N, Horiguchi Y, Matsuda M, Sugimoto N. Clostridium perfringens enterotoxin utilizes two structurally related membrane proteins as functional receptors in vivo. J Biol Chem. 1997;272:26652–58. doi: 10.1074/jbc.272.42.26652. [DOI] [PubMed] [Google Scholar]
- 16.Robertson S, Smedley JG, III, McClane BA. Identification of a claudin-4 residue important for mediating the host cell binding and action of Clostridium perfringens enterotoxin. Infect Immun. 2010;78:505–17. doi: 10.1128/IAI.00778-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Robertson SL, Smedley JG, 3rd, Singh U, Chakrabarti G, Van Itallie CM, Anderson JM, et al. Compositional and stoichiometric analysis of Clostridium perfringens enterotoxin complexes in Caco-2 cells and claudin 4 fibroblast transfectants. Cell Microbiol. 2007;9:2734–55. doi: 10.1111/j.1462-5822.2007.00994.x. [DOI] [PubMed] [Google Scholar]
- 18.Shrestha A, McClane BA. Human claudin-8 and -14 are receptors capable of conveying the cytotoxic effects of Clostridium perfringens enterotoxin. mBio. 2013:4. doi: 10.1128/mBio.00594-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Veshnyakova A, Piontek J, Protze J, Waziri N, Heise I, Krause G. Mechanism of Clostridium perfringens enterotoxin interaction with claudin-3/-4 protein suggests structural modifications of the toxin to target specific claudins. J Biol Chem. 2012;287:1698–708. doi: 10.1074/jbc.M111.312165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wieckowski EU, Wnek AP, McClane BA. Evidence that an ~50kDa mammalian plasma membrane protein with receptor-like properties mediates the amphiphilicity of specifically-bound Clostridium perfringens enterotoxin. J Biol Chem. 1994;269:10838–48. [PubMed] [Google Scholar]
- 21.Singh U, Van Itallie CM, Mitic LL, Anderson JM, McClane BA. CaCo-2 cells treated with Clostridium perfringens enterotoxin form multiple large complex species, one of which contains the tight junction protein occludin. J Biol Chem. 2000;275:18407–17. doi: 10.1074/jbc.M001530200. [DOI] [PubMed] [Google Scholar]
- 22.Smedley JG, 3rd, Uzal FA, McClane BA. Identification of a prepore large-complex stage in the mechanism of action of Clostridium perfringens enterotoxin. Infect Immun. 2007;75:2381–90. doi: 10.1128/IAI.01737-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen J, Theoret JR, Shrestha A, Smedley JG, 3rd, McClane BA. Cysteine scanning mutagenesis supports the importance of Clostridium perfringens enterotoxin amino acids 80–106 for membrane insertion and pore formation. Infect Immun. 2012;80:4078–88. doi: 10.1128/IAI.00069-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chakrabarti G, McClane BA. The importance of calcium influx, calpain, and calmodulin for the activation of CaCo-2 cell death pathways by Clostridium perfringens enterotoxin. Cell Microbiol. 2005;7:129–46. doi: 10.1111/j.1462-5822.2004.00442.x. [DOI] [PubMed] [Google Scholar]
- 25.Chakrabarti G, Zhou X, McClane BA. Death pathways activated in CaCo-2 cells by Clostridium perfringens enterotoxin. Infect Immun. 2003;71:4260–70. doi: 10.1128/IAI.71.8.4260-4270.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Caserta JARS, Saputo J, Shrestha A, McClane BA, Uzal FA. Development and application of a mouse intestinal loop model to study the in vivo action of Clostridium perfringens enterotoxin. Infect Immun. 2011;79:3020–7. doi: 10.1128/IAI.01342-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bos J, Smithee L, McClane B, Distefano RF, Uzal F, Songer JG, et al. Fatal necrotizing colitis following a foodborne outbreak of enterotoxigenic Clostridium perfringens type A infection. Clin Infect Dis. 2005;40:E78–E83. doi: 10.1086/429829. [DOI] [PubMed] [Google Scholar]
- 28.CDC. Fatal Foodborne Clostridium perfringens Illness at a State Psychiatric Hospital — Louisiana, 2010. MMWR. 2012;61:605–08. [PubMed] [Google Scholar]
