Highlights
-
•
We modeled the mitochondrial disease MELAS by generating patient-specific iPS cells.
-
•
MELAS-iPS cells show a wide variety of heteroplasmy levels.
-
•
MELAS-iPS cells with high heteroplasmy levels showed impaired complex I activity.
Abbreviations: bFGF, basic fibroblast growth factor; EB, embryoid body; ES, embryonic stem; iPSCs, induced pluripotent stem cells; KSR, Knock-out Serum Replacement; MEF, mouse embryonic fibroblast; MELAS, mitochondrial myopathy, encephalomyopathy, lactic acidosis, and stroke-like episodes; mtDNA, mitochondrial DNA; OXPHOS, oxidative phosphorylation system
Keywords: MELAS, iPS cell, Mitochondrial disease, Disease modeling
Abstract
Mitochondrial diseases are heterogeneous disorders, caused by mitochondrial dysfunction. Mitochondria are not regulated solely by nuclear genomic DNA but by mitochondrial DNA. It is difficult to develop effective therapies for mitochondrial disease because of the lack of mitochondrial disease models. Mitochondrial myopathy, encephalomyopathy, lactic acidosis, and stroke-like episodes (MELAS) is one of the major mitochondrial diseases. The aim of this study was to generate MELAS-specific induced pluripotent stem cells (iPSCs) and to demonstrate that MELAS-iPSCs can be models for mitochondrial disease. We successfully established iPSCs from the primary MELAS-fibroblasts carrying 77.7% of m.3243A>G heteroplasmy. MELAS-iPSC lines ranged from 3.6% to 99.4% of m.3243A>G heteroplasmy levels. The enzymatic activities of mitochondrial respiratory complexes indicated that MELAS-iPSC-derived fibroblasts with high heteroplasmy levels showed a deficiency of complex I activity but MELAS-iPSC-derived fibroblasts with low heteroplasmy levels showed normal complex I activity. Our data indicate that MELAS-iPSCs can be models for MELAS but we should carefully select MELAS-iPSCs with appropriate heteroplasmy levels and respiratory functions for mitochondrial disease modeling.
1. Introduction
Mitochondrial diseases are genetically and clinically heterogeneous disorders, commonly caused by mitochondrial dysfunction [1,2]. Clinically, they involve multiple tissues although they mainly affect nervous system, skeletal muscle, and cardiac muscle, all of which are rich in mitochondria. Each cell in a human body has thousands of mitochondrial DNA (mtDNA) copies, and the expression and severity of mitochondrial disease symptoms depend on the levels of mtDNA mutation-specific heteroplasmy (mixture of wild-type and mutated mtDNA) [3].
Mitochondrial myopathy, encephalomyopathy, lactic acidosis, and stroke-like episodes (MELAS) is one of the major clinical subgroups of the mitochondrial diseases and is caused by a single base replacement in the mtDNA [4]. The majority (80%) of MELAS patients possess an A to G substitution at mtDNA nucleotide position 3243, denoted as m.3243A>G [5]. MELAS presents a broad clinical spectrum involving seizures, stroke-like episodes, mental disorders, sensorineural deafness, progressive muscle weakness, cardiomyopathy, and other organ symptoms. The estimated threshold proportion of m.3243A>G heteroplasmy sufficient to impair mitochondrial respiratory function in MELAS is 90% in cellular systems [6] and 70% in muscle fibers [7]. Accumulating evidences help us understand the pathogenesis, but there is no effective therapy for mitochondrial diseases. Available treatments serve only to alleviate symptoms and slightly delay disease progression. In addition, the development of new potential treatments are complicated due to the rarity of patients with mitochondrial disease and the lack of disease models. Transmitochondrial cybrid cell lines have been used as artificial mitochondrial disease models to identify relationships between the proportion of mutant mtDNA and impaired biochemical functions in mitochondria [8,9]. However, human cell models are needed to sufficiently understand the cellular pathophysiology in various mitochondrial diseases.
The generation of induced pluripotent stem cells (iPSCs) from patients with genetic disorders holds enormous promise for understanding the pathogenesis of complex diseases [10,11]. Specifically, disease-specific iPSCs have opened new possibilities for generating continuous supplies of human diseased cells for basic research and drug screening. Most genetic diseases are affected by genomic DNA mutation but mitochondrial diseases are mostly affected by mtDNA mutation. It remains unknown whether mitochondrial disease model could also be generated by patient-specific iPSCs. The present study sought to demonstrate the generation of disease-specific iPSCs from a patient with MELAS carrying m.3243A>G heteroplasmy for disease modeling. We also evaluated the mitochondrial genotype-phenotype correlation between mtDNA heteroplasmy and the mitochondrial oxidative phosphorylation system (OXPHOS).
