Abstract
The ability of HIV to establish long-lived latent infection is mainly due to transcriptional silencing of viral genome in resting memory T lymphocytes. Here, we show that new semi-synthetic ingenol esters reactivate latent HIV reservoirs. Amongst the tested compounds, 3-caproyl-ingenol (ING B) was more potent in reactivating latent HIV than known activators such as SAHA, ingenol 3,20-dibenzoate, TNF-α, PMA and HMBA. ING B activated PKC isoforms followed by NF-κB nuclear translocation. As virus reactivation is dependent on intact NF-κB binding sites in the LTR promoter region ING B, we have shown that. ING B was able to reactivate virus transcription in primary HIV-infected resting cells up to 12 fold and up to 25 fold in combination with SAHA. Additionally, ING B promoted up-regulation of P-TEFb subunits CDK9/Cyclin T1. The role of ING B on promoting both transcription initiation and elongation makes this compound a strong candidate for an anti-HIV latency drug combined with suppressive HAART.
Keywords: HIV, Latency, Ingenol, PKC, NF-kB, Resting cells, P-TEFb
Introduction
The eradication of HIV by current anti-retroviral therapies in infected individuals is not accomplished due to the establishment of viral latency during early stages of infection. HIV establishes long-lived latent infection in resting memory T lymphocytes and other non-dividing cells mainly due to transcriptional silencing (Finzi et al., 1997; Geeraert et al., 2008). Despite undetectable viral load in patients treated with potent antiretroviral therapy, proviral genomes remain in the latent reservoirs rendering the total clearance of HIV an unattainable goal at present (Finzi et al., 1997; Pierson et al., 2000; Yang et al., 2009). Hence, the search for a cure is one of the most challenging and rewarding areas of HIV/AIDS research (Geeraert et al., 2008; Johnston, 2010).
Multiple mechanisms contribute to the maintenance of HIV latency, including integration of the proviral DNA in transcriptionally inactive sites, Tat trans-activation, histone modifications or unavailability of cellular transcription factors (Ganesh et al., 2003; Ishida et al., 2006; Jones and Peterlin, 1994; Mbonye and Karn, 2014; Williams et al., 2006). Additionally, post-transcriptional mechanisms affecting the export or translation of HIV mRNAs can also block HIV expression during latency (Huang et al., 2007; Karn and Stoltzfus, 2012; Lassen et al., 2006). HIV activation and replication is highly dependent on T lymphocyte activation status, relying on the availability of host transcription factors, such as NF-κB, NFAT and AP1, which bind to HIV-1 LTR triggering virus RNA transcription, and others such as P-TEFb that act in transcriptional elongation (Bartholomeeusen et al., 2013; Contreras et al., 2012, 2007; González et al., 2001; Khalaf et al., 2010; Kinoshita et al., 1997; Mbonye and Karn, 2014; Sgarbanti et al., 2008; Torgerson et al., 1998; Yang et al., 1999). Protein kinase C (PKC) activation followed by NF-κB nuclear translocation is an important step required for latent HIV reactivation.
The PKC family comprises a large number of serine-threonine kinases dependent on the hydrolysis of phosphatidyl-inositol-4, 5-bisphosphate (PIP2) for activation. PIP2 is hydrolyzed in diacyl glycerol (DAG) and inositol-1,4,5-trisphosphate (IP3), promoting calcium release from endoplasmic reticulum. This step is important to activate PKC calcium dependent isoforms, such as PKC α, βI, βII, and γ. DAG binds with high affinity to cis-rich zinc finger domains in conventional and novel PKC isoforms promoting their activation (Spitaler and Cantrell, 2004; Steinberg, 2008). In T lymphocytes, active PKC-θ and PKC-γ stimulate IKK-dependent phosphorylation and degradation of IκB-α (a NF-κB cellular inhibitor). Free NF-κB is then able to translocate to the nucleus and bind to NF-κB responsive promoters stimulating transcription of several cellular genes, as well as binding to specific sites in the LTR region triggering HIV-transcription (Spitaler and Cantrell, 2004; Steinberg, 2008; Williams et al., 2004; Yang et al., 1999).
Several compounds have been investigated as potential candidates for elimination of latent HIV-reservoirs. These, in association with HAART therapy could potentially eradicate HIV from infected patients. Therapeutically, anti-latency agents would be given intermittently or for a limited period of time to purge reservoirs of dormant HIV. In this scenario, PKC agonists are potential targets worth exploring (Richman et al., 2009). The phorbol esther PMA and prostratin reactivate latent HIV in lymphoid and myeloid cells (Spitaler and Cantrell, 2004; McKernan et al., 2012; Van Lint et al., 2013; Williams et al., 2004). In a pilot clinical trial, bryostatin (another modulator of PKC isoforms) has been used to reactivate HIV from latency (Van Lint et al., 2013).
Histone deacetylase inhibitors (HDACi) have been also tested as HIV reactivator compounds. SAHA (suberoylanilide hydroxamic acid) is a histone deacetylases inhibitor that can disrupt HIV-1 latency in vitro (Archin et al., 2012, 2009; Liu et al., 2006) and in HIV positive patients submitted to HAART combined with 400 mg of SAHA (Archin et al., 2012). Introduction of yet another HDACi; valproic acid (VPA), was envisioned to flush out the latent virus from these reservoirs within few years, but VPA in combination with HAART failed to deplete latent HIV reservoir sufficiently (Routy et al., 2012).
Some compounds are able to disrupt HIV latency activating the transcriptional elongation factor b (P-TEFb). This cellular factor can form two different complexes: an active one, composed by cyclin-dependent kinase 9 (CDK9) and cyclin T1 (Cyc T1) and an inactive complex, which in addition to CDK9 and Cyc T1 also contains the inhibitory protein HEXIM 1 or 2 and the 7SK small nuclear RNA, amongst other proteins (Cho et al., 2010; Contreras et al., 2009, 2007). Productive transcriptional elongation requires hyper-phosphorylation of RNA polymerase II C-terminal domain (CTD), which is accomplished by the CDK9 subunit of active P-TEFb (Cho et al., 2010). The HMBA (hexamethylene bisacetamide) transiently activates the PI3K/Akt pathway, leading to the phosphorylation of HEXIM1 and the subsequent release of active P-TEFb, which then stimulates HIV transcription and reactivation of the latent HIV reservoir (Contreras et al., 2007). SAHA can also disrupt HIV-1 latency in vitro and in HAART treated HIV-positive patients (Archin et al., 2012, 2009; Liu et al., 2006) by transiently turning on the PI3K/Akt pathway promoting P-TEFb activation (Contreras et al., 2009; Liu et al., 2006). In resting primary CD4+ T cells, where levels of P-TEFb are lower, the most potent HDACi, SAHA, has minimal effects. In contrast, when these cells are treated with a PKC agonist, bryostatin 1, which increased levels of P-TEFb, then SAHA once again, reactivated HIV. In this way, HDACis, which can reactivate HIV, work via the release of free P-TEFb from the 7SK snRNP (Bartholomeeusen et al., 2013).
While multiple transcriptional regulatory mechanisms for HIV-1 latency have been described in the context of progressive epigenetic silencing and maintenance, recent reports suggested that productive infection is positively correlated with cellular activation and NF-κB activity (Dahabieh et al., 2014). Many natural compounds are currently been screened for their antiviral properties and some have been reported as possible candidates for clinical tests. These include terpenoids, polyphenols and phorbol esters (Fujiwara et al., 1998; Jassbi, 2006; Salatino et al., 2007). The diterpene ingenol is a secondary metabolite of Euphorbiacea, a large family of succulent plants that commonly grow in semi-desertic areas of Brazil. Ingenol derivates can inhibit HIV reverse transcription and down modulate CD4 and CXCR4 surface expression by activating PKC (Ersvaer et al., 2010; Fujiwara et al., 1998; Hong et al., 2011; Lee et al., 2010; Warrilow et al., 2006). Additionally, ingenol 3-angelate has anti-tumor activity and activates responses mediated by PKC pathway, thus being considered a PKC agonist (Ersvaer et al., 2010; Lee et al., 2010). Another semi-synthetic derivative, ingenol 3,20-dibenzoate, displays trombopoietic activity and also activates PKC pathways (Racke et al., 2012). In this article, in an attempt to obtain less toxic compounds retaining anti-latency activities, we synthesized new ingenol derivates through chemical substitutions in the polyhydroxylated southern region of ingenol structure, which apparently is critical for the activity over HIV transcription. The most promising molecule isolated here, 3-caproyl-ingenol (ingenol B, ING B), promotes PKC activation and NF-κB nuclear translocation, activating transcription of B and C HIV-1 subtypes. Finally, ING B induces higher levels of reactivation when compared to TNF-α, PMA, SAHA and HMBA, both in established HIV latency models and in HIV infected primary resting cells.
Results
Screening of ingenol derivates with anti-HIV latency activity
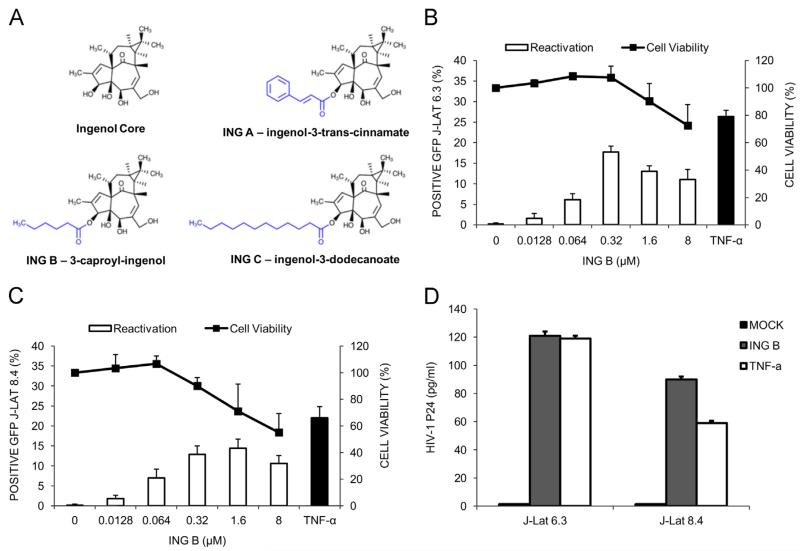
Euphorbia tirucalli latex contains a complex mixture of ingenol esters. They are mostly esters of dodecatrienic and dodecatetraenic acids attached at various hydroxyl groups. Alkaline hydrolysis cleaved the ester bonds producing free ingenol, which was then isolated in a single chromatographic step. Subsequently, selective esterification at C-3 position produced three new esters of ingenol; trans-cinnamate (ING A), caprate (ING B), and myristate (ING C) (Fig. 1A and S1). The primary reason for choosing these ester groups was to explore initial structure-activity relationship for various 3-acyl-ingenols for their ability to reactivate latent HIV-1. We used the J-Lat cell line (clones 6.3 and 8.4), which are derived T cells that harbor a transcriptionally silent HIV-GFP proviral genome as a HIV latency model (Jordan et al., 2003).