- 29.Li J, McClane BA. A novel small acid soluble protein variant is important for spore resistance of most Clostridium perfringens food poisoning isolates. PLoS Pathog. 2008;4:e1000056. doi: 10.1371/journal.ppat.1000056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li J, Paredes-Sabja D, Sarker MR, McClane BA. Further charactrization of Clostridium perfringens small acid soluble protein-4 (Ssp4) properties and expression. PLoS One. 2009;4:e6249. doi: 10.1371/journal.pone.0006249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li J, McClane BA. Evaluating the involvement of alternative sigma factors SigF and SigG in Clostridium perfringens sporulation and enterotoxin synthesis. Infect Immun. 2010;78:4286–93. doi: 10.1128/IAI.00528-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harry KH, Zhou R, Kroos L, Melville SB. Sporulation and Enterotoxin (CPE) synthesis are controlled by the sporulation-specific factors SigE and SigK in Clostridium perfringens. J Bacteriol. 2009;191:2728–42. doi: 10.1128/JB.01839-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li J, Chen J, Vidal JE, McClane BA. The Agr-like quorum-sensing system regulates sporulation and production of enterotoxin and beta2 toxin by Clostridium perfringens type A non-food-borne human gastrointestinal disease strain F5603. Infect Immun. 2011;79:2451–9. doi: 10.1128/IAI.00169-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Varga J, Stirewalt VL, Melville SB. The CcpA protein is necessary for efficient sporulation and enterotoxin gene (cpe) regulation in Clostridium perfingens. J Bacteriol. 2004;186:5221–29. doi: 10.1128/JB.186.16.5221-5229.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ohtani K, Hirakawa H, Paredes-Sabja D, Tashiro K, Kuhara S, Sarker MR, et al. Unique regulatory mechanism of sporulation and enterotoxin production in Clostridium perfringens. J Bacteriol. 2013;195:2931–6. doi: 10.1128/JB.02152-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miyamoto K, Fisher DJ, Li J, Sayeed S, Akimoto S, McClane BA. Complete sequencing and diversity analysis of the enterotoxin-encoding plasmids in Clostridium perfringens type A non-food-borne human gastrointestinal disease isolates. J Bacteriol. 2006;188:1585–98. doi: 10.1128/JB.188.4.1585-1598.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brynestad S, Sarker MR, McClane BA, Granum PE, Rood JI. The enterotoxin (CPE) plasmid from Clostridium perfringens is conjugative. Infect Immun. 2001;69:3483–87. doi: 10.1128/IAI.69.5.3483-3487.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li J, Sayeed S, McClane BA. Prevalence of enterotoxigenic Clostridium perfringens isolates in Pittsburgh (Pennsylvania) area soils and home kitchens. Appl Environ Microbiol. 2007;73:7218–24. doi: 10.1128/AEM.01075-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ma M, Li J, McClane BA. Genotypic and phenotypic characterization of Clostridium perfringens isolates from Darmbrand cases in post-World War II Germany. Infect Immun. 2012;80:4354–63. doi: 10.1128/IAI.00818-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gurjar A, Li J, McClane BA. Characterization of toxin plasmids in Clostridium perfringens type C isolates. Infect Immun. 2010;78:4860–69. doi: 10.1128/IAI.00715-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sayeed S, Li J, McClane BA. Virulence plasmid diversity in Clostridium perfringens type D isolates. Infect Immun. 2007;75:2391–98. doi: 10.1128/IAI.02014-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Billington SJ, Wieckowski EU, Sarker MR, Bueschel D, Songer JG, McClane BA. Clostridium perfringens type E animal enteritis isolates with highly conserved, silent enterotoxin sequences. Infect Immun. 1998;66:4531–36. doi: 10.1128/iai.66.9.4531-4536.1998. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 43.Li J, Miyamoto K, McClane BA. Comparison of virulence plasmids among Clostridium perfringens type E isolates. Infect Immun. 2007;75:1811–19. doi: 10.1128/IAI.01981-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miyamoto K, Yumine N, Mimura K, Nagahama M, Li J, McClane BA, et al. Identification of novel Clostridium perfringens type E strains that carry an iota toxin plasmid with a functional enterotoxin gene. PLoS One. 2011;6:e20376. doi: 10.1371/journal.pone.0020376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Keyburn AL, Boyce JD, Vaz P, TLB, Ford ME, Parker D, et al. NetB, a new toxin that is associated with avian necrotic enteritis caused by Clostridium perfringens. PLoS Pathog. 2008;4:e26. doi: 10.1371/journal.ppat.0040026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Keyburn AL, Yan XX, Bannam TL, Van Immerseel F, Rood JI, Moore RJ. Association between avian necrotic enteritis and Clostridium perfringens strains expressing NetB toxin. BMC Vet Res. 2010;41:21. doi: 10.1051/vetres/2009069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cooper KK, Songer JG, Uzal FA. Diagnosing clostridial enteric disease in poultry. J Vet Diagn Invest. 2013;25:314–27. doi: 10.1177/1040638713483468. [DOI] [PubMed] [Google Scholar]
