Abstract
The CAG repeat expansion in the Huntington's disease gene HTT extends a polyglutamine tract in mutant huntingtin that enhances its ability to facilitate polycomb repressive complex 2 (PRC2). To gain insight into this dominant gain of function, we mapped histone modifications genome-wide across an isogenic panel of mouse embryonic stem cell (ESC) and neuronal progenitor cell (NPC) lines, comparing the effects of Htt null and different size Htt CAG mutations. We found that Htt is required in ESC for the proper deposition of histone H3K27me3 at a subset of ‘bivalent’ loci but in NPC it is needed at ‘bivalent’ loci for both the proper maintenance and the appropriate removal of this mark. In contrast, Htt CAG size, though changing histone H3K27me3, is prominently associated with altered histone H3K4me3 at ‘active’ loci. The sets of ESC and NPC genes with altered histone marks delineated by the lack of huntingtin or the presence of mutant huntingtin, though distinct, are enriched in similar pathways with apoptosis specifically highlighted for the CAG mutation. Thus, the manner by which huntingtin function facilitates PRC2 may afford mutant huntingtin with multiple opportunities to impinge upon the broader machinery that orchestrates developmentally appropriate chromatin status.
Introduction
Huntington's disease (HD) is a dominantly inherited neurodegenerative disorder characterized by the prominent loss of medium size spiny neurons in the caudate and putamen (1,2). Discovery of the HD mutation revealed an unstable expanded CAG trinucleotide repeat, of more than ∼35 units, in exon 1 of HTT (previously HD) (3). The mutation triggers a truly dominant disease process that leads to the onset of diagnostic motor signs in a manner that is inversely correlated with the size of only the expanded HTT CAG repeat (4), although both the mutant and normal range repeat HTT alleles are expressed from conception (5–8).
The key features of the HD mutation (true dominance and progressivity with CAG size of the expanded repeat) provide genetic criteria with which to delineate the mutational mechanism and its earliest consequences. Furthermore, the occurrence of polyglutamine-encoding CAG-expansion mutations in unrelated genes that cause eight different inherited neurodegenerative disorders strongly implies a disease mechanism whose specificity entails an effect of the expanded polyglutamine tract on the existing function of the protein in which it is embedded (9–11). Therefore, although other scenarios are possible, the most parsimonious explanation of the genetic findings is that its expanded polyglutamine tract endows full-length mutant huntingtin with some novel property that is related to the protein's normal function as a HEAT/HEAT-like domain α-helical scaffold molecule (12–14).
To provide a genetic framework for focused biochemical and molecular studies, we and others have created precisely targeted mutations in the mouse HD gene homolog Htt (previously Hdh). Phenotypes associated with inactivating Htt mutations disclose inherent normal huntingtin functions (15–18), whereas phenotypes that worsen with the size of expanded Htt CAG alleles, which precisely replicate the HD mutation, reveal dominant effects of full-length mutant huntingtin that fulfill the HD genetic criteria (18–25). Studies with these mutant Htt alleles have revealed that huntingtin function is required for proper embryonic development and have established that Htt CAG-expansion alleles can bypass blocks early in embryonic (15–17) and fetal (18) development imposed by huntingtin deficiency, thereby arguing against loss of function (simple or dominant-negative) mechanisms for the HD mutation. In support of a simple gain of existing function, extending its polyglutamine tract enhances huntingtin's function as a facilitator for the chromatin regulator polycomb repressive complex 2 (PRC2) (14), comprising Eed (Extra sex combs—Esc) (26), Suz12 (Suppressor of Zeste 12—Su(z)12), Ezh2 (Enhancer of Zeste E(z) (27) and RbAp48/Caf1/Nurf55. Full-length wild-type huntingtin protein is able to physically interact with Ezh2 and Suz12 PRC2 core component in the nucleus of embryoid bodies (EBs) and in vitro, suggesting its function as a PRC2 facilitator is, at least in part, mediated by this interaction (14). EBs developing from embryonic stem cells (ESCs) that lack huntingtin exhibit inefficient deposition of the histone H3K27me3 mark, whereas EBs with Htt CAG-expansion display mildly increased histone H3K27me3 (14). Moreover, purified full-length human huntingtin stimulates, progressively with its polyglutamine tract size, the histone H3K27me3 catalytic activity of reconstituted PRC2 (14).
Using an approach that compares the members of an isogenic Htt knock-out and Htt CAG knock-in mutant ESC panel (23), it has been shown recently that full-length mutant huntingtin subtly alters the developmental fate of lineage committed neuronal progenitor cells (NPC) in a qualitatively different manner than does lack of huntingtin. One study demonstrates that neurons derived from CAG-expanded NPC have an increased propensity to undergo apoptosis, whereas NPC that lack huntingtin tend to give rise to glial cells in a slightly, but significantly higher proportion than wild-type Htt NPC (28). Another study finds that CAG knock-in NPC have unaltered multi-lineage potential, but premature neuronal differentiation, whereas Htt null NPC exhibit increased cell death and altered lineage potential (29). Based upon these observations, we hypothesized distinctly altered molecular states for huntingtin-null and mutant huntingtin expressing cells at earlier developmental stages. To delineate these states, while gaining molecular insights into huntingtin’'s facilitation of PRC2 activity and the effects of extending its polyglutamine tract, we have mapped histone modifications genome-wide across all of the members of the isogenic Htt null and Htt CAG repeat knock-in ESC series and lineage committed NPC derived from them.
Results
Lack of huntingtin or presence of mutant huntingtin do not alter PRC2 expression
Our Htt mutation ESC panel is composed of six pluripotent cell lines (or their derivative cell types) that were created to be analyzed together as a series of isogenic samples that enable the discovery and direct comparison of the effects of targeted inactivation of both copies of Htt (Htt null cells, dKO) with the more subtle effects of targeted mutations that progressively expand a CAG stretch in one allele (Htt CAG knock-in cells) (23). The panel comprises the parental ESC line with wild-type Htt alleles, expressing only wild-type huntingtin, the Htt null double knock-out Hdhex4/5/ex4/5 ESC line, which lacks huntingtin, and a set of four heterozygous Htt CAG repeat knock-in HdhQ20/7, HdhQ50/7, HdhQ91/7 and HdhQ111/7 ESC lines, that express both wild-type huntingtin and mutant huntingtin with expanded polyglutamine tracts of 20, 50, 91 and 111 residues, respectively (Fig. 2C) (23).
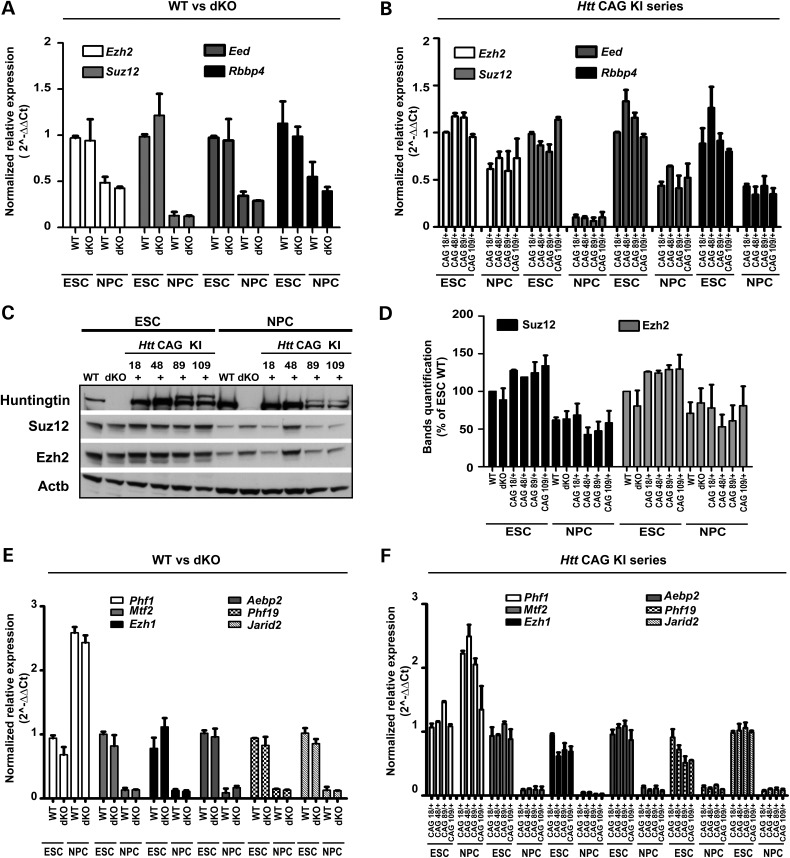
Figure 2.
PRC2 core and accessory factors are similar in across the Htt allelic series. (A and B) Bar graphs plotting normalized relative mRNA levels of genes encoding PRC2 core subunits (Ezh2, Suz12, Eed and Rbbp4/RbAp48) determined by RT-qPCR amplification assays for wild-type (WT) and Htt null (dKO) ESC and NPC lines and (B) for the heterozygous Htt CAG knock-in HdhQ20/7, HdhQ50/7, HdhQ91/7, HdhQ111/7 (CAG 18/+, 48/+, 89/+ 109/+) ESC and NPC lines. (C) Immunoblot analyses of wild-type (WT) and Htt null (dKO) and HdhQ20/7, HdhQ50/7, HdhQ91/7, HdhQ111/7 (CAG 18/+, 48/+, 89/+ 109/+) ESC and NPC lines. The band of normal mouse huntingtin (7 glutamine tract) is absent in Htt null cells and across the CAG knock-in series the normal mouse huntingtin band and the more slowly migrating mutant huntingtin bands are progressively separated with increasing size of the latter's polyglutamine tract (comprising 20, 50, 91 and 111 glutamines, respectively). The bands of Ezh2 and Suz12 and β-actin (Actb) are detected in all of the lines. (D) Quantification of Esh2 and Suz12 immunoreactive bands relative to β-actin using ImageJ software. The mean values determined in two independent biological replicates are plotted. Error bars represent the standard deviations from the mean. (E and F) Bar graphs plotting normalized relative mRNA levels of genes encoding PRC2-associated factors (Phf1, Mtf2, Ezh1, Aebp2, Phf19, Jarid2) determined by RT-qPCR amplification assays for wild-type (WT) and Htt null (dKO) ESC and NPC lines and (F) for the heterozygous Htt CAG knock-in HdhQ20/7, HdhQ50/7, HdhQ91/7, HdhQ111/7 (CAG 18/+, 48/+, 89/+ 109/+) ESC and NPC lines. Error bars represent standard deviations from the mean for two biological replicates and two technical replicates.
