Abstract
White matter abnormalities have been reported in premanifest Huntington's disease (HD) subjects before overt striatal neuronal loss, but whether the white matter changes represent a necessary step towards further pathology and the underlying mechanism of these changes remains unknown. Here, we characterized a novel knock-in mouse model that expresses mouse HD gene homolog (Hdh) with extended CAG repeat- HdhQ250, which was derived from the selective breeding of HdhQ150 mice. HdhQ250 mice manifest an accelerated and robust phenotype compared with its parent line. HdhQ250 mice exhibit progressive motor deficits, reduction in striatal and cortical volume, accumulation of mutant huntingtin aggregation, decreased levels of DARPP32 and BDNF and altered striatal metabolites. The abnormalities detected in this mouse model are reminiscent of several aspects of human HD. In addition, disturbed myelination was evident in postnatal Day 14 HdhQ250 mouse brain, including reduced levels of myelin regulatory factor and myelin basic protein, and decreased numbers of myelinated axons in the corpus callosum. Thinner myelin sheaths, indicated by increased G-ratio of myelin, were also detected in the corpus callosum of adult HdhQ250 mice. Moreover, proliferation of oligodendrocyte precursor cells is altered by mutant huntingtin both in vitro and in vivo. Our data indicate that this model is suitable for understanding comprehensive pathogenesis of HD in white matter and gray matter as well as developing therapeutics for HD.
Introduction
Huntington's disease (HD) is a dominantly inherited neurodegenerative disorder marked by involuntary movements, cognitive impairment and psychiatric abnormalities. HD is caused by polyglutamine-encoding expanded CAG repeats (1), and increasing polyglutamine length correlates with earlier onset of disease (2). Although HD pathology shows massive degeneration of neurons, non-neuronal cells also play critical role in the pathogenic process (3). Indeed, altered white matter morphology has been found in the corpus callosum in premanifest HD, and impaired white matter integrity appears to be an early event and related to neuronal loss and disease progression (4–11). HD postmortem brain showed increased progenitor cell proliferation (12) indicating the regenerative potential, and HD brain also exhibits increased proliferation of oligodendrocyte precursor cells (OPCs) (13), suggesting a possible homeostatic response to oligodendrocyte degeneration or a generic reactive response to mutant huntingtin protein (HTT). A recent study indicates that oligodendrocyte lineage cells are the most vulnerable population among non-neuronal cells in the human HD striatum (14). The selective degeneration of myelinated projection neurons with sparing of interneurons in the basal ganglia of HD patients further suggests that a defect of myelination may be involved in HD pathogenesis (15).
The underlying mechanisms of white matter abnormalities remain largely unexplored. Whether white matter changes represent a necessary step towards further pathology or a response to neuronal degeneration is a key question. The temporal dynamics of white matter changes become critical to answer this question. Therefore, development of genetic animal models that recapitulate salient genetic, behavioral and neuropathological features of HD is required; valuable mouse models, recapitulating genetics, pathology and behavioral features of HD, appears an important tool for us to unravel cellular logic and elucidate molecular mechanisms underlying the disease pathogenesis. In fact, many clues to the mechanism have come from the analysis of mouse lines engineered to express additional copies of partial or full-length versions of mutant huntingtin (HTT) (16,17). Determining the molecular cause of pathology in transgenic mouse models requires caution, however, since transgene location and differences between transgene expression and the endogenous gene might cause abnormalities not related to HD in human (18). Knock-in (KI) models in the appropriate genomic context are, in some respects, the most representative models. Nonetheless, most existing HD KI models have later onset of behavioral abnormalities and neuropathological features than do transgenic models (19,20). The usefulness of these KI models in studying pathogenesis and testing drug efficacy is limited by their delayed and mild phenotype.
