Key Points
Acquired mutations of myeloid-related genes are present in a proportion of AA patients.
Somatic mutations in AA predict higher risk of transformation to MDS.
Abstract
The distinction between acquired aplastic anemia (AA) and hypocellular myelodysplastic syndrome (hMDS) is often difficult, especially nonsevere AA. We postulated that somatic mutations are present in a subset of AA, and predict malignant transformation. From our database, we identified 150 AA patients with no morphological evidence of MDS, who had stored bone marrow (BM) and constitutional DNA. We excluded Fanconi anemia, mutations of telomere maintenance, and a family history of BM failure (BMF) or cancer. The initial cohort of 57 patients was screened for 835 known genes associated with BMF and myeloid cancer; a second cohort of 93 patients was screened for mutations in ASXL1, DNMT3A, BCOR, TET2, and MPL. Somatic mutations were detected in 19% of AA, and included ASXL1 (n = 12), DNMT3A (n = 8) and BCOR (n = 6). Patients with somatic mutations had a longer disease duration (37 vs 8 months, P < .04), and shorter telomere lengths (median length, 0.9 vs 1.1, P < .001), compared with patients without mutations. Somatic mutations in AA patients with a disease duration of >6 months were associated with a 40% risk of transformation to MDS (P < .0002). Nearly one-fifth of AA patients harbor mutations in genes typically seen in myeloid malignancies that predicted for later transformation to MDS.
Introduction
Acquired aplastic anemia (AA) is an immune-mediated disorder characterized by quantitative defects in the hematopoietic stem cell compartment.1-4 Evolution to myelodysplastic syndrome (MDS) or acute myeloid leukemia (AML) occurs in up to 15% of AA patients, especially in those not attaining complete response following treatment with immunosuppressive therapy (IST).5
The most frequent and often most difficult differential diagnosis of AA, especially the nonsevere subtype of AA, is hypocellular MDS (hMDS). The age of onset of AA varies widely with 2 peaks, 1 in adolescents and young adults and another around 60 years of age,6,7 although the much more frequent occurrence of MDS in older patients often results in uncertainty about the diagnosis being hMDS rather than AA. Morphologic differences between the 2 conditions may be subtle, and often there are too few cells to be sure of morphologic characterization. The lack of established diagnostic criteria to distinguish these 2 entities is a major reason for this inconsistency.8 However, the significance of abnormal cytogenetic clones seen in around 10% to 15% of patients with AA is controversial and may not indicate a diagnosis of established MDS.9 Some abnormal clones, for example +810 and del13q,11 are associated with a good response to IST. Furthermore, detection of single nucleotide polymorphism array (SNP-A) karyotype abnormalities may identify those AA patients who are at risk of clonal evolution.12
The principal aim of this study was to examine a large cohort of AA patients to determine whether a subgroup of AA patients, especially those with less severe disease, had hMDS rather than AA, based on the presence of acquired somatic mutations that typically occur in MDS and whether this would help predict those at higher risk of later transformation to MDS/AML. This would be important for treatment decisions, especially for consideration of allogeneic transplant at an earlier stage before transformation to high-risk MDS or AML.
To determine the presence of acquired somatic mutations in acquired AA, we screened 150 patients who had no evidence of Fanconi anemia and no detectable mutations of the telomere maintenance genes (TERC, TINF2, and TERT) by using targeted high-throughput DNA sequencing. We found acquired somatic mutations in 20% of AA patients, and this predicted for evolution to MDS/AML.
Materials and methods
Patients
We screened the King’s College Hospital adult bone marrow failure (BMF) database of 345 patients with idiopathic BMF; 150 had stored bone marrow (BM) and skin or buccal mucosa samples. Informed written consent was obtained in accordance with National Research Ethics Protocol (NRES 08/H0906/94-KCLPRO29) and the Declaration of Helsinki. On the basis of peripheral blood and BM criteria, patients were classified as nonsevere AA (NSAA; n = 65), severe AA (SAA; n = 51), very severe AA (VSAA; n = 23),13 AA/paroxysmal nocturnal hemoglobinuria (PNH; n = 6), pure red cell aplasia (n = 4), or acquired amegakaryocytic thrombocytopenia (n = 1). All AA patients fulfilled the criteria for diagnosis of AA.3,14 The sequencing was done less than 6 months from diagnosis in 63 patients (median, 1.4 months; range, 0 to 6 months) and beyond 6 months in 87 patients (median, 44.3 months; range, 6.1 to 249 months) (Table 1). The median follow-up from diagnosis was 46 months (range, 1 to 360 months).
Table 1.
Features of patients with AA with somatic mutations and wild-type, based on disease duration
| Disease duration* | No. of patients | Mutations (n = 29) | Wild-type (n = 131) | P |
|---|---|---|---|---|
| <6 months | 63 | 9 | 54 | |
| Transformation to MDS | 3 | 3 | <.03 | |
| Median mutant allele burden <10% | 7 | NA | ||
| >6 months | 87 | 20 | 67 | |
| Transformation to MDS | 8 | 3 | <.0002 | |
| Median mutant allele burden <10% | 4 | NA |
NA, not applicable.
Time interval from initial diagnosis of AA (first presentation) to sampling date for patients with median mutant allele burden (proportion of sequence reads harboring mutation) less than 10% (ie, subclonal mutations). The data include both patients evaluated in the initial cohort and in the second focused cohort; the breakdown for second focused cohort is provided in supplemental Table 6.