2. Materials and methods
2.1. Human iPSC generation
The iPSCs were established as described previously [11], using lentiviral vectors to introduce mouse solute carrier family 7, a member 1 (Slc7a1) gene encoding the ecotropic retrovirus receptor. Transfectants were plated at 2 × 105 cells per 60-mm dish, and the next day, OCT4, SOX2, KLF4, and MYC were introduced by retroviral transduction. After another 24 h, the virus-containing medium was aspirated and the cell culture continued under fibroblast conditions. Thereafter those cells were cultured under iPSC culture condition. Six days later, the cells were harvested and plated at 5 × 104 cells per 100-mm dish for another 20 days of culturing. At day 25, embryonic stem (ES) cell-like colonies were mechanically dissociated and transferred to a 24-well plate containing adhered mouse embryonic fibroblast (MEF) feeder cells. The isolation and use of patient somatic cells was approved by the Institutional Review Board, NCNP (21-9-10) and the Ethics Committee of Keio University (approval No. 20-92-5), and conducted under the Declaration of Helsinki. Patient information was encoded to protect privacy, and written informed consent obtained. The MELAS-iPSC line was maintained in the undifferentiated state on irradiated MEF feeder cells in DMEM/F12 medium (Invitrogen, CA, USA) supplemented with 20% Knock-out Serum Replacement (KSR) (Invitrogen), 1 mM l-glutamine (Invitrogen), 1 mM nonessential amino acids (Sigma–Aldrich, MO, USA), 0.1 mM β-mercaptoethanol, and 4 ng/ml basic fibroblast growth factor (bFGF). The iPSC culture medium was changed daily, and the cells were passaged every 5–6 days. We generated iPSCs from donor MELAS-fibroblasts multiple times and named iPSC clone in same dish as same alphabet.
2.2. Teratoma formation
To confirm pluripotency in vivo, teratoma formation was assessed in accordance with the Institutional Animal Care and Use Committee of Keio University. Approximately 1–2 × 106 iPSCs were injected into the testis of anesthetized immune-compromised NOD-SCID mice (CREA-Japan, Tokyo, Japan). At 10–12 weeks after the injection, mice were euthanized and the teratomas were excised, fixed overnight in formalin, embedded in paraffin, and analyzed by haematoxylin-eosin staining [12]. The mice were anesthetized using a mixture of ketamine (50 mg/kg), xylazine (10 mg/kg), and chlorpromazine (1.25 mg/kg). The adequacy of anesthesia was monitored by heart rate, muscle relaxation, and the loss of sensory reflex responses, i.e., nonresponsive to tail pinching. The investigation conforms with the Guide for the Care and Use of Laboratory Animals published by the US National Institutes of Health (publications number 23–80 revised in 2011).
2.3. Immunofluorescence
The immunostaining was performed using the following primary antibodies and reagents: anti-OCT4 (sc-5279, Santa Cruz, CA, USA), anti-NANOG (RCAB0003P, ReproCELL, Yokohama, Japan), anti-SSEA 4 (MAB4304, Millipore), anti-Tra1-60 (MAB4360, Millipore, MA, USA), anti-human smooth muscle actin monoclonal antibody (Agilent Technologies, CA, USA), goat anti-human SOX17 polyclonal antibody (R&D Systems, MN, USA), rabbit anti-Nestin polyclonal antibody (Sigma), or Prolyl 4-hydroxylase beta (PH4B) (Acris Antibodies GmbH, Herford, Germany), together with 4′,6′-diamidino-2-phenylindole (DAPI; ThermoFisher SCIENTIFIC, MA, USA) [13]. Signal was detected using a conventional fluorescence laser microscope (BZ-9000, KEYENCE, Osaka, Japan) equipped with a color charge-coupled device camera (BZ-9000, KEYENCE).
2.4. mtDNA mutation analysis
Mutation ratios of m.3243A>G were determined as follows. Extracted DNA (1 ng) was used for quantitative PCR with the TaqMan Universal PCR Master Mix kit (ThermoFisher SCIENTIFIC) according to the manufacturer’s instructions. A sequence detection system (ABI PRISM 7900HT; Applied Biosystems) was used, and a calibration curve was created using several known copy numbers of plasmid containing the amplified mtDNA fragments for either wild-type or mutant sequences. Primers and allele-specific TaqMan probes used were as follows: forward primer (5′-CCACCCAAGAACAGGGTTTG-3′), reverse primer (5′-GGAATTGAACCTCTGACTGTAAAGTTT-3′), wild-type allele-specific TaqMan probe (5′-AGATGGCAGAGCCCGGTA-3′; with VIC-fluorogenic probe at the 5′ terminus), mutant allele-specific TaqMan probe (5′-AGATGGCAGGGCCCGGTA-3′; with FAM-fluorogenic probe at the 5′ terminus).
