Abstract
Wild waterfowl and their habitats are the main reservoirs of influenza A virus (IAV) mainly during the breeding season and prior to migration. This study describes the molecular characterization of an IAV isolated from 240 water samples of a small wetland during non-breeding season of migratory wild ducks in the State of Mexico, Mexico. The results showed that the virus belongs to the H4N2 subtype and each of its eight segments of the viral genome has similarity to IAV isolated from ducks in North America. This study suggests that IAV can be isolated from small wetland during non-breeding season of migrating waterfowl.
Keywords: influenzavirus A, migrating waterfowl, water, wetland
It is known that influenza A virus (IAV) is maintained in anatids and their habitat, because of 2 factors: the transmission of the virus between birds and its persistence in the environment [2]. The transmission occurs among multiple species and bird populations that share the same habitat through the fecal-oral route, which pollutes wetlands [5, 12]. Most studies of IAV ecology have been conducted in birds in tundra and taiga wetlands, where the water freezes [11]. However, wild aquatic birds’ habitats in the central high plateau of Mexico depend on variable precipitation. There is limited and small habitat for local or migrating aquatic birds compared to the other regions. Even so, a low pathogenic IAV subtype H7N3 has been detected in a migratory cinnamon teal (Anas cyanoptera) during hunting activities in wetlands of central high plateau [3].
During the particular winter stay of migratory wild waterfowl in 2011, there was maximum density of birds and high level of water in a small wetland (~3ha) in the county of Toluca in the State of Mexico, and this wetland belongs to central high plateau, which allowed the survey of IAV in water.
The sampled water (1.5 ml) was gathered from the top 5 cm of the surface using 2 ml transfer pipettes and stored in 1.7 ml eppendorf tubes in triplicate, at each of the 8 sites identified where wild ducks were mostly located. Samples were kept at 4°C and transported to the laboratory for immediate processing. The sampling was performed 10 times from October 2010 to March 2011 at a time interval of approximately 21 days, resulting in a total of 240 samples collected and processed.
Water samples were centrifuged at 12,000 × g for 3 min. The supernatant (~1.3 ml) was transferred to 1.7 ml new eppendorf tubes, and 13 µl of PBS with antibiotic-antimycotic (penicillin G sodium salt 10,000 IU/ml, streptomycin 10,000 µg/ml and amphotericin B 25 mg/ml) (Gibco, Carlsbad, CA, U.S.A.) was added; samples were allowed to stand for 2 hr at room temperature before inoculating in 4 specific pathogen-free 9-day-old chick embryos with 200 µl of each sample via the allantoic cavity [10]. Allantoic fluid was harvested and evaluated by the hemagglutination technique. Positive samples were frozen at −75°C and sent to the Laboratory of the Mexico-United States Commission for the Prevention of Foot and Mouth Disease and Other Exotic Animal Diseases (CPA), to identify virus subtype (A/environment/Mexico/CPA-04740/2011).
To amplify each segment, we used primers previously reported [6], and we design other primers for sequencing the genome with the Primer3 program version 0.4.0 [9] and corroborated by the NetPrimer program [8] based on publicly available sequences from GenBank. Primers are upon requested.
Viral RNA was extracted from allantoic fluid (stored at −75°C) using the kit PureLink Viral RNA/DNA Mini Kit (Invitrogen, Carlsbad, CA, U.S.A.), according to the supplier’s specifications. Each of the 8 segments of the viral genome was amplified using the kit Qiagen® OneStep RT-PCR Kit (Qiagen, Hilden, Germany), with primers designed for each segment. PCR products were purified using the QIAquick Gel Extraction Kit (Qiagen). Purified products were used for sequencing reactions for each segment by chain termination method using the BigDye® Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, U.S.A.), and the reactions were read using an ABI 3130 sequencer (Applied Biosystems). The obtained sequences were aligned and edited to build consensus sequences for each segment using the software MEGA version 5.05 [13].