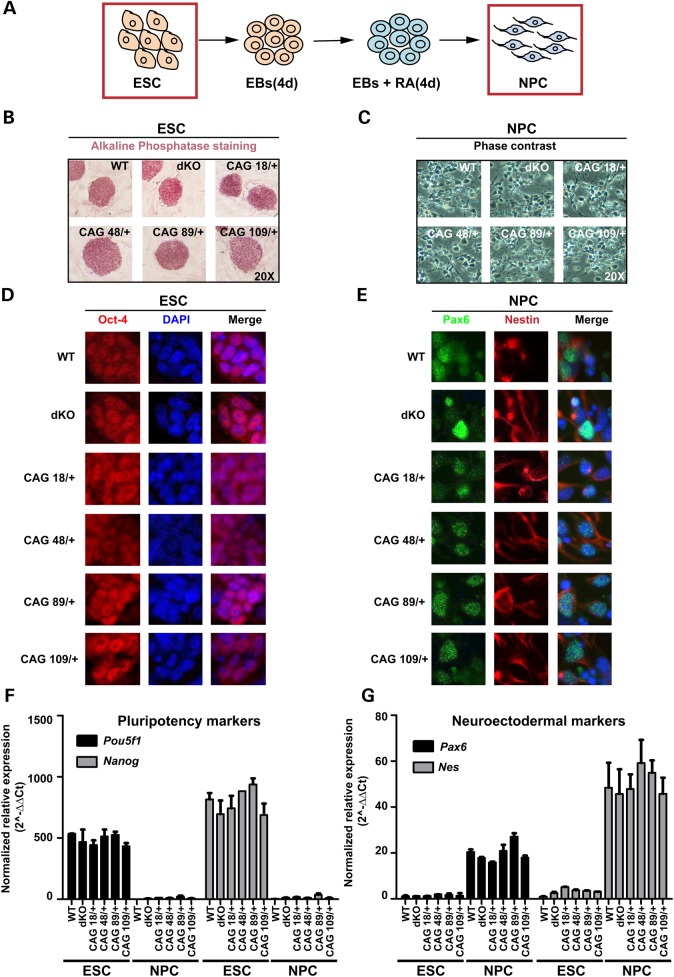
The distinctly altered fates of Htt null and Htt CAG knock-in neuronal cells do not reflect overt differences at the pluripotent stem cell or neuronal lineage committed progenitor cell stage. All six members of the panel exhibit appropriate stage-specific morphology (Fig. 1A–C) and expression of canonical markers, such as alkaline phosphatase, Oct-4 and Nanog in ESC (Fig. 1B, D and F) and Pax6 and Nestin in retinoic acid-induced NPC (Fig. 1C, E and G). Thus, neither the lack of huntingtin nor the presence of mutant huntingtin dramatically compromises either pluripotency or the induced transition to NPC. The specific Htt genotype also does not obviously alter the expression of Eed, Ezh2, Suz12 and Rbbp4 or of genes encoding the PRC2-associated factors (Phf1, Mtf2, Jarid2, Aebp2 and Phf19) and non-canonical H3K27 methyltransferase (Ezh1). The steady-state mRNA levels for these genes are not distinguished by Htt genotype at either developmental stage (Fig. 2A, B, E and F) and immunoblot analysis disclosed similar levels of Ezh2 and Suz12 in wild-type, Htt-null and Htt CAG knock-in cells (Fig. 2C and D). Since inactivation of a core member can destabilize and decrease levels of the complex (30,31), these results imply that the stability of PRC2 is not greatly altered by the absence of huntingtin or by the presence of mutant huntingtin.
Figure 1.
Members of the isogenic Htt null and Htt CAG-expansion ESC and NPC panels exhibit similar stage-appropriate morphological and molecular characteristics. (A) Schematic representation of the protocol by which mouse embryonic stem cells (ESC) develop into neuronal progenitor cells (NPC) through EBs and timed addition of retinoic acid (RA). (B and C) Phase contrast micrographs of wild-type (WT), Htt null Hdhex4/5/ex4/5 (dKO) and heterozygous Htt CAG knock-in HdhQ20/7, HdhQ50/7 HdhQ91/7 and HdhQ111/7 (CAG 18/+, 48/+, 89/+, 109/+) ESC lines showing colonies stained for alkaline phosphatase and the NPC lines derived from them displaying appropriate morphology with neurite extensions. (D and E) Images of cells, with DAPI stained nuclei to show proper Oct-4 expression in Htt wild-type, Htt null and Htt CAG knock-in ESC colonies and appropriate expression of Pax6 and Nestin neuroectodermal markers in the NPC for each genotype. (F and G) Bar graphs plot relative normalized mRNA expression levels of pluripotency marker genes Pou5f1 and Nanog encoding Oct-4 and Nanog and neuroectodermal marker genes Pax6 and Nes encoding Pax6 and Nestin as determined by RT-qPCR amplification assays. Error bars represent standard deviations from the mean of two biological and two technical replicates.
Genome-wide ChIP-seq and RNA-seq across the members of the ESC and NPC series
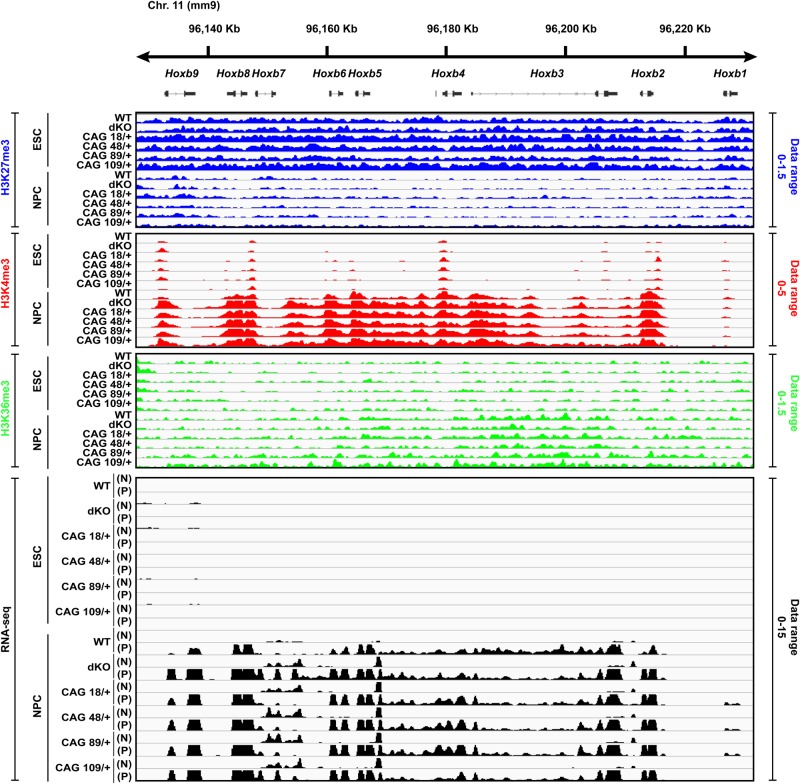
These findings are consistent with the proposed role for huntingtin as a PRC2-facilitator rather than a core member of the complex (14,32). Therefore, reasoning that the molecular consequences of the absence of huntingtin and the potentially milder effects of mutant huntingtin on PRC2 function would become evident from unbiased genome-wide chromatin mapping, we performed histone H3K27me3, H3K4me3 and H3K36me3 ChIP-seq and RNA-seq analyses across the members of the isogenic Htt ESC and NPC series (Supplementary Material, Table S1). To gauge the quality of the biological replicates formed by the six ESC datasets and six NPC datasets, we performed extensive QC analyses, which demonstrate a high degree of similarity both across the Htt genotypes for each stage and with previously reported datasets (see Materials and Methods and Supplementary Material, Figs S1A, B and S2A, B). As an example, during the transition of ESC to NPC the Hoxb cluster is expected to show loss of histone H3K27me3 and increases of histone H3K4me3 at transcriptional start site (TSS) and histone H3K36me3 over the gene body with increasing RNA expression (33). All six Htt genotypes display comparable, stage-appropriate histone H3K27me3, H3K4me3 and H3K36me3 marks and RNA profiles across this dynamically regulated gene cluster (Fig. 3).
Figure 3.
Hoxb cluster illustrating genome-wide ChIP-seq and RNA-seq analyses. A snapshot of the IGV (http://broadinstitute.org) genome browser view at the location of the developmentally regulated Hoxb cluster (mouse chromosome 11qD) shows the ChIP-seq library-size normalized reads density (see Materials and Methods) for histone H3K27me3, histone H3K4me3 and histone H3K36me3 across the Htt wild-type, Htt null and the four Htt CAG knock-in ESC lines and for the NPC derived from them. Also shown are the RNA-seq reads density with the + strand (P) and – strand (N) indicated. Library-size normalized reads density data range for each histone modification and RNA-seq datasets are indicated on the right side of the tracks. For all six genotypes, after neural induction, the level of histone H3K27me3 at the gene TSSs is decreased with increased enrichment of histone H3K4me3 that is concomitant with RNA expression and enrichment of histone H3K36me3 across the gene bodies, thereby indicating comparable pluripotency and neural differentiation status for the members of the isogenic panel. Additional QC results for the ChIP-seq and RNA-seq datasets are provided in Supplementary Material, Figures S1, S2A and B.