- 48.Van Immerseel F, Rood JI, Moore RJ, Titball RW. Rethinking our understanding of the pathogenesis of necrotic enteritis in chickens. Trends Microbiol. 2009;17:32–6. doi: 10.1016/j.tim.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 49.Skinner JT, Bauer S, Young V, Pauling G, Wilson J. An economic analysis of the impact of subclinical (mild) necrotic enteritis in broiler chickens. Avian Dis. 2010;54:1237–40. doi: 10.1637/9399-052110-Reg.1. [DOI] [PubMed] [Google Scholar]
- 50.Keyburn AL, Sheedy SA, Ford ME, Williamson MM, Awad MM, Rood JI, et al. Alpha-toxin of Clostridium perfringens is not an essential virulence factor in necrotic enteritis in chickens. Infect Immun. 2006;74:6496–500. doi: 10.1128/IAI.00806-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chalmers G, Bruce HL, Hunter DB, Parreira VR, Kulkarni RR, Jiang YF, et al. Multilocus sequence typing analysis of Clostridium perfringens isolates from necrotic enteritis outbreaks in broiler chicken populations. J Clin Microbiol. 2008;46:3957–64. doi: 10.1128/JCM.01548-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Johansson A, Aspan A, Kaldhusdal M, Engstrom BE. Genetic diversity and prevalence of netB in Clostridium perfringens isolated from a broiler flock affected by mild necrotic enteritis. Vet Microbiol. 2010;144:87–92. doi: 10.1016/j.vetmic.2009.12.017. [DOI] [PubMed] [Google Scholar]
- 53.Martin TG, Smyth JA. NetB, a pore-forming toxin from necrotic enteritis strains of Clostridium perfringens. Vet Microbiol. 2009;136:202–5. doi: 10.1016/j.vetmic.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 54.Keyburn AL, Bannam TL, Moore RJ, Rood JI. NetB, a Pore-Forming Toxin from Necrotic Enteritis Strains of Clostridium perfringens. Toxins. 2010;2:1913–27. doi: 10.3390/toxins2071913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Savva CG, Fernandes da Costa SP, Bokori-Brown M, Naylor CE, Cole AR, Moss DS, et al. Molecular architecture and functional analysis of NetB, a pore-forming toxin from Clostridium perfringens. J Biol Chem. 2013;288(5):3512–22. doi: 10.1074/jbc.M112.430223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yan X, Porter CJ, Hardy SP, Steer D, Smith AI, Quinsey NS, et al. Structural and functional analysis of the pore-forming toxin NetB from Clostridium perfringens. mBio. 2013;5:e00019–13. doi: 10.1128/mBio.00019-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cheung JK, Keyburn AL, Carter G, Lanckriet A, Van Immerseel F, Moore R, et al. The VirSR two-component signal transduction system regulates NetB toxin production in Clostridium perfringens. Infect Immun. 2010;78:3064–72. doi: 10.1128/IAI.00123-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ohtani K, Yuan Y, Hassan S, Wang R, Wang Y, Shimizu T. Virulence gene regulation by the agr system in Clostridium perfringens. J Bacteriol. 2009;191:3919–27. doi: 10.1128/JB.01455-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vidal JE, Ma M, Saputo J, Garcia J, Uzal FA, McClane BA. Evidence that the Agr-like quorum sensing system regulates the toxin production, cytotoxicity and pathogenicity of Clostridium perfringens type C isolate CN3685. Mol Microbiol. 2012;83:179–194. doi: 10.1111/j.1365-2958.2011.07925.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lepp D, Roxas B, Parreira V, Marri P, Rosey E, Gong J, et al. Identification of novel pathogenicity loci in Clostridium perfringens strains that cause avian necrotic enteritis. PLoS One. 2010;5:e10795. doi: 10.1371/journal.pone.0010795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bannam T, Yan X, Harrison P, Seemann T, Keyburn A, Stubenrauch C, et al. Necrotic enteritis-derived Clostridium perfringens strain with three closely related independently conjugative toxin and antibiotic plasmids. mBio. 2011;2:e00190–11. doi: 10.1128/mBio.00190-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Parreira VR, Costa M, Eikmeyer F, Blom J, Prescott JF. Sequence of two plasmids from Clostridium perfringens chicken necrotic enteritis isolates and comparison with C. perfringens conjugative plasmids. PLoS One. 2012;7:e49753. doi: 10.1371/journal.pone.0049753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gibert M, Jolivet-Reynaud C, Popoff MR. Beta2 toxin, a novel toxin produced by Clostridium perfringens. Gene. 1997;203:65–73. doi: 10.1016/s0378-1119(97)00493-9. [DOI] [PubMed] [Google Scholar]