Fig. 1.
Ingenol derivate promotes HIV transcription and virus production. J-Lat cells 6.3 and 8.4 were used as a model of HIV latency. (A) Schematic representation of the novel ingenol ester derivates from Euphorbia tirucalli. (B) and (C) ING B activates HIV transcription in J-Lat cells, clones 6.3 and 8.4, respectively. Cells were exposed to increasing concentrations of ING B and virus activation was evaluated by GFP expression using flow cytometer 24 h after treatment (empty bars). Cytotoxicity was measured using Cell Titer Blue reagent five days post ING B treatment (black lines) and expressed as viability (%). TNF-α (20 ng/ml) was used as positive control (black bars). Error bars represent standard deviation of three independent experiments. (D) ING B treatment promotes HIV production. HIV viral particles were evaluated in the supernatants of J-Lat cells (clones 6.3 and 8.4) exposed to ING B for 48 h by ELISA (HIV-1 p24 Antigen). TNF-α (20 ng/ml) was used as positive control. Mock stands for supernatant of non-treated cells. Error bars represent standard deviation of three experiments.
Cells were exposed to different ingenol derivates and HIV transcriptional reactivation was evaluated by flow cytometry measuring GFP expression 24, 48 and 72 h after treatment. In an initial screening, the original ingenol molecule (ING) activated up to 35% of J-Lat 6.3 cells at 8 μM; however, only 20% of the cells were viable at this concentration (Fig. S1A). The modifications in the C-3 position of the ING core produced the derivates ING A and ING C (Fig. 1A). Both of them activated HIV transcription in about 20% of latent cells at 0.32 μM. However, at this concentration these compounds induced significant cytotoxicity (Fig. S1, panels B and D). ING B ester at the same concentration (0.32 μM) promoted 20% of HIV activation in the J-Lat 6.3 and 15% of J-Lat 8.4 cells with 100% of cell viability in both cases (Fig. 1B and C) and no major differences were observed for the different exposure times suggesting that 24 h treatment was sufficient to promote HIV transcription (Fig. S1C). The EC50 for ING B activation was calculated for both J-lat clones (J-Lat 6.3, EC50=0.13 μM and J-Lat 8.4, EC50=0.16 μM) with no differences between these two cells.
All these compounds were compared with TNF-α activation levels as positive control (Fig. 1B and C, black bars). To confirm the production of HIV particles by ING B treated cells, their supernatant was collected after 48 hours and levels of p24 antigen were measured by ELISA. ING B stimulated virus production to the same extent as the positive control TNF-α in both J-Lat 6.3 and 8.4 clones(Fig. 1D). These results suggested that ING B was able to reactivate transcription of a latent HIV provirus and induced production of viral particles. Based on these results we proceeded with further characterization of this derivate at 0.32–1 μM concentration.
ING B promotes PKC activation and NF-kB translocation to the nucleus
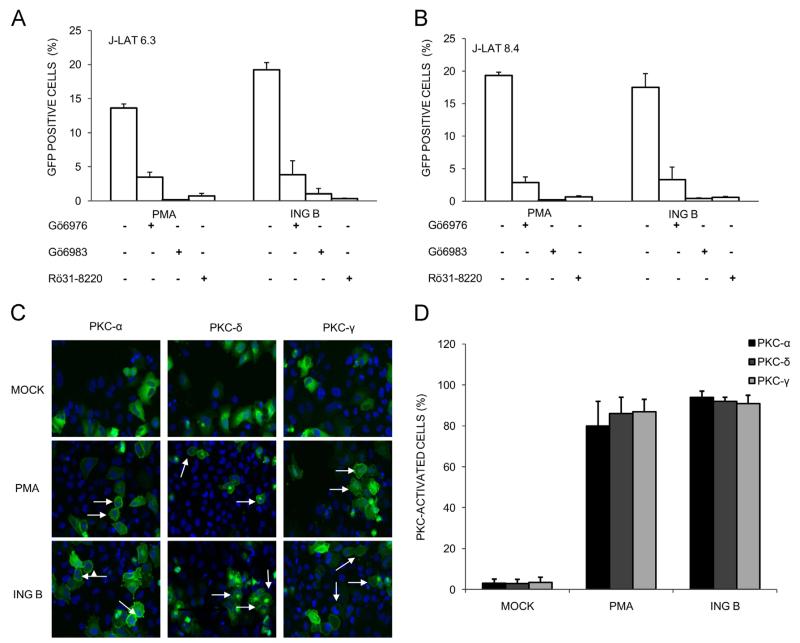
To evaluate if ING B ester activates PKC, J-Lat cells were pre-treated with known PKC inhibitors (Gö 6976, Gö 6983 and Ro-31-8220) and then treated either with PMA, a potent PKC activator, or with ING B. A similar pattern of HIV activation was found for both PMA and ING B, suggesting that the latter effect is indeed mediated by PKC activation in both clones of J-Lat cells (Fig. 2, panels A and B). Only 3.8%, 1.0% and 0.3% of cells were reactivated by ING B when J-Lat clone 6.3 was pretreated with PKC inhibitors Gö 6976, Gö 6983 and Ro-31–8220, respectively (Fig. 2A). Similar results were obtained with the same PKC inhibitors and ING B treatment in J-Lat clone 8.4 (Fig. 2B) suggesting that PKC activity is modulated by ING B. Neither cytotoxic effects nor HIV-GFP expression were observed in controls treated only with PKC inhibitors.
Fig. 2.
HIV reactivation addressed by ING B is mediated by PKC activation. J-Lat cells were pre-treated with different PKC inhibitors at 1 μM for 24 h (Gö 6976, Gö 6983 and Ro-31-8220) before ING B addition (0.32 μM). GFP expression was evaluated 24 h post ING B treatment by flow cytometer. PMA (1 μM) was used as positive control of PKC dependent HIV reactivation. (A) HIV-GFP expression in J-Lat 6.3 cells in the presence of PKC inhibitors. (B) HIV-GFP expression in J-Lat 8.4 cells in the presence of PKC inhibitors. (C) Analysis of cellular localization of HeLa cells expressing different GFP-tagged PKC isoforms by confocal microscopy. HeLa cells were transfected with expressing vectors coding PKC-α (left columns), PKC-δ (central columns) and PKC-γ (right columns) and treated with PMA (1 μM) or ING B (0.32 μM) for 10 min. Untreated cells are labeled as Mock. Cell nuclei are labeled with DAPI (blue color) and PKC-GFP are labeled in green. White arrows are highlighting the sub-membrane location when PKC isoforms are activated. (D) PKC sub-membrane localization was quantified in 200 GFP positive cells for each treatment (Mock non-treated, PMA and ING B). PKC-α, PKC-δ and PKC-γ are represented by black, dark gray and gray bars, respectively. Standard deviation is shown at the top of the bars.
Upon activation, PKCs are translocated to the plasma membrane by RACK proteins (membrane-bound receptor for activated protein kinase C proteins) (Mochly-Rosen and Gordon, 1998). In order to verify if ING B is able to promote PKC translocation to the plasma membrane, HeLa cells expressing PKC isoforms fused to GFP were treated with ING B or PMA (positive control). Confocal fluorescence microscopy analysis revealed a diffuse cytoplasmic PKC-GFP distribution in untreated cells (Fig. 2C, upper panels). However, 10 min of ING B or PMA treatment were sufficient to induce translocation of PKCα-GFP, PKCδ-GFP and PKCγ-GFP to the plasma membrane (Fig. 2C, middle and bottom panels). Quantification analysis showed that 80% of the cells presented PKC activation in the presence of ING B and PMA (Fig. 2D). This profile of PKC-GFP sub-membrane localization was maintained up to 120 min after ING B treatment (Fig. S2). Together, these data show that ING B acts by promoting PKC activation.
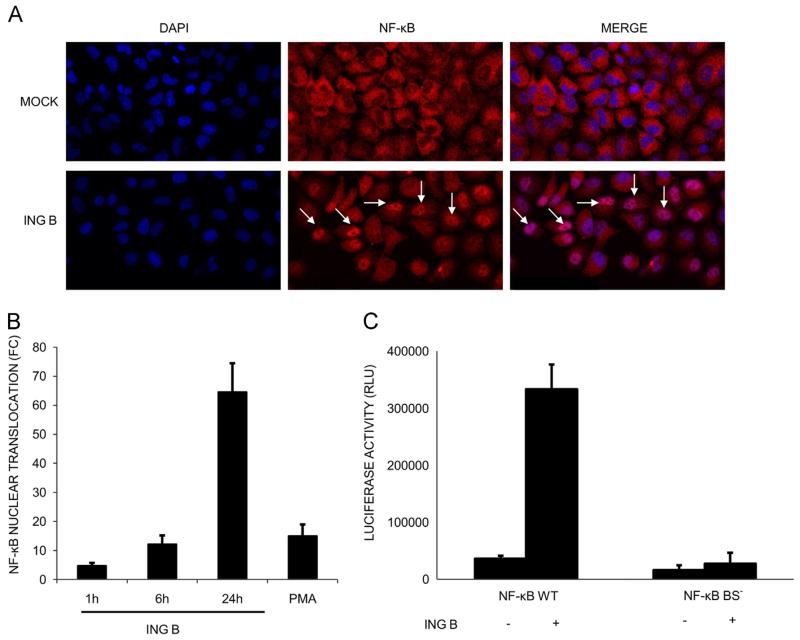
As mentioned before, PKC activation leads to nuclear translocation of the transcription factor NF-κB due to induction of IKK degradation in the proteasome (McKernan et al., 2012; Spitaler and Cantrell, 2004; Williams et al., 2004). The effect of ING B treatment on nuclear translocation of NF-κB was addressed by two different approaches. First, using confocal microscopy, the presence of this transcription factor inside the nucleus of treated HeLa cells was evaluated using anti-p65 antibodies (Fig. 3A). Levels of NF-κB translocation were quantified using the Translocation module of MetaXpress software of a High Content Screening confocal microscope (Molecular Devices, Inc). ING B treatment for 24 h induced the translocation of NF-κB in 65% of the cells analyzed; reaching higher values than the positive control with PMA treatment at the same time point (24 h) (Fig. 3B).
Fig. 3.
HIV transcriptional activation by ING B is dependent on NF-κB activation. (A) ING B promotes NF-kB translocation to the nucleus. HeLa cells were treated with ING B (1 μM) for different time points. After that, the cells were fixed and submitted to immunofluorescence using anti-NF-κB antibodies (red color) and DAPI staining (blue color). Cells treated with ING B are presented in lower panel and untreated cells (mock) in upper panel. White arrows highlight NF-κB nuclear translocation. (B) Quantification of NF-κB nuclear translocation in HeLa cells, expressed as fold change (FC) at different time points: 1, 6 and 24 h after ING B treatment (1 μM). PMA (1 μM) was used as positive control (24 h treatment). The fold change was calculated using the average of internalized NF-κB cells of each treatment in comparison to the untreated cells (Mock). 800 HeLa cells were scored and these are results of three independent experiments. (C) Transcription activation by ING B is dependent on intact NF-kB binding site. Jurkat cells were transfected with pBlue3′LTR-Luc (NF-κB WT) and with the construct pBlue3′LTR NF-κB MUT-Luc (NF-κB BS−) harboring mutations at NF-κB binding sites that prevent its interaction. After transfections, cells were treated with ING B (0.32 μM). Luciferase assays were performed 24 h post-treatment.