2.5. mtDNA copy number analysis
mtDNA copy number was determined as follows. Extracted DNA (1 ng) was used as template for quantitative PCR with Power SYBR Green PCR Master Mix kit (ThermoFisher SCIENTIFIC) according to the manufacturer’s instructions. A Sequence detection system (ABI PRISM 7900HT; Applied Biosystems) was used, and the threshold cycle number for mtDNA gene (MT-CYB) and for nuclear DNA gene (FBXO15) were adopted for ΔΔCt-based relative quantification. Primers used were as follows: MT-CYB forward primer (5′-TGCAACTATAGCAACAGCCTTCA-3′), MT-CYB reverse primer (5′-GAACTAGGTCTGTCCCAATGTATGG-3′), FBXO15 forward primer (5′-GCCAGGAGGTCTTCGCTGTA-3′), FBXO15 reverse primer (5′-AATGCACGGCTAGGGTCAAA-3′).
2.6. In vitro differentiation
iPSCs were harvested using 1 mg/ml collagenase IV (Invitrogen), and then colonies were transferred to ultralow attachment plates (Corning, NY, USA) in iPSC culture medium without bFGF. After 8 days, aggregated cells (embryoid body (EB)) were plated onto gelatin-coated tissue culture dishes. The cells were incubated at 37 °C in 5% CO2 and the medium was replaced every other day for 8–10 days.
2.7. Fibroblast differentiation from iPSCs
Cells were harvested using 1 mg/ml collagenase IV (Invitrogen), and then transferred to ultra-low attachment plates (Corning) in differentiation medium (DMEM/F12 supplemented with 20% FBS, 1 mM l-glutamine, 1 mM nonessential amino acids, and 0.1 mM β-mercaptoethanol), as described previously [11]. The EBs were incubated at 37 °C in 5% CO2, and the medium was replaced every 3–4 days for 8–10 days. EBs were plated onto gelatin-coated tissue culture dishes. After 7–10 days, differentiated fibroblast-like cells were seen and these were expanded by culturing in DMEM medium with 10% FBS and serially passaged with trypsin for at least six passages.
2.8. Cell growth
20 wells of a 24 well tissue culture plate with MEF feeder cells were seeded with approximately 20,000 iPSCs/well. 24 h later, 4 wells of MEF feeder cells and 4 wells of iPSCs with MEF feeder cells were trypsinized to a single cell suspension and counted. The eight counts for the MEF feeder cells were averaged and subtracted from the average of the eight counts that corresponded to those wells with iPSCs. This value was used as a baseline (Day 0) for the doubling time assay. This procedure was repeated with 4 wells of iPSCs in the condition of regular iPSC culture.
2.9. Chromosome karyotyping
iPSCs treated with colcemid solution (60 ng/ml) were cultured for 5 h at 37 °C to stop cell growth in metaphase. These cells were then incubated in 0.075 M KCl solution for 10 min at room temperature, followed by fixation in Carnoy’s solution. Fixed cells were sent to Nihon Gene Research Laboratories Inc. (Sendai, Japan) for analysis.