- 64.Petit L, Gilbert M, Popoff M. Clostridium perfringens: toxinotype and genotype. Trends Microbiol. 1999;7:104–10. doi: 10.1016/s0966-842x(98)01430-9. [DOI] [PubMed] [Google Scholar]
- 65.Manteca C, Daube G, Jauniaux T, Linden A, Pirson V, Detilleux J, et al. The role of Clostridium perfringens beta2-toxin in bovine enterotoxaemia? Vet Microbiol. 2002;86:191–202. doi: 10.1016/S0378-1135(02)00008-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Waters M, Savoie A, Garmory HS, Bueschel D, Popoff MR, Songer JG, et al. Genotyping and phenotyping of beta2-toxigenic Clostridium perfringens fecal isolates associated with gastrointestinal diseases in piglets. J Clin Microbiol. 2003;41:3584–91. doi: 10.1128/JCM.41.8.3584-3591.2003. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 67.Ohtani K, Kawsar HI, Okumura K, Hayashi H, Shimizu T. The VirR/VirS regulatory cascade affects transcription of plasmid-encoded putative virulence genes in Clostridium perfringens. FEMS Microbiol Lett. 2003;222:137–41. doi: 10.1016/S0378-1097(03)00255-6. [DOI] [PubMed] [Google Scholar]
- 68.Chen J, McClane BA. Role of the agr-Like quorum-sensing system in regulating toxin production by Clostridium perfringens type B strains CN1793 and CN1795. Infect Immun. 2012;80:3008–17. doi: 10.1128/IAI.00438-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sayeed S, Li J, McClane BA. Characterization of virulence plasmid diversity among Clostridium perfringens type B isolates. Infect Immun. 2010;78:495–504. doi: 10.1128/IAI.00838-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Miyamoto K, Li J, Sayeed S, Akimoto S, McClane BA. Sequencing and diversity analyses reveal extensive similarities between some epsilon-toxin-encoding plasmids and the pCPF5603 Clostridium perfringens enterotoxin plasmid. J Bacteriol. 2008;190:7178–88. doi: 10.1128/JB.00939-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yonogi S, Matsuda S, Kawai T, Yoda T, Harada T, Kumeda Y, et al. BEC, a novel enterotoxin of Clostridium perfringens found in human clinical isolates from acute gastroenteritis outbreaks. Infect Immun. 2014;82:2390–9. doi: 10.1128/IAI.01759-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shimizu T, Ohtani K, Hirakawa H, Ohshima K, Yamashita A, Shiba T, et al. Complete genome sequence of Clostridium perfringens, an anaerobic flesh-eater. Proc Natl Acad Sci. 2002;99:996–1001. doi: 10.1073/pnas.022493799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Songer JG. Clostridial enteric diseases of domestic animals. Clin Microbiol Rev. 1996;9:216–34. doi: 10.1128/cmr.9.2.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Songer JG, Uzal FA. Diagnosis of Clostridium perfringens intestinal infections in sheep and goats. J Vet Diagn Invest. 2008;20:253–65. doi: 10.1177/104063870802000301. [DOI] [PubMed] [Google Scholar]
- 75.Uzal FA. Diagnosis of Clostridium perfringens intestinal infections in sheep and goats. Anaerobe. 2004;10:135–43. doi: 10.1016/j.anaerobe.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 76.Uzal FA, Songer G. Diagnosis of Clostridium perfringens intestinal infections in sheep and goat. J Vet Diag Invest. 2008;20:253–65. doi: 10.1177/104063870802000301. [DOI] [PubMed] [Google Scholar]