SNP-A karyotyping
Genotyping was performed on DNA from BM mononuclear cells by using the Affymetrix 250K or SNP6 platform as described previously.15
Measurement of relative telomere length
The telomere length was assessed on the genomic DNA by a modified Cawthon’s multiplex quantitative polymerase chain reaction and reported as telomere to single copy gene (T/S) ratio16 (see supplemental Methods available on the Blood Web site).
Sequencing
For cohort 1 (n = 57), a custom panel of 835 gene exons (supplemental Table 1) were targeted via Agilent SureSelect technology and sequenced by using Illumina 100 base-pair paired-end sequencing on the HiSeq2000 sequencing system. Variant analysis was performed on patient BM/skin paired samples by using SAMtools17 and VarScan18 software following alignment and realignment/recalibration with Burrows-Wheeler Aligner and Genome Analysis Toolkit software, respectively. Mean base coverage across the custom regions was 190-640X per sample for >95% of samples.
For cohort 2 (n = 93), Illumina Nextera-amplicon sequencing was also used in high-depth screens across genes found to be mutated in the initial screen (mean base coverage >1000X for 90% of samples). Independent sequencing experiments were used to validate all mutations and confirm their acquired nature by their absence in paired constitutional DNA in the majority of patients (supplemental Methods).
Results
Clinical data
The median age was 44 years (range, 17 to 84 years). The ratio of males to females was 0.9 (71:79). Median disease duration from diagnosis to sample date was 10 months (range, 0 to 246 months). The demographic details, clinical characteristics, treatments received, and survival status are provided in Table 2, supplemental Data, and supplemental Table 2. High sensitivity flow detected granulocyte PNH clone in 65 patients (43%), large (>50%), moderate (10% to 50%), and subclinical (<10%) in 15, 14, and 36 patients, respectively (Figure 1).
Table 2.
Clinical features of patients with AA stratified by the presence/absence of non-PIGA somatic mutations
| Somatic mutation (n = 29) | No somatic mutation (n = 121) | P | |||
|---|---|---|---|---|---|
| No. | % | No. | % | ||
| Sex | .5 | ||||
| Male | 16 | 55 | 55 | 46 | |
| Female | 13 | 45 | 66 | 54 | |
| Age at diagnosis, years | .4 | ||||
| Median | 44 | 41 | |||
| Range | 27-83 | 11-87 | |||
| Age at sampling, years | .2 | ||||
| Median | 47 | 43 | |||
| Range | 28-83 | 17-87 | |||
| Disease duration, months* | .04 | ||||
| Median | 37 | 8 | |||
| Range | 0-305 | 0-605 | |||
| Severity of AA | .7 | ||||
| Very severe | 2 | 7 | 21 | 17 | |
| Severe | 10 | 35 | 41 | 34 | |
| Nonsevere | 16 | 55 | 49 | 41 | |
| Other† | 1 | 3 | 10 | 8 | |
| PNH clone | .3 | ||||
| None | 18 | 62 | 67 | 55 | |
| Subclinical | 5 | 17 | 31 | 26 | |
| Moderate | 1 | 3 | 13 | 11 | |
| Large | 5 | 17 | 10 | 8 | |
| MDS/AML evolution | .001 | ||||
| Yes | 11 | 38 | 6 | 5 | |
| No | 18 | 62 | 115 | 94 | |
| IST | .5 | ||||
| Yes | 22 | 76 | 85 | 70 | |
| No | 7 | 24 | 36 | 30 | |
Time interval between the initial diagnosis and date of sampling.
Included pure red cell aplasia, acquired amegakaryocytic thrombocytopenia, and AA with hemolytic PNH.
Figure 1.
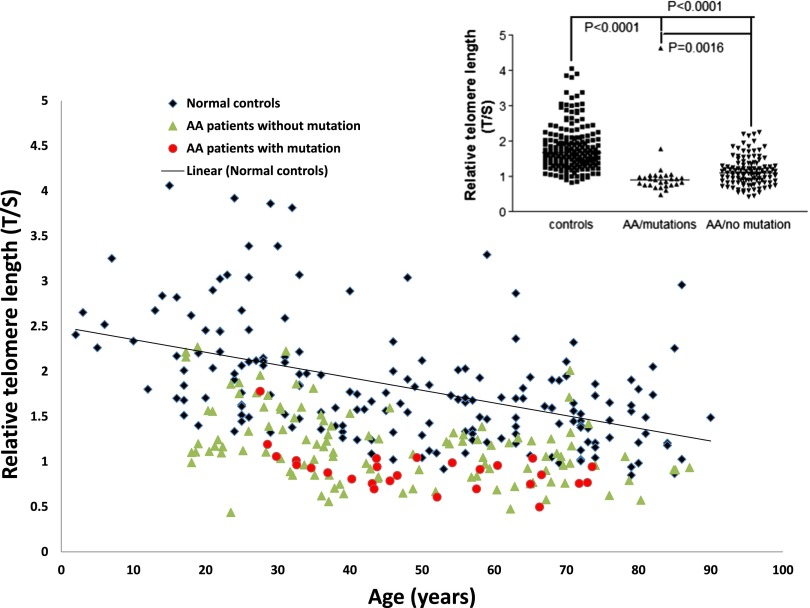
Telomere lengths of patients in the study cohort segregated into different subgroups as compared with healthy age-matched controls. The groups are AA patients with no acquired mutations (green triangles) and AA patients with somatic mutations (red circles). The trend line for normal controls (black rhomboid) is also shown. The relative telomere lengths of normal controls and AA patients with or without somatic mutations is shown in the figure inset.