Htt null mutation but not Htt CAG expansion predominantly affects histone H3K27me3
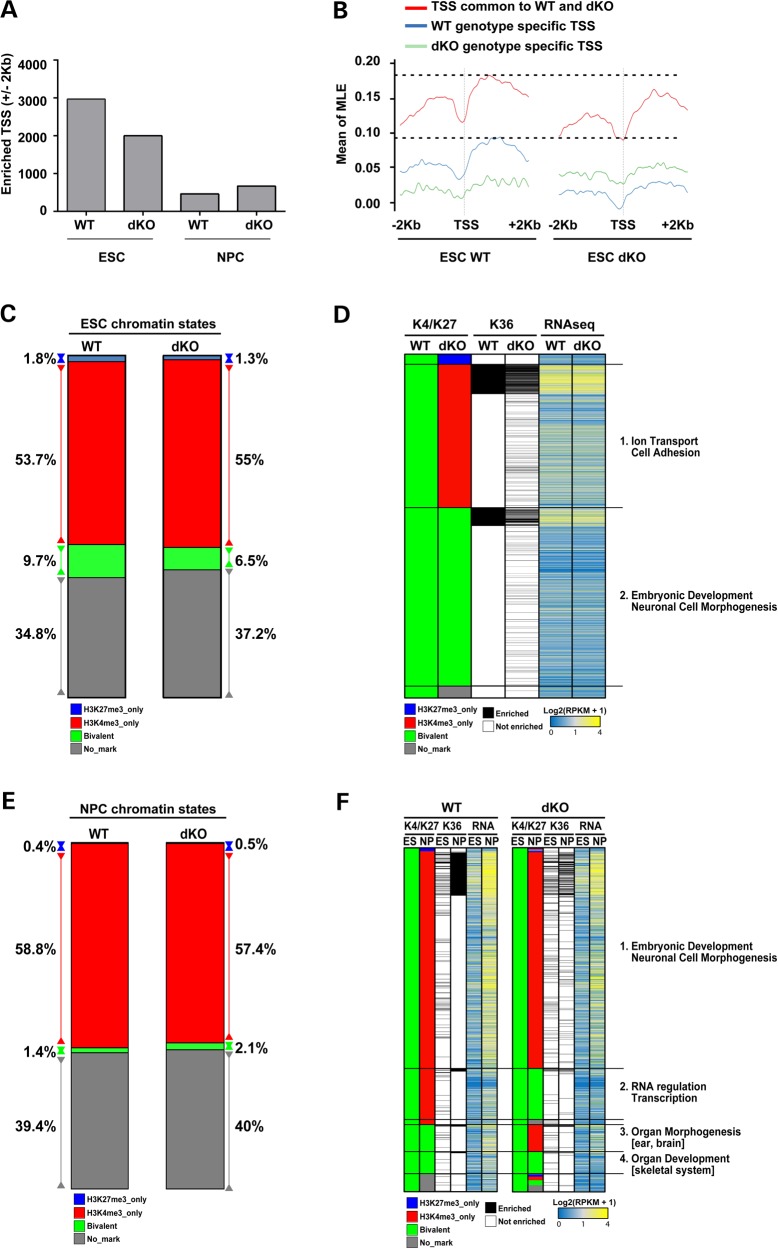
We then performed comparative quantitative analysis of the chromatin marks at the TSSs of genes genome-wide to determine the global consequences of the absence of huntingtin, by comparing wild-type versus Htt null cells, and of lengthening mutant huntingtin's polyglutamine tract by examining the four Htt CAG knock-in cell lines. The number of TSS enriched over input control for histone H3K4me3 and the number of genes with histone H3K36me3 over the gene body is not greatly affected by the Htt null genotype or by the Htt CAG-expansion alleles at either the ESC or the NPC developmental stages (Supplementary Material, Fig. S2C and D). However, using the most stringent threshold (Threshold 4) (see Materials and Methods), and as confirmed by analysis at more relaxed thresholds (Thresholds 1–3) (data not shown), the numbers of histone H3K27me3 enriched TSS are significantly decreased by one-third (4.47 × 10−79, χ2 test) in Htt null ESC and, seemingly incongruously, are nearly doubled (1.23 × 10−22, χ2 test) in Htt null NPC (Fig. 4A), compared with their wild-type counterparts. This mark is not consistently changed across the CAG knock-in ESC or NPC series (Supplementary Material, Fig. S2C and D), thereby revealing that mutant huntingtin does not mimic the loss of huntingtin function. Furthermore, we analyzed metagene profiles plotting the average ChIP-seq enrichment signals (input normalized ChIP enrichment) for three different groups of TSS presenting H3K27me3 enrichment in (a) both wild-type and Htt null ESC, (b) wild-type ESC only and (c) Htt null ESC only. The striking decrease in TSS histone H3K27me3 enrichment in Htt null ESC, which is most evident from the metagene profiles for genes with this mark in wild-type ESC (WT genotype-specific TSS) but is also noticeable at genes with this mark in both wild-type and Htt null ESC (TSS common to WT and dKO) (Fig. 4B), is consistent with huntingtin function assisting PRC2 in the deposition of histone H3K27 trimethyl mark. However, the observation of a significant excess of histone H3K27me3 enriched genes in Htt null NPC implies a further, distinct role for huntingtin in the proper resolution of this mark during neuronal cell differentiation.
Figure 4.
Htt null mutation predominantly affects histone H3K27me3 at ‘bivalent’ loci in ESC and NPC. (A) Bar graph of the total number of TSS with histone H3K27me3 enrichment (scaled ChIP read counts over input control using threshold 4 as described in Materials and Methods) in a region of ±2 kb around the TSS, for the Htt wild-type (WT) and Htt null (dKO) ESC and NPC lines. Supplementary Material, Figure S3C and D presents the TSS data for all of the other assessed histone marks for all of the members of the isogenic Htt allelic ESC and NPC series. (B) Metagene profiles displaying the average of TSS histone H3K27me3 enrichment (scaled ChIP read counts over input control exceeding threshold 3—see also Materials and Methods) in a region of ±2 kb around the TSS in Htt wild-type and Htt null ESC. TSS enriched for H3K27me3 in both genotypes are depicted in red. TSS enriched only in wild-type are given in blue. TSS enriched only in Htt null ES are depicted in green. Y axis shows mean of smoothed maximum likelihood enrichment estimates (MLE) for the groups of genes in each category (red, blue and green). (C) Bar plot depicting the fraction (as percentage) of the total TSS (N = 30 489) analyzed that are classified as ‘repressed’ (histone H3K27me3 only), ‘active’ (histone H3K4me3 only) or ‘bivalent’ (histone H3K27me3 and histone H3K4me3) for Htt wild-type (WT) and Htt null (dKO) ESC lines. (D) Composite heatmap plotting (in rows) the 2949 loci with TSS classified in Htt wild-type (WT) ESC as ‘bivalent’ to illustrate their chromatin status in Htt null (dKO) ESC. The adjacent columns show the corresponding histone H3K36me3 enrichment calculated over the gene body and the RNA-seq expression levels as Log2(RPKM+1) values. The major GO terms highlighted by pathways analyses for the subsets of loci with Htt null sensitive TSS enrichment (Category 1) and Htt null insensitive TSS enrichment (Category 2) are given (further details on the categories are provided in the Results). (E) Bar plot depicting the fraction (as percentage) of the total TSS (N = 30 489) analyzed that are classified as ‘repressed’ (histone H3K27me3 only), ‘active’ (histone H3K4me3 only) or ‘bivalent’ (histone H3K27me3 and histone H3K4me3) for Htt wild-type (WT) and Htt null (dKO) NPC lines. (F) Composite heatmap plotting (in rows) the 1525 TSS that are classified as ‘bivalent’ in both Htt wild-type (WT) ESC and Htt null (dKO) ESC with the adjacent column indicating their chromatin status in their cognate NPC line. The corresponding paired ESC and NPC histone H3K36me3 enrichment and RNA expression levels, as Log2(RPKM+1) values, are in the adjacent columns. The major GO terms highlighted by pathways analyses for the subsets of loci with Htt null insensitive TSS enrichment (Categories 1 and 4) or Htt null sensitive TSS enrichment (Categories 2 and 3) are given (further details on the categories are provided in the Results). Supplementary Material, Figure S3 reports ChIP-qPCR confirmation of selected Htt null TSS changes in ESC and NPC.
The absence of huntingtin predominantly affects histone H3K27me3 at ‘bivalent’ loci
To understand the impact of the Htt null genotype, we utilized an approach pioneered by Mikkelsen et al. (34) to generate ESC and NPC chromatin maps. The marked TSSs are classified as transcriptionally ‘repressed’ (histone H3K27me3 only) or ‘active’ (histone H3K4me3 only) or are classified as ‘bivalent’ (histone H3K27me3 and histone H3K4me3) and poised for transcription. At the ESC stage, the fraction of TSS in each category for wild-type parental ESC is similar to that reported in the literature (34) and, as reported, only ∼8% of genes in the ‘bivalent’ category are expressed, as judged by RNA-seq analysis (>2RPKM) and histone H3K36me3 enrichment across the gene body (34) (Supplementary Material, Table S2).
By comparison, the latter ‘bivalent’ chromatin class is predominantly affected by the absence of huntingtin. In Htt null ESC, although the proportions of TSS in each category are similar to wild-type ESC (Fig. 4C), nearly 42% of the TSS classified as ‘bivalent’ in wild-type ESC are instead classified as ‘active’ in HTT null ESC, such that 1229 TSS display the histone H3K4me3 mark but lack histone H3K27me3 (Fig. 4D and Supplementary Material, Table S2), consistent with inefficient PRC2 deposition of H3K27me3 in the absence of huntingtin. About 14% of these abnormally ‘active’ category genes exhibit an increase (>1.5-fold) in RNA expression in Htt null ESC (Supplementary Material, Table S2), which is consistent with the previous report showing the increased expression of only a small fraction of the genes with decreased histone H3K27me3 in Eed-null ESC (35,36).
In contrast, Htt null NPC exhibit a relative excess of TSS in the ‘bivalent’ category, although the fractions of TSS in the other chromatin categories resemble those of wild-type NPC (Fig. 4E). In wild-type NPC, the majority of loci with TSS classified as ‘bivalent’ at the ESC stage have lost the histone H3K27me3 mark and are found in the ‘active’ category (Fig. 4F). However, a small fraction (7.9%) of TSS that are properly ‘bivalent’ in Htt null ESC are inappropriately resolved to ‘active’ loci in Htt null NPC (Fig. 4F and Supplementary Material, Table S2), thereby implying that lack of huntingtin impairs the efficiency with which histone H3K27me3 is maintained at 121 poised loci. In addition, a larger proportion (17.6%) of the TSS that are appropriately ‘bivalent’ in Htt null ESC inappropriately retain the histone H3K27me3 mark and remain aberrantly ‘bivalent’ in Htt null NPC (Fig. 4F and Supplementary Material, Table S2), as confirmed by ChIP-qPCR analysis of selected loci (Supplementary Material, Fig. S3). This reveals impaired removal of the PRC2-deposited mark in the absence of huntingtin at 223 ‘bivalent’ TSS and highlights an unexpected role for huntingtin in the process by which histone H3K27me3 is removed from promoters of genes that are destined to be expressed during the transition to neuronal lineage fate.
Mutant huntingtin mainly affects histone H3K4me3 and gene expression at ‘active’ loci
To uncover the mild effects of the CAG-expansion mutation, which must satisfy the genetic parameters of the HD mechanism (dominance and progressivity with CAG size), we performed continuous analysis across the Htt CAG knock-in ESC ChIP-seq and NPC ChIP-seq datasets to disclose marks at TSS whose enrichment is progressively increased or decreased with increasing size of mutant huntingtin's polyglutamine tract [see Fig. 5A and B for visual examples of integrative genomics viewer (IGV) snapshots of H3K27me3, H3K4me3]. Across the ESC lines, CAG-progressive enrichment is found at 360 of the histone H3K27me3 marked TSS (6.05%) and 962 of the histone H3K4me3 TSS (4.21%), as confirmed by ChIP-qPCR analysis of selected loci (Supplementary Material, Fig. S4A–H). At the NPC stage, CAG-associated enrichment is detected at 110 histone H3K27me3 marked TSS (3.91%) and 933 histone H3K4me3 decorated TSS (5.09%), as validated by ChIP-qPCR analysis for selected genes with decreases in histone H3K4me3 (Supplementary Material, Fig. S4I and J).