- 77.Fernandez-Miyakawa ME, Fisher DJ, Poon R, Sayeed S, Adams V, Rood JI, et al. Both epsilon-toxin and beta-toxin are important for the lethal properties of Clostridium perfringens type B isolates in the mouse intravenous injection model. Infect Immun. 2007;75:1443–52. doi: 10.1128/IAI.01672-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fernandez-Miyakawa ME, Fisher DJ, Poon R, Sayeed S, Adams V, Rood JI, et al. Both epsilon-toxin and beta-toxin are important for the lethal properties of Clostridium perfringens type B isolates in the mouse intravenous injection model. Infect Immun. 2007;75:1443–52. doi: 10.1128/IAI.01672-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gill DM. Bacterial toxins: a table of lethal amounts. Microbiol Rev. 1982;46:86–94. doi: 10.1128/mr.46.1.86-94.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Miclard J, Jaggi M, Sutter E, Wyder M, Grabscheid B, Posthaus H. Clostridium perfringens beta-toxin targets endothelial cells in necrotizing enteritis in piglets. Vet Microbiol. 2009;137:320–5. doi: 10.1016/j.vetmic.2009.01.025. [DOI] [PubMed] [Google Scholar]
- 81.Nagahama M, Hayashi S, Morimitsu S, Sakurai J. Biological activities and pore formation of Clostridium perfringens beta toxin in HL 60 cells. J Biol Chem. 2003;278:36934–41. doi: 10.1074/jbc.M306562200. [DOI] [PubMed] [Google Scholar]
- 82.Steinthorsdottir V, Halldorsson H, Andresson OS. Clostridium perfringens beta-toxin forms multimeric transmembrane pores in human endothelial cells. Microb Pathog. 2000;28:45–50. doi: 10.1006/mpat.1999.0323. [DOI] [PubMed] [Google Scholar]
- 83.Autheman D, Wyder M, Popoff M, D’Herde K, Christen S, Posthaus H. Clostridium perfringens beta-toxin induces necrostatin-inhibitable, calpain-dependent necrosis in primary porcine endothelial cells. PLoS One. 2013;8:e64644. doi: 10.1371/journal.pone.0064644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hunter SEC, Clarke IN, Kelley DC, Titball RW. Cloning and neucleotide sequencing of the Clostridium perfringens epsilon-toxin gene and its expression in Escherichia coli. Infect Immun. 1992;60:102–10. doi: 10.1128/iai.60.1.102-110.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Minami J, Katayama S, Matsushita O, Matsushita C, Okabe A. Lambda-toxin of Clostridium perfringens activates the precursor of epsilon-toxin by releasing its N- and C-terminal peptides. Microbiol Immun. 1997;41:527–35. doi: 10.1111/j.1348-0421.1997.tb01888.x. [DOI] [PubMed] [Google Scholar]
- 86.Miyata S, Matsushita O, Minami J, Katayama S, Shimamoto S, Okabe A. Cleavage of a C-terminal peptide is essential for heptamerization of Clostridium perfringens epsilon-toxin in the synaptosomal membrane. J Biol Chem. 2001;276:13778–83. doi: 10.1074/jbc.M011527200. [DOI] [PubMed] [Google Scholar]
- 87.Worthington RW, Mulders MS. Physical changes in the epsilon prototoxin molecule of Clostridium perfringens during enzymatic activation. Infect Immun. 1977;18:549–51. doi: 10.1128/iai.18.2.549-551.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hambrook JL, Lindsay CD, Hughes N. Morphological alterations in MDCK cells induced by exposure to Clostridium perfringens epsilon-toxin. Biochem Soc Trans. 1995;23:44S. doi: 10.1042/bst023044s. [DOI] [PubMed] [Google Scholar]
- 89.Robertson SL, Li J, Uzal FA, McClane BA. Evidence for a prepore stage in the action of Clostridium perfringens epsilon toxin. PLoS One. 2011;6:e22053. doi: 10.1371/journal.pone.0022053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Popoff MR. Epsilon toxin: a fascinating pore-forming toxin. FEBS J. 2011;278:4602–15. doi: 10.1111/j.1742-4658.2011.08145.x. [DOI] [PubMed] [Google Scholar]
- 91.Garcia JP, Adams V, Beingesser J, Hughes ML, Poon R, Lyras D, et al. Epsilon toxin is essential for the virulence of Clostridium perfringens type D infection in sheep, goats and mice. Infect Immun. 2013;81:2405–2414. doi: 10.1128/IAI.00238-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Knapp O, Maier E, Benz R, Geny B, Popoff MR. Identification of the channel-forming domain of Clostridium perfringens Epsilon-toxin (ETX) Biochim Biophys Acta. 2009;1788:2584–93. doi: 10.1016/j.bbamem.2009.09.020. [DOI] [PubMed] [Google Scholar]
- 93.Chen J, Rood JI, McClane BA. Epsilon toxin production by Clostridium perfringens type D strain CN3718 is dependent upon the agr operon but not the VirS/VirR. mBio. 2011;2:e00275–300275–11. doi: 10.1128/mBio.00275-11. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 94.Hughes ML, Poon R, Adams V, Sayeed S, Saputo J, Uzal FA, et al. Epsilon-toxin plasmids of Clostridium perfringens type D are conjugative. J Bacteriol. 2007;189:7531–8. doi: 10.1128/JB.00767-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Songer JG, Uzal FA. Clostridial enteric infections in pigs. J Vet Diag. 2005;17:528–36. doi: 10.1177/104063870501700602. [DOI] [PubMed] [Google Scholar]