The second approach involved the use of LTR-Luciferase reporter assays to quantify NF-κB nuclear function. HIV transcription is sensitive to NF-κB activation as its promoter carries multiple copies of the NF-κB recognition sequence. HIV-LTR subtype B region has two NF-κB binding sites defined by the conserved sequence GGGACTTTCC (Bachu et al., 2012; Naghavi et al., 1999). To verify if HIV transcriptional activation by ING B is mediated by NF-κB binding to the LTR promoter, Jurkat cells were transfected with pBlue3′LTR-Luc (WT) and pBlue3′LTR NF-κB MUT-Luc (NF-κB BS−) that express the luciferase reporter gene controlled by intact and mutated NF-κB binding sites, respectively (Dong et al., 2000). Transfected cells were treated with ING B and transcription activation was evaluated by luciferase activity. ING B treatment induced a 10 fold increase in luciferase expression in cells containing the intact pBlue3′LTR-Luc (WT) plasmid (Fig. 3C). As expected, no difference was observed in the luciferase expression under the control of mutagenized NF-κB binding sites (NF-κB BS−), suggesting that transcriptional activation promoted by ING B is dependent on intact NF-κB binding site of HIV LTR (Fig. 3C).
Our results show that ING B treatment activates PKC signaling and NF-κB translocation to the nucleus, activating in turn transcription from the HIV LTR promoter.
LTR from different subtypes of HIV-1 are sensitive to ING B
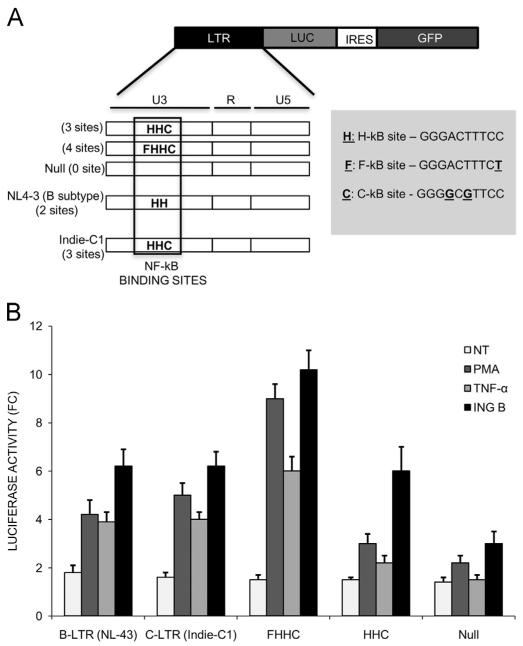
The LTR promoter region of HIV subtype C contains an extra NF-κB binding site (GGGGCGTTCC) that differs from the classical sequence (GGGACTTTCC). Indeed, subtype C of Indian origin contains a fourth NF-κB binding site (GGGACTTTCT) that has been associated with higher viral loads in these patients (Bachu et al., 2012; Dong et al., 2000). In order to check if ING B can activate virus transcription in the context of both subtype B and C LTRs, Jurkat cells were transfected with luciferase expression vectors under the control of LTR promoters from classical subtype B, harboring two NF-κB binding sites, and from subtype C, containing 3 or 4 NF-κB binding sites.
The two classical NF-kB binding sites were nominated as H, and the third and fourth NF-κB binding site found in Indian HIV strains were identified as C and F, respectively (Fig. 4A). All these versions of the NF-κB binding site are functional, as described before (Bachu et al., 2012). The cells were treated with different transcription activators (ING B, TNF-α and PMA) and the levels of luciferase expression compared to non-treated cells. All the treatments induced luciferase expression regardless the subtype origin of the LTR regions (Fig. 4B). ING B induced higher levels of luciferase expression compared to TNF-α and PMA in all conditions (Fig. 4B, compare black with gray bars). Importantly, the LTR region of subtype C harboring the forth extra NF-κB binding site (FHHC) was more responsive to ING B treatment than LTR promoters carrying less NF-κB binding sites, reinforcing the previous results that transcription activation promoted by this compound is mediated by NF-κB binding.
Fig. 4.
Non-B subtype LTR promoters are also activated by ING B. (A) Schematic representation of B and C subtype LTR enhancers used. The numbers of NF-κB binding sites are highlighted in the figure. The constructs were denominated Null (without NF-κB binding site), B-LTR (with two canonical NF-κB binding sites), C-LTR and HHC (both with three NF-κB binding sites) and FHHC-LTR (with four NF-κB binding sites). All of these LTR regions described were cloned in the same backbone vector expressing gaussia luciferase. (B) ING B activates luciferase expression controlled by LTR regions from subtype B and C of HIV-1. Luciferase activity in fold change of Jurkat cells transfected with the plasmids cited above and treated with three different activator compounds for 24 h: PMA (1 μM), TNF-α (20 ng/ml) and ING B (0.32 μM). NT stands for non-treated control cells and is represented by white bars.
These results suggest that ING B elicits activation of HIV transcription regardless of HIV-LTR subtype and hence could be used to tackle latency from reservoirs formed by different HIV subtypes.
ING B is the most potent anti-HIV latency drug
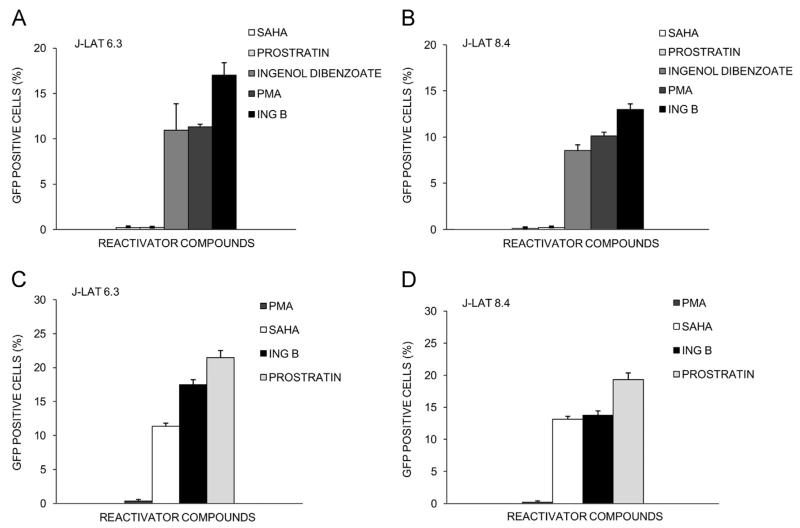
We compared the ability of ING B to other previously described HIV activators in their ability to reactivate HIV from latency. Initially, J-Lat 6.3 and 8.4 were treated with PMA, prostratin, SAHA and ingenol 3,20-dibenzoate at the minimum concentration of ING B capable to produce HIV activation without cytotoxic effects (0.32 μM). ING B was a superior activator reaching higher levels of HIV transcription in both clones of J-Lat cells 6.3 (Fig. 5A) and 8.4 (Fig. 5B). SAHA and prostratin had no effect on HIV transcription at 0.32 μM (Fig. 5A and B). In J-Lat 6.3 cells, ING B activates up to 18–20% of the cells, while ingenol 3,20-dibenzoate and PMA activate 12% of the cells. In J-Lat 8.4 cells, ING B activates up to 14% of the cells while ingenol 3,20-dibenzoate and PMA activate only 8% and 10%, respectively.
Fig. 5.
ING B compares favorably to other known compounds in HIV reactivation potential. J-Lat cells were treated with different transcriptional activators (ING B, PMA, prostratin, SAHA and ingenol 3,20-dibenzoate) for 24 h. First, J-Lat 6.3 cells (A) and J-Lat 8.4 cells (B) were incubated with each compounds (PMA, prostratin, SAHA and ingenol 3,20-dibenzoate) at 0.32 μM, which is the best concentration of ING B described and percentage of GFP positive cells was determined by FACS. Next, J-Lat 6.3 cells (C) and J-Lat 8.4 cells (D) were incubated with optimal concentrations of each compound (PMA, 20 nM; prostratin, 10 μM; SAHA, 10 μM and ING B, 0.32 μM) and percentage of GFP positive cells were also determined by FACS. Error bars represent the standard deviation of triplicate experiments.
Since SAHA and prostratin did not activate HIV transcription at 0.32 μM, cells were treated with the optimal concentrations described for these inhibitors (Contreras et al., 2009; Williams et al., 2004; Wu et al., 2005). Therefore the reactivation levels of ING B at 0.32 μM were compared to the obtained using higher concentrations of PMA (20 nM), prostratin (10 μM) and SAHA (10 μM) in both clones, J-Lat 6.3 (Fig. 5C) and 8.4 (Fig. 5D). ING B activated 30% more cells than SAHA in J-Lat 6.3 cells (Fig. 5C) and the same levels in J-Lat 8.4 cells (Fig. 5D). Here, prostratin treatment reactivated a higher number of cells from latency compared to ING B in both cell lines, when used at higher concentrations (10 μM).
These results suggest that ING B is a superior candidate to promote HIV transcription compared to other HIV activators such as ingenol 3,20-dibenzoate, SAHA and PMA.
ING B activates HIV transcription in infected primary resting CD4+ T cells
To expand the results of ING B activation to primary cells, the levels of transcription activation were evaluated in peripheral blood mononuclear cells (PBMC) isolated from healthy donors expressing the luciferase reporter gene under LTR control (pBlue3′LTR-Luc). The ING B treatment promoted potent activation of luciferase expression in PBMCs from all tested donors (Fig. S3A). Since PKC activators such as PMA and prostratin are able to mediate cellular activation, we investigated the effects of ING B in primary cells activation measuring CD38, HLA-DR and CD69 markers in human lymphocytes. Three different activator compounds were used at 1 μM concentration (ING B, PMA and prostratin). All of them were able to activate primary cells at this concentration after 48 h induction (Fig. S3B). ING B activated primary cells at a higher level than prostratin evidenced by the induction of early and late activation markers (Fig. S3B). On the other hand, PMA demonstrated the highest activation levels at this concentration when compared to ING B and prostratin.