2.10. Mitochondorial enzymatic activity measurement
Enzymatic activities for mitochondrial respiratory complexes were analyzed as described elsewhere [14] with modifications. In brief, after culturing the harvested cells were resuspended in isolation buffer (pH 7.4) containing 210 mM mannitol, 70 mM sucrose, 1 mM EGTA, and 5 mM HEPES, and then homogenized on ice. Cell lysates were centrifuged to isolate mitochondrial proteins, which were quantified by Bradford assay, with a calibration curve created using several known concentrations of BSA. All samples were measured in triplicate using a spectrophotometric multi-well plate reader (SPECTROstar Nano; BMG Labtech). Citrate synthase (CS) activity was used for normalization of respiratory complexes activities in each sample. For measuring complex I activity, reaction buffer (pH 7.4; containing 50 mM Tris–HCl, 250 mM sucrose, 1 mM EDTA, 5 μg/ml antimycin A, 2 mM KCN, 10 μM decylubiquinone (DQ), and 50 μM NADH) was added and the mixture was incubated at 37 °C. Isolated mitochondrial proteins (1 μg) were then added to monitor the time-dependent spectrum alterations at 340 nm. For complex II activity, reaction buffer (pH 7.4; containing 50 mM potassium phosphate, 5 μg/ml rotenone, 5 μg/ml antimycin A, 2 mM KCN, 20 mM succinate, 50 μM 2,6-dichlorophenolindophenol (DCPIP), and 50 μM DQ) was added and the mixture was incubated at 37 °C. Isolated mitochondrial proteins (1 μg) were then added to monitor the time-dependent spectrum alterations at 600 nm. For complex III activity, reaction buffer (pH 7.4; containing 50 mM Tris–HCl, 250 mM sucrose, 1 mM EDTA, 2 mM KCN, 50 μM cytochrome c, and 50 μM decylubiquinol [reduced form of DQ]) was added and the mixture was incubated at 37 °C. Isolated mitochondrial proteins (1 μg) were then added to monitor the time-dependent spectrum alterations at 550 nm. For complex IV activity, reaction buffer (pH 7.4; containing 10 mM potassium phosphate and 25 μM ferrocytochrome c [reduced form of cytochrome c]) was added and the mixture was incubated at 37 °C. Isolated mitochondrial proteins (1 μg) were then added to monitor the time-dependent spectrum alterations at 550 nm. For citrate synthase activity, reaction buffer (pH 8.0; containing 125 mM Tris–HCl, 300 μM acetyl-CoA, 100 μM 5,5′-dithiobis [2-nitrobenzoic acid] (DTNB), and 500 μM oxaloacetate) was added and the mixture was incubated at 37 °C. Isolated mitochondrial proteins (1 μg) were then added to monitor the time-dependent spectrum alterations at 415 nm.
2.11. Statistical analysis
All results are expressed as mean ± SD of three independent experiments. The measurements were statistically analyzed using Student’s t-test for comparing two groups.
3. Results
3.1. iPSC generation from a patient with MELAS
Primary fibroblasts were derived from skin biopsy samples from a MELAS patient. The primary MELAS fibroblasts carrying 77.7% of m.3243A>G. Retroviral vectors carrying OCT4, SOX2, KLF4, and MYC were infected into those fibroblasts. After 3–4 weeks, compact embryonic stem (ES)-cell-like colonies emerged, and these were clonally expanded to create 20 stable iPSC lines. All iPSC clones expressed pluripotent markers OCT4, Tra1-60, NANOG, and SSEA4 (Fig. 1A). We also confirmed the in vitro differentiation capability of these MELAS-iPSCs. Embryoid bodies (EBs) were generated from each MELAS-iPSC line and plated on gelatin-coated dishes on day 7. A few days later, the EBs were immunostained for Nestin (ectoderm marker), αSMA (mesoderm marker), and SOX17 (endoderm marker). Each iPSC lines tested similarly generated EBs and contained derivatives of all three germ layers (Fig. 1B). Next we examined the multipotency of MELAS-iPSC, based on teratoma formation which involved tissues derived from all three germ layers (Fig. 1C). Karyotype and growth rate are also examined in representative MELAS-iPSC lines (Fig. 1D and E).
Fig. 1.
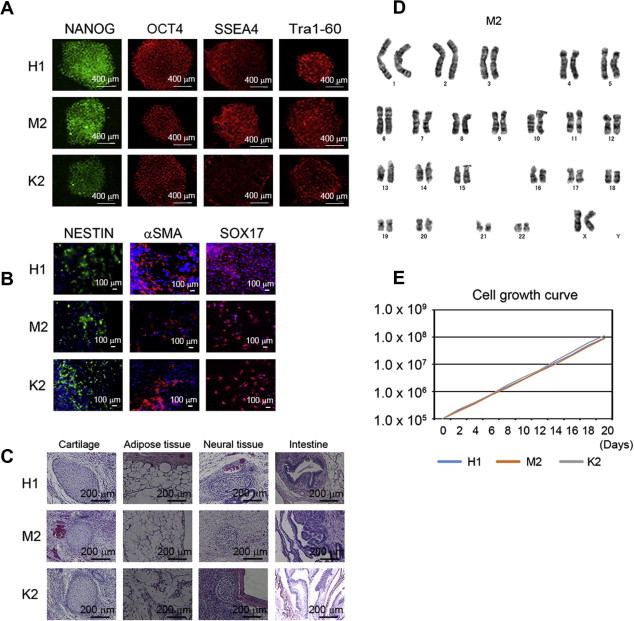
(A) Immunostaining for pluripotency and surface markers (NANOG, OCT4, SSEA4, and Tra1-60) in representative MELAS-iPSCs (H1, M2, and K2). Scale bars represent 500 μm. (B) Immunofluorescence staining for Nestin (ectodermal marker), αSMA (mesodermal marker), and Sox17 (endodermal marker) in representative MELAS-iPSC-derived differentiated cells (H1, M2, and K2). (C) Microscopic observation of teratoma sections in representative MELAS-iPSCs (H1, M2, and K2), showing tissue structures resembling gut (endoderm), cartilage (mesoderm), adipose (mesoderm), and neural tissue (ectoderm). (D) Normal karyotype of representative MELAS-iPSCs (M2). (E) Cell growth curves of representative MELAS-iPSCs (H1, M2, and K2).