- 96.Johnson S, Gerding DN. Enterotoxemic Infections. In: Rood JI, McClane BA, Songer JG, Titball RW, editors. The Clostridia: Molecular Biology and Pathogenesis. Academic Press; London: 1997. pp. 117–40. [Google Scholar]
- 97.Lawrence GW. The pathogenesis of enteritis necroticans. In: Rood JI, McClane BA, Songer JG, Titball RW, editors. The Clostridia: Molecular Genetics and Pathogenesis. Academic Press; London: 1997. pp. 198–207. [Google Scholar]
- 98.Petrillo TM, Beck-Sague CM, Songer JG, Abramowsky C, Fortenberry JD, Meacham L, et al. Enteritis necroticans (pigbel) in a diabetic child. N Engl J Med. 2000;342:1250–3. doi: 10.1056/NEJM200004273421704. [DOI] [PubMed] [Google Scholar]
- 99.Gui L, Subramony C, Fratkin J, Hughson M. Fatal enteritis necroticans (pigbel) in a diabetic adult. Mod Pathol. 2002;15:66–70. doi: 10.1038/modpathol.3880491. [DOI] [PubMed] [Google Scholar]
- 100.Fisher DJ, Fernandez-Miyakawa ME, Sayeed S, Poon R, Adams V, Rood JI, et al. Dissecting the contributions of Clostridium perfringens type C toxins to lethality in the mouse intravenous injection model. Infect Immun. 2006;74:5200–10. doi: 10.1128/IAI.00534-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sayeed S, Uzal FA, Fisher DJ, Saputo J, Vidal JE, Chen Y, et al. Beta toxin is essential for the intestinal virulence of Clostridium perfringens type C disease isolate CN3685 in a rabbit ileal loop model. Mol Microbiol. 2008;67:15–30. doi: 10.1111/j.1365-2958.2007.06007.x. [DOI] [PubMed] [Google Scholar]
- 102.Vidal JE, McClane BA, Saputo J, Parker J, Uzal FA. Effects of Clostridium perfringens beta-toxin on the rabbit small intestine and colon. Infect Immun. 2008;76:4396–404. doi: 10.1128/IAI.00547-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Skjelkvale R, Duncan CL. Enterotoxin formation by different toxigenic types of Clostridium perfringens. Infect Immun. 1975;11:563–75. doi: 10.1128/iai.11.3.563-575.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ma M, Gurjar A, Theoret JR, Garcia JP, Beingesser J, Freedman JC, et al. Synergistic effects of Clostridium perfringens enterotoxin and beta toxin in rabbit small intestinal loops. Infect Immun. 2014;82:2958–70. doi: 10.1128/IAI.01848-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Shimizu T, Ba-Thein W, Tamaki M, Hayashi H. The virR gene, a member of a class of two-component response regulators, regulates the production of perfringolysin O, collagenase, and hemagglutinin in Clostridium perfringens. J Bacteriol. 1994;176:1616–23. doi: 10.1128/jb.176.6.1616-1623.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lyristis M, Bryant AE, Sloan J, Awad MM, Nisbet IT, Stevens DL, et al. Identification and molecular analysis of a locus that regulates extracellular toxin production in Clostridium perfringens. Mol Microbiol. 1994;12:761–77. doi: 10.1111/j.1365-2958.1994.tb01063.x. [DOI] [PubMed] [Google Scholar]
- 107.Vidal JE, Ohtani K, Shimizu T, McClane BA. Contact with enterocyte-like Caco-2 cells induces rapid upregulation of toxin production by Clostridium perfringens type C isolates. Cell Microbiol. 2009;11:363–9. doi: 10.1111/j.1462-5822.2009.01332.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ma M, Vidal J, Saputo J, McClane BA, Uzal F. The VirS/VirR two-component system regulates the anaerobic cytotoxicity, intestinal pathogenicity, and enterotoxemic lethality of Clostridium perfringens type C isolate CN3685. mBio. 2011;2:e00338–10. doi: 10.1128/mBio.00338-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Li J, Ma M, Sarker MR, McClane BA. CodY is a global regulator of virulence-associated properties for Clostridium perfringens type D strain CN3718. mBio. 2013;4:e00770–13. doi: 10.1128/mBio.00770-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sakurai J, Nagahama M, Oda M, Tsuge H, Kobayashi K. Clostridium perfringens iota-toxin: structure and function. Toxins. 2009;1:208–28. doi: 10.3390/toxins1020208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Stiles BG, Wigelsworth DJ, Popoff MR, Barth H. Clostridial binary toxins: iota and c2 family portraits. Front Cell Infect Microbiol. 2011;1:11. doi: 10.3389/fcimb.2011.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Aktories K, Schwan C, Papatheodorou P, Lang AE. Bidirectional attack on the actin cytoskeleton. Bacterial protein toxins causing polymerization or depolymerization of actin. Toxicon. 2012;60:572–81. doi: 10.1016/j.toxicon.2012.04.338. [DOI] [PubMed] [Google Scholar]