The nucleotide sequences of the eight segments obtained in this study were submitted to the GenBank nucleotide sequences database with the accession numbers: Polymerase basic protein 2 (PB2) [KF648921]; polymerase basic protein 1 (PB1) [KF648922]; polymerase acid protein (PA) [KF648923]; hemagglutinin (HA) [KF648924]; nucleocapsid protein (NP) [KF648925]; neuraminidase (NA) [KF648926]; matrix protein (M) [KF648927]; and non-structural protein (NS) [KF648928]. Each of the nucleotide sequences of the 8 segments was compared using multiple sequence alignment tools available through the NCBI BLAST software. The nucleotide sequences of the segments encoding the HA and NA genes were used for phylogenetic analysis corresponding to isolates with >86% and >91% similarity, respectively, from North America and Eurasia-Oceania. Alignments were performed using the “ClustalW multiple alignment” tool of the MEGA version 5.05 software package. Additionally, genetic distance was calculated using Kimura’s 2-parameter method. Phylogenetic trees were constructed with H4 viruses and N2 viruses, respectively, using the Neighbor-Joining algorithm based on genetic distances using 1,000 bootstrap replicates. The translation of the nucleotide sequences into a deduced amino acid sequence was performed using the ExPASy Translate Tool [4]. The sequence data were aligned using also the “ClustalW multiple alignment” in the MEGA version 5.05 software [13].
The virus was isolated on February 28, 2011 from 1 of 240 water samples processed during the coldest time of the winter and 3 weeks after congregation of birds prior to migration (~3,000 individuals), which differ with previous studies documented by other authors [12]. At the time of the present study, the water temperature was ranging from ~6 to 16°C and pH from 7.0 to 7.8, and these conditions are known to maintain virus infectivity [2, 12], but the virus was isolated when temperature was around 11–14°C.