Here, we report a KI model derived from the HdhQ150 KI line (16) which has ∼250 CAG/polyQ repeats (HdhQ250). These heterozygous mice exhibit accelerated behavioral and pathological phenotypes compared with those in the parental HdhQ150 line and other recently characterized heterozygous KI mouse models (21–23). In addition, HdhQ250 mice display abnormal myelination and sustained white matter abnormalities in the corpus callosum. The OPCs exhibit enhanced proliferation in adult HdhQ250 mouse brain, but numbers of mature oligodendrocytes are dramatically less in postnatal Day 14 (P14) HdhQ250 brain than those in age-matched littermate control mouse brain. To our knowledge, this is the first KI HD mouse model which demonstrated abnormal myelination during the critical myelin development period and sustained white matter pathology in adulthood. Furthermore, we reveal that mutant huntingtin directly alters the proliferation property of cultured OPCs. Although this mouse model demonstrates many pathological aspects of HD, it should be noted that these mice express a mouse HD gene homolog (Hdh), under the control of murine promoters and genomic regulatory elements, and hence there are subtle differences at the levels of Hdh genomic DNA and protein sequences between KI mice and HD patients. Inherent differences between humans and mice must be considered in the search for efficacious treatments for HD, nevertheless the phenotypes displayed in this mouse model suggest that HdhQ250 mouse model is valuable for preclinical HD research. Our findings provide new insight into understanding disease pathogenesis and development of therapeutics for HD.
Results
Mutant huntingtin expression, body weight and motor deficits in HdhQ250 mice
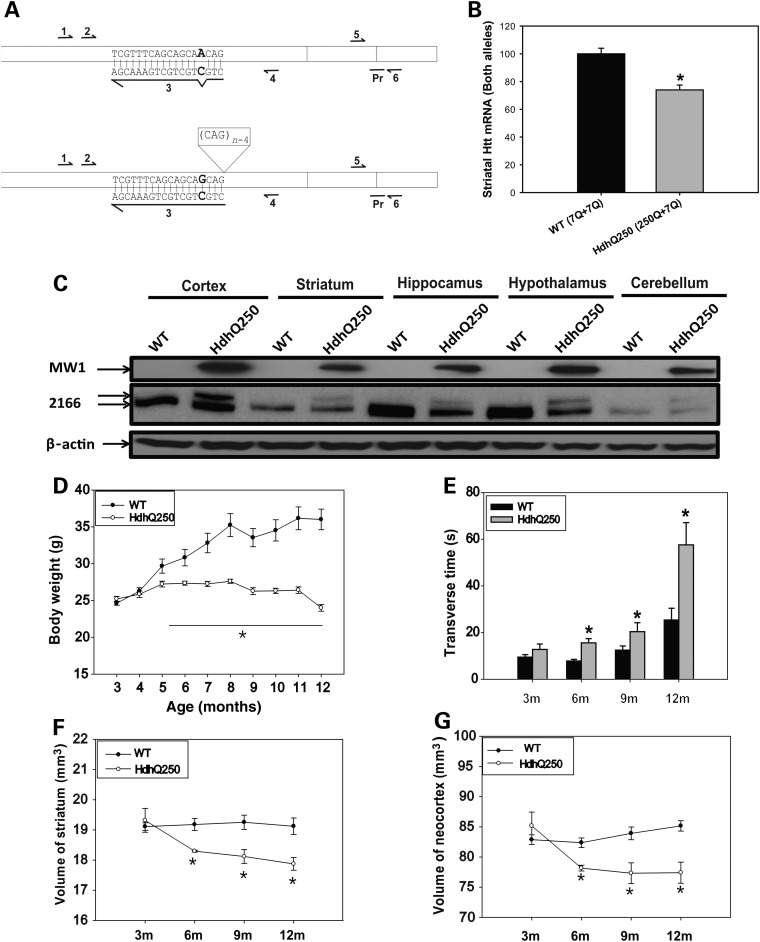
In previous reports of parent line HdhQ150 and extended HdhQ200 line, mutant huntingtin mRNA expression levels were reduced to about 61–68% of wild-type (WT) levels in the striatum (13,21). Therefore, we quantified mRNA levels of Hdh in HdhQ250 mice by allele specific huntingtin mRNA assay (see detail in Materials and Methods), our results indicates that the mRNA levels of mutant Hdh allele were significantly lower than those of WT allele (Fig. 1A and B). Western blot analysis indicated that mutant HTT was expressed in different brain regions, including cerebral cortex, striatum, hippocampus, hypothalamus and cerebellum as described in HdhQ150 and HdhQ200 line (16,21) (Fig. 1C).
Figure 1.