Metaphase, fluorescence in situ hybridization, and molecular karyotyping
The metaphase cytogenetic karyotype was normal in 112 patients; in 12 patients, monosomy 7 (n = 3), trisomy 8 (n = 2), loss of Y (n = 2), gain of Y (n = 1), del(13)(q12q21) (n = 1), trisomy 15 (n = 1), add(6)(p22) (n = 1), and del(10)(q22q24) (n = 1) were detected. Karyotype was not available in 7 patients. Fluorescence in situ hybridization for chromosomes 5, 7, and 8 in patients for whom metaphase cytogenetic karyotyping failed (n = 19) revealed 1 patient with trisomy 8 (supplemental Table 3). SNP-A karyotyping was performed in 43 patients, and new additional aberrations were identified in 3 patients: uniparental disomy 6p25.3-21.1, gain of 11q24.3-25, and uniparental disomy of both 15q23-25.1 and 18q21.3-21.1 (supplemental Figure 1 and supplemental Data).
Telomere length
The telomere length was shorter for patients with AA (n = 142) compared with healthy control participants (n = 182) (Figure 1 and supplemental Table 3). AA patients without somatic mutations had significantly longer telomere length (median T/S, 1.1) compared with patients with acquired mutations (median T/S, 0.9; P = .0016).
Somatic mutations are frequent in AA
Excluding PIGA mutations, 29 (19%) of 150 patients had 32 somatic mutations (Table 3). The mutations detected were ASXL1 (n = 12), DNMT3A (n = 8), BCOR (n = 6), SRSF2 (n = 1), U2AF1 (n = 1), TET2 (n = 1), MPL (n = 1), IKZF1 (n = 1), and ERBB2 (n = 1). Mutations were confirmed to be acquired by their absence in constitutional DNA. The frequency of mutations was not different between the initial cohort (12 [21%] of 57) and the second focused cohort (17 [18%] of 93), with similar incidence of mutations of ASXL1, DNMT3A, and BCOR (supplemental Data).
Table 3.
Details of all the somatic mutations in the study
| UPN | Gene | Mutant allele burden (%) | Variant class | Nucleotide and protein change | Constitutional DNA |
|---|---|---|---|---|---|
| 2* | ASXL1 | 30 | Frameshift insertion | c.1927_1928insG:p.G643fs | Skin |
| 2* | DNMT3A | 42 | Nonsynonymous SNV | c.C1540G:p.L514V | Skin |
| 2* | ERBB2 | 44 | Nonsynonymous SNV | c.G922A:p.V308M | Skin |
| 5* | TET2 | 5 | Stopgain SNV | c.C3100T:p.Q1034X | Skin |
| 6* | ASXL1 | 38 | Stopgain SNV | c.C2242T:p.Q748X | Buccal |
| 10* | SRSF2 | 43 | Nonsynonymous SNV | c.C284T:p.P95L | Buccal |
| 16* | ASXL1 | 23 | Frameshift insertion | c.2469_2470insAG:p.L823fs | Skin |
| 18* | DNMT3A | 31 | Nonsynonymous SNV | c.C2644T:p.R882C | Skin |
| 19* | IKZF1 | 14 | Nonsynonymous SNV | c.C640G:p.H214D | Skin |
| 21* | BCOR | 5 | Stopgain SNV | c.C526T:p.Q176X | Buccal |
| 29* | ASXL1 | 41 | Stopgain SNV | c.G4068A:p.W1356X | Skin |
| 33* | BCOR | 68 | Stopgain SNV | c.G4832A:p.W1611X | Skin |
| 40* | ASXL1 | 31 | Nonframeshift deletion | c.2894_2896del:p.965_966del | Buccal |
| 46* | MPL | 10 | Nonsynonymous SNV | c.G1544T:p.W515L | Buccal |
| 64 | DNMT3A | 47 | Nonsynonymous SNV | c.C2644T:p.R882C | Skin |
| 66 | ASXL1 | 37 | Frameshift deletion | c.2433delT:p.N811fs | Skin |
| 67 | U2AF1 | 19 | Nonsynonymous SNV | c.C101A:p.S34Y | Skin |
| 69 | ASXL1 | 34 | Stopgain SNV | c.C2077T:p.R693X | Buccal |
| 70 | ASXL1 | 2 | Stopgain SNV | c.G2026T:p.E676X | Buccal |
| 70 | BCOR | 14 | Stopgain SNV | c.T912G:p.Y304X | Buccal |
| 73 | BCOR | 6 | Frameshift insertion | c.4834_4835insC:p.L1612fs | Skin |
| 79 | ASXL1 | 36 | Stopgain SNV | c.G2026T:p.E676X | Buccal |
| 81 | ASXL1 | 3 | Stopgain SNV | c.T2324G:p.L775X | Skin |
| 88 | ASXL1 | 7 | Frameshift deletion | c.2126delC:p.A709fs | Skin |
| 93 | DNMT3A | 8 | Stopgain SNV | C2311T:p.R771X | Skin |
| 94 | BCOR | 30 | Splice site | splice site c.3052-2A>G | Skin |
| 97 | DNMT3A | 7 | Nonsynonymous SNV | c.C2644T:p.R882C | Buccal |
| 107 | ASXL1 | 30 | Stopgain SNV | c.T2468G:p.L823X | Buccal |
| 129 | DNMT3A | 5 | Nonsynonymous SNV | c.G2207A:p.R736H | Skin |
| 130 | DNMT3A | 5 | Nonsynonymous SNV | c.G2645A:p.R882H | Skin |
| 140 | BCOR | 5 | Frameshift deletion | c.4760delC:p.P1587fs | Buccal |
| 142 | DNMT3A | 1.5 | Nonsynonymous SNV | c.C2644T:p.R882C | Buccal |
c, nucleotide; g, genomic sequence; p, protein sequence; SNV, single nucleotide variant.