The genome analysis of the virus consists of eight segments with a total of 13,583 bp, including: PB2 (2,341 bp), PB1 (2,341 bp), PA (2,233 bp), HA (1,738 bp), NP (1,560 bp), NA (1,453 bp), M (1,027 bp) and NS (890 bp).
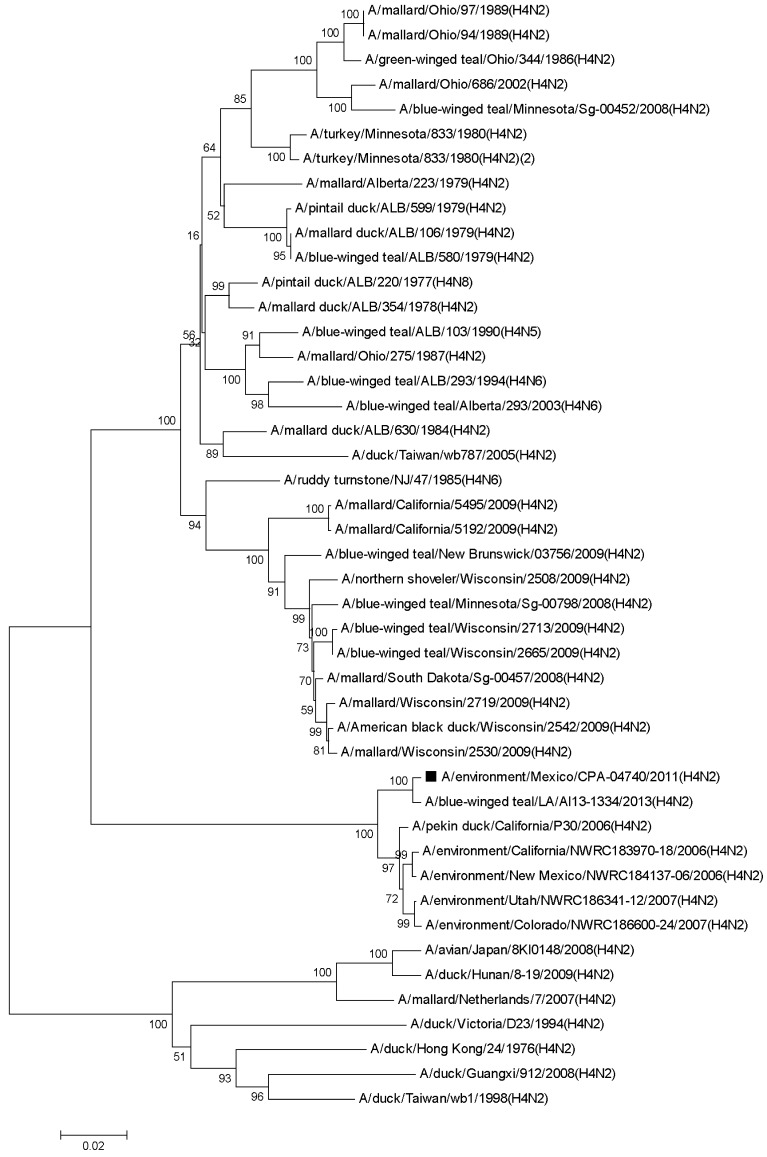
We found that HA gene of the virus showed 99% similarity to the homologous sequence of the virus A/blue-winged teal/LA/AI13-1334/2013 (H4N2) [KJ413565.1] and 98% similarity to the viruses A/pekin duck/California/P30/2006 (H4N2) [CY053821.1], A/environment/California/NWRC183970-18/2006 (H4N2) [CY122499.1] and A/environment/New Mexico/NWRC184137-06/2006 (H4N2) [CY122503.1], except for other isolates of the H4 subtype from North American wild waterfowl and environmental samples, collected between 1975 and 2009 with similarity <97% (Fig. 1).
Fig. 1.
Phylogenetic tree of the HA gene of A/environment/Mexico/CPA-04740/2011 (H4N2) [■] Influenza A virus isolated from a wetland. The phylogenetic tree was constructed with the Neighbor-Joining method. The percentages of replicate trees in which the associated taxa clustered together in a bootstrap test (1,000 replicates) are shown next to the branches. The evolutionary distances were computed using the Kimura 2-parameter method. The analysis involved 45 nucleotide sequences. Evolutionary analyses were conducted in MEGA5.05.
The genetic distance of the HA gene sequence was closest to the group of sequences from North America with ≤0.18 (≤252 nucleotides changes), while it was ≥0.18 (≥259 nucleotides changes) to Euroasia-Oceania sequences, according to the Kimura 2 parameter model.
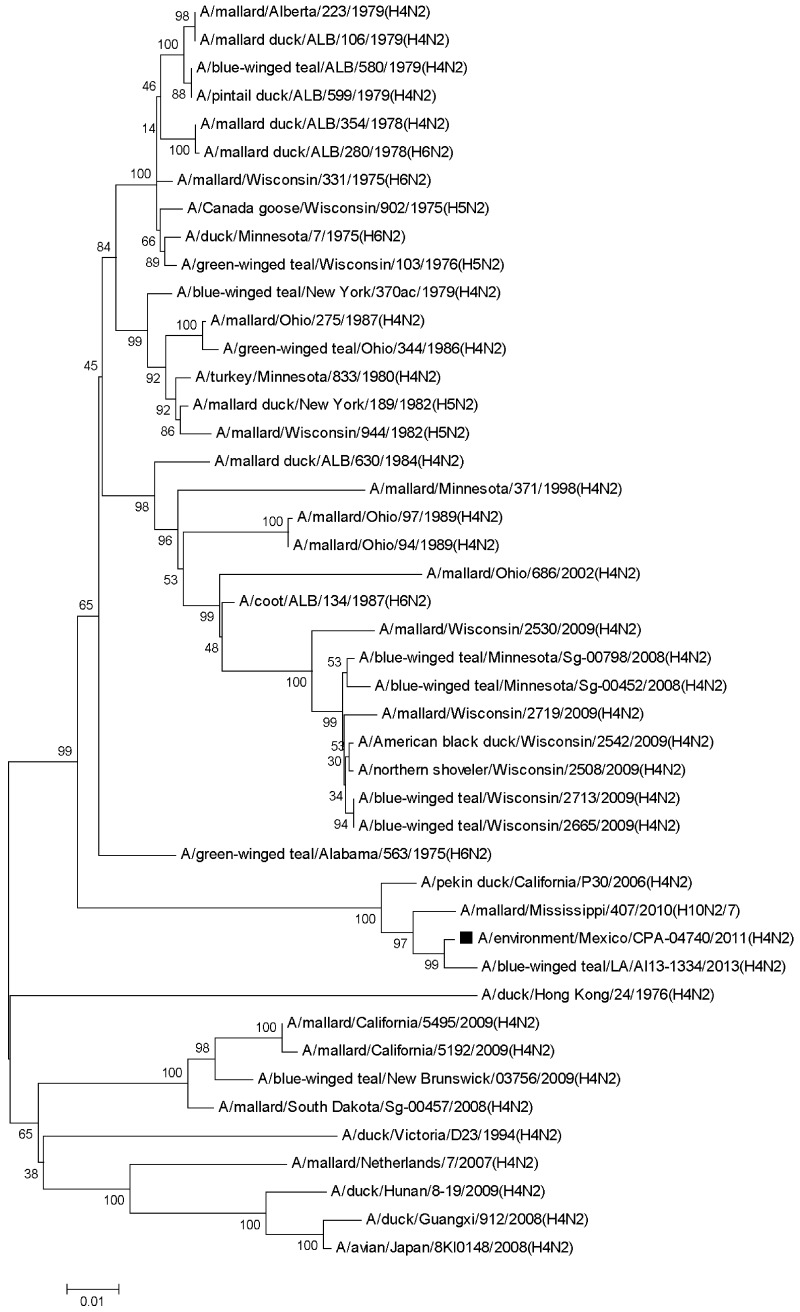