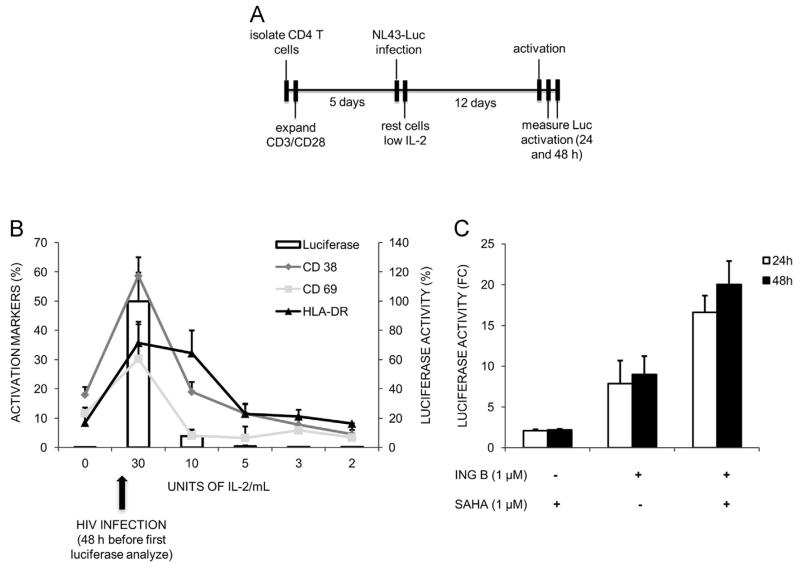
Resting CD4+ T lymphocytes are the body reservoir of transcriptionally silent HIV in infected individuals, hence these cells are the best model to evaluate ING B ability to reactivate virus from latency (Finzi et al., 1997; Geeraert et al., 2008;Bartholomeeusen et al., 2013). Naïve CD4+ T cells were isolated, activated with anti-CD3/CD28 and infected with a pseudotyped HIV-1 harboring a luciferase reporter gene. This virus is defective in the envelope gene and restricted to a single cycle of replication, which allows the virus to integrate in the cellular genome and establish latency. After infection, cells were allowed to return to the resting state by gradually decreasing IL-2 concentration (30– 2 U/ml) over the course of two weeks. The entire experiment is depicted in the Fig. 6A. The resting state of the cells was verified by detection of activator markers (CD38, CD69 and HLA-DR) by FACS and virus latency was evaluated by measuring luciferase activity during all IL-2 reduction points (Fig. 6B).
Fig. 6.
ING B reactivates latent HIV-1 in primary CD4+ T cells. Naïve CD4+ T cells were isolated from three different healthy donors. CD4+ T cells were activated and infected with an envelope defective HIV-1 virus and returned to a resting state by gradually reducing IL-2 concentration (30–2 U/ml) in the medium. Luciferase activity and levels of activator markers CD38, CD69 and HLA-DR were measured before and at intermediate time points after infection. After resting, cells were stimulated with SAHA, ING B or both. Luciferase activity was measured 24 and 48 h later. (A) Chronological representation of the entire experiment. (B) Average levels of CD38, CD69, HLA-DR and luciferase activity at different time points are indicated. Error bars indicate the standard error of triplicate experiments. Black arrow below the graph indicates the point that HIV infection was performed. (C) Reactivation levels of latent HIV-1 in the resting CD4+ T cells. ING B (1 μM) and SAHA (1 μM) were used alone or in combination as indicated. Luciferase activity was analyzed 24 and 48 h after activation (white and black bars respectively) and is shown as fold change relative to non-treated cells (FC). Error bars indicate the standard deviation of triplicate experiments.
HIV latency was established in all replicates, as confirmed by the low levels of luciferase expression and reduced activation markers (CD69, CD38 and HLA-DR) at the lowest concentration of IL-2 (2 U/ml) (Fig. 6B). At this point cells were treated with ING B, SAHA or both and the levels of HIV transcription were evaluated by luciferase expression 24 and 48 h post-treatment (Fig. 6C). The level of HIV transcription activation was higher with ING B treatment (up to 12-fold) than with SAHA (Fig. 6C). No differences of luciferase expression were observed 24 or 48 h post-treatment either with SAHA or ING B. Indeed, higher levels of HIV activation were observed when ING B was combined with SAHA (20-fold), suggesting an additive effect between these different classes of HIV activators (Fig. 6C).
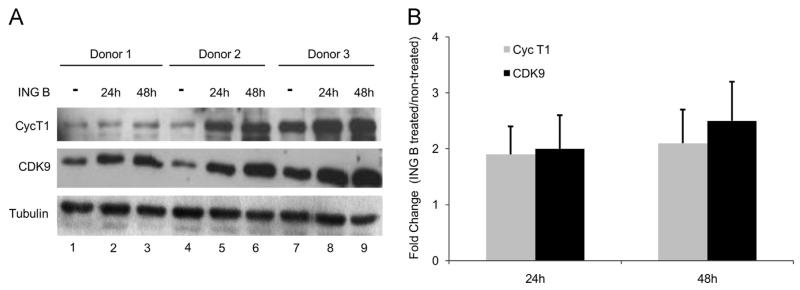
Several reports suggested that transcription initiation promoted by PKC activation and NF-κB binding is not sufficient for HIV activation, indicating that transcription elongation is also required. It is well known that HIV active transcription depends on P-TEFb activation. In order to verify if ING B exerts some influence on the expression of P-TEFb components, naïve T CD4+ cells were isolated from different healthy blood donors and treated with 1 μM of ING B. The levels of P-TEFb components (CDK9 and Cyclin T1) were evaluated by immunoblotting. We observed an increase in protein levels of both P-TEFb components (CDK9 and Cyclin T1) in the T CD4+ cells isolated from all donors and treated with ING B (Fig. 7A). The quantification analysis showed the increase of CDK9 and Cyclin T1 protein levels (up to 2-fold) in ING B treated T CD4+ compared with non-treated cells (Fig. 7B). The same analysis was performed with the latent models of HIV (J-Lat cells), revealing that CDK9 and Cyclin T1 protein levels also increase in the ING B treated cells (Fig. S4A and B).
Fig. 7.
ING B induces higher levels of P-TEFb components in human primary cells. Naïve T CD4+ cells were isolated from three different healthy donors and treated with ING B (1 μM) for 24 or 48 h. After these periods, P-TEFb components (Cyc T1 and CDK9) expression was analyzed. (A) Cyc T1 (top panel) and CDK9 (middle panel) proteins levels were evaluated by immunoblotting of cell lysates from non-treated (mock) and ING B treated cells for 24 and 48 h. Tubulin (bottom panel) was used as loading control. Non-treated cells are presented in the lines 1, 4 and 7. (B) Quantification of the immunoblots from the three donors is shown as fold change (treated/non-treated cells ratio).
To validate our results with another ingenol derivate compound already described as PKC activator, we performed the same experiments with ingenol 3,20-dibenzoate (DBZ) (Racke et al., 2012). T CD4+ resting HIV infected cells were treated with DBZ, SAHA or HMBA alone or in combination. Ingenol 3,20-dibenzoate was combined with SAHA and HMBA at different concentrations (Fig. S5A and B). The treatment with ingenol 3,20-dibenzoate (DBZ) at 20 nM induced the HIV activation at higher levels than SAHA or HMBA at different concentrations (Fig. S5A and B). However, ING B induced even higher levels of HIV activation (up to 12-fold) than ingenol 3,20-dibenzoate (6-fold). The ingenol 3,20-dibenzoate combined with HMBA induced higher levels of HIV transcription if compared with the same combination with SAHA (Fig. S5A and B).
The same HIV infected resting cells described above were treated with ingenol 3,20-dibenzoate and the levels of P-TEFb components were also evaluated by immunoblotting using anti-bodies anti-cyclin T1 and CDK9 (Fig. S5C and D). The results indicate that ingenol 3,20-dibenzoate induced a higher expression of both P-TEFb components (Cyclin T1 and CDK9).
We thus concluded that ING B is able to activate virus transcription in HIV reservoirs by promoting both transcription initiation, mediated by NF-kB nuclear translocation, and increasing of levels of P-TEFb components, suggesting potential usefulness in HIV/AIDS cure strategies.
Discussion
In this work we addressed the ability of three new semi-synthetic ingenol esters, ING A, ING B, and ING C, to reactivate HIV latency. Chemical modifications at the C-3 position of ingenol produced ING B, an ester with the best reactivation/cytotoxicity profile. This new compound activates the PKC pathway by promoting NF-kB internalization into the nucleus and increasing HIV transcription, which is dependent on NF-kB binding to LTR promoter. ING B was also efficient in reactivating HIV transcription directly in resting T CD4+ cells as primary model of HIV latency. Ingenol derivates also induced the protein levels of P-TEFb components (Cyclin T1 and CDK9) that could explain the additive effect found in activation of HIV when cells were treated with ingenol in the presence of either SAHA or HMBA.
There are several publications describing anti-cancer activities of ingenol esters and the HIV transcription activation properties of this class of drugs (Ersvaer et al., 2010; Fujiwara et al., 1998; Hong et al., 2011; Lee et al., 2010; Warrilow et al., 2006). Ingenol-3-angelate (referred to as PEP005) is derived from the plant Euphorbia peplus and is currently in clinical trials for eradicating basal cell carcinoma, actinic keratosis (a disease associated with photo-damaged skin that may lead to invasive squamous cell carcinoma) and squamous cell carcinoma (SCC) in situ by topical application (Ersvaer et al., 2010; Li et al., 2010.; Song et al., 2013). Ingenol-3-angelate also shows pro-apoptotic effects in several malignant cells, including melanoma cell lines and primary human acute myelogenous leukemia cells (Ersvaer et al., 2010; Li et al., 2010; Olsnes et al., 2009). Similar to ING B, PEP005 is a broad range activator of the classical (α, β, γ) and novel (δ, ε, ν, θ) protein kinase C isoenzymes (Ersvaer et al., 2010) and is capable to activate NF-κB (Olsnes et al., 2009). The results with PEP005 are very promising and this drug is FDA approved for the topical treatment of actinic keratoses (AK). Ingenol-3-angelate also inhibits HIV infection by downmodulating CD4 and CXCR4 receptors and enhances HIV-1 replication in chronically infected cells, reiterating the clinical application of ingenol compounds to HIV cure (Ersvaer et al., 2010; Hong et al., 2011; Olsnes et al., 2009). Other important medical effects obtained using ingenol derivates such as ingenol mebuate and ingenol 3,20-dibenzoate have been published (Aditya and Gupta, 2013; Racke et al., 2012). Our comparison of ING B with the latter compound showed that ING B was more efficient in activating HIV from latency than ingenol 3,20-dibenzoate.