Figure 5.
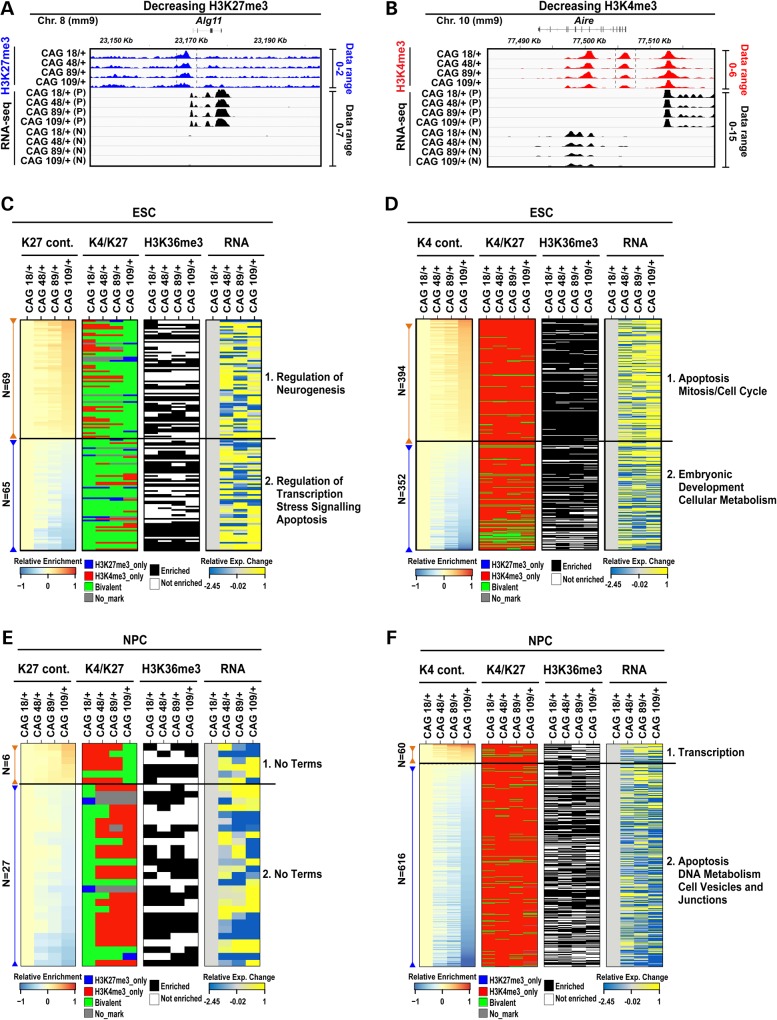
Htt CAG expansion yields pleiotropic chromatin changes in ESC and NPC. Two illustrative IGV snapshots at two genomic locations presenting decreasing H3K27me3 (A) and decreasing H3K4me3 (B) enrichments show the ChIP-seq library-size normalized reads (RPKM) for histone H3K27me3 or histone H3K4me3 across the four Htt CAG knock-in ESC lines. Vertical dashed lines indicate the ±2 kb region around the TSS considered for the evaluation of the histone modifications enrichment. Also shown are the RNA-seq reads (RPKM) with the + strand (P) and – strand (N) indicated. Library-size normalized RPKM data range for each histone modification and RNA-seq datasets are indicated on the right side of the tracks. The level of histone H3K27me3 and histone H3K4me3 at the specific TSS is decreased with increasing Htt CAG size and this change is concomitant with RNA expression alterations: specifically, increasing RNA levels were observed following histone H3K27me3 decreased enrichment (A panel, Alg11 gene), while decreasing RNA levels were identified following decreased histone H3K4me3 enrichment (B panel, Aire gene). Composite heatmaps plotting the loci (rows) with TSS that exhibit continuously (cont.) increasing (brown) or decreasing (blue) levels of histone H3K27me3 enrichment (C and E) or histone H3K4me3 enrichment (D and F) in the CAG knock-in HdhQ20/7, HdhQ50/7, HdhQ91/7, HdhQ111/7 (CAG 18/+, 48/+, 89/+, 109/+) ESC (C and D) and NPC lines (E and F), displayed by ranking of their library-size normalized ChIP enrichment relative to that of the CAG 18/+ ESC line (enrichment score). The respective chromatin status of each TSS by histone code classification as ‘active’, ‘bivalent’ or ‘repressed’ and the respective enrichment of histone H3K36me3 and RNA-seq expression levels are plotted in the adjacent sets of columns. The loci chosen exhibit at least >1 RPKM in at least one Htt CAG knock-in sample. RNA-seq data are reported as relative log2(RPKM+1) ratio of over CAG 18/+ ESC line and standardized by dividing these values over standard deviation across the knock-in samples. The major GO terms highlighted by pathway analyses for the subsets of loci with the TSS histone mark enrichment that increases (Category 1) or decreases (Category 2) with Htt CAG size are given (further details on the categories are provided in the Results). Supplementary Material, Figure S4 reports ChIP-qPCR confirmation of progressive TSS enrichment with Htt CAG size.
From the HD criteria-conforming histone marks that are located at the TSS of genes for which RNA is reliably detected (>1 RPKM), we generated ESC and NPC chromatin maps displayed as HEAT maps in Figure 5C–F (Supplementary Material, Table S3), which reveal apparently pleiotropic effects of mutant huntingtin. In pluripotent stem cells, most genes with progressively enriched histone H3K27me3 (increases and decreases) are in the ‘bivalent’ class (Fig. 5C) (Supplementary Material, Table S3). Notably, regardless of their TSS histone H3K27me3 level, the majority of all of the conforming ‘bivalent’ genes are not reliably expressed (<1 RPKM), as found for ‘bivalent’ genes in wild-type ESC, and, therefore, did not meet the criteria of this analysis. In contrast, most genes with progressive histone H3K4me3 TSS levels (increases and decreases) are in the ‘active’ category and are expressed (>1 RPKM) (Supplementary Material, Table S3) (Fig. 5D). Interestingly, genes with increasing histone H3K4me3 enrichment exhibit concomitantly increasing RNA levels (Pearson's R ≥ 0.5), indicating an effect of mutant huntingtin on gene regulation in pluripotent cells.
At the NPC stage, most of the TSS with altered histone H3K27me3 levels are those that remained ‘bivalent’ in HdhQ20/7 NPC but with increasing Htt CAG-size tend to exhibit decreasing enrichment and, thereby, tend to be classified as ‘active’ in the NPC lines with longer repeats (HdhQ91/7, HdhQ111/7) (Fig. 5E and Supplementary Material, Table S3). However, the predominant progressive changes are in histone H3K4me3 enrichment where the bulk of the conforming TSS exhibit decreasing levels of this mark at ‘active’ chromatin genes, with concomitantly decreased RNA expression (Pearson's correlation coefficient R ≤ −0.5) that discloses an effect of mutant huntingtin on gene regulation in mutant huntingtin expressing neuronal lineage cells (Fig. 5F).
Chromatin changes due to mutant huntingtin gain of function define distinct processes
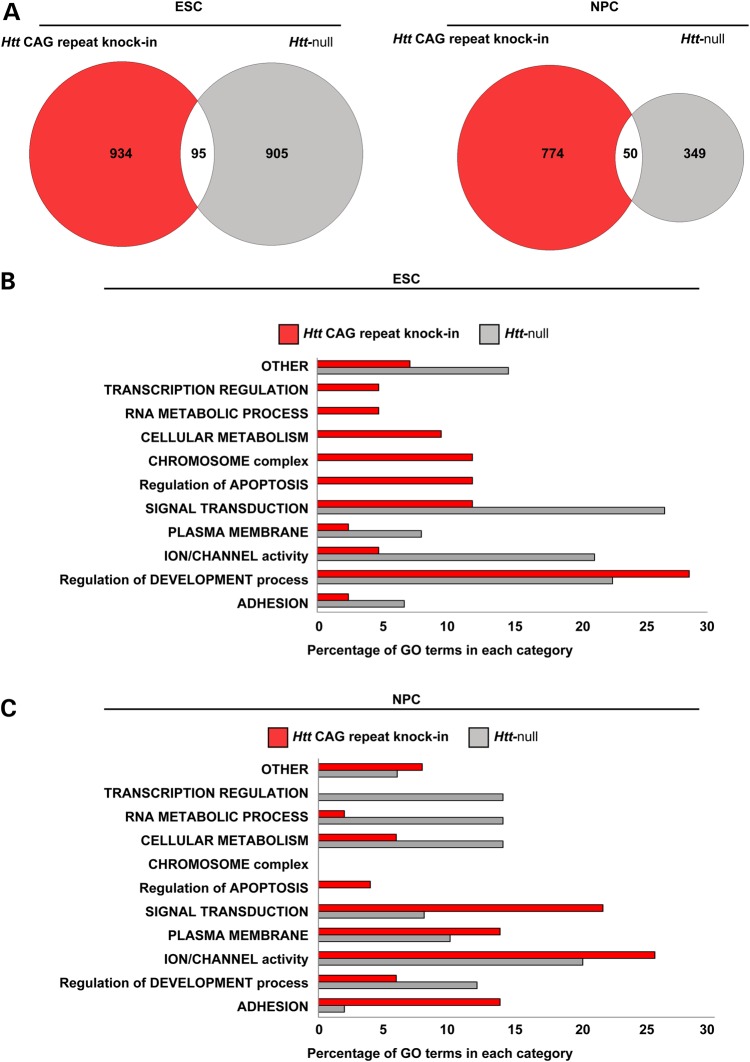
The polyglutamine expansion mutation is expected to endow mutant huntingtin with a gain of function that is related to its existing normal huntingtin function. Therefore, although the dominant Htt CAG mutation is expected to target different genes than the recessive Htt null inactivating mutation, the gain of function should be reflected in the uniquely altered chromatin states of ESC and NPC that express the mutant protein when compared with the chromatin states conferred by the lack of huntingtin normal function. Comparison of the Htt CAG-delineated ESC and NPC gene sets (TSS marks fulfilling the HD genetic criteria of showing a trend across CAG repeat length) (Supplementary Material, Table S4) with the Htt null-defined ESC and NPC gene sets (TSS marks differing from wild-type) (Supplementary Material, Table S4) reveals that the overwhelming majority of loci (∼90%) are unique to one genetic paradigm (Fig. 6A), commensurate with the Htt CAG repeat conferring a gain of new function rather than a loss of huntingtin normal function. Pathways enrichment analyses were then utilized to determine if the altered chromatin states delineated by sets of Htt CAG expansion- and Htt null-targeted loci might highlight cellular processes that are similar or dissimilar, such that the latter may denote mutant huntingtin gains of function.