The NA gene of the virus presented 99% identity to the homologous sequences of the mixed virus A/mallard/Mississippi/407/2010 (H10N2,7) [CY097754.1] and the virus A/blue-winged teal/LA/AI13-1334/2013 (H4N2) [KJ413567.1], and also, it was observed 98% identity to the virus A/pekin duck/California/P30/2006 (H4N2) [CY053823.1]. Most of the remaining analyzed sequences corresponded to viruses isolated of the N2 subtype from North American wild ducks, collected between 1975 and 2009 with similarity <92%. The genetic distance of the NA gene sequence of the virus was closest to the sequences from North America with ≤0.15 (≤176 nucleotides changes), while it was ≥0.14 (≥172 nucleotides changes) to Eurasia-Oceania sequences according to the Kimura 2 parameter model (Fig. 2).
Fig. 2.
Phylogenetic tree of the NA gene of A/environment/Mexico/CPA-04740/2011 (H4N2) [■] Influenza A virus isolated from a wetland. The phylogenetic tree was constructed with the Neighbor-Joining method. The percentages of replicate trees in which the associated taxa clustered together in a bootstrap test (1,000 replicates) are shown next to the branches. The evolutionary distances were computed using the Kimura 2-parameter method. The analysis involved 45 nucleotide sequences. Evolutionary analyses were conducted in MEGA5.05.
The remaining six segments, PB2, PB1, PA, NP, M and NS genes, were closely related to North American wild ducks viruses, principally of the H6N5, H4N6, H3N5, H2N3, H6N1 and H3N7 subtypes, which correspond to the Pacific and Mississippi migratory routes, respectively.
At protein level, the HA of the virus had the amino acid sequence PEKTARGLF at the cleavage site, and its NA has no deletions in the stalk region, which is a feature of non-poultry adapted IAV [7]. The receptor binding sites, especially Q239 and G241, were similar among these sequences, suggesting that viruses would preferentially bind to 2,3-linked sialic acid receptors which are predominant in avian species [1].