Indicates patients in the initial cohort (UPN 1 through UPN 57); the correlation with PIGA mutation is illustrated in supplemental Table 4.
The median mutant allele burden (proportion of sequence reads harboring mutation) was 20% (range, 1.5% to 68%). All mutations were heterozygous except for a hemizygous BCOR mutation (male patient). Eleven patients had a mutation load of <10% (small clones), specifically DNMT3A (n = 5), ASXL1 (n = 3), BCOR (n = 2), and TET2 (n = 1). These small clones were frequently seen in patients with shorter disease duration (7 [77%] of 9) compared with those AA patient samples >6 months from diagnosis (4 [20%] of 20; P < .01; Table 1). Mutations were missense (n = 12), nonsense (n = 12), frameshift (n = 6), nonframeshift deletion (n = 1), and splice site changes (n = 1).
Somatic mutations were more frequent in SAA and NSAA (n = 26) compared with VSAA (n = 2), but the difference was not significant (21% vs 9%; P = .2) (Figure 2). All patients with mutations had a diagnosis of AA at the time of the study, with various disease severity and remission status following IST. The median age of patients with somatic mutations was older than that of patients without somatic mutations (47 vs 43 years; P < .2) and, similarly, the duration of AA was longer in patients with mutations (37 vs 8 months; P < .04). Response rate to IST, either partial or complete, was not different between patients with or without mutations.
Figure 2.
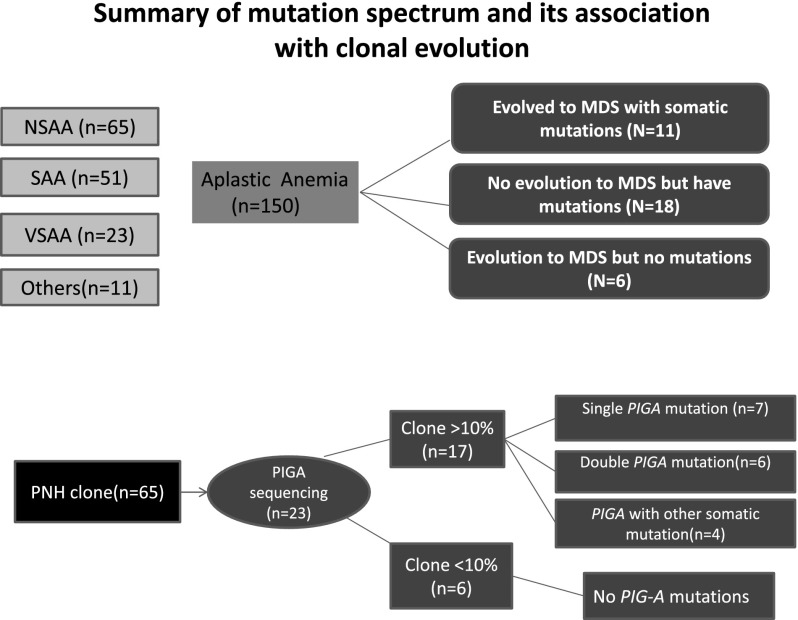
Schematic representation of patients in the study cohort based on severity of AA, presence of PNH clone, and presence of somatic mutations with or without transformation to MDS. The detailed data on PIGA mutations are provided in the supplemental Data. Only patients in the first cohort (n = 57) were screened for PIGA mutations, and 23 of 57 patients had a PNH clone detected by flow cytometry.
Somatic mutations of ASXL1 was the most frequent abnormality, present in 8% of patients (n = 12). Although there was no predilection for a particular hot spot within exon 12, all mutations were heterozygous and were predicted to have a deleterious effect on protein structure (stop or frameshift). The commonly reported mutation of G646fs was seen in 1 patient, and most other mutations have been previously reported in MDS/AML.19 The median allele burden for ASXL1 mutations was 31% (range, 2% to 41%); only 3 patients had <10% clone. A majority of ASXL1 mutations were isolated except for 2 with coexisting DNMT3A and BCOR, respectively.
DNMT3A mutations were seen in 8 patients (5%), with the more frequently reported R882 (R882C and R882H) mutation seen in 5 patients. All were missense and heterozygous mutations with median allele burden of 7% (range, 1% to 47%). Clone sizes were <10% in 62% (5 of 8) of DNMT3A mutants.
BCOR mutations were detected in 6 patients (4%) and were nonsense (n = 3), frameshift (n = 2), and splice site (n = 1) variations. Median allele burden was 10% (range, 5% to 68%). Notably, all patients with BCOR mutation also had a PNH clone (2 large; 4 subclinical). Of interest, unique patient number (UPN) 33 presented with VSAA with monosomy 7 but with disappearance of the −7 clone after antithymocyte globulin; this patient acquired a large PNH clone along with a BCOR mutation at study entry. Additionally, the mutant allele burden was similar for both PIGA and BCOR, suggesting their presence in the same clone.