We demonstrated that the ING B action mechanism is based on the activation of both classes of PKC isoforms: calcium dependent and independent. Gö 6976 is a PKC inhibitor specific for Ca2+ dependent (PKC-α and β) and Gö 6983 targets both classes (PKC-α, PKC-β, PKC-γ, PKC-δ and PKC-ζ), similar to Ro-31-8220 that inhibits PKC-α, PKC-β, PKC-ε and PKC-γ. All these inhibitors block ING B activation, suggesting that this compound is a broad range PKC agonist. These results were confirmed by confocal microscopy that showed that ING B treatment promotes translocation of calcium dependent and independent PKC isoforms (PKC-α, PKC-γ and PKC-δ) to the cytoplasmic membrane. PKC activation leads to NF-κB nuclear translocation and this cellular factor appears to be the key factor required for latent provirus reactivation (Bachu et al., 2012; Spitaler and Cantrell, 2004; Steinberg, 2008; Torgerson et al., 1998; Van Lint et al., 2013; Williams et al., 2004; Dahabieh et al., 2014). We observed that constructs harboring deficient NF-κB binding sites in the HIV-LTR are not responsive to ING B activation. Indeed, ING B was able to induce NF-κB nuclear translocation regardless of HIV infection. Overall, LTR regions of HIV subtype C presenting duplications of NF-kB binding sites were more responsive to ING B transcription activation, if compared with HIV LTR from subtype B viruses. These are promising and innovative results concerning anti-latency agent activity in non-B HIV subtypes. Our results suggest that HIV subtype C viruses can potentially be more responsive to transcription activators compounds that target NF-kB interaction. This aspect should be considered in latency studies, since subtype C accounts for the majority of worldwide infections (Rossenkhan et al., 2013).
The variation in the number of transcription factor-responsive elements in the LTR promoter has been shown to differentially regulate viral transcription and replication in a dose-dependent manner upon exposure of HIV-infected cells to cytokines, such as TNF-α (Montano et al., 2000; Opijnen et al., 2004). It is known that a single NF-κB binding site is present in the promoter of HIV-1 subtype E (HIV-1E), yet subtype B contains two, with subtype C containing at least three, but as many as four, NF-κB consensus sequences (Bachu et al., 2012; Roof et al., 2002; Rossenkhan et al., 2013). Moreover, Tat transactivation potential from different sub-types is not uniform: for example, subtype E and C Tat are strong mediators of LTR transcription compared to subtype B, and this is thought to be due to a higher affinity for the TAR hairpin (Desfosses et al., 2005). Tat half-life from subtype E is about twice as long as that from subtypes B and C, which may be a compensatory mechanism for the reduction in NF-κB binding sites (Desfosses et al., 2005; Rossenkhan et al., 2013). All these reports support the fact that prolonged high viremia has been shown in a subset of patients during primary HIV-1 subtype C infection. It is during this primary phase of infection that the reservoir of latently infected, resting CD4+ T cells is established and anti-latency potential candidates should be considered to restore complete viral gene expression in subtype C infected patients.
ING B activates virus transcription in our ex vivo model using primary human resting CD4+ T lymphocytes infected with HIV-1. This result was even more striking when ING B was combined with SAHA, suggesting an additive effect between these compounds. Additive and synergistic activation generally occurs when molecules act in different activation pathways. There are some descriptions about synergism among reactivator compounds leading to higher levels of gene expression when used together (Remoli et al., 2012). It has been demonstrated that histone deacetylase inhibitor and PKC agonist can work synergistically as shown by SAHA and prostratin (Burnett et al., 2010; Reuse et al., 2009). We hypothesized that PKC activation mediated by ING B in addition with HDACi (SAHA) effects lead to a more robust activation of viral transcription. Nevertheless, some activator compounds have failed to reactivate HIV in vivo (Bullen et al., 2014). One effective single agent able to disrupt HIV latency in vivo was the PKC agonist bryostatin-1, which is probably too toxic for clinical use (Bullen et al., 2014). Overall, apparent clinical side effects of latency-reversing agents such as PKC agonists administration to patients include respiratory distress syndrome, hypotension, and other toxicities due to the release of pro-inflammatory cytokines via non specific cellular activation (Maioli and Fortino, 2004; McKernan et al., 2012; Strair et al., 2002). The search for other compounds that reactivate latent HIV, independently of action pathway, and that can also be safely administered to patients must proceed. Further progress may depend on finding safe and active combinations of latency-reversing agents (Bullen et al., 2014). Although our findings demonstrated safe cytotoxicity profile in vitro and ex vivo models with ING B we did not address toxicity in animal models or for clinical treatment.
Additionally, both ING B and ingenol 3,20-dibenzoate could induce differential expression of P-TEFb subunits, such as CDK9 and Cyc T1. Interestingly, we and others observed little effect on HIV latency of HDAC inhibition in primary resting CD4+ cells (Friedman et al., 2011; Sahu and Cloyd, 2011; Tyagi et al., 2010). In these cells, levels of P-TEFb are particularly low due to actions of specific miRNAs and NF-90, which block the translation of CycT1 mRNA (Bartholomeeusen et al., 2013; Hoque et al., 2011). Since primary resting cells contain insignificant levels of P-TEFb, these had to be increased first with PKC agonists before effects of the most potent HDACi, SAHA, could be observed. The effects of ING B on P-TEFb could explain the results of additive effects of ING B with SAHA to reactivate HIV from latency. This combination doubled virus transcription compared to ING B treatment alone and a ten-fold induction was detected compared to the induction obtained with SAHA alone. These compounds could be administrated together or sequentially to lower chromatin condensation and release P-TEFb from its inactive complex. We envisage that if ingenol is given first, it would also lead to P-TEFb synthesis and release, boosting SAHA’s effect on HIV reactivation. It is important to point out that the additive effect was not observed with the close related molecule ingenol 3,20-dibenzoate, indicating that it is not a feature shared by all ingenol compounds.
Conclusions
In our quest for an effective agent capable of reactivating latent HIV reservoirs, we found a promising lead: 3-caproyl-ingenol (ING B). HIV latency reactivators could be applied intermittently to the patients undergoing anti-retroviral therapy. Based on the results presented here, it would be expected that several rounds of treatments with ING B would result either in complete elimination of HIV or decrease in viral load to the point at which the immune system could complete the elimination of this pathogen. Several aspects favor ING B use as follows: (a) there would be abundant source of ingenol since the Brazilian Euphorbia tirucalli is easily grown in plantations; (b) ingenol derivatives have already been used therapeutically in humans to tackle diseases (Aditya and Gupta, 2013; Ersvaer et al., 2010) and their potential in HIV activation has been largely described (Fujiwara et al., 1998; Hong et al., 2011; Spitaler and Cantrell, 2004; Steinberg, 2008; Van Lint et al., 2013); (c) we obtained a proof of principle that ING B does reactivate latent HIV ex vivo. Additionally, this has also been proven in a study with Rhesus macaque model where it was shown that treatment with ING B could activate SIV mac 251 latency in Rhesus monkeys by oral administration and had an excellent toxicity profile (manuscript in preparation). All these in vivo properties in non-human primate models added to the molecular mechanisms of reactivation presented here, strongly recommends 3-caproyl-ingenol (ingenol B, ING B) as a great candidate for future clinical trials for HIV eradication.
Materials and methods
Cell lines
Jurkat, J-Lat 6.3, J-Lat 8.4 and Hella cells were obtained from National Institute of Health (NIH). Primary cells were isolated from healthy blood donors from Hemorio, Rio de Janeiro, Brazil. Jurkat, J-Lat 6.3, J-Lat 8.4 and primary cells were grown in RPMI 1640 medium containing penicillin (100 IU/ml), streptomycin (100 g/ml), and 10% fetal bovine serum at 37°C with 5% CO2. Hella cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) containing the same supplements and conditions.
Reagents
The natural compound ingenol-3-dodecatrienoates extracted from Euphorbiaceae and the derivate esters Ingenol A, B and C were isolated by Kyolab Laboratories (São Paulo, Brazil). Ingenol 3,20-dibenzoate, PMA, prostatin and the PKC inhibitors Gö 6976, Gö 6983 and Ro-31–8220 were obtained from Santa Cruz Biotechnology. SAHA was obtained from SelleckChem (Houston, USA). All these compounds were diluted to specific concentrations in DMEM or RPMI, and the final concentration of solvent (DMSO) was always less than 1%. We used 20 ng/ml of TNF-α (eBiosciences) as positive control of reactivation. Levels of P-TEFb components were evaluated by immunoblotting using specific antibodies anti-CDK9 and anti-Cyc T1 (Santa Cruz Biotechnologies, cat. number 484 and 10750, respectively). Anti-tubulin (Abcam, cat. number 56676) was used to normalize proteins level.
Latency reactivation and cell viability assays
J-Lat cells (clones 6.3 and 8.4) were used as HIV-1 latency model. These cells harbor a HIV-GFP latent proviral clone integrated in the genome (Jordan et al., 2003). The level of reactivation was measured by flow cytometry. Before expansion, cells were treated with different concentrations of each compound, ING (parent molecule), Ingenol A, B and C and ingenol 3,20-dibenzoate, separately at different times. Cells (2 × 105) were fixed with paraformaldehyde (2%) for 30 min at 4 °C followed by PBS washing. In the experiments involving PKC inhibitors, 1 μM of each compound was separately added in the cell culture 24 h before the treatment with ING B (0.32 μM). TNF-α (20 ng/ml) was used as positive control. Flow cytometry analyses were performed 24 h after ING B addition. Cell viability assays were performed using Cell Titer Blue (Promega) following manufacturer’s instructions. Briefly, J-Lat 6.3 and 8.4 (104 cells) were plated and treated with each compound at different concentrations. Cell Titer Blue reagent was added five days post-treatment and followed by overnight incubation. Absorbance levels were evaluated by spectrophotometer at 595 nm.
HIV-1 p24 antigen ELISA
J-Lat cells 6.3 and 8.4 were treated with ING B (0.32 μM) for 48 h. TNF-α (20 ng/ml) was used as positive control to particle release, for the same time of induction (48 h). Virus production was evaluated by ELISA HIV-1 p24 antigen detection (Zeptometrix) from culture supernatants. Absorbance levels were evaluated at 450 nm using a spectrophotometer.
Plasmids and site directed mutagenesis
The luciferase expressing vector (pBlue3′ LTR-LUC B) was obtained from NIH. This vector expresses firefly luciferase gene under HIV-1 LTR control. Site directed mutagenesis at the two NF-κB binding site of this same plasmid was performed using Quick Change Lighting Multi Site Directed Mutagenesis (Stratagene), following manufacturer’s instructions. The primers NF-κB MutF - 5′- CGA GCT TGC TAC AAG CGA CTT TCC GCT GGC GAC TTT CCA GGG AGG −3′ and NF-κB MutR - 5′- CCT CCC TGG AAA GTC GCC AGC GGA AAG TCG CTT GTA GCA AGC TCG −3′ were designed to perform the site directed mutagenesis. This pair of primers changed the second guanine at both NF-κB binding sites into LTR promoter to a cytosine, resulting in the plasmid designed here as pBlue3′LTR NF-κB MUT-Luc (NF-κB BS−) lacking functional NF-κB binding sites.
Transfections, ING B induction and luciferase assays
Jurkat cells (5 × 105) were transiently transfected with 15 μg of plamids pBlue3′LTR-Luc wild type (WT) and pBlue3′LTR NF-κB MUT-Luc (NF-κB BS−) using the Neon Transfection System (Life Technologies) according manufacturer’s instructions. After transfections, cells were equally separated in two groups: treated and non-treated with 0.32 μM of ING B. Luciferase assay was performed 24 h after ING B induction. Cell lysates were obtained using Luciferase Reporter Lysis Buffer (Promega) and the luciferase activity measured using GloMax luminometer (Promega).