HdhQ250 knock-in mice exhibit robust phenotypes. (A) Primers used for allele specific qRT–PCR. Boxes represent the first 3 exons of mouse HTT mRNA for wild-type (upper) and mutant (lower). Arrows represent primer binding site regions for PCR. Allele specific PCR uses primer 4 for reverse transcription, primers 1 and 4 to generate copy number controls and primers 2 and 3 for Sybr Green based qRT–PCR. Sequence of primer 3 and its binding site are shown with bold-lettered sequence indicating site of PCR primer mismatch with wild-type cDNA. Taqman primers 5 and 6 and probe (Pr) spanning junction of exon 2 and 3 are also shown and have been described in the Material and Methods. Genotyping primers 1 and 4 were also used to generate DNA quantity standards for allele specific HTT qRT–PCR. Primer 4 alone was used for reverse transcription for allele specific qRT–PCR. (B) Striatal HTT mRNA levels of 20-week-old mice as determined qRT–PCR assays. Error bars indicate standard error of the mean and asterisks show level of statistical certainty as determined by one-way ANOVA with Tukey–Kramer Test. *P < 0.01. n = 12 WT mice and 9 HD mice. (C) Western blot of mutant huntingtin (HTT) expression in different brain regions detected by MW1 antibody which recognizes mutant HTT, and MAB2166 (2166) which recognizes both wild-type HTT (lower band) and mutant HTT (upper band). (D) Body weight was recorded from 3 through 12 months of age in male HdhQ250 mice and their littermate control wild-type (WT) mice. n = 10. *P < 0.05 compared with the values of WT mice by repeated-measures ANOVA. (E) Mice were trained and tested on the balance beam (5-mm square beam), and transverse time on the beam was recorded. HdhQ250 mice exhibited progressive motor deficits indicated by longer transverse time on the beam along with age. n = 10, *P < 0.05 compared with the values of age- and gender-matched WT mice by standard Student's t-tests. (F and G) Volumes of the striatum (F) and neocortex (G) were measured by in vivo structural MRI at indicated ages. n = 6, *P < 0.05 compared with the volume of gender-matched WT control mice by repeated-measures ANOVA.
HdhQ250 mice displayed significantly lower body weight than their WT littermates. HdhQ250 mice gained body weight normally up to 4 months, stopped gaining body weight and then lost body weight (Fig. 1D). The body weight loss was also shown in HD patients (24). This effect on body weight is not seen in either heterozygous HdhQ150 (16) or HdhQ200 line (21), a first indication that HdhQ250 mice have more robust phenotype.
To assess motor function, we evaluated the time which mice spent to cross a 5-mm square shaped balance beam, HdhQ250 mice exhibited significant motor deficit at 6 months of age, indicated by longer time spent to cross the beam and motor deficits of HdhQ250 mice progressed along with the age (Fig. 1E). We also performed other behavioral tests, including open-field activity, the accelerating rotarod test and 11-mm round balance beam (Supplementary Material, Fig. S1). Our results indicated that 5-mm balance beam test was the most sensitive measure in the HdhQ250 mice. This compares to heterozygous HdhQ150 mice which did not show significant motor deficits on the balance beam until 25 months (16). For each motor abnormality tested in HdhQ250 mice showed accelerated phenotypes compared with the HdhQ150 line.
Mutant huntingtin causes selective reduction of brain volumes and protein aggregation in HdhQ250 mice
Although mutant HTT is expressed ubiquitously, the neuropathology in HD is selective, in which robust atrophy is seen in the striatum and to some extent in the cortex, and extends to other brain regions with disease progression. In order to determine whether the HdhQ250 mouse model demonstrates selective neuropathological changes, we performed in vivo longitudinal structural MRI whole brain scans from 3- to 12-month-old HdhQ250 mice. At 3 months of age, no differences in regional volumes were detected between HdhQ250 mice and WT mice. By 6 months of age, we detected selective and significant reductions in the striatum and neocortex of HdhQ250 heterozygous mice among 17 brain regions that we analyzed (Table 1). Acceleration of brain atrophy was most robust between 3 and 6 months of age, and then amplitude of striatal and neocortical volume reduction slow down (Fig. 1F and G). Our results indicate that HdhQ250 mice have an accelerated phenotype than that in HdhQ150 and HdhQ200 lines in terms of brain volume changes. Striatal volume does not change in heterozygous HdhQ200 mice even by 80 weeks (21). These results suggest that preventive preclinical trials starting at or before 3 months of age might provide better protection than those starting at later age in these mice.
Table 1.