Only 2 patients had RNA splicing gene mutations SRSF2 and U2AF1, and no mutations of SF3B1 or ZRSR2 were seen. Evolution to high-risk MDS was observed in 1 patient with U2AF1 mutation. A small TET2 mutant clone (5%) was identified in 1 patient who evolved to florid hMDS.
Morphologic progression to MDS/AML
Progression to MDS was seen in 17 patients (11%) over median disease duration of 86 months (range, 9 to 260 months) and, with the exception of 2 patients, all others had received adenine-thymine-guanine–based IST. Two patients who evolved to MDS subsequently progressed to AML. The median time to progression to MDS from date of sampling was 12 months (range, 2 to 50 months). The cytogenetic karyotype at the time of evolution was normal (n = 10), monosomy 7 (n = 4), trisomy 15 (n = 1), trisomy 8 (n = 1), and complex karyotype (n = 1). None of the patients had sequential samples analyzed to understand whether some patients acquired novel mutations during the natural history of the disease. However, the median follow-up was similar between patients with or without mutations; the median time of follow-up for the nonmutant patients was 23 months (range, 4 to 87 months) from sequencing and a median of 43 months (range, 4 to 224 months) from the time of diagnosis.
Of the 17 patients with morphologic progression to MDS, 11 (65%) had somatic mutations detected at a median of 12 months prior to onset of MDS. Somatic mutations were not detectable in the other 6 patients who progressed to MDS, although 2 patients (UPN 56 and UPN 75) underwent allogeneic transplantation, which precluded adequate long-term follow-up for monitoring the natural course of the disease.
AA patients with somatic mutations were more likely to evolve to MDS (11 [38%] of 29), compared with those without mutations (6 [6%] of 121; P < .001). For those patients with a somatic mutation, there was a highly significant effect of disease duration of AA on the risk of MDS transformation (Table 1). For patients with disease duration of AA of >6 months, the presence of a somatic mutation was associated with a 40% risk of MDS transformation compared with a 4.5% risk without a somatic mutation (P < .0002). A shorter disease duration was observed in patients with mutations without MDS compared with those with both mutations and morphologic evidence of MDS (52 vs 81 months; P < .4). The median clone size was also smaller (10%) in the former group compared with 31% in those with morphologic and molecular evidence of MDS.
Seven of 12 patients with ASXL1 mutations showed progression to MDS. Of the 3 patients with low-level (<10%) ASXL1 mutations, 2 progressed to hMDS. All 3 patients with a large DNMT3A clone showed progression to MDS, with monosomy 7 in 2 patients. None of the other 5 patients with DNMT3A subclones (<10%) showed progression. None of the patients with isolated BCOR mutations showed disease progression; 1 patient with a coexistent low-level ASXL1 clone evolved to MDS.
All 4 patients with monosomy 7 in our cohort had either ASXL1 (n = 3) and/or DNMT3A (n = 2) mutations and evolved to MDS (supplemental Results). Overall, 35 (24%) of AA patients had morphologic evidence of evolution to MDS (n = 6), mutations typically seen in myeloid malignancies (n = 18), or a combination of both (n = 11).
Discussion
We have identified a subgroup (19%) of AA patients who at the time of sequencing had no morphologic evidence of MDS but who did have somatic mutations in genes associated with hematologic malignancies, especially MDS/AML.20-24 The genes frequently affected were ASXL1, DNMT3A, and BCOR. Although we undertook a targeted sequencing of 835 candidate genes involved in DNA maintenance, epigenetic regulation, cell growth, and those implicated in other cancers, we found no novel genes to be mutated, except a single patient with an ERBB2 mutation. The frequency of mutations was not different between the initial cohort (12 [21%] of 57) and the second focused cohort (17 [18%] of 93), with a similar incidence of mutations of ASXL1, DNMT3A, and BCOR. Although we postulated the presence of mutations in novel genes and undertook a targeted sequencing of a large number of candidate genes, we found no novel genes (except ERBB2) to be mutated, at least at a higher allele frequency of >5%. Hence, a larger second cohort (n = 103) was analyzed for only those genes mutated in the initial cohort plus splicing factors rather than a further comprehensive screen.
AA patients with somatic mutations had longer disease duration and were more likely to progress to MDS. The effect of AA disease duration on the risk of MDS transformation was especially striking; it was significantly increased (40%) in those patients with a disease duration of >6 months compared with those without a somatic mutation (4.5%; P < .0002). Somatic mutations were detected at a median of 1 year prior to emergence of morphologic features of MDS. However, the time interval between initial diagnosis and date of sampling was significantly longer in the patients with mutations (with a median of 37 months vs less than 8 months for the group with no mutations). The longer disease duration prior to sequencing may have skewed the results because clones would have a longer time to gain clonal dominance and become detected. Patients with a longer time interval between diagnosis and sequencing would have received more therapy, which could also have skewed the results and selected for the emergence of clones. Furthermore, there was a lack of uniform follow-up time, but in a very large study evaluating a rare disease, a median follow-up of 4 years may be regarded as reasonable. Nevertheless, on the basis of our results, future studies with sequential analyses of somatic mutations in AA are now indicated.