Figure 6.
Comparison of the biological states forecast by the chromatin landscapes of Htt CAG knock-in and Htt null ESC and NPC. (A) Venn diagrams showing the modest intersection of the two genome-wide gene sets that are delineated either by histone H3K27me3 and histone H3K4me3 enrichment that conforms to HD genetic criteria in Htt CAG knock-in ESC and NPC lines (red) or, alternatively, by enrichment levels that are changed in Htt null ESC and NPC lines compared with their wild-type counterparts (gray). (B) Bar graph summarizing the results of DAVID pathways analysis for the Htt CAG- and Htt null-delineated ESC gene sets (red and gray, respectively), such that the proportion of significant Biological Process, Molecular Functions, Cellular Component and KEGG terms in any given general category is plotted as percentage of the total for each dataset. (C) Bar graph is same as in B, but for NPC samples.
Huntingtin normal function was evaluated by separately analyzing each of the main subsets of ‘bivalent’ loci for the Htt null ESC (Categories 1–2 in Fig. 4D) and NPC (Categories 1–4 in Fig. 4F) as listed in Supplementary Material, Table S4 as well. As expected, those not changed by the absence of huntingtin (ESC Category 2; NPC Categories 1 and 4 in Fig. 4D and F) are over-represented in processes that denote ‘embryonic development’ and ‘neuronal cell morphogenesis'. This finding supports the concept that genes in the ‘bivalent’ chromatin class anticipate development in the pluripotent state and are involved in the commitment to the neuronal cell lineage fate in NPC (34), thereby confirming the normal developmental potential of Htt null cells. However, this potential is subtly altered. The subsets of ‘bivalent’ genes that are improperly marked in Htt null pluripotent and neuronal fated cells (ESC Category 1; NPC Categories 2 and 3 in Fig. 4D and F) indicate deviations from normalcy in ‘ion transport’, ‘cell adhesion’ and ‘regulation of RNA/transcription’ or in ‘forebrain and inner ear morphogenesis', respectively.
Mutant huntingtin function was assessed by analyzing each of the main subsets of Htt CAG-conforming ESC and NPC loci (Supplementary Material, Table S4), as indicated beside the heatmaps for each histone mark in Figure 5C–F. At the pluripotent stage, the gene sets highlight processes that imply altered development (‘regulation of neurogenesis' and ‘embryonic development and metabolism’) or altered cellular homeostasis (‘regulation of transcription’, ‘stress signaling’, ‘cell cycle’, ‘apoptosis'). Similarly, at the NPC stage, the conforming gene sets are enriched in processes that imply altered signaling (‘transcription’, ‘membrane vesicle and junctions'), with processes that indicate ‘DNA metabolism’ and ‘apoptosis', thereby denoting anticipated stress and cell death.
These observations strongly imply that mutant huntingtin or the lack of huntingtin may differentially alter the status of pluripotent and neuronal progenitor cells. However, the cell states highlighted by selected subsets of genes may be misleading because unselected TSS changes do not contribute to the pathways analysis and genes from each genetic paradigm may be differentially enriched in multiple processes within the same general category. Therefore, we broadened the pathways analyses to compare the cell states forecast by all ESC and NPC loci whose TSS histone marks conformed to the HD genetic criteria (Supplementary Material, Table S4) and for all ESC and NPC loci whose TSS chromatin states are altered in the absence of huntingtin (Supplementary Material, Table S4). Then, we grouped the resulting significant cellular processes, molecular functions, cellular components and KEGG pathways (Supplementary Material, Table S4) into general categories, such that the relative proportions of the processes in each category reveal the evident similarities and differences between the genetic paradigms (Fig. 6B). Overall, at both the pluripotent and lineage restricted developmental stages, the presence of mutant huntingtin and the lack of huntingtin produce cell states that are to a similar extent enriched for many different processes that are under the general categories that denote embryonic and neuronal development (‘adhesion’, ‘membrane’, ‘ion channel’, ‘signal transduction’). However, the chromatin states of mutant huntingtin pluripotent stem cells and neuronal progenitor cells uniquely highlight categories with processes that denote cell stress and cell death (‘chromosome’, ‘cell cycle’, ‘apoptosis'). Therefore, mutant huntingtin does not replicate the loss of huntingtin function but instead forecasts uniquely altered developmental potential of progenitor cells that predicts reduced fitness and an enhanced readiness for cell death.
Discussion
The key genetic parameters of the mechanism by which the expanded HTT CAG repeat triggers the HD disease process (true dominance, progressivity with CAG size) and a unique specificity that is provided by the gene product itself, point to a gain of mutant huntingtin function. Employing a comparative strategy that utilizes these HD genetic criteria, we evaluated an initial hypothesis suggested by previous studies: that expanding mutant huntingtin's polyglutamine tract may simply enhance its ability to stimulate PRC2, thereby altering the chromatin landscapes of pluripotent and lineage restricted progenitor cells in a predictable manner that provides insights into the distinct fates of neurons that express mutant huntingtin or lack huntingtin function. However, while huntingtin's role as PRC2-facilitator is evident under some circumstances, the simple hypothesis does not fully explain all of the chromatin states produced by Htt inactivation. Nor does it readily account for the multiple distinct chromatin states that are associated with mutant huntingtin in Htt CAG knock-in cells, which uniquely forecast reduced viability at the earliest stages of development.
The view of huntingtin as a PRC2-facilitator is supported by two findings in cells that lack huntingtin: dramatically decreased H3K27me3 at TSS of developmentally poised ‘bivalent’ loci in Htt null ESC and the failure of some TSS appropriately marked in Htt null ESC to remain properly ‘bivalent’ at the NPC stage, which implies that huntingtin is needed to efficiently maintain the trimethyl group deposited by PRC2. The effect of huntingtin on facilitating the deposition of H3K27me3 could be achieved through the physical interaction between full-length huntingtin and the canonical members of PRC2 complex (14) such as Ezh2 or Suz12, however, an effect on Ezh1, the other H3 methyltransferase with a partially redundant trimethylation activity on lysine 27 (37,38), cannot be excluded and this point warrants additional investigations. Conversely, huntingtin must also serve to facilitate the removal of this mark, perhaps by a histone H3K27me3 demethylase such as Utx (39,40) because during the transition of ESC to NPC the histone H3K27me3 mark at ‘bivalent’ loci is inappropriately retained in the absence of huntingtin. In order to better understand how huntingtin interplays with PRC2 activity during neuronal development, additional investigation to reveal potential variations in PRC2 subunit occupancy in the absence of huntingtin will be useful. Moreover, a close comparison between H3K27me3 signature in PRC2 knock-down or conditional knock-out model systems compared with huntingtin null will provide additional hints to dissect the PRC2-huntingtin functional interaction.
Furthermore, since huntingtin normal function provides the platform for mutant huntingtin's novel ‘gains', a refined hypothesis that invokes simple increases in the two aforementioned functions would readily explain only two of the multiple outcomes of mutant huntingtin that satisfy the HD genetic criteria: the increases in histone H3K27me3 at a subset of ‘bivalent’ TSS in Htt CAG knock-in ESC lines and the decreases in histone H3K27me3 at a subset of ‘bivalent’ loci at the NPC stage. A fundamentally updated hypothesis of huntingtin function that fully accounts for the pleiotropy conferred by extending its polyglutamine tract in mutant huntingtin should also explain the other outcomes that we detected: (1) decreases in histone H3K27me3 observed at ‘bivalent’ TSS in the Htt CAG knock-in ESC lines, (2) increases and decreases in the histone H3K4me3 mark, catalyzed by Mll methyltransferase and removed by Jarid1/Kdm5 family demethylases (39,40), observed at ‘active’ loci in the Htt CAG knock-in ESC lines and (3) decreases in histone H3K4me3 levels that are found at ‘active’ loci in the Htt CAG knock-in the NPC lines.
Though huntingtin may separately modulate multiple discrete chromatin complexes, a more parsimonious hypothesis is that huntingtin may fine-tune PRC2 in some long-range manner when the complex is embedded within the larger machinery that coordinates both the balance of ‘activation’ and ‘repressive’ histone marks and the other layers of regulatory factors required for developmentally appropriate gene silencing and activation. There is growing precedent for large factories and for long-range regulation. For example, super-complexes containing histone methyltransferases and demethylases can coordinately deposit one mark while removing the opposing mark (41). The ‘repressive complexes' PRC2 and polycomb-like proteins (Ring6a/MBLR) coexist with histone H3K4me3 demethylase (31,42) and ‘activating complexes' containing MLL2 or MLL4 histone H3K4me3 methyltransferases also contain the histone H3K27me3 demethylase UTX (39,40). In ESC, Jarid1b histone H3K4me3 demethylase is found at TSS of genes encoding developmental regulators, together with PRC2 (43). Furthermore, dramatically expanding its chromatin locations, PRC2 polycomb group proteins detected at ‘active’ chromatin in association with nascent transcripts suggest a model for PRC2 surveillance that ‘samples' TSS, such that PRC2 becomes catalytically active upon binding of a transcriptional repressor (44,45). Thus, assessing the occupancy of activating and repressing DNA binding proteins may help to characterize the effects of mutant huntingtin on gene regulation in ESC and NPC. In addition, at a higher order of structural organization, polycomb group proteins and trithorax group proteins, with insulator proteins such as CTCF, orchestrate large scale ‘repressed’ or ‘active’ chromatin loops that ensure coordinated silencing or expression, respectively, of genes on the same and different chromosomes, as befits a given cell state (46–48).
In this view, huntingtin may primarily influence some long-range interaction that fine-tunes the efficiency with which PRC2 catalyzes histone H3K27me3 to maintain this mark at ‘bivalent’ TSS in ESC. Post neural induction, this long-range function may also serve to fine-tune the catalytic efficiency of the associated histone H3K27me3 demethylase that removes the trimethyl group at ‘bivalent’ loci that are becoming transcriptionally active, while fine-tuning the efficiency with which PRC2 maintains proper histone H3K27me3 levels at other ‘bivalent’ loci that should remain ‘bivalent’ at the NPC stage. The hypothesized long-range PRC2 fine-tuning role for huntingtin would present multiple opportunities for mutant huntingtin gains of function: both simple increases in its aforementioned existing huntingtin activities and the gains of new function at TSS of ‘bivalent’ and transcriptionally ‘active’ genes. For example, the aberrant fine-tuning conferred by mutant huntingtin, which increases its ‘normal’ huntingtin activities, may also relax the constraints in ESC that keep some poised ‘bivalent’ loci from becoming excessively ‘active’ or ‘repressed’ and certain ‘active’ genes from being exuberantly transcribed. After neural induction the aberrant fine-tuning may relax the constraints that keep transcribed ‘active’ genes in NPC from becoming excessively ‘repressed’.