3.2. The variation of mitochondria heteroplasmy in MELAS-iPSCs
By quantitative real-time PCR, the m.3243A>G heteroplasmy levels in MELAS-iPSC lines ranged from 3.6% (line D1) to 99.4% (line G1), and 14 MELAS-iPSC lines had a mutant heteroplasmy level of over 80% (Fig. 2). These results indicated that MELAS-iPSC lines with a wide variety of m.3243A>G contents (<5% to >95%) could be established from the same patient. Nevertheless, those MELAS-iPSC lines showed typical iPSC morphology (Fig. 1A), differentiation capability into three different germ layer-derived tissues (Fig. 1B and C) and typical growth rates of human iPSCs (Fig. 1E). During reprogramming of the fibroblasts to iPSCs, mitochondria assumed an apparently “quiescent” state: a round-shaped morphology with poor cristae structure, less active OXPHOS, and reduced mtDNA copies [15]. Undifferentiated human iPSCs predominantly produce energy by anaerobic glycolysis, and this could explain why MELAS-iPSC lines carrying a higher proportion of m.3243A>G, even over its pathogenic threshold level, were also stably established.
Fig. 2.
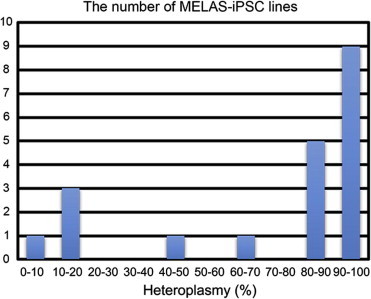
Histogram of the number of MELAS-iPSC lines with different heteroplasmy levels.
3.3. Maintenance of heteroplasmy levels after long-term culture and fibroblast differentiation
To examine the influences of long-term iPSC culture on m.3243A>G content, we compared m.3243A>G heteroplasmy levels between cells from passage 10 and passage 40 using fifteen MELAS-iPSC lines. The heteroplasmy levels were increased after long-term culture in iPSC with low heteroplasmy levels (C1 and K2) (Fig. 3A). But no apparent differences in m.3243A>G heteroplasmy levels were observed among iPSC with high heteroplasmy levels after long-term culture (Fig. 3A). We also examined the mtDNA copy number between cells from passage 10 and passage 40 using eleven MELAS-iPSC lines, which showed that mtDNA copy numbers dynamically changed after passage (Fig. 3B). We didn’t observe the relation between the iPSC clones with high heteroplasmy and those with low heteroplasmy. Next we asked if the level of heteroplasmy could be maintained during differentiation of the MELAS-iPSCs into fibroblasts. Ten MELAS-iPSC lines were enzymatically harvested and transferred to be cultured on low attachment dishes with the differentiation medium to form EBs. EBs were plated on gelatin-coated dishes at day 7. After five passages, the EBs differentiated to uniform fibroblasts (Fig. 3C). The purities of fibroblasts were approximately over 95%. Interestingly, evaluation of the change in m.3243A>G heteroplasmy levels upon differentiation by quantitative real-time PCR measurements revealed that the heteroplasmy levels were similar after fibroblast differentiation in each MELS-iPSC line (Fig. 3D).
Fig. 3.

(A) Heteroplasmy levels in 15 MELAS-iPSC lines at passage 10 and 40. (B) mtDNA/nuclearDNA (nDNA) copy number in 11 MELAS-iPSC lines at passage 10 and 40. (C) Immunostaining for fibroblast marker (P4HB) in representative MELAS-iPSC-derived fibroblasts. (D) Heteroplasmy levels in 10 MELAS-iPSC lines and iPSC-derived fibroblasts.