This study suggests that IAV can be isolated from water during non-breeding season in migratory aquatic birds that stay in an area of ~3ha. Further studies on physical and chemical water conditions in situ are needed to understand virus ecology and their maintenance at different latitudes.
Acknowledgments
The molecular characterization of this study was funded by the project PAPIIT IN216212, while viral isolation in the wetlands was funded by the collaborative project DGVS/SEMARNAT-FMVZ/UNAM/20556-1061-14-IX-07.
REFERENCES
- 1.Bateman A. C., Busch M. G., Karasin A. I., Bovin N., Olsen C. W.2008. Amino acid 226 in the hemagglutinin of H4N6 influenza virus determines binding affinity for a 2,6-linked sialic acid and infectivity levels in primary swine and human respiratory epithelial cells. J. Virol. 82: 8204–8209. doi: 10.1128/JVI.00718-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown J. D., Goekjian G., Poulson R., Valeika S., Stallknecht E. D.2009. Avian influenza virus in water: Infectivity is dependent on pH, salinity and temperature. Vet. Microbiol. 136: 20–26. doi: 10.1016/j.vetmic.2008.10.027 [DOI] [PubMed] [Google Scholar]
- 3.Cuevas-Domínguez E., González-Guzmán S., Quintana-López J., Loza-Rubio E., González-Rebeles C., García-Espinosa G.2009. Detección de orthomyxovirus H7N3 en anátidos del Estado de México. Red. Vet.,http://www.veterinaria.org/revistas/redvet/n040409.html. Accessed March 2012.
- 4.Expert Protein Aalysis System. 2012. Translate Tool Swiss Institute of Bioinformatics, http://web.expasy.org/translate/. Accessed August 2012.
- 5.Hinshaw V. S., Webster R. G., Turner B.1979. Water-borne transmission of influenza A viruses? Intervirology 11: 66–68. doi: 10.1159/000149014 [DOI] [PubMed] [Google Scholar]
- 6.Hoffmann E., Stech J., Guan Y., Webster R. G., Perez D. R.2001. Universal primer set for the full-length amplification of all influenza A viruses. Arch. Virol. 146: 2275–2289. doi: 10.1007/s007050170002 [DOI] [PubMed] [Google Scholar]
- 7.Matrosovich M., Zhou N., Kawaoka Y., Webster R.1999. The surface glycoproteins of H5 influenza viruses isolated from humans, chickens, and wild aquatic birds have distinguishable properties. J. Virol. 73: 1146–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Premier Biosoft. 2012. NetPrimer Primer Analysis Software, http://www.premierbiosoft.com/servlet/com.pbi.crm.clientside.FreeToolLoginServlet#. Accessed March 2012.
- 9.Rozen S., Skaletsky J. H.2000. Primer3 on the www for general users and for biologist programmers, http://frodo.wi.mit.edu/. Accessed March 2012. [DOI] [PubMed]
- 10.Senne D. A.2008. Virus propagation in embrionating eggs. pp. 204–208. In: A Laboratory Manual for the Isolation, Identification and Characterization of Avian Pathogens, 5th ed. (Dufour-Zavala, L., Swayne, D. E., Glisson, J. R., Pearson, J. E., Reed, W. M., Jackwood, M. W. and Woolcock, P. R. eds.), American Association of Avian Pathologists, Athens. [Google Scholar]
- 11.Shoham D., Jahangir A., Ruenphet S., Takehara K.2012. Persistence of avian influenza viruses in various artificially frozen environmental water types. Influenza Res. Treat.. doi: 10.1155/2012/912326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stallknecht D. E., Brown J. D.2007. Wild birds and the epidemiology of avian influenza. J. Wildl. Dis. 43: S15–S20. [DOI] [PubMed] [Google Scholar]
- 13.Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S.2011. MEGA5: Molecular Evolutionary Genetics Analysis using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Method, http://www.megasoftware.net/. Accessed June 2012. [DOI] [PMC free article] [PubMed]