The distinctly altered chromatin states of the pluripotent and neuronal lineage committed progenitor cells that we find to be associated with either the presence of mutant huntingtin or the lack of normal huntingtin function in each case consistently predict distinct subtly altered developmental status' that anticipate the different fates for the differentiated mature cell types that are derived from them, as reported by Conforti et al. (28). It is now evident that the expression of mutant huntingtin confers from the earliest stages of development an altered cellular homeostasis that forecasts reduced viability. The specific biological processes and molecular functions that are highlighted by pathways enrichment analyses of the different chromatin states produced by the Htt null and Htt CAG-expansion mutations now provide a framework for investigating the molecular basis of huntingtin's role in neuronal development and the dominant impact of expanding its polyglutamine tract.
Materials and Methods
Cell culture
The isogenic panel of wild-type, Htt null Hdhex4/5/ex4/5 and heterozygous Htt CAG knock-in HdhQ20/7, HdhQ50/7 HdhQ91/7 and HdhQ111/7 mouse ES cell lines are grown as described previously (15,18,22–24). Differentiation to neuronal lineage committed progenitor cells was performed essentially as described in (49). Briefly, ES cells were maintained on feeder layers of γ-irradiated mouse embryonic fibroblasts (Global Stem Sciences) or on gelatin-coated plates (1% gelatin solution; Millipore). ES cell media contained Knock-Out D-MEM (Invitrogen), 15% FBS (Hyclone), 50 I.U/ml of penicillin, 50 mg/ml of streptomycin (Invitrogen), 0.2 mm GlutaMax (Invitrogen), 0.1 mm non-essential aminoacids (Invitrogen), 0.1 mm 2-mercaptoethanol (Sigma) and 1000 U/ml of leukemia inhibitory factor (LIF) (Millipore).
For neural progenitors differentiation, ESCs were deprived of feeder cells for four passages, then 3 × (106) cells were used for formation of EBs. EBs were grown in non-adherent bacterial dishes (Greiner, Germany) for 8 days. Retinoic acid (SIGMA, 5 uM) was added from day 4 to day 8 and medium was changed every other day. Subsequently, EBs were dissociated by trypsin digestion and plated on Poly-ornithine (SIGMA) and laminin (SIGMA) coated plates. Two hours after plating, neural committed cells were collected for different analyses.
RNA isolation, reverse transcription and quantitative PCR
RNA was extracted from cell lines by using TRIzol reagent (Invitrogen) and following manufacturer's instructions. All RNA samples were subjected to DNAse I treatment (Ambion). One microgram of RNA was subjected to retro-transcription using iScript cDNA Synthesis Kit (Bio-Rad) accordingly to the vendor's instructions. Quantitative RT-PCR was performed by using SYBER-Green PCR Master Mix and 480 Light Cycler Detection System (Roche). TATA-binding protein (Tbp) and glyceraldehyde 3-phosphate dehydrogenase (Gapdh) were used as housekeeping genes through different samples and conditions. Primers were chosen by using the Universal ProbeLibrary Assay Design Center from Roche Applied Science. Primer sequences used in this work are available upon request. The amplified transcripts were quantified using 2−(▵▵Ct) method and the differences in gene expression were presented as normalized fold expression. All the results are presented as average ± standard deviation of two biological and two technical duplicates.
Imaging and immunocytofluorescence
For immunofluorescence experiments cells were fixed in 4% paraformaldehyde (Tousimis Research). Cell were then treated with 0.1 m glycine in PBS and permeabilized with 0.1% Triton X-100. After washing and blocking with 0.2% BSA, 1% normal goat serum, and 0.1% Triton X-100 in PBS (blocking solution), cells were incubated with the indicated antibodies diluted in blocking solution. Cells were probed with fluorescent secondary antibodies (Molecular Probes). For nuclear staining, 1 g/mL 4’,6-diamidino-2-phenylindole (DAPI) staining was used. Cells were washed and mounted with Vectashield mounting medium (Vector Laboratories).
All images were collected using an epifluorescence microscope (Zeiss). The following antibodies were used for this study: anti-Pax6 (1:500) [monoclonal antibody, developed by Dr Atsushi Kawakami (Yokohama, Japan) was obtained from the Developmental Studies Hybridoma Bank, created by the NICHD of the NIH and maintained at The University of Iowa, Department of Biology, Iowa City, IA 52 242], anti-Oct4 (1:500; Millipore), anti-Nestin (1:1000; Millipore).
Alkaline phosphatase staining was performed using ‘Alkaline Phosphatase Detection Kit’ from Millipore Corporation accordingly to the vendor's instructions.
Protein extraction and immunoblot analysis
Protein extracts were prepared from PBS-washed cell pellets by RIPA (Boston Bio-Products) lysis buffer and protease inhibitor mixture (Roche). The lysates were then sonicated with A Bioruptor sonicator (Diagenode), cleared by centrifugation at 14 000g for 30 min and the supernatants collected. The protein concentration was determined using the Bio-Rad (detergent compatible) protein assay. Fifty micrograms (hundred for Htt) of protein extract were mixed with SDS-loading buffer, boiled for 5 min and subjected to 6 or 10% SDS–PAGE. After electrophoresis, the proteins were transferred to nitrocellulose membranes (Schleicher and Schuell) and incubated for 30 min in a blocking solution containing 5% non-fat powdered milk in TBS-T (50 mm Tris–HCl, 150 mm NaCl, pH 7.4, 0.1% Tween-20). The membranes were probed overnight at 4°C with the primary antibody Htt-mAb2166 (Millipore, 1:500), Ezh2 (Millipore, 1:2000), Suz12 (Abcam, 1:1000) and β-actin (SIGMA, 1:5000). After TBS-T washes, the blots were incubated for 1 h at room temperature with horseradish peroxidase-conjugated secondary antibodies. The membranes were then processed using an ECL chemiluminescence substrate kit (Pierce) and exposed to autoradiographic film (Hyperfilm ECL; Amersham Bioscience).
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) was performed using the protocol described by (50) with minor modifications. Briefly, ∼40–60 million cells were fixed with 1% formaldehyde, washed with ice-cold PBS, harvested, pelleted and directly resuspended in SDS lysis buffer (50 mm Tris–HCl [pH8.1], 1% SDS, 10 mm EDTA) or stored at −80C for future use. Sonication of the samples was accomplished by using a Bioruptor sonicator (Diagenode), then, shared-chromatin was centrifuged at 13 000g for 10 min and diluted 10-fold in ChIP dilution buffer (16.7 mm Tris–HCl [pH 8.1], 167 mm NaCl, 0.01% SDS, 1.1% Triton X-100, 1.2 mm EDTA). After removing A control aliquot (INPUT), each sample was incubated at 4°C overnight with antibodies of interest: histone H3K27me3 (ABE44 or 07-449, Millipore), histone H3K4me3 (07-473, Millipore) and histone H3K36me3 (Ab9050, Abcam). Chromatin–Antibody complexes were precipitated with Dynabeads Protein A beads (Invitrogen) and washed sequentially with low-salt (20 mm Tris–HCl [pH 8.1], 150 mm NaCl, 0.1% SDS, 1% Triton X-100, 2 mm EDTA), high-salt (20 mm Tris–HCl [pH 8.1], 500 mm NaCl, 0.1%SDS, 1% Triton X-100, 2 mm EDTA), LiCl (10 mm Tris–HCl [pH 8.1], 0.25 M LiCl, 1% NP40, 1% sodium deoxycholate,1 mm EDTA), and TE wash buffers (10 mm Tris–HCl [pH 8.0], 1 mm EDTA). Immunoprecipitated chromatin was eluted in elution buffer (TE plus 1% SDS, 150 mm NaCl, 5 mm DTT), de-crosslinked at 65°C for 8 h (or overnight), and treated with proteinase K. The DNA was treated with RNase and purified with A MinElute Kit (Qiagen). Quantification of ChIP and INPUT DNA was done using Qubit 2.0 Fluorometer system (Invitrogen). ChIP enrichments were assessed by quantitative PCR analysis on 480 Light Cycler Detection System (Roche) with 200–400 pg of ChIP DNA and an equal amount of un-enriched INPUT DNA. Enrichments were calculated from 2 biological replicates. A gene desert region (Chr.6: 120 258 500-120 259 000) (GD) and the β-actin (Actb) promoter, commercially available from Active Motif, were used as negative controls. Error bars in the graphs represent standard error of the mean.
Helicos library preparation and single-molecule sequencing
Nanogram quantities of DNA (cDNA) were sequenced on an Helicoscope single molecule sequencer (Helicos Bioscience, Cambridge, MA, USA) obtaining an average of 8 million successful reads, consisting of 30–35 bases each (Supplementary Material, Table S1). Poly-A tailing and Helicos sequencing of ChIP fragments (300–500 bp) was performed as described in (51).
ChIP-seq data processing
The sequenced reads were filtered based on quality scores and aligned to the Mus musculus genome (mm9 assembly) using the basic processing pipeline available in the Helisphere tools suite by Helicos (http://sourceforge.net/projects/openhelisphere/). Default parameters were used, except for ‘globalAmbig = none’ to retain only uniquely aligned reads.
Read density tracks for visual inspection of data were generated using Gaussian smoothing of the 5′-end mapping positions, normalized by the number of uniquely mapped reads (millions). A 200 bp bandwidth was used for smoothing to browse data and 5′-end mapping positions were shifted by the average sequencing fragment size estimated from using cross correlation analysis of histone H3K4me3 samples for each cell type, as implemented in the SPP R package (52).
We detected ChIP-enriched regions by comparing scaled ChIP and input control read counts to see if their ratio exceeded that expected from a Poisson process, using get.broad.enrichment.cluster function in the SPP R package with a sliding window of 1 kb (default). The clusters of significant windows with Z-score > 3 (default) [for Hdh KI ChIP-seq analysis, see Figure 5 and Supplementary Material, Table S3] or >4 (more stringent) [for WT versus Htt null ChIP-seq comparison, see Figure 4A, C–F and Supplementary Material, Table S2], were determined as enriched regions. Raw sequencing data are available in the SRA database (http://www.ncbi.nlm.nih.gov/sra/) with accession number PRJNA252361. A summary of samples and aligned reads is presented in Supplementary Material, Table S1.