PKC activation and confocal microscopy
Three different plasmids coding for human PKC isoforms fused with GFP reporter (PKCα-GFP, PKCδ-GFP and PKCγ-GFP) were gently provided by Dr. Morgan Huse (Memorial Sloan-Katterin Cancer Center, NY, USA). HeLa cells (2 × 104) were grown in Black 96 Well Cell Culture Microplate (Corning) at 70–80% of confluence. Afterwards, transient transfection was performed with human PKCα-GFP, PKCδ-GFP and PKCγ-GFP plasmids using Fugene HD (Promega) according manufacturer’s instructions. ING B (1 μM) was added 24 h post transfection and the cells were fixed with paraformaldehyde (2%) at different times. Positive control was performed using PMA (1 μM). The cells were washed with PBS and submitted to DAPI staining. Finally, the sub-cellular localization of PKC-GFP was evaluated using a High Content Screening confocal microscope (Molecular Devices, Inc). GFP-positive cells (200 per well) were counted using ImageJ software and the relative percentage of PKC-activated cells were measured for each PKC isoform.
NF-κB nuclear translocation
HeLa cells (2 × 104) were plated in Black 96 Well Cell Culture Microplate (Corning). Cells were treated with 1 μM of PMA (positive control) or ING B (1 μM). After induction, cells were fixed with paraformaldehyde (2%) at different times and immunofluorescense assay was performed to endogenous subunits of NF-kB using anti-p65 (1:300) (Santa Cruz Biotechnologies, cat. number 109). Nuclear staining was performed using DAPI. Finally, NF-κB nuclear translocation was analyzed using the Translocator Module of MetaXpress software of the High Content Screening confocal microscope (Molecular Devices, Inc). Each treatment was performed three times and about 800 cells in average were analyzed per treatment. In order to detect NF-kB internalization the cells were scored using DAPI staining and the NF-kB visualized using Alexa Fluors 594 Donkey anti-Rabbit IgG (1:1000) secondary antibody (Life Technologies, cat. number A-21207).
Transcription activation of reporter genes controlled by B and C LTR subtypes
Four different constructs derivate from the dual-reporter vector pLTR-sLuc-IRES-EGFP that simultaneously expresses two different reporter genes, secreted Gaussia luciferase (sLuc) and enhanced green fluorescent protein (EGFP), were gently provided for Dr. Udaykumar Ranga (HIV-AIDS Laboratory, Molecular Biology and Genetics Unit, Jawaharlal Nehru Centre for Advanced Scientific Research, Bengaluru, India). The first construct comprises a typical B LTR, and was called B-LTR (NL-43). The second one comprises a typical C LTR derivate for a HIV subtype C from India, described here as C-LTR (Indie1). Typically, LTR from HIV C-subtype comprises three NF-κB binding sites. A third construct comprises a C-LTR with four NF-κB binding sites was designated as FHHC-LTR. The forth one was designated null mutant been a LTR carrying no NF-κB binding site. All constructs were previously described (Bachu et al., 2012). Jurkat cells were transfected with all these constructions separately by electroporation. Six hours post transfections the cells were separated in two groups (treated or not-treated) and submitted to the following treatments: PMA (1 μM), TNF-α (20 ng/ml) or ING B (0.32 μM). The cells lysates were obtained 24 h post treatment and the luciferase activity evaluated using the Gaussia Luciferase kit (New England BioLabs) (Bachu et al., 2012).
Transcription activation in primary cells
Buffy coat from different healthy adults were obtained from Hemorio Blood Bank, Rio de Janeiro, Brazil. PBMC was isolated using Ficoll Paque Plus (GE Lifesciences), according to manufacturer’s instructions. Cells were grown in RPMI 1640 medium containing penicillin (100 IU/ml), streptomycin (100 g/ml), and 10% fetal bovine serum at 37 °C with 5% CO2. Isolated PBMCs from three different donors were separately transfected with pBlue3′TR-Luciferase B (WT) using Neon Transfection System (Life Technologies) according manufacturer’s instructions, using the following conditions: 12 μg of plasmid were mixed to 106 cells and after that, cells were exposed to one pulse with 20 ms of width and voltage of 2,150 V. After transfections, cells were separated in two groups: non-treated and treated with ING B (1 μM). Luciferase assays were performed 24 h after treatments, as described above.
Activation markers analyses
For analysis of cellular activation marker expression, enrichment T CD4+ cells from human PBMCs were incubated with 1 μM of each compound (ING B, PMA and prostratin) at 37 °C for 48 h. After incubation, cells were washed with PBS and incubated for 20 min at room temperature with antibodies against CD69-Pacific Blue (clone FN50 – BD Biosciences), CD38-FITC (clone AT-1 – Stem Cell Technologies), and HLA-DR-Qdot 605 (clone TÜ36 – Life Technologies). Activation markers were evaluated on a LSR Fortessa cytometer (BD Biosciences). Data analysis was performed using FlowJo software (Tree Star).
Preparation, infection, resting and reactivation of primary CD4+ T cells
Buffy coat from different healthy adults were obtained from Hemorio Blood Bank, Rio de Janeiro, Brazil. PBMC was isolated using Ficoll Paque Plus (GE Lifesciences), according to manufacturer’s instructions. Naïve CD4+ T cells were purified using the Dynabeads® Untouched™ Human CD4 T Cells kit (Invitrogen) according to the manufacturer’s protocol. CD4+ T cells were maintained in RPMI, 10% FBS, 30 U/ml IL-2 at 37 °C with 5% CO2 for 24 h. After this period, cells were activated using anti-CD3/anti-CD28 Dynabeads (Invitrogen) and maintained for 5 days in RPMI with the same conditions above described. Purity and activation levels were determined by FACS analysis using specific antibodies against CD4, CD69 and HLA-DR (BD Biosciences). Activated CD4+T cells were infected by spin inoculation using a luciferase reporter virus pseudotyped with HIV envelope proteins (pNL4.3ΔenvΔNef-Luc). To produce this pseudotyped virus HEK-293T cells were transfected by Fugene HD (Promega) using the pNL4.3ΔenvΔNef-Luc and pEnvHIV at 1:3 proportion. The pEnvHIV plasmid was kindly provided by Dr. Kotaro Shirakawa. The virus was previously titrated in Jurkat cells and we used about 200 ng of p24 to infect the Naïve CD4+ T cells. Cells were reverted to a resting state by gradually decreasing the amount of IL-2 in the medium to 2 U/ml over the period of 12 days. Luciferase activity was measured over this period until they reached a background plateau. After this period, reactivation treatment with the following drugs alone or conjugated in different concentrations was performed for 24 and 48 h: ING B (1 μM), SAHA (330 and 1000 mM), HMBA (330 and 3000 μM) and ingenol 3,20 dibenzoate (20 nM). HIV lantency reactivation was detected by luciferase activity as previously described.
Supplementary Material
Acknowledgments
We thank Dr. Morgan Huse and Dr. Kotaro Shirakawa to gently provide the human PKC-GFP plasmids (α, δ and γ) and pEnvHIV, respectively. We also thank Dr. Ana Lucia Moraes Giannini for English review. This work was supported by research grants from Brazilian agencies CNPq (Grant nos. 480861/2011-0, 405283/2013-0) and FAPERJ (Grant no. E-26/111.267/2013) and Diego Pandeló José was supported by CNPq fellowship. We acknowledge funding support to UR from DST, India toward the Indo-Brazil collaboration.
Footnotes
Appendix A. Supporting information
Supplementary data associated with this article can be found in the online version at http://dx.doi.org/10.1016/j.virol.2014.05.033.