HdhQ250 mice display selective brain atrophy
| Regions | WT (mm3) | HdhQ250 (mm3) |
|---|---|---|
| Whole brain | 436.0 ± 4.0 | 430.2 ± 1.5 |
| Accumbens Nu | 1.76 ± 0.01 | 1.64 ± 0.06 |
| Amygdala | 6.34 ± 0.12 | 6.46 ± 0.05 |
| Anterior commissure | 1.25 ± 0.02 | 1.18 ± 0.02 |
| Striatum | 19.18 ± 0.20 | 18.30 ± 0.03* |
| Corpus collasum | 13.30 ± 0.22 | 13.01 ± 0.05 |
| Cerebellum | 50.33 ± 0.92 | 53.34 ± 0.25 |
| Fimbria | 2.11 ± 0.06 | 2.25 ± 0.14 |
| Fornix | 0.08 ± 0.00 | 0.08 ± 0.00 |
| Hippocampus | 22.54 ± 0.29 | 23.07 ± 0.41 |
| Hypothalamus | 11.73 ± 0.20 | 11.97 ± 0.11 |
| Internal capsule | 4.86 ± 0.12 | 4.83 ± 0.02 |
| Lateral globus pallidus | 1.96 ± 0.04 | 1.91 ± 0.01 |
| Neocortex | 82.37 ± 0.80 | 78.16 ± 0.48* |
| Periform cortex | 2.79 ± 0.07 | 2.72 ± 0.02 |
| Thalamus | 18.68 ± 0.28 | 18.15 ± 0.12 |
| Lateral ventricle | 9.90 ± 0.68 | 11.89 ± 0.97 |
In vivo MRI quantification data in 15 different brain regions and lateral ventricle size in 6-month-old HdhQ250 mice and age-matched WT controls. Note the striatum and neocortex regions exhibit significant reduced volume while other regions have not shown difference in 6-month-old HdhQ250 mice. Data are mean ± SEM, n = 6
*P < 0.05 compared with the volume of age-matched WT control mice by standard Student's t-test.
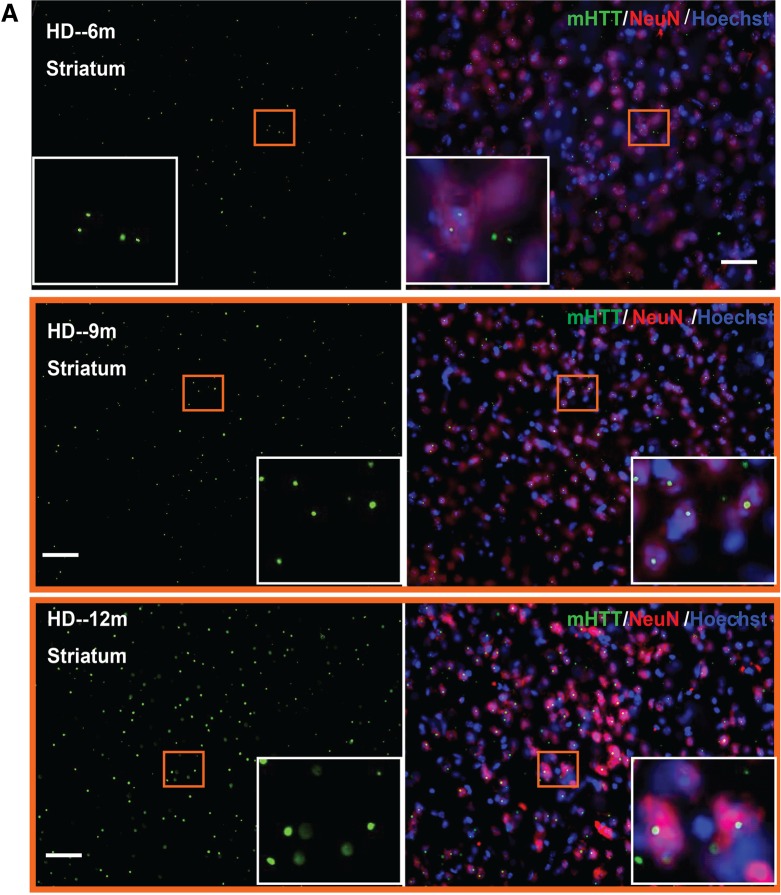
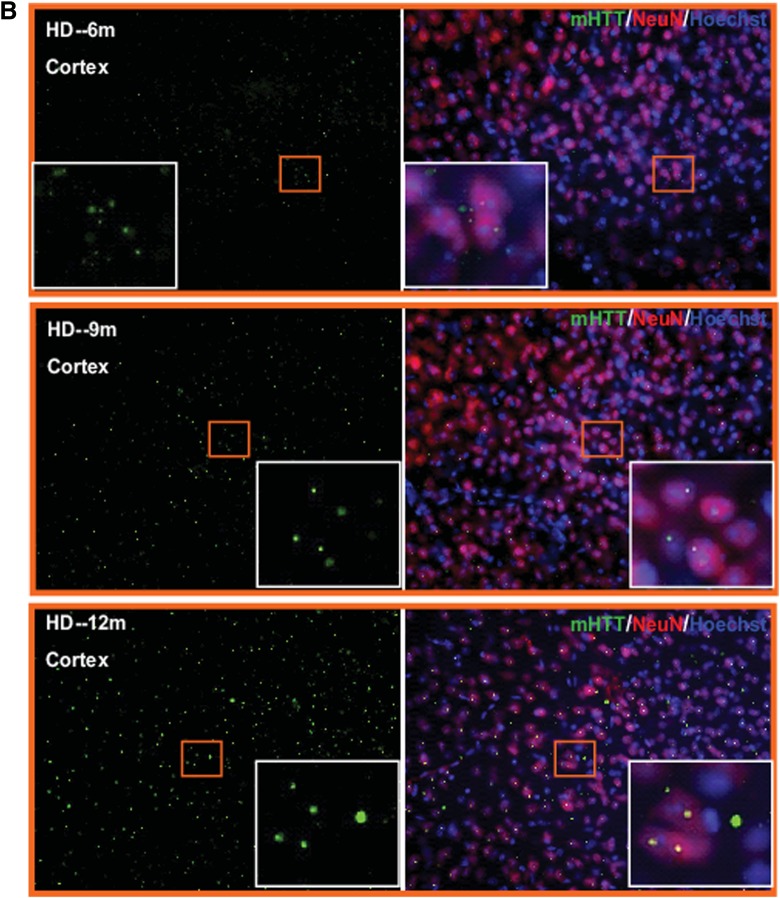