- 113.Barth H, Stiles BG. Binary actin-ADP-ribosylating toxins and their use as molecular Trojan horses for drug delivery into eukaryotic cells. Curr Med Chem. 2008;15:459–69. doi: 10.2174/092986708783503195. [DOI] [PubMed] [Google Scholar]
- 114.Gibert M, Petit L, Raffestin S, Okabe A, Popoff MR. Clostridium perfringens iota-toxin requires activation of both binding and enzymatic components for cytopathic activity. Infect Immun. 2000;68:3848–53. doi: 10.1128/iai.68.7.3848-3853.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Papatheodorou P, Carette JE, Bell GW, Schwan C, Guttenberg G, Brummelkamp TR, et al. Lipolysis-stimulated lipoprotein receptor (LSR) is the host receptor for the binary toxin Clostridium difficile transferase (CDT) Pro Nat Acad Sci. 2011;108:16422–7. doi: 10.1073/pnas.1109772108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wigelsworth DJ, Ruthel G, Schnell L, Herrlich P, Blonder J, Veenstra TD, et al. CD44 promotes intoxication by the clostridial iota-family toxins. PLoS One. 2012;7:e51356. doi: 10.1371/journal.pone.0051356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hale ML, Marvaud JC, Popoff MR, Stiles BG. Detergent-resistant membrane microdomains facilitate Ib oligomer formation and biological activity of Clostridium perfringens iota-toxin. Infect Immun. 2004;72:2186–93. doi: 10.1128/IAI.72.4.2186-2193.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Marvaud JC, Stiles BG, Chenal A, Gillet D, Gibert M, Smith LA, et al. Clostridium perfringens iota toxin. Mapping of the Ia domain involved in docking with Ib and cellular internalization. J Biol Chem. 2002;277:43659–66. doi: 10.1074/jbc.M207828200. [DOI] [PubMed] [Google Scholar]
- 119.Nagahama M, Yamaguchi A, Hagiyama T, Ohkubo N, Kobayashi K, Sakurai J. Binding and internalization of Clostridium perfringens iota-toxin in lipid rafts. Infect Immun. 2004;72:3267–75. doi: 10.1128/IAI.72.6.3267-3275.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Gibert M, Monier MN, Ruez R, Hale ML, Stiles BG, Benmerah A, et al. Endocytosis and toxicity of clostridial binary toxins depend on a clathrin-independent pathway regulated by Rho-GDI. Cell Microbiol. 2011;13:154–70. doi: 10.1111/j.1462-5822.2010.01527.x. [DOI] [PubMed] [Google Scholar]
- 121.Miyamoto K, Chakrabarti G, Morino Y, McClane BA. Organization of the plasmid cpe locus of Clostridium perfringens type A isolates. Infect Immun. 2002;70:4261–72. doi: 10.1128/IAI.70.8.4261-4272.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Li J, Adams V, Bannam TL, Miyamoto K, Garcia JP, Uzal FA, et al. Toxin plasmids of Clostridium perfringens. MMBR. 2013;77:208–33. doi: 10.1128/MMBR.00062-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Bantwal R, Bannam TL, Porter CJ, Quinsey NS, Lyras D, Adams V, et al. The peptidoglycan hydrolase TcpG is required for efficient conjugative transfer of pCW3 in Clostridium perfringens. Plasmid. 2012;67:139–47. doi: 10.1016/j.plasmid.2011.12.016. [DOI] [PubMed] [Google Scholar]
- 124.Parsons JA, Bannam TL, Devenish RJ, Rood JI. TcpA, an FtsK/SpoIIIE homolog, is essential for transfer of the conjugative plasmid pCW3 in Clostridium perfringens. J Bacteriol. 2007;189:7782–90. doi: 10.1128/JB.00783-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Porter CJ, Bantwal R, Bannam TL, Rosado CJ, Pearce MC, Adams V, et al. The conjugation protein TcpC from Clostridium perfringens is structurally related to the type IV secretion system protein VirB8 from Gram-negative bacteria. Mol Microbiol. 2012;83:275–88. doi: 10.1111/j.1365-2958.2011.07930.x. [DOI] [PubMed] [Google Scholar]