Public ChIP-seq data
Publicly available and previously published ChIP-seq datasets obtained with Helicos sequencing technology were processed as above described. Public ChIP-seq datasets obtained with Illumina sequencing instead were aligned using Bowtie (53), to mm9 reference genome. The parameters of alignments were -n 2 -l 28 -E 70 -m 1 for unique mapping. The subsequent analyses steps were analogous to those used for Helicos ChIP-seq data.
Hdh KI ChIP-seq data analysis
To enable a discovery strategy based on the continuous relationship between CAG size and phenotype, we utilized a correlation approach to distinguish loci that may specifically be affected by CAG mutation. This approach also did not rely on classification of loci by combination of histone mark, but evaluated histone H3K4me3 and histone H3K27me3 separately, as we could not anticipate whether or not a specific category would be affected. We quantified the enrichment values for ChIP-seq datasets of the CAG knock-in genotypes in ESC and NPC, evaluating the total reads number in a region of ±2 kb centered on TSS. The enrichment values for each promoter region in a specific dataset were then selected to present monotonic increases or decreases with CAG size thereby assessing quantitative differences in ChIP enrichment with CAG expansion. In order to discriminate between real ChIP enrichment and fluctuation of the background, we additionally filtered ChIP enrichment for being higher than 60 normalized reads in the analyzed window (±2 kb) in Q20, the lowest CAG-expanded allele (for monotonically decreasing promoters) or Q111, the highest CAG-expanded allele (for monotonically increasing promoters). The ChIP enrichment values obtained using these parameters were further sorted based on the degree of variance among the different CAG expansions in order to rank the changes from the top to the least most affected by CAG size.
RNA-seq data processing
RNA-sequence (RNA-seq) data were obtained with strand-specific RNA-seq protocol for Helicos sequencing technology. Sequencing reads were quality filtered and aligned to reference RefSeq transcripts sequences for the mm9 genome (RefSeq build date 29 June 2012) using the DGE analysis pipeline of Helicos software (Helisphere), which uses the RMC algorithm to assign read counts to transcripts (54). Default parameters were used, except for Global.rpkm = True, to obtain read counts normalized by gene length (RPKM).
Strand-specific read density tracks for visual inspection of data were generated using Gaussian smoothing of the 5′-end mapping positions, normalized by the number of uniquely mapped reads (millions) using the SPP R package. A 200 bp bandwidth was used for smoothing and 5′-end mapping positions were shifted by half (≈15 bp) of the average read length in each sample.
Raw sequencing data are available in the SRA database (http://www.ncbi.nlm.nih.gov/sra/) with accession number PRJNA252362. A summary of samples and aligned reads is presented in Supplementary Material, Table S1.
Gene annotations
RefSeq annotations tables were retrieved from UCSC Genome Browser (mm9 genome version—build date 29 June 2012). RefSeq transcript annotations were downloaded from the UCSC genome browser. For each RefSeq transcript a window spanning 2 kb upstream and 2 kb downstream the annotated TSS was considered. This window was truncated downstream of the TSS if the RefSeq transcript was <2 kb long, and was also truncated upstream of the TSS if another RefSeq transcript ending on the same genomic strand was located <2 kb upstream. Each RefSeq TSS was annotated as associated with a chromatin mark (histone H3K27me3 or histone H3K4me3) if any overlap existed between the ChIP-seq peak the TSS window. Instead, histone H3K36me3 enrichment regions were compared for overlap with gene body regions (between annotated 5′ and 3′ ends). Depending on the analysis, these overlaps were also summarized at the gene level, as indicated in specific results.
In the Venn diagrams (Supplementary Material, Fig. S2A and B), the lists of chromatin mark-associated genes/TSSs were compared across different ChIP-seq targets. Venn diagrams with proportional overlapping areas were plotted using the Vennerable R package.
Metagene profiles
To obtain an estimation of binding distribution near genes we plotted metagene profiles (Fig. 4B and Supplementary Material, Fig. S1B). These plots show average ChIP-seq enrichment signal over a large number of genes. For each RefSeq transcript a window of −2 kb upstream from the annotated 5′-end and 2 kb downstream from the 3′-end was considered (or −5 kb/+2 kb for Supplementary Material, Fig. S2B). RefSeq transcripts overlapping each other in the range of such windows were discarded to avoid interference in ChIP-seq signal distribution. Only genes 2.5 kb or longer were used. Scaling of genes was achieved by splitting smoothed profiles into 200 bins for each gene and flanking regions, then computing the average for each bin in individual genes. Read density was then log2 transformed, after adding a pseudocount of 1 to each bin to avoid log transformation of zero values. The value of the Nth bin in the metagene profile is then the average of the log2 read density in the Nth bins across the gene set. The regions outside the gene body are given in real scale, as well as the first and last 500 bp in the gene body.
Accession numbers
The sequencing data were deposited in the Sequence Read Archive under accession numbers PRJNA252361 and PRJNA252362 for ChIP-seq and RNA-seq data, respectively.
Pathway analysis
To gain insight into the functional roles of genes associated with a particular chromatin mark in ESC or NPC, we relied on DAVID (55). In identifying enriched functional terms, we selected those with nominal P-value < 0.05. DAVID was used with Mus musculus background. Enriched terms included biological processes (BP), molecular functions (MF), cellular component (CC) and KEGGS pathways. A table summarizing pathways analysis (BP only) results for single categories (related to Figs 4D, F and 5C–F) and for general analysis (BP, MF, CC and KEGGS) (related to Fig. 6B) is presented in Supplementary Material, Table S4. To assess commonality between significant pathways in Htt null and CAG knock-in cells, the top 25 knock-out significant terms (top 25 BP, top 25 MF, top 25 CC and top 25 KEGGS pathways) and the top 25 terms in the CAG knock-in data were compared. Broad categories of main cellular processes grouping significant terms from BP, MF, CC and KEGGS pathways were hand-annotated and. The percentage of significant terms was plotted for Htt null and Htt CAG knock-in genotypes (23).
Supplementary Material
Funding
This work was supported by CHDI Foundation Inc.; and the Huntington's Society of Canada; and National Institutes of Health/National Institute of Neurological Disorders and Stroke (R01 NS32765 to M.E.M., R01 NS079651 to I.S.S.).
Supplementary Material
Acknowledgements
We are grateful to the MacDonald, Gusella, Seong, Wheeler and Talkowski laboratory members for helpful discussions. We thank Eric Vallabh Minikel for helping with statistical analyses, Jayalakshmi Mysore and Tammy Gillis for technical support. Pax6 monoclonal antibody, developed by Dr Atsushi Kawakami (Yokohama, Japan) was obtained from the Developmental Studies Hybridoma Bank, created by the NICHD of the NIH and maintained at The University of Iowa, Department of Biology, Iowa City, IA 52242.
Conflict of Interest statement. F.O. was an employee of Helicos BioSciences.
References
- 1.Rosas H.D., Koroshetz W.J., Chen Y.I., Skeuse C., Vangel M., Cudkowicz M.E., Caplan K., Marek K., Seidman L.J., Makris N., et al. (2003) Evidence for more widespread cerebral pathology in early HD: an MRI-based morphometric analysis. Neurology, 60, 1615–1620. [DOI] [PubMed] [Google Scholar]
- 2.Vonsattel J.P., Myers R.H., Stevens T.J., Ferrante R.J., Bird E.D., Richardson E.P., Jr. (1985) Neuropathological classification of Huntington’s disease. J. Neuropathol. Exp. Neurol., 44, 559–577. [DOI] [PubMed] [Google Scholar]
- 3.1993) A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. The Huntington’s Disease Collaborative Research Group. Cell, 72, 971–983. [DOI] [PubMed] [Google Scholar]
- 4.Lee J.M., Ramos E.M., Lee J.H., Gillis T., Mysore J.S., Hayden M.R., Warby S.C., Morrison P., Nance M., Ross C.A., et al. (2012) CAG repeat expansion in Huntington disease determines age at onset in a fully dominant fashion. Neurology, 78, 690–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.An M.C., Zhang N., Scott G., Montoro D., Wittkop T., Mooney S., Melov S., Ellerby L.M. (2012) Genetic correction of Huntington’s disease phenotypes in induced pluripotent stem cells. Cell Stem Cell, 11, 253–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feyeux M., Bourgois-Rocha F., Redfern A., Giles P., Lefort N., Aubert S., Bonnefond C., Bugi A., Ruiz M., Deglon N., et al. (2012) Early transcriptional changes linked to naturally occurring Huntington’s disease mutations in neural derivatives of human embryonic stem cells. Hum. Mol. Genet., 21, 3883–3895. [DOI] [PubMed] [Google Scholar]
- 7.Niclis J., Trounson A.O., Dottori M., Ellisdon A., Bottomley S.P., Verlinsky Y., Cram D. (2009) Human embryonic stem cell models of Huntington disease. Reprod. Biomed. Online, 19, 106–113. [DOI] [PubMed] [Google Scholar]
- 8.Consortium H.D.i. (2012) Induced pluripotent stem cells from patients with Huntington’s disease show CAG-repeat-expansion-associated phenotypes. Cell Stem Cell, 11, 264–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gusella J.F., MacDonald M.E. (2006) Huntington's disease: seeing the pathogenic process through a genetic lens. Trends Biochem. Sci., 31, 533–540. [DOI] [PubMed] [Google Scholar]
- 10.Gusella J., MacDonald M. (2002) No post-genetics era in human disease research. Nat. Rev. Genet., 3, 72–79. [DOI] [PubMed] [Google Scholar]
- 11.Orr H.T., Zoghbi H.Y. (2007) Trinucleotide repeat disorders. Annu. Rev. Neurosci., 30, 575–621. [DOI] [PubMed] [Google Scholar]
- 12.Andrade M.A., Bork P. (1995) HEAT repeats in the Huntington’s disease protein. Nat. Genet., 11, 115–116. [DOI] [PubMed] [Google Scholar]
- 13.MacDonald M.E. (2003) Huntingtin: alive and well and working in middle management. Sci. STKE, 2003, pe48. [DOI] [PubMed] [Google Scholar]