The nonsevere subtype of AA is often difficult to distinguish morphologically from hMDS. We found a trend for more frequent somatic mutations in SAA and NSAA compared with VSAA, but larger numbers of patients are needed to explore this further. Although classified as AA at the time of sampling, the existence of myeloid-associated gene mutations provides evidence for the prior existence or emergence of a clone. Furthermore, transformation to MDS/AML was seen more frequently in patients with somatic mutations compared with those without mutations. Careful morphologic evaluation of features of dysplasia in the peripheral blood and BM in patients with mutations needs to be undertaken to confirm conclusive evidence for MDS, which has not only diagnostic but also therapeutic consequences. A future systematic and in-depth sequencing screen in acquired AA at the time of diagnosis and sequentially after IST could therefore provide invaluable tools for clinical evaluation.
The diversity of somatic mutations seen in acquired marrow failure highlights the complexity of pathogenetic mechanisms underlying this immune-mediated disease. Response to IST seen in patients with clonal markers is not surprising because a variable proportion of MDS patients with specific disease characteristics respond to IST.25-27 However, an association of shorter telomeres with somatic mutations may predict an increased risk of future relapse of AA after IST in addition to higher clonal transformation rates, as has been previously reported.28 This strengthens the argument for earlier hematopoietic stem cell transplantation rather than continued exposure to IST.
Overall, 24% of AA patients in our study showed either morphologic evidence of MDS or had mutations typically seen in myeloid malignancies, providing a combined higher detection rate for MDS on incorporation of a broader targeted genetic screen.
The lack of somatic mutation in a minority of patients (6%) with subsequent morphologic evidence of MDS raises the possibility of mutations in small subclones below the limit of detection at the time of sequencing or mutations in genes not included in our panel, for example, the recently described SETBP129,30 and STAG2.31 Additionally, the lack of progression in the presence of somatic mutations could be explained by small disease clones and the need for longer follow-up to evaluate clonal expansion and disease progression combined with a need to comprehensively evaluate early signs of morphologic dysplasia. Indeed, this was true of DNMT3A mutations, because none of the very small disease clones (below 10% mutant allele burden in 5 patients) showed progression to MDS, although longer follow-up is needed. Conversely, a patient with ASXL1 and DNMT3A mutations progressed to MDS with monosomy 7 at 50 months after the sampling date. Additionally, clone size correlated with disease duration, because small clones were more likely to be present in patients sequenced <6 months from diagnosis. This is suggestive of a model of disease evolution that directly reflects clonal expansion of a defined disease clone over time.
We detected a significant number of disease clones with a relatively low mutant allele burden more frequently in early disease stage. Although dilution with blood could contribute to this finding as a result of depleted BM cell numbers, 1 explanation is that we are dealing with small subpopulations of disease clones at an early stage in the progression to MDS. Alternatively, the mutant clones might be preexisting small clones that expand as an effect of immune-mediated suppression of hematopoiesis and/or treatment. These small clones, if not present in the founding clone, might need other cooperating mutations for progression to MDS. This is 1 plausible explanation for the lack of progression to MDS seen in all 5 patients with low-level (<10%) DNMT3A clones. Others have also recently identified clonal mutations by deep sequencing in AA,32 and in contrast to our study, identified only small clones. A sequential evaluation of low-level clones in the future would reveal the clonal architecture of AA with variable clonal flux in response to immune suppression.
Values for mutation load are also potentially difficult to interpret, because we do not know whether the DNA sequenced was derived exclusively from myeloid cells. DNA was obtained from BM mononuclear cells, and patients with AA often have a prominent lymphocyte population in their BM. However, we demonstrated the absence of such mutations in constitutional DNA (skin), and thus it is highly unlikely that lymphocytes harbor such abnormalities. The retrospective aspect of this study precludes us from analyzing all the cellular compartments in details, although as discussed above, we have recently published the role of regulatory T cells and T-effector subsets in immune-mediated AA.4 The ideal control for AA patients with somatic mutations would involve sequencing of age-matched individuals with normal hematopoiesis.
Recently, whole-exome sequencing has revealed age-associated mutations in seemingly healthy individuals.31 Furthermore, TET2 mutations specifically have been detected as small clones in healthy elderly females with an associated clonal hematopoiesis,33 and in MDS, a higher incidence of this mutation was observed with increased age.15 One possibility here is that these age-related disease clones in relatively small clonal subpopulations is a predilection (founder) stage that requires subsequent cooperating mutations for clonal expansion and disease.31 Thus, the detection of both small and relatively large disease clone populations in this study may indicate different stages of clonal evolution rather than normal aging. Furthermore, detection of small DNMT3A mutant clones with no morphologic evidence of MDS might represent the early stages of disease or an intermediary state that may require acquisition of further cooperating mutations.
Clinical observations of dyskeratosis congenita and mouse telomerase knockout experiments indicate that critical shortening of telomeres causes chromosome instability, tumor formation, and cancer progression.34,35 Our work shows that short telomeres correlated well with acquired somatic mutations, possibly implying clonal evolution as a result of hematopoietic stress. The stromal microenvironment and the inflammatory milieu could also potentially favor selection of mutant clones.
It is interesting to note that the majority of the mutations detected in our cohort are genes involved in epigenetic regulation of DNA transcription.36-38 Gene expression profiling of patients with ASXL1 mutation has previously demonstrated an upregulation of immune response pathway compared with patients with wild-type ASXL1.19 The detection of ASXL1, DNMTA, BCOR, and TET2 mutations in our cohort coupled with published expression data provides a role for the potential association and cooperation between mutations in epigenetic regulators and immune-mediated BMF.