3.4. The enzymatic activity of mitochondrial respiratory complexes in MELAS-iPSC derived fibroblasts
To further characterize the potential for disease modeling, we examined the enzymatic activities of mitochondrial respiratory complexes I, II, III, and IV in MELAS-iPSC-differentiated fibroblasts from the four MELAS-iPSC lines (K1, H1, C11, and M2) and in primary skin fibroblasts derived from a donor patient, and compared them with skin fibroblasts of ten healthy persons (normalized as 100%). Enzymatic activities for mitochondrial respiratory complexes were analyzed as described elsewhere [14] with modifications. Mitochondrial heteroplasmy levels at the time of enzymatic analysis in the donor and iPSC-derived fibroblasts (K1, H1, C11, and M2) were 75%, 64%, 70%, 81%, and 100%, respectively. To normalize for content of mitochondria, each enzymatic activity was normalized against citrate synthase (CS) activity (Fig. 4A) and complex II (Fig. 4B) [16]. The complex II is regulated not by mtDNA but by nuclear DNA. The difference in the complex II would not be directly affected by mtDNA mutation. The normalization by complex II can minimize the effect of mitochondrial DNA mutation. The enzymatic activities of complex I were decreased in MELAS-iPSC lines H1, C11, and M2, and in the MELAS-donor fibroblasts (Fig. 4A and B). However, no decrease in enzymatic activity was observed for complexes II, III, and IV in the MELAS-iPSC-derived fibroblasts, despite the possibility that these complexes might be induced by the overall tRNA functional defect. In MELAS patients, complex I deficiency is the most common feature of mitochondrial enzymatic activity. A defect in the taurine modification of mutant tRNALeu(UUR) causes reduction in mitochondrial translation of the ND6 gene, which encodes a component of respiratory chain complex I. On the contrary, complexes II, III, and IV are not affected in MELAS patients [17]. Complexes II and IV normalized against CS activity were increased in M2 (Fig. 4A), which would be due to functional complementation for the decreased activity of complex I, but complex IV normalized against complex II didn’t show significant difference (Fig. 4B). These results indicate that the pathogenic threshold levels of m.3243A>G were recapitulated in MELAS-iPSC-derived fibroblasts from a donor patient.
Fig. 4.
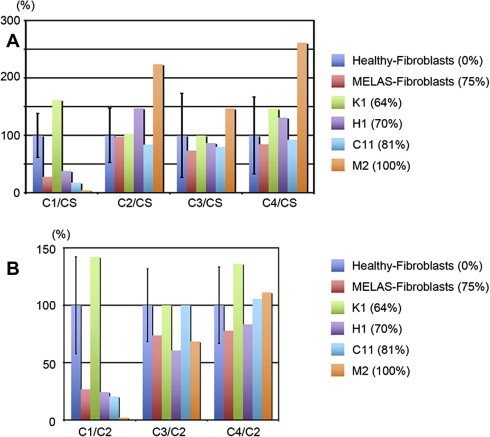
Enzymatic activity analysis for mitochondrial respiratory complexes in healthy-fibroblasts, donor MELAS-fibroblasts and 4 MELAS-iPSC-derived fibroblasts, normalized against citrate synthase (CS) activity (A) and complex II (B).
4. Discussion
In this study, we successfully generated disease-specific iPSCs from a MELAS patient carrying m.3243A>G heteroplasmy, and our MELAS-iPSC lines showed a wide range of m.3243A>G content (<5% to >95%) suitable for possible applications in disease modeling and drug screening. Mitochondria is the only organelle which contains their own DNA besides the nuclear DNA in mammals. Mitochondrial DNA is exclusively maternally transmitted during mammalian fertilization. mtDNA can expand clonally during mtDNA replication in somatic cells and mtDNA mutation would randomly segregate during cell division and result in daughter cells that exhibit low or high levels of heteroplasmy in somatic cells [18]. In this study, we examined the levels of heteroplasmy of mtDNA during reprogramming, iPSC maintenance and iPSC differentiation, and the enzymatic activities of mitochondrial respiratory complexes in iPSC-derived fibroblasts.