Previous studies of mutant HTT aggregation in mouse models expressing full-length mutant HTT revealed early and selective diffuse nuclear accumulation of aggregated mutant HTT in the striatum and cortex (25–30). To determine the mutant HTT aggregation patterns in HdhQ250 mouse brains, we performed immunohistochemistry with an anti-HTT antibody (N-18) in 3-, 6-, 9-, 12-month-old HdhQ250 brain. We found no detectable mutant HTT aggregates in 3-month-old HdhQ250 mouse brain (data not shown). Nuclear accumulations of mutant HTT aggregates were detected at 6, 9 and 12 months of age, and the numbers of aggregates increased dramatically with disease progression (Fig. 2A and B). Aggregation of mutant HTT is a pathological hallmark of HD (31–33). In adult-onset HD patients, mutant HTT aggregation patterns consist predominantly of large aggregates in the neuropil, with only a small percentage of aggregates present in the nucleus. Interestingly, this nuclear versus cytoplasmic distribution of mutant HTT aggregates in the cortex is opposite to that in juvenile-onset HD patients, in whom nuclear inclusions are the predominant species (31). Additionally, more large aggregates are observed in the cortex of both juvenile- and adult-onset HD brains compared with those in the striatum (31,32).
Figure 2.
Immunohistochemistry staining with anti-HTT antibody (N-18, Santa Crutz) of 6-, 9-, and 12-month-old HdhQ250 mouse striatum (A) or cerebral cortex (B). Note that there is progressively increased mutant HTT aggregation with age in HdhQ250 mice. Green color represents mHTT staining, red color represents NeuN staining and blue color represents nuclear staining. The insert box showing the enlarge pictures for the indicated square area. Scale bar = 50 µm.