We have identified clonal hematopoiesis in a fifth of AA patients and specific gene mutations associated with transformation to MDS. The presence of mutated clones could indicate clonal myelopoiesis like that in MDS or selective adaptation in the context of immune surveillance/subversion secondary to AA or the normal aging process. In conclusion, large prospective studies with sequential analysis of somatic mutations are now indicated to more accurately determine the impact and dynamics of mutations in predicting the risk of evolution to MDS and AML prior to onset of frank morphologic dysplasia.
Acknowledgments
The authors thank Nigel B. Westwood, Rajani Chelliah, and Swaibu Mugambwa for assisting with sample processing and tissue separation.
This work was supported by the Leukaemia Lymphoma Research (LLR) Fund (United Kingdom), by LLR and the British Society for Haematology (A.G.K.), and by King’s College London (the King's College Haemato-oncology Tissue Bank).
Footnotes
The online version of this article contains a data supplement.
There is an Inside Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: A.G.K., A.E.S., J.J., J.C.W.M., and G.J.M. contributed to the conception and design of the study; A.E.S., J.J., and S.M. performed sequencing; J.J. performed telomere length measurement; A.M.M. and J.G. performed SNP-A karyotyping; B.C. performed metaphase cytogenetics and fluorescence in situ hybridization analysis; A.G.K., S.G., J.C.W.M., and G.J.M. provided clinical samples; and all authors contributed to the analysis and interpretation of data.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Ghulam J. Mufti, King’s College London, Department of Haematological Medicine, The Rayne Institute, 123 Coldharbour Ln, London, SE5 9NU, United Kingdom; e-mail: ghulam.mufti@kcl.ac.uk.
References
- 1.Young NS, Calado RT, Scheinberg P. Current concepts in the pathophysiology and treatment of aplastic anemia. Blood. 2006;108(8):2509–2519. doi: 10.1182/blood-2006-03-010777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Scheinberg P, Young NS. How I treat acquired aplastic anemia. Blood. 2012;120(6):1185–1196. doi: 10.1182/blood-2011-12-274019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marsh JC, Ball SE, Cavenagh J, et al. British Committee for Standards in Haematology. Guidelines for the diagnosis and management of aplastic anaemia. Br J Haematol. 2009;147(1):43–70. doi: 10.1111/j.1365-2141.2009.07842.x. [DOI] [PubMed] [Google Scholar]
- 4.Kordasti S, Marsh J, Al-Khan S, et al. Functional characterization of CD4+ T cells in aplastic anemia. Blood. 2012;119(9):2033–2043. doi: 10.1182/blood-2011-08-368308. [DOI] [PubMed] [Google Scholar]
- 5.Afable MG, 2nd, Tiu RV, Maciejewski JP. Clonal evolution in aplastic anemia. Hematology Am Soc Hematol Educ Program. 2011;2011:90-55. [DOI] [PubMed]
- 6.Young NS, Kaufman DW. The epidemiology of acquired aplastic anemia. Haematologica. 2008;93(4):489–492. doi: 10.3324/haematol.12855. [DOI] [PubMed] [Google Scholar]
- 7.Montané E, Ibáñez L, Vidal X, et al. Catalan Group for Study of Agranulocytosis and Aplastic Anemia. Epidemiology of aplastic anemia: a prospective multicenter study. Haematologica. 2008;93(4):518–523. doi: 10.3324/haematol.12020. [DOI] [PubMed] [Google Scholar]
- 8.Bennett JM, Orazi A. Diagnostic criteria to distinguish hypocellular acute myeloid leukemia from hypocellular myelodysplastic syndromes and aplastic anemia: recommendations for a standardized approach. Haematologica. 2009;94(2):264–268. doi: 10.3324/haematol.13755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gupta V, Brooker C, Tooze JA, et al. Clinical relevance of cytogenetic abnormalities at diagnosis of acquired aplastic anaemia in adults. Br J Haematol. 2006;134(1):95–99. doi: 10.1111/j.1365-2141.2006.06105.x. [DOI] [PubMed] [Google Scholar]
- 10.Maciejewski JP, Risitano A, Sloand EM, Nunez O, Young NS. Distinct clinical outcomes for cytogenetic abnormalities evolving from aplastic anemia. Blood. 2002;99(9):3129–3135. doi: 10.1182/blood.v99.9.3129. [DOI] [PubMed] [Google Scholar]
- 11.Hosokawa K, Katagiri T, Sugimori N, et al. Favorable outcome of patients who have 13q deletion: a suggestion for revision of the WHO ‘MDS-U’ designation. Haematologica. 2012;97(12):1845–1849. doi: 10.3324/haematol.2011.061127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Afable MG, II, Wlodarski M, Makishima H, et al. SNP array-based karyotyping: differences and similarities between aplastic anemia and hypocellular myelodysplastic syndromes. Blood. 2011;117(25):6876–6884. doi: 10.1182/blood-2010-11-314393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Camitta BM, Rappeport JM, Parkman R, Nathan DG. Selection of patients for bone marrow transplantation in severe aplastic anemia. Blood. 1975;45(3):355–363. [PubMed] [Google Scholar]
- 14.[No authors listed] Incidence of aplastic anemia: the relevance of diagnostic criteria. By the International Agranulocytosis and Aplastic Anemia Study. Blood. 1987;70(6):1718–1721. [PubMed] [Google Scholar]