- 126.Steen JA, Bannam TL, Teng WL, Devenish RJ, Rood JI. The putative coupling protein TcpA interacts with other pCW3-encoded proteins to form an essential part of the conjugation complex. J Bacteriol. 2009;191:2926–33. doi: 10.1128/JB.00032-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Low HH, Gubellini F, Rivera-Calzada A, Braun N, Connery S, Dujeancourt A, et al. Structure of a type IV secretion system. Nature. 2014;508:550–3. doi: 10.1038/nature13081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Guglielmini J, Neron B, Abby SS, Garcillan-Barcia MP, de la Cruz F, Rocha EP. Key components of the eight classes of type IV secretion systems involved in bacterial conjugation or protein secretion. Nucleic Acids Res. 2014;42:5715–27. doi: 10.1093/nar/gku194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Bannam TL, Teng WL, Bulach D, Lyras D, Rood JI. Functional identification of conjugation and replication regions of the tetracycline resistance plasmid pCW3 from Clostridium perfringens. J Bacteriol. 2006;188:4942–51. doi: 10.1128/JB.00298-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Bhatty M, Laverde Gomez JA, Christie PJ. The expanding bacterial type IV secretion lexicon. Res Microbiol. 2013;164:620–39. doi: 10.1016/j.resmic.2013.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Goessweiner-Mohr N, Arends K, Keller W, Grohmann E. Conjugative type IV secretion systems in Gram-positive bacteria. Plasmid. 2013;70:289–302. doi: 10.1016/j.plasmid.2013.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Teng WL, Bannam TL, Parsons JA, Rood JI. Functional characterization and localization of the TcpH conjugation protein from Clostridium perfringens. J Bacteriol. 2008;190:5075–86. doi: 10.1128/JB.00386-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Goessweiner-Mohr N, Grumet L, Pavkov-Keller T, Birner-Gruenberger R, Grohmann E, Keller W. Crystallization and preliminary structure determination of the transfer protein TraM from the Gram-positive conjugative plasmid pIP501. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2013;69:178–83. doi: 10.1107/S1744309113000134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Alvarez-Martinez CE, Christie PJ. Biological diversity of prokaryotic type IV secretion systems. MMBR. 2009;73:775–808. doi: 10.1128/MMBR.00023-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Guglielmini J, de la Cruz F, Rocha EP. Evolution of conjugation and type IV secretion systems. Mol Biol Evol. 2013;30:315–31. doi: 10.1093/molbev/mss221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Gomis-Ruth FX, Sola M, de la Cruz F, Coll M. Coupling factors in macromolecular type-IV secretion machineries. Curr Pharm Des. 2004;10:1551–65. doi: 10.2174/1381612043384817. [DOI] [PubMed] [Google Scholar]
- 137.Garcillan-Barcia MP, Francia MV, de la Cruz F. The diversity of conjugative relaxases and its application in plasmid classification. FEMS Microbiol Rev. 2009;33:657–87. doi: 10.1111/j.1574-6976.2009.00168.x. [DOI] [PubMed] [Google Scholar]
- 138.de la Cruz F, Frost LS, Meyer RJ, Zechner EL. Conjugative DNA metabolism in Gram-negative bacteria. FEMS Microbiol Rev. 2010;34:18–40. doi: 10.1111/j.1574-6976.2009.00195.x. [DOI] [PubMed] [Google Scholar]
- 139.Ebersbach G, Gerdes K. Plasmid segregation mechanisms. Annu Rev Genet. 2005;39:453–79. doi: 10.1146/annurev.genet.38.072902.091252. [DOI] [PubMed] [Google Scholar]
- 140.Salje J, Gayathri P, Lowe J. The ParMRC system: molecular mechanisms of plasmid segregation by actin-like filaments. Nat Rev Microbiol. 2010;8:683–92. doi: 10.1038/nrmicro2425. [DOI] [PubMed] [Google Scholar]
- 141.Adams V, Li J, Wisniewski JA, Uzal FA, Moore RJ, McClane BA, et al. Virulence plasmids of spore-forming bacteria. Microbiol Spectr. 2014 doi: 10.1128/microbiolspec.PLAS-0024-2014. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.del Solar G, Giraldo R, Ruiz-Echevarria MJ, Espinosa M, Diaz-Orejas R. Replication and control of circular bacterial plasmids. MMBR. 1998;62:434–64. doi: 10.1128/mmbr.62.2.434-464.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Ionesco H. Transferable tetracycline resistance in “Clostridium difficile” (author’s transl) Ann Microbiol (Paris) 1980;131A:171–9. [PubMed] [Google Scholar]
- 144.Lyras D, Storie C, Huggins AS, Crellin PK, Bannam TL, Rood JI. Chloramphenicol resistance in Clostridium difficile is encoded on Tn4453 transposons that are closely related to Tn4451 from Clostridium perfringens. Antimicrob Agents Chemother. 1998;42:1563–7. doi: 10.1128/aac.42.7.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]