Hdhq250 mice exhibit decreased levels of DARPP32 and brain-derived neurotrophic factor
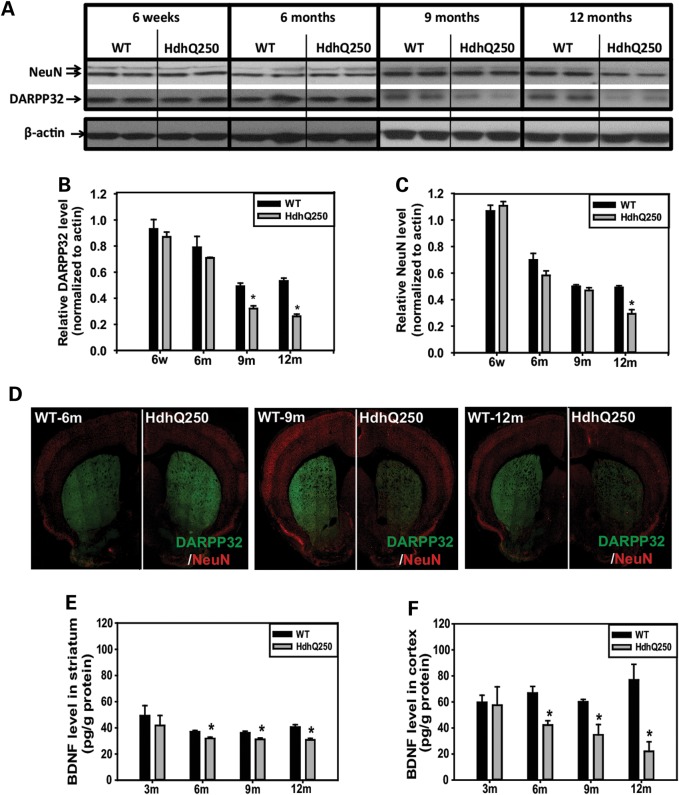
Dopamine and adenosine 3′,5′-monophosphate-regulated phosphoprotein of 32 kDa (DARPP32) is a marker of striatal medium spiny neurons that are most vulnerable and selectively degenerated in HD. Using both western blot analysis and immunofluorescent staining, we found that DARPP32 protein levels decreased progressively in the striatum of HdhQ250 mice before pan-neuronal marker NeuN levels decreased in this region (Fig. 3A and D), consistently suggesting that the medium spiny neurons are more sensitive to mutant HTT.
Figure 3.
HdhQ250 mutation causes progressive reduction of striatal DARPP32 and BDNF levels in cortex and striatum. (A) Representative western blots of DARPP32 and NeuN in the striatum of male HdhQ250 mice and WT controls at indicated ages. (B and C) Quantification of DARPP32 (B) and NeuN (C) western blots from WT and HdhQ250 mice at indicated ages. n = 4, all male mice, *P < 0.05 compared with the levels of age-matched WT control mice by standard Student's t-tests. (D) Immunostaining of DARPP32 in the brain sections of indicated genotypes and ages. Note progressive reduction of DARPP32 staining in the striatum of HdhQ250 mice. Red fluorescent staining, NeuN; green fluorescent staining, for DARPP32. (E and F) BDNF protein levels were measured by ELISA in the cortex (E) and striatum (F) at indicated ages in male HdhQ250 (HD) mice and wild-type (WT) littermate controls. n = 4, *P < 0.05 compared with the volume of age-matched WT control mice by standard Student's t-tests.
The cascade of events initiated by the mutant HTT which are responsible for the selective vulnerability of the striatum in HD remains unknown. Evidence suggests that deficits in brain-derived neurotrophic factor (BDNF) supply could play an important role in the pathogenesis. BDNF is needed for the survival of striatal medium-sized spiny neurons. It is produced by the cortex and delivered via the corticostriatal afferent. Both the loss of the beneficial function of normal huntingtin on BDNF expression and BDNF vesicle axonal transport could be involved in the selective vulnerability observed in HD. It has been reported that both cortical BDNF expression and transportation to the striatum are affected in HD (34,35). Therefore, we measured BDNF protein levels in the cortex and striatum of 3-, 6-, 9-, 12-month-old HdhQ250 mice and their littermate controls. The levels of BDNF protein were significantly reduced in the cortex and striatum of HdhQ250 mice at 6-, 9- and 12-month-old age (Fig. 3E and F), indicating that the HdhQ250 model displays neurotrophin deficit similar to that found in HD patients.