References
- Aditya S, Gupta S. Ingenol mebutate: a novel topical drug for actinic keratosis. Indian Dermatol. Online J. 2013;4:246–249. doi: 10.4103/2229-5178.115538. http://dx.doi.org/10.4103/2229-5178.115538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archin NM, Espeseth A, Parker D, Cheema M, Hazuda D, Margolis DM. Expression of latent HIV induced by the potent HDAC inhibitor suberoylanilide hydroxamic acid. AIDS Res. Hum. Retrovir. 2009;25:207–212. doi: 10.1089/aid.2008.0191. http://dx.doi.org/10.1089/aid.2008.0191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archin NM, Liberty a L., Kashuba a D., Choudhary SK, Kuruc JD, Crooks a M., Parker DC, Anderson EM, Kearney MF, Strain MC, Richman DD, Hudgens MG, Bosch RJ, Coffin JM, Eron JJ, Hazuda DJ, Margolis DM. Administration of vorinostat disrupts HIV-1 latency in patients on antiretroviral therapy. Nature. 2012;487:482–485. doi: 10.1038/nature11286. http://dx.doi.org/10.1038/nature11286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachu M, Yalla S, Asokan M, Verma A, Neogi U, Sharma S, Murali RV, Mukthey AB, Bhatt R, Chatterjee S, Rajan RE, Cheedarla N, Yadavalli VS, Mahadevan A, Shankar SK, Rajagopalan N, Shet A, Saravanan S, Balakrishnan P, Solomon S, Vajpayee M, Satish KS, Kundu TK, Jeang K-T, Ranga U. Multiple NF-κB sites in HIV-1 subtype C long terminal repeat confer superior magnitude of transcription and thereby the enhanced viral predominance. J. Biol. Chem. 2012;287:44714–44735. doi: 10.1074/jbc.M112.397158. http://dx.doi.org/10.1074/jbc.M112.397158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartholomeeusen K, Fujinaga K, Xiang Y, Peterlin BM. Histone deacetylase inhibitors (HDACis) that release the positive transcription elongation factor b (P-TEFb) from its inhibitory complex also activate HIV transcription. J. Biol. Chem. 2013;288:14400–14407. doi: 10.1074/jbc.M113.464834. http://dx.doi.org/10.1074/jbc.M113.464834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullen CK, Laird GM, Durand CM, Siliciano JD, Siliciano RF. New ex vivo approaches distinguish effective and ineffective single agents for reversing HIV-1 latency in vivo. Nat. Med. 2014;20:425–429. doi: 10.1038/nm.3489. http://dx.doi.org/10.1038/nm.3489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnett JC, Lim K-I, Calafi A, Rossi JJ, Schaffer DV, Arkin AP. Combinatorial latency reactivation for HIV-1 subtypes and variants. J. Virol. 2010;84:5958–5974. doi: 10.1128/JVI.00161-10. http://dx.doi.org/10.1128/JVI.00161-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho W-K, Jang MK, Huang K, Pise-Masison C. a, Brady JN. Human T-lymphotropic virus type 1 Tax protein complexes with P-TEFb and competes for Brd4 and 7SK snRNP/HEXIM1 binding. J. Virol. 2010;84:12801–12809. doi: 10.1128/JVI.00943-10. http://dx.doi.org/10.1128/JVI.00943-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras X, Barboric M, Lenasi T, Peterlin BM. HMBA releases P-TEFb from HEXIM1 and 7SK snRNA via PI3K/Akt and activates HIV transcription. PLoS Pathog. 2007;3:1459–1469. doi: 10.1371/journal.ppat.0030146. http://dx.doi.org/10.1371/journal.ppat.0030146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras X, Mzoughi O, Gaston F, Peterlin MB, Bahraoui E. Protein kinase C-delta regulates HIV-1 replication at an early post-entry step in macrophages. Retrovirology. 2012;9:37. doi: 10.1186/1742-4690-9-37. http://dx.doi.org/10.1186/1742-4690-9-37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras X, Schweneker M, Chen C-S, McCune JM, Deeks SG, Martin J, Peterlin BM. Suberoylanilide hydroxamic acid reactivates HIV from latently infected cells. J. Biol. Chem. 2009;284:6782–6789. doi: 10.1074/jbc.M807898200. http://dx.doi.org/10.1074/jbc.M807898200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahabieh MS, Ooms M, Brumme C, Taylor J, Harrigan PR, Simon V, Sadowski I. Direct non-productive HIV-1 infection in a T-cell line is driven by cellular activation state and NFκB. Retrovirology. 2014;11:17. doi: 10.1186/1742-4690-11-17. http://dx.doi.org/10.1186/1742-4690-11-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desfosses Y, Solis M, Sun Q, Grandvaux N, Van Lint C, Burny A, Gatignol A, Wainberg MA, Lin R, Hiscott J. Regulation of human immunodeficiency virus type 1 gene expression by clade-specific tat proteins. J. Virol. 2005;79:9180–9191. doi: 10.1128/JVI.79.14.9180-9191.2005. http://dx.doi.org/10.1128/JVI.79.14.9180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Q, Kelkar S, Xiao Y, Joshi-Barve S, McClain CJ, Barve SS. Ethanol enhances TNF-alpha-inducible NFkappaB activation and HIV-1-LTR transcription in CD4+ Jurkat T lymphocytes. J. Lab. Clin. Med. 2000;136:333–343. doi: 10.1067/mlc.2000.110104. http://dx.doi.org/10.1067/mlc.2000.110104 [DOI] [PubMed] [Google Scholar]
- Ersvaer E, Kittang AO, Hampson P, Sand K, Gjertsen BT, Lord JM, Bruserud O. The protein kinase C agonist PEP005 (ingenol 3-angelate) in the treatment of human cancer: a balance between efficacy and toxicity. Toxins (Basel) 2010;2:174–194. doi: 10.3390/toxins2010174. http://dx.doi.org/10.3390/toxins2010174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finzi D, Hermankova M, Pierson T, Carruth LM, Buck C, Chaisson RE, Quinn TC, Chadwick K, Margolick J, Brookmeyer R, Gallant J, Markowitz M, Ho DD, Richman DD, Siliciano RF. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science. 1997;278:1295–1300. doi: 10.1126/science.278.5341.1295. http://dx.doi.org/10.1126/science.278.5341.1295 [DOI] [PubMed] [Google Scholar]
- Friedman J, Cho W-K, Chu CK, Keedy KS, Archin NM, Margolis DM, Karn J. Epigenetic silencing of HIV-1 by the histone H3 lysine 27 methyltransferase enhancer of Zeste 2. J. Virol. 2011;85:9078–9089. doi: 10.1128/JVI.00836-11. http://dx.doi.org/10.1128/JVI.00836-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara M, Okamoto M, Ijichi K, Tokuhisa K, Hanasaki Y, Katsuura K, Uemura D, Shigeta S, Konno K, Yokota T, Baba M. Upregulation of HIV-1 replication in chronically infected cells by ingenol derivatives Brief Report. Arch. Virol. 1998;143:2003–2010. doi: 10.1007/s007050050436. [DOI] [PubMed] [Google Scholar]
- Ganesh L, Burstein E, Guha-Niyogi A, Louder MK, Mascola JR, Klomp LWJ, Wijmenga C, Duckett CS, Nabel GJ. The gene product Murr1 restricts HIV-1 replication in resting CD4+ lymphocytes. Nature. 2003;426:853–857. doi: 10.1038/nature02171. http://dx.doi.org/10.1038/nature02171 [DOI] [PubMed] [Google Scholar]
- Geeraert L, Kraus G, Pomerantz RJ. Hide-and-seek: the challenge of viral persistence in HIV-1 infection. Annu. Rev. Med. 2008;59:487–501. doi: 10.1146/annurev.med.59.062806.123001. http://dx.doi.org/10.1146/annurev.med.59.062806.123001 [DOI] [PubMed] [Google Scholar]
- González E, Punzón C, González M, Fresno M. HIV-1 tat inhibits IL-2 gene transcription through qualitative and quantitative alterations of the cooperative Rel/AP1 complex bound to the CD28RE/AP1 composite element of the IL-2 promoter. J. Immunol. 2001;166:4560–4569. doi: 10.4049/jimmunol.166.7.4560. [DOI] [PubMed] [Google Scholar]
- Hong K-J, Lee HS, Kim Y-S, Kim SS. Ingenol protects human T cells from HIV-1 infection. Osong Public Heal. Res. Perspect. 2011;2:109–114. doi: 10.1016/j.phrp.2011.07.001. http://dx.doi.org/10.1016/j.phrp.2011.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoque M, Shamanna RA, Guan D, Pe’ery T, Mathews MB. HIV-1 replication and latency are regulated by translational control of cyclin T1. J. Mol. Biol. 2011;410:917–932. doi: 10.1016/j.jmb.2011.03.060. http://dx.doi.org/10.1016/j.jmb.2011.03.060.HIV-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Wang F, Argyris E, Chen K, Liang Z, Tian H, Huang W, Squires K, Verlinghieri G, Zhang H. Cellular microRNAs contribute to HIV-1 latency in resting primary CD4+ T lymphocytes. Nat. Med. 2007;13:1241–1247. doi: 10.1038/nm1639. http://dx.doi.org/10.1038/nm1639 [DOI] [PubMed] [Google Scholar]
- Ishida T, Hamano A, Koiwa T, Watanabe T. 5′ long terminal repeat (LTR)-selective methylation of latently infected HIV-1 provirus that is demethylated by reactivation signals. Retrovirology. 2006;3:69. doi: 10.1186/1742-4690-3-69. http://dx.doi.org/10.1186/1742-4690-3-69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jassbi AR. Chemistry and biological activity of secondary metabolites in Euphorbia from Iran. Phytochemistry. 2006;67:1977–1984. doi: 10.1016/j.phytochem.2006.06.030. http://dx.doi.org/10.1016/j.phytochem.2006.06.030 [DOI] [PubMed] [Google Scholar]
- Johnston R. HIV cure: controversy, consensus, and a consortium. AIDS Res. Hum. Retrovir. 2010;26:943–946. doi: 10.1089/aid.2010.0087. http://dx.doi.org/10.1089/aid.2010.0087 [DOI] [PubMed] [Google Scholar]
- Jones KA, Peterlin BM. Control of RNA initiation and elongation at the HIV-1 promoter. Annu. Rev. Biochem. 1994;63:717–743. doi: 10.1146/annurev.bi.63.070194.003441. [DOI] [PubMed] [Google Scholar]
- Jordan A, Bisgrove D, Verdin E. HIV reproducibly establishes a latent infection after acute infection of T cells in vitro. EMBO J. 2003;22:1868–1877. doi: 10.1093/emboj/cdg188. http://dx.doi.org/10.1093/emboj/cdg188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karn J, Stoltzfus CM. Regulation of HIV-1 gene expression. Cold Spring Harb. Perspect. Med. 2012;2:1–17. doi: 10.1101/cshperspect.a006916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalaf H, Jass J, Olsson P-E. Differential cytokine regulation by NF-kappaB and AP-1 in Jurkat T-cells. BMC Immunol. 2010;11:26. doi: 10.1186/1471-2172-11-26. http://dx.doi.org/10.1186/1471-2172-11-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita S, Su L, Amano M, Timmerman L. a, Kaneshima H, Nolan GP. The T cell activation factor NF-ATc positively regulates HIV-1 replication and gene expression in T cells. Immunity. 1997;6:235–244. doi: 10.1016/s1074-7613(00)80326-x. [DOI] [PubMed] [Google Scholar]
- Lassen KG, Ramyar KX, Bailey JR, Zhou Y, Siliciano RF. Nuclear retention of multiply spliced HIV-1 RNA in resting CD4+ T cells. PLoS Pathog. 2006;2:650–661. doi: 10.1371/journal.ppat.0020068. http://dx.doi.org/10.1371/journal.ppat.0020068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W-Y, Hampson P, Coulthard L, Ali F, Salmon M, Lord JM, Scheel-Toellner D. Novel antileukemic compound ingenol 3-angelate inhibits T cell apoptosis by activating protein kinase Ctheta. J. Biol. Chem. 2010;285:23889–23898. doi: 10.1074/jbc.M109.041962. http://dx.doi.org/10.1074/jbc.M109.041962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Shukla S, Lee A, Garfield SH, Maloney DJ, Suresh V, Yuspa SH. The skin cancer chemotherapeutic agent ingenol-3-angelate (PEP005) is a substrate for the epidermal multidrug transporter (ABCB1) and targets tumor vasculature. Cancer Res. 2010;70:4509–4519. doi: 10.1158/0008-5472.CAN-09-4303. http://dx.doi.org/10.1158/0008-5472.CAN-09-4303.The [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Denlinger CE, Rundall BK, Smith PW, Jones DR. Suberoylanilide hydroxamic acid induces Akt-mediated phosphorylation of p300, which promotes acetylation and transcriptional activation of RelA/p65. J. Biol. Chem. 2006;281:31359–31368. doi: 10.1074/jbc.M604478200. http://dx.doi.org/10.1074/jbc.M604478200 [DOI] [PubMed] [Google Scholar]