Disease modeling using disease-specific iPSCs will shed light on the unknown disease pathogenesis and drug screening. Specifically, patient-specific iPSCs harboring m.3243A>G were recently generated from two diabetic patients [19] and examined for m.3243A>G content, but not mitochondrial function. In that study, the parent fibroblasts contained small amounts of mutated mtDNA. Disease-specific iPSCs carrying different mutant mtDNAs have also been reported: m.13513G>A and a 2501 base-pair deletion in mtDNA [20,21]. Those reports mostly argued about heteroplasmy levels and cellular phenotypes such as cell growth and differentiation ability. Another recent report also showed the generation of MELAS-iPSCs harboring m.3243A>G [22]. They showed the expression of mtDNA encoded genes related to mitochondrial respiratory complexes I was decreased in MELAS-iPSC derived neurons. Mitochondrial diseases can be critically manifested by mitochondrial function and are heterogenic disorders partly due to the proportion of mtDNA mutation and/or absolute number of mtDNA copies. In terms of mitochondrial disease modeling, we have to carefully evaluate both heteroplasmy levels in iPSCs and mitochondrial function. Our patient-derived MELAS-iPSCs carrying a range of m.3243A>G levels could therefore serve as cellular disease models for facilitating mitochondrial medicine.
Cell replacement therapy for diseased organs is one of the attractive applications of patient-specific iPSCs, although such therapy could be problematic for mitochondrial disease due to the multiple organs affected. Specific organs such as skeletal muscle, heart, and nervous system, were seriously affected in many cases. However, the scarcity of therapeutic options thus far for these patients also raises the potential value of cell replacement therapy. In our study, we demonstrate that some MELAS-iPSC lines have extremely low levels of heteroplasmy, meaning that we can obtain isogenic iPSCs with normal mitochondrial respiratory function from the one patient. This indicates the potential for future therapeutic applications in mitochondrial diseases without genetic correction by using these isogenic iPSC-derived target cell types in autologous cell replacement therapy without concern about immunological rejection.
Author contributions
Conceived and designed the experiments: S.Y. Performed the experiments: M.K., H.H., S.Y., T.S., T.E., S.T., Y.K., A.T., S.O., H.H., D.K., A.K., M.T., S.K., T.Y., G.Y., M.S., C.M., N.H., and Y.S. Analyzed the data: M.K., H.H., S.Y., Y.G., and K.F. Wrote the paper: M.K., H.H., and S.Y.
Acknowledgements
This study was supported in part by research grants from the Ministry of Education, Culture, Sports, Science, and Technology, Health Labour Sciences Research Grant, the New Energy and Industrial Technology Development Organization, Japan, the Program for Promotion of Fundamental Studies in Health Science of the National Institute of Biomedical Innovation, Japan Science and Technology Agency, Research Center Network for Realization of Regenerative Medicine “The Program for Intractable Diseases Research utilizing Disease-specific iPS cells”, The Nakatomi Foundation, Japan Heart Foundation/Novartis Grant for Research Award on Molecular and Cellular Cardiology SENSHIN Medical Research Foundation, Kimura Memorial Heart Foundation Research Grant, Japan Intractable Diseases Research Foundation, Japan, and The Cell Science Research Foundation, The Tokyo Biochemical Research Foundation, Suzuken Memorial Foundation, and by Core Research for Evolutional Science and Technology, Japan Science and Technology Agency, Japan.
References
- 1.Wallace D.C. Mitochondrial diseases in man and mouse. Science. 1999;283:1482–1488. doi: 10.1126/science.283.5407.1482. [DOI] [PubMed] [Google Scholar]
- 2.Taylor R.W., Turnbull D.M. Mitochondrial DNA mutations in human disease. Nat. Rev. Genet. 2005;6:389–402. doi: 10.1038/nrg1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gropman A.L. Diagnosis and treatment of childhood mitochondrial diseases. Curr. Neurol. Neurosci. Rep. 2001;1:185–194. doi: 10.1007/s11910-001-0015-9. [DOI] [PubMed] [Google Scholar]
- 4.Kobayashi Y. Respiration-deficient cells are caused by a single point mutation in the mitochondrial tRNA-Leu (UUR) gene in mitochondrial myopathy, encephalopathy, lactic acidosis, and strokelike episodes (MELAS) Am. J. Hum. Genet. 1991;49:590–599. [PMC free article] [PubMed] [Google Scholar]