- 15.Smith AE, Mohamedali AM, Kulasekararaj A, et al. Next-generation sequencing of the TET2 gene in 355 MDS and CMML patients reveals low-abundance mutant clones with early origins, but indicates no definite prognostic value. Blood. 2010;116(19):3923–3932. doi: 10.1182/blood-2010-03-274704. [DOI] [PubMed] [Google Scholar]
- 16.Cawthon RM. Telomere length measurement by a novel monochrome multiplex quantitative PCR method. Nucleic Acids Res. 2009;37(3):e21. doi: 10.1093/nar/gkn1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li H, Handsaker B, Wysoker A, et al. 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koboldt DC, Zhang Q, Larson DE, et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22(3):568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thol F, Friesen I, Damm F, et al. Prognostic significance of ASXL1 mutations in patients with myelodysplastic syndromes. J Clin Oncol. 2011;29(18):2499–2506. doi: 10.1200/JCO.2010.33.4938. [DOI] [PubMed] [Google Scholar]
- 20.Bejar R, Stevenson K, Abdel-Wahab O, et al. Clinical effect of point mutations in myelodysplastic syndromes. N Engl J Med. 2011;364(26):2496–2506. doi: 10.1056/NEJMoa1013343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yoshida K, Sanada M, Shiraishi Y, et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011;478(7367):64–69. doi: 10.1038/nature10496. [DOI] [PubMed] [Google Scholar]
- 22.Patel JP, Gönen M, Figueroa ME, et al. Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N Engl J Med. 2012;366(12):1079–1089. doi: 10.1056/NEJMoa1112304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cancer Genome Atlas Research Network. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368(22):2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grossmann V, Tiacci E, Holmes AB, et al. Whole-exome sequencing identifies somatic mutations of BCOR in acute myeloid leukemia with normal karyotype. Blood. 2011;118(23):6153–6163. doi: 10.1182/blood-2011-07-365320. [DOI] [PubMed] [Google Scholar]
- 25.Sloand EM, Wu CO, Greenberg P, Young N, Barrett J. Factors affecting response and survival in patients with myelodysplasia treated with immunosuppressive therapy. J Clin Oncol. 2008;26(15):2505–2511. doi: 10.1200/JCO.2007.11.9214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Passweg JR, Giagounidis AA, Simcock M, et al. Immunosuppressive therapy for patients with myelodysplastic syndrome: a prospective randomized multicenter phase III trial comparing antithymocyte globulin plus cyclosporine with best supportive care—SAKK 33/99. J Clin Oncol. 2011;29(3):303–309. doi: 10.1200/JCO.2010.31.2686. [DOI] [PubMed] [Google Scholar]
- 27.Lim ZY, Killick S, Germing U, et al. Low IPSS score and bone marrow hypocellularity in MDS patients predict hematological responses to antithymocyte globulin. Leukemia. 2007;21(7):1436–1441. doi: 10.1038/sj.leu.2404747. [DOI] [PubMed] [Google Scholar]
- 28.Scheinberg P, Cooper JN, Sloand EM, Wu CO, Calado RT, Young NS. Association of telomere length of peripheral blood leukocytes with hematopoietic relapse, malignant transformation, and survival in severe aplastic anemia. JAMA. 2010;304(12):1358–1364. doi: 10.1001/jama.2010.1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Piazza R, Valletta S, Winkelmann N, et al. Recurrent SETBP1 mutations in atypical chronic myeloid leukemia. Nat Genet. 2013;45(1):18–24. doi: 10.1038/ng.2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Damm F, Itzykson R, Kosmider O, et al. SETBP1 mutations in 658 patients with myelodysplastic syndromes, chronic myelomonocytic leukemia and secondary acute myeloid leukemias. Leukemia. 2013;27(6):1401–1403. doi: 10.1038/leu.2013.35. [DOI] [PubMed] [Google Scholar]
- 31.Welch JS, Ley TJ, Link DC, et al. The origin and evolution of mutations in acute myeloid leukemia. Cell. 2012;150(2):264–278. doi: 10.1016/j.cell.2012.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lane AA, Odejide O, Kopp N, et al. Low frequency clonal mutations recoverable by deep sequencing in patients with aplastic anemia. Leukemia. 2013;27(4):968–971. doi: 10.1038/leu.2013.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Busque L, Patel JP, Figueroa ME, et al. Recurrent somatic TET2 mutations in normal elderly individuals with clonal hematopoiesis. Nat Genet. 2012;44(11):1179–1181. doi: 10.1038/ng.2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alter BP, Giri N, Savage SA, Rosenberg PS. Cancer in dyskeratosis congenita. Blood. 2009;113(26):6549–6557. doi: 10.1182/blood-2008-12-192880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blasco MA, Lee HW, Hande MP, et al. Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell. 1997;91(1):25–34. doi: 10.1016/s0092-8674(01)80006-4. [DOI] [PubMed] [Google Scholar]
- 36.Ley TJ, Ding L, Walter MJ, et al. DNMT3A mutations in acute myeloid leukemia. N Engl J Med. 2010;363(25):2424–2433. doi: 10.1056/NEJMoa1005143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mardis ER, Ding L, Dooling DJ, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med. 2009;361(11):1058–1066. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Delhommeau F, Dupont S, Della Valle V, et al. Mutation in TET2 in myeloid cancers. N Engl J Med. 2009;360(22):2289–2301. doi: 10.1056/NEJMoa0810069. [DOI] [PubMed] [Google Scholar]