- Maioli E, Fortino V. Protein kinase C: a target for anticancer drugs? Endocr. Relat. Cancer. 2004;11:161–162. doi: 10.1677/erc.0.0110161. [DOI] [PubMed] [Google Scholar]
- Mbonye U, Karn J. Transcriptional control of HIV latency: cellular signaling pathways, epigenetics, happenstance and the hope for a cure. Virology. 2014:454–455C. 328–339. doi: 10.1016/j.virol.2014.02.008. http://dx.doi.org/10.1016/j.virol.2014.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKernan LN, Momjian D, Kulkosky J. Protein Kinase C: one pathway towards the eradication of latent HIV-1 reservoirs. Adv. Virol. 2012;2012:1–8. doi: 10.1155/2012/805347. http://dx.doi.org/10.1155/2012/805347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochly-Rosen D, Gordon AS. Anchoring proteins for protein kinase C: a means for isozyme selectivity. FASEB J. 1998;12:35–42. [PubMed] [Google Scholar]
- Montano M. a., Nixon CP, Ndung’u T, Bussmann H, Novitsky V. a., Dickman D, Essex M. Elevated tumor necrosis factor-alpha activation of human immunodeficiency virus type 1 subtype C in Southern Africa is associated with an NF-kappaB enhancer gain-of-function. J. Infect. Dis. 2000;181:76–81. doi: 10.1086/315185. http://dx.doi.org/10.1086/315185 [DOI] [PubMed] [Google Scholar]
- Naghavi MH, Schwartz S, Sönnerborg a., Vahlne a. Long terminal repeat promoter/enhancer activity of different subtypes of HIV type 1. AIDS Res. Hum. Retrovir. 1999;15:1293–1303. doi: 10.1089/088922299310197. http://dx.doi.org/10.1089/088922299310197 [DOI] [PubMed] [Google Scholar]
- Olsnes AM, Ersvaer E, Ryningen A, Paulsen K, Hampson P, Lord JM, Gjertsen BT, Kristoffersen EK, Bruserud Ø. The protein kinase C agonist PEP005 increases NF-kappaB expression, induces differentiation and increases constitutive chemokine release by primary acute myeloid leukaemia cells. Br. J. Haematol. 2009;145:761–774. doi: 10.1111/j.1365-2141.2009.07691.x. http://dx.doi.org/10.1111/j.1365-2141.2009.07691.x [DOI] [PubMed] [Google Scholar]
- Van Opijnen T, Jeeninga RE, Boerlijst MC, Pollakis GP, Zetterberg V, Salminen M, Berkhout B. Human immunodeficiency virus type 1 subtypes have a distinct long terminal repeat that determines the replication rate in a host-cell-specific manner. J. Virol. 2004;78:3675–3683. doi: 10.1128/JVI.78.7.3675-3683.2004. http://dx.doi.org/10.1128/JVI.78.7.3675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierson T, Hoffman TL, Blankson J, Finzi D, Chadwick K, Margolick JB, Buck C, Siliciano JD, Doms RW, Siliciano RF. Characterization of Chemokine receptor utilization of viruses in the latent reservoir for human immunodeficiency virus type 1. J. Virol. 2000;74:7824–7833. doi: 10.1128/jvi.74.17.7824-7833.2000. http://dx.doi.org/10.1128/JVI.74.17.7824-7833.2000.Updated [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racke FK, Baird M, Barth RF, Huo T, Yang W, Gupta N, Weldon M, Rutledge H. Unique in vitro and in vivo thrombopoietic activities of ingenol 3,20 dibenzoate, a Ca(+ +)-independent protein kinase C isoform agonist. PLoS One. 2012;7:e51059. doi: 10.1371/journal.pone.0051059. http://dx.doi.org/10.1371/journal.pone.0051059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remoli AL, Marsili G, Battistini A, Sgarbanti M. The development of immune-modulating compounds to disrupt HIV latency. Cytokine Growth Factor Rev. 2012;23:159–172. doi: 10.1016/j.cytogfr.2012.05.003. http://dx.doi.org/10.1016/j.cytogfr.2012.05.003 [DOI] [PubMed] [Google Scholar]
- Reuse S, Calao M, Kabeya K, Guiguen A, Gatot J-S, Quivy V, Vanhulle C, Lamine A, Vaira D, Demonte D, Martinelli V, Veithen E, Cherrier T, Avettand V, Poutrel S, Piette J, de Launoit Y, Moutschen M, Burny A, Rouzioux C, De Wit S, Herbein G, Rohr O, Collette Y, Lambotte O, Clumeck N, Van Lint C. Synergistic activation of HIV-1 expression by deacetylase inhibitors and prostratin: implications for treatment of latent infection. PLoS One. 2009;4:e6093. doi: 10.1371/journal.pone.0006093. http://dx.doi.org/10.1371/journal.pone.0006093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richman DD, Margolis DM, Delaney M, Greene WC, Hazuda D, Pomerantz RJ. The challenge of finding a cure for HIV infection can we do better than HAART? Science. 2009;323:1304–1307. doi: 10.1126/science.1165706. [DOI] [PubMed] [Google Scholar]
- Roof P, Ricci M, Genin P, Montano M. a, Essex M, Wainberg M. a, Gatignol A, Hiscott J. Differential regulation of HIV-1 clade-specific B, C, and E long terminal repeats by NF-kappaB and the Tat transactivator. Virology. 2002;296:77–83. doi: 10.1006/viro.2001.1397. http://dx.doi.org/10.1006/viro.2001.1397 [DOI] [PubMed] [Google Scholar]
- Rossenkhan R, MacLeod IJ, Sebunya TK, Castro-Nallar E, McLane MF, Musonda R, Gashe B. a, Novitsky V, Essex M. tat Exon 1 exhibits functional diversity during HIV-1 subtype C primary infection. J. Virol. 2013;87:5732–5745. doi: 10.1128/JVI.03297-12. http://dx.doi.org/10.1128/JVI.03297-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Routy JP, Tremblay CL, Angel JB, Trottier B, Rouleau D, Baril JG, Harris M, Trottier S, Singer J, Chomont N, Sékaly RP, Boulassel MR. Valproic acid in association with highly active antiretroviral therapy for reducing systemic HIV-1 reservoirs: results from a multicentre randomized clinical study. HIV Med. 2012;13:291–296. doi: 10.1111/j.1468-1293.2011.00975.x. http://dx.doi.org/10.1111/j.1468-1293.2011.00975.x [DOI] [PubMed] [Google Scholar]
- Sahu GK, Cloyd MW. Latent HIV in primary T lymphocytes is unresponsive to histone deacetylase inhibitors. Virol. J. 2011;8:400–406. doi: 10.1186/1743-422X-8-400. http://dx.doi.org/10.1186/1743-422X-8-400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salatino A, Salatino MLF, Negri G. Traditional uses, chemistry and pharmacology of Croton species (Euphorbiaceae) J. Braz. Chem. Soc. 2007;18:11–33. [Google Scholar]
- Sgarbanti M, Remoli AL, Marsili G, Ridolfi B, Borsetti A, Perrotti E, Orsatti R, Ilari R, Sernicola L, Stellacci E, Ensoli B, Battistini A. IRF-1 is required for full NF-kappaB transcriptional activity at the human immunodeficiency virus type 1 long terminal repeat enhancer. J. Virol. 2008;82:3632–3641. doi: 10.1128/JVI.00599-07. http://dx.doi.org/10.1128/JVI.00599-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X, Lopez-Campistrous A, Sun L, Dower N. a, Kedei N, Yang J, Kelsey JS, Lewin NE, Esch TE, Blumberg PM, Stone JC. RasGRPs are targets of the anti-cancer agent ingenol-3-angelate. PLoS One. 2013;8:e72331. doi: 10.1371/journal.pone.0072331. http://dx.doi.org/10.1371/journal.pone.0072331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitaler M, Cantrell D. a. Protein kinase C and beyond. Nat. Immunol. 2004;5:785–790. doi: 10.1038/ni1097. http://dx.doi.org/10.1038/ni1097 [DOI] [PubMed] [Google Scholar]
- Steinberg SF. Structural basis of protein Kinase C isoform function. Physiol. Rev. 2008;88:1341–1378. doi: 10.1152/physrev.00034.2007. http://dx.doi.org/10.1152/physrev.00034.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strair RK, Schaar D, Goodell L, Aisner J, Chin K, Eid J, Senzon R, Cui XX, Han ZT, Knox B, Rabson AB, Chang R, Conney A. Administration of a Phorbol ester to patients with hematological malignancies: preliminary results from a phase I clinical trial of 12-O-tetradecanoylphorbol-13-acetate. Clin Cancer Res. 2002:2512–2518. Administration of a Phorbol Ester to Patients with Hematological Malignancies: Preli. [PubMed] [Google Scholar]
- Torgerson TR, Colosia a D., Donahue JP, Lin YZ, Hawiger J. Regulation of NF-kappa B, AP-1, NFAT, and STAT1 nuclear import in T lymphocytes by noninvasive delivery of peptide carrying the nuclear localization sequence of NF-kappa B p50. J. Immunol. 1998;161:6084–6092. [PubMed] [Google Scholar]
- Tyagi M, Pearson RJ, Karn J. Establishment of HIV latency in primary CD4+ cells is due to epigenetic transcriptional silencing and P-TEFb restriction. J. Virol. 2010;84:6425–6437. doi: 10.1128/JVI.01519-09. http://dx.doi.org/10.1128/JVI.01519-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Lint C, Bouchat S, Marcello A. HIV-1 transcription and latency: an update. Retrovirology. 2013;10:67. doi: 10.1186/1742-4690-10-67. http://dx.doi.org/10.1186/1742-4690-10-67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warrilow D, Gardner J, Darnell G. a, Suhrbier A, Harrich D. HIV type 1 inhibition by protein kinase C modulatory compounds. AIDS Res. Hum. Retrovir. 2006;22:854–864. doi: 10.1089/aid.2006.22.854. http://dx.doi.org/10.1089/aid.2006.22.854 [DOI] [PubMed] [Google Scholar]
- Williams SA, Chen L-F, Kwon H, Fenard D, Bisgrove D, Verdin E, Greene WC. Prostratin antagonizes HIV latency by activating NF-kappaB. J. Biol. Chem. 2004;279:42008–42017. doi: 10.1074/jbc.M402124200. http://dx.doi.org/10.1074/jbc.M402124200 [DOI] [PubMed] [Google Scholar]
- Williams SA, Chen L-F, Kwon H, Ruiz-Jarabo CM, Verdin E, Greene WC. NF-kappaB p50 promotes HIV latency through HDAC recruitment and repression of transcriptional initiation. EMBO J. 2006;25:139–149. doi: 10.1038/sj.emboj.7600900. http://dx.doi.org/10.1038/sj.emboj.7600900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Liu F, Zhou X, Cheng Z, Yang X, Xiao H, Chen Q, Cai K. Effect of protein kinase C on proliferation and apoptosis of T lymphocytes in idiopathic thrombocytopenic purpura children. Cell. Mol. Immunol. 2005;2:197–203. [PubMed] [Google Scholar]
- Yang H, Xing S, Shan L, Connell KO, Dinoso J, Shen A, Zhou Y, Shrum CK, Han Y, Liu JO, Zhang H, Margolick JB, Siliciano RF. Small-molecule screening using a human primary cell model of HIV latency identifies compounds that reverse latency without cellular activation. J. Clin. Invest. 2009;119:3473–3486. doi: 10.1172/JCI39199. http://dx.doi.org/10.1172/JCI39199DS1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Chen Y, Gabuzda D. ERK MAP kinase links cytokine signals to activation of latent HIV-1 infection by stimulating a cooperative interaction of AP-1 and NF-kappa B. J. Biol. Chem. 1999;274:27981–27988. doi: 10.1074/jbc.274.39.27981. http://dx.doi.org/10.1074/jbc.274.39.27981 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.