- 14.Seong I.S., Woda J.M., Song J.J., Lloret A., Abeyrathne P.D., Woo C.J., Gregory G., Lee J.M., Wheeler V.C., Walz T., et al. (2010) Huntingtin facilitates polycomb repressive complex 2. Hum. Mol. Genet., 19, 573–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duyao M.P., Auerbach A.B., Ryan A., Persichetti F., Barnes G.T., McNeil S.M., Ge P., Vonsattel J.P., Gusella J.F., Joyner A.L., et al. (1995) Inactivation of the mouse Huntington's disease gene homolog Hdh. Science, 269, 407–410. [DOI] [PubMed] [Google Scholar]
- 16.Nasir J., Floresco S.B., O'Kusky J.R., Diewert V.M., Richman J.M., Zeisler J., Borowski A., Marth J.D., Phillips A.G., Hayden M.R. (1995) Targeted disruption of the Huntington’s disease gene results in embryonic lethality and behavioral and morphological changes in heterozygotes. Cell, 81, 811–823. [DOI] [PubMed] [Google Scholar]
- 17.Zeitlin S., Liu J.P., Chapman D.L., Papaioannou V.E., Efstratiadis A. (1995) Increased apoptosis and early embryonic lethality in mice nullizygous for the Huntington's disease gene homologue. Nat. Genet., 11, 155–163. [DOI] [PubMed] [Google Scholar]
- 18.White J.K., Auerbach W., Duyao M.P., Vonsattel J.P., Gusella J.F., Joyner A.L., MacDonald M.E. (1997) Huntingtin is required for neurogenesis and is not impaired by the Huntington's disease CAG expansion. Nat. Genet., 17, 404–410. [DOI] [PubMed] [Google Scholar]
- 19.Lin C.H., Tallaksen-Greene S., Chien W.M., Cearley J.A., Jackson W.S., Crouse A.B., Ren S., Li X.J., Albin R.L., Detloff P.J. (2001) Neurological abnormalities in a knock-in mouse model of Huntington's disease. Hum. mol. Genet., 10, 137–144. [DOI] [PubMed] [Google Scholar]
- 20.Shelbourne P., Coote E., Dadak S., Cobb S.R. (2007) Normal electrical properties of hippocampal neurons modelling early Huntington disease pathogenesis. Brain Res., 1139, 226–234. [DOI] [PubMed] [Google Scholar]
- 21.Joshi P.R., Wu N.P., Andre V.M., Cummings D.M., Cepeda C., Joyce J.A., Carroll J.B., Leavitt B.R., Hayden M.R., Levine M.S., et al. (2009) Age-dependent alterations of corticostriatal activity in the YAC128 mouse model of Huntington disease. The Journal of Neuroscience: the Official Journal of the Society for Neuroscience, 29, 2414–2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Auerbach W., Hurlbert M.S., Hilditch-Maguire P., Wadghiri Y.Z., Wheeler V.C., Cohen S.I., Joyner A.L., MacDonald M.E., Turnbull D.H. (2001) The HD mutation causes progressive lethal neurological disease in mice expressing reduced levels of huntingtin. Hum. Mol. Genet., 10, 2515–2523. [DOI] [PubMed] [Google Scholar]
- 23.Jacobsen J.C., Gregory G.C., Woda J.M., Thompson M.N., Coser K.R., Murthy V., Kohane I.S., Gusella J.F., Seong I.S., MacDonald M.E., et al. (2011) HD CAG-correlated gene expression changes support a simple dominant gain of function. Hum. Mol. Genet., 20, 2846–2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wheeler V.C., Auerbach W., White J.K., Srinidhi J., Auerbach A., Ryan A., Duyao M.P., Vrbanac V., Weaver M., Gusella J.F., et al. (1999) Length-dependent gametic CAG repeat instability in the Huntington's disease knock-in mouse. Hum. mol. Genet., 8, 115–122. [DOI] [PubMed] [Google Scholar]
- 25.Hickey M.A., Kosmalska A., Enayati J., Cohen R., Zeitlin S., Levine M.S., Chesselet M.F. (2008) Extensive early motor and non-motor behavioral deficits are followed by striatal neuronal loss in knock-in Huntington's disease mice. Neuroscience, 157, 280–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jones C.A., Ng J., Peterson A.J., Morgan K., Simon J., Jones R.S. (1998) The Drosophila esc and E(z) proteins are direct partners in polycomb group-mediated repression. Mol. Cell. Biol., 18, 2825–2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Czermin B., Melfi R., McCabe D., Seitz V., Imhof A., Pirrotta V. (2002) Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell, 111, 185–196. [DOI] [PubMed] [Google Scholar]
- 28.Conforti P., Camnasio S., Mutti C., Valenza M., Thompson M., Fossale E., Zeitlin S., MacDonald M.E., Zuccato C., Cattaneo E. (2013) Lack of huntingtin promotes neural stem cells differentiation into glial cells while neurons expressing huntingtin with expanded polyglutamine tracts undergo cell death. Neurobiol. Dis., 50, 160–170. [DOI] [PubMed] [Google Scholar]
- 29.Nguyen G.D., Gokhan S., Molero A.E., Mehler M.F. (2013) Selective roles of normal and mutant huntingtin in neural induction and early neurogenesis. PLoS ONE, 8, e64368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pasini D., Bracken A.P., Jensen M.R., Lazzerini Denchi E., Helin K. (2004) Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. EMBO J., 23, 4061–4071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pasini D., Hansen K.H., Christensen J., Agger K., Cloos P.A., Helin K. (2008) Coordinated regulation of transcriptional repression by the RBP2 H3K4 demethylase and Polycomb-Repressive Complex 2. Genes Dev., 22, 1345–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Woda J.M., Calzonetti T., Hilditch-Maguire P., Duyao M.P., Conlon R.A., MacDonald M.E. (2005) Inactivation of the Huntington's disease gene (Hdh) impairs anterior streak formation and early patterning of the mouse embryo. BMC Dev. Biol., 5, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chambeyron S., Bickmore W.A. (2004) Chromatin decondensation and nuclear reorganization of the HoxB locus upon induction of transcription. Genes Dev., 18, 1119–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mikkelsen T.S., Ku M., Jaffe D.B., Issac B., Lieberman E., Giannoukos G., Alvarez P., Brockman W., Kim T.K., Koche R.P., et al. (2007) Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature, 448, 553–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boyer L.A., Plath K., Zeitlinger J., Brambrink T., Medeiros L.A., Lee T.I., Levine S.S., Wernig M., Tajonar A., Ray M.K., et al. (2006) Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature, 441, 349–353. [DOI] [PubMed] [Google Scholar]
- 36.Ku M., Koche R.P., Rheinbay E., Mendenhall E.M., Endoh M., Mikkelsen T.S., Presser A., Nusbaum C., Xie X., Chi A.S., et al. (2008) Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet., 4, e1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ezhkova E., Lien W.H., Stokes N., Pasolli H.A., Silva J.M., Fuchs E. (2011) EZH1 and EZH2 cogovern histone H3K27 trimethylation and are essential for hair follicle homeostasis and wound repair. Genes Dev., 25, 485–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Margueron R., Li G., Sarma K., Blais A., Zavadil J., Woodcock C.L., Dynlacht B.D., Reinberg D. (2008) Ezh1 and Ezh2 maintain repressive chromatin through different mechanisms. Mol. Cell, 32, 503–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Agger K., Cloos P.A., Christensen J., Pasini D., Rose S., Rappsilber J., Issaeva I., Canaani E., Salcini A.E., Helin K. (2007) UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature, 449, 731–734. [DOI] [PubMed] [Google Scholar]
- 40.Kim J.H., Sharma A., Dhar S.S., Lee S.H., Gu B., Chan C.H., Lin H.K., Lee M.G. (2014) UTX and MLL4 coordinately regulate transcriptional programs for cell proliferation and invasiveness in breast cancer cells. Cancer Res., 74, 1705–1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cloos P.A., Christensen J., Agger K., Helin K. (2008) Erasing the methyl mark: histone demethylases at the center of cellular differentiation and disease. Genes Dev., 22, 1115–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee M.G., Norman J., Shilatifard A., Shiekhattar R. (2007) Physical and functional association of a trimethyl H3K4 demethylase and Ring6a/MBLR, a polycomb-like protein. Cell, 128, 877–887. [DOI] [PubMed] [Google Scholar]
- 43.Schmitz S.U., Albert M., Malatesta M., Morey L., Johansen J.V., Bak M., Tommerup N., Abarrategui I., Helin K. (2011) Jarid1b targets genes regulating development and is involved in neural differentiation. EMBO J., 30, 4586–4600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Davidovich C., Zheng L., Goodrich K.J., Cech T.R. (2013) Promiscuous RNA binding by Polycomb repressive complex 2. Nat. Struct. Mol. Biol., 20, 1250–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kaneko S., Son J., Shen S.S., Reinberg D., Bonasio R. (2013) PRC2 binds active promoters and contacts nascent RNAs in embryonic stem cells. Nat. Struct. Mol. Biol., 20, 1258–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Follmer N.E., Wani A.H., Francis N.J. (2012) A polycomb group protein is retained at specific sites on chromatin in mitosis. PLoS Genet., 8, e1003135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Herold M., Bartkuhn M., Renkawitz R. (2012) CTCF: insights into insulator function during development. Development, 139, 1045–1057. [DOI] [PubMed] [Google Scholar]
- 48.Pirrotta V., Li H.B. (2012) A view of nuclear polycomb bodies. Curr. Opin. Genet. Dev., 22, 101–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bibel M., Richter J., Lacroix E., Barde Y.A. (2007) Generation of a defined and uniform population of CNS progenitors and neurons from mouse embryonic stem cells. Nat. Protoc., 2, 1034–1043. [DOI] [PubMed] [Google Scholar]
- 50.Bernstein B.E., Kamal M., Lindblad-Toh K., Bekiranov S., Bailey D.K., Huebert D.J., McMahon S., Karlsson E.K., Kulbokas E.J., 3rd, Gingeras T.R., et al. (2005) Genomic maps and comparative analysis of histone modifications in human and mouse. Cell, 120, 169–181. [DOI] [PubMed] [Google Scholar]
- 51.Goren A., Ozsolak F., Shoresh N., Ku M., Adli M., Hart C., Gymrek M., Zuk O., Regev A., Milos P.M., et al. (2010) Chromatin profiling by directly sequencing small quantities of immunoprecipitated DNA. Nat. Methods, 7, 47–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kharchenko P.V., Tolstorukov M.Y., Park P.J. (2008) Design and analysis of ChIP-seq experiments for DNA-binding proteins. Nat. Biotechnol., 26, 1351–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Langmead B., Trapnell C., Pop M., Salzberg S.L. (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol., 10, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lipson D., Raz T., Kieu A., Jones D.R., Giladi E., Thayer E., Thompson J.F., Letovsky S., Milos P., Causey M. (2009) Quantification of the yeast transcriptome by single-molecule sequencing. Nat. Biotechnol., 27, 652–658. [DOI] [PubMed] [Google Scholar]
- 55.Huang da W., Sherman B.T., Lempicki R.A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc., 4, 44–57. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.