- 5.Goto Y., Nonaka I., Horai S. A mutation in the tRNA(Leu)(UUR) gene associated with the MELAS subgroup of mitochondrial encephalomyopathies. Nature. 1990;348:651–653. doi: 10.1038/348651a0. [DOI] [PubMed] [Google Scholar]
- 6.Attardi G., Yoneda M., Chomyn A. Complementation and segregation behavior of disease-causing mitochondrial DNA mutations in cellular model systems. Biochim. Biophys. Acta. 1995;1271:241–248. doi: 10.1016/0925-4439(95)00034-2. [DOI] [PubMed] [Google Scholar]
- 7.Jeppesen T.D., Schwartz M., Olsen D.B., Vissing J. Oxidative capacity correlates with muscle mutation load in mitochondrial myopathy. Ann. Neurol. 2003;54:86–92. doi: 10.1002/ana.10594. [DOI] [PubMed] [Google Scholar]
- 8.Yoneda M., Chomyn A., Martinuzzi A., Hurko O., Attardi G. Marked replicative advantage of human mtDNA carrying a point mutation that causes the MELAS encephalomyopathy. Proc. Natl. Acad. Sci. 1992;89:11164–11168. doi: 10.1073/pnas.89.23.11164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garrido-Maraver J. Screening of effective pharmacological treatments for MELAS syndrome using yeasts, fibroblasts and cybrid models of the disease. Br. J. Pharmacol. 2012;167:1311–1328. doi: 10.1111/j.1476-5381.2012.02086.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tanaka A. Endothelin-1 induces myofibrillar disarray and contractile vector variability in hypertrophic cardiomyopathy-induced pluripotent stem cell-derived cardiomyocytes. J. Am. Heart Assoc. 2014;3 doi: 10.1161/JAHA.114.001263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Egashira T. Disease characterization using LQTS-specific induced pluripotent stem cells. Cardiovasc. Res. 2012;95:419–429. doi: 10.1093/cvr/cvs206. [DOI] [PubMed] [Google Scholar]
- 12.Seki T. Generation of induced pluripotent stem cells from human terminally differentiated circulating T cells. Cell Stem Cell. 2010;7:11–14. doi: 10.1016/j.stem.2010.06.003. [DOI] [PubMed] [Google Scholar]
- 13.Seki T., Yuasa S., Fukuda K. Generation of induced pluripotent stem cells from a small amount of human peripheral blood using a combination of activated T cells and Sendai virus. Nat. Protoc. 2012;7:718–728. doi: 10.1038/nprot.2012.015. [DOI] [PubMed] [Google Scholar]
- 14.Trounce I.A., Kim Y.L., Jun A.S., Wallace D.C. Assessment of mitochondrial oxidative phosphorylation in patient muscle biopsies, lymphoblasts, and transmitochondrial cell lines. Methods Enzymol. 1996;264:484–509. doi: 10.1016/s0076-6879(96)64044-0. [DOI] [PubMed] [Google Scholar]
- 15.Prigione A., Fauler B., Lurz R., Lehrach H., Adjaye J. The senescence-related mitochondrial/oxidative stress pathway is repressed in human induced pluripotent stem cells. Stem Cells. 2010;28:721–733. doi: 10.1002/stem.404. [DOI] [PubMed] [Google Scholar]
- 16.Gellerich F.N. Mitochondrial respiratory rates and activities of respiratory chain complexes correlate linearly with heteroplasmy of deleted mtDNA without threshold and independently of deletion size. Bioenergetics. 2002;1556:41–52. doi: 10.1016/s0005-2728(02)00305-5. [DOI] [PubMed] [Google Scholar]
- 17.Suzuki T., Nagao A., Suzuki T. Human mitochondrial tRNAs: biogenesis, function, structural aspects, and diseases. Annu. Rev. Genet. 2011;45:299–329. doi: 10.1146/annurev-genet-110410-132531. [DOI] [PubMed] [Google Scholar]
- 18.Lagouge M., Larsson N.G. The role of mitochondrial DNA mutations and free radicals in disease and ageing. J. Intern. Med. 2013;273:529–543. doi: 10.1111/joim.12055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fujikura J. Induced pluripotent stem cells generated from diabetic patients with mitochondrial DNA A3243G mutation. Diabetologia. 2012;55:1689–1698. doi: 10.1007/s00125-012-2508-2. [DOI] [PubMed] [Google Scholar]
- 20.Folmes C.D.L. Disease-causing mitochondrial heteroplasmy segregated within induced pluripotent stem cell clones derived from a patient with MELAS. Stem Cells. 2013;31:1298–1308. doi: 10.1002/stem.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cherry A.B.C. Induced pluripotent stem cells with a mitochondrial DNA deletion. Stem Cells. 2013;31:1287–1297. doi: 10.1002/stem.1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hamalainen R.H. Tissue- and cell-type-specific manifestations of heteroplasmic mtDNA 3243A>G mutation in human induced pluripotent stem cell-derived disease model. Proc. Natl. Acad. Sci. USA. 2013;110:E3622–E3630. doi: 10.1073/pnas.1311660110. [DOI] [PMC free article] [PubMed] [Google Scholar]