Altered striatal metabolites detected by magnetic resonance spectroscopy in HdhQ250 mice
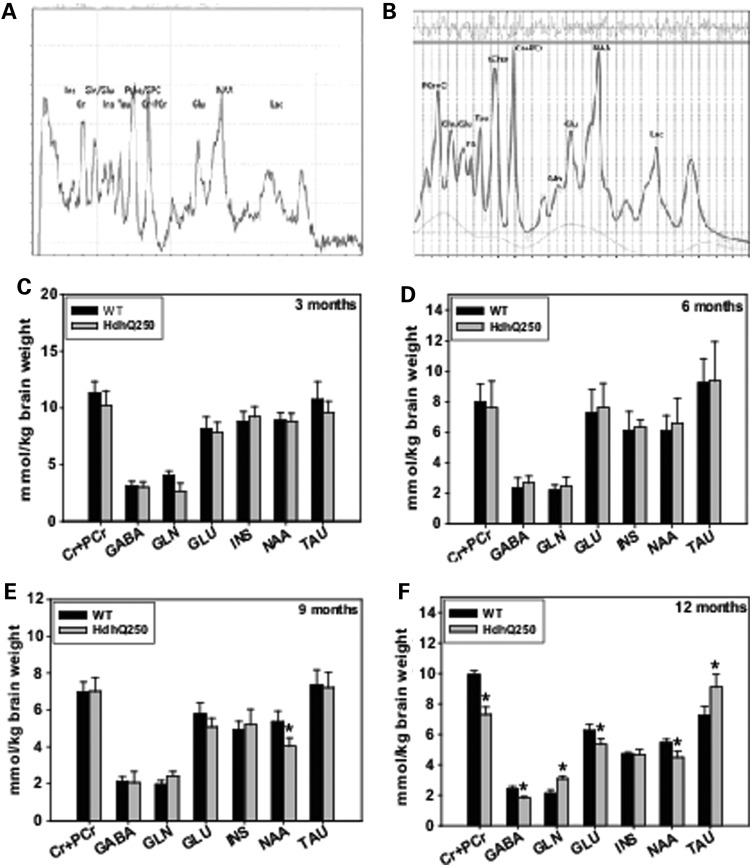
Magnetic resonance spectroscopy (MRS) allows non-invasive measurements of the concentrations of brain metabolites, several of which are involved in brain energy metabolism. Impaired energy production and increased energy demand are evident in HD (36). To determine whether MRS measures can be alternative non-invasive biomarkers used in the preclinical trials with HdhQ250 mice, we employed proton MRS (1H MRS) measurements of in vivo neurochemical profiles together with principal component analysis to determine whether we could distinguish HdhQ250 mice from WT control mice by differences in metabolites levels. We are able to measure eight metabolites in the striatum of mice reliably (Fig. 4A). Using LC Model software (37), we quantified the concentrations of these metabolites (Fig. 4B).
Figure 4.
HdhQ250 mutation caused altered striatal metabolites. (A) In vivo 1H MR spectrum of the HD mouse striatum. The spectrum was obtained by the standard PRESS sequence (TE = 15 ms, TR = 5 s, 256 averages). After initial scout image, a 3 × 3 × 3 mm3 voxel was placed in the mouse striatum. First- and second-order shim coil currents were adjusted using FASTMAP. The line-width of water was ∼15 Hz after adjustment. (B) Analysis of the 1H MR spectrum from an HD mouse using LC Model software with unsuppressed water as internal reference. The basis set was generated by numerical simulation from the pulse sequence acquisition parameters and the literature chemical shift and coupling constant parameters. Adjustment of the LC Model quantification was made with respect to the water relaxation time and concentration. Metabolite values with an LC Model fit of a Cramér–Rao lower bound (CRLB) above 20% were excluded. (C–F) Quantification of striatal metabolites in HdhQ250 mice and their littermate controls at indicated ages. n = 6. *P < 0.05 compared with the values of age-matched wild-type (WT) mice by standard Student's t-tests.
We started assessing metabolite levels in the striatum of 3-month-old mice, and measured the metabolite concentrations every 3 months longitudinally thereafter. At 3 months and 6 months, there are no significant differences of these metabolites between HdhQ250 mice and WT mice (Fig. 4C and D). By 9 months of age, HdhQ250 mice displayed lower levels of N-acetylaspartate (NAA) in striatum than did those in WT control (Fig. 4E). At 12 months of age, HdhQ250 mice displayed significantly altered metabolites in the striatum, including decreased levels in gamma-aminobutyric acid (GABA), glutamate (Glu), NAA, Creatine + PCr and increased Glutamine (Gln) and taurine (Tau) levels (Fig. 4F).
Developmental delay of myelination and sustained white matter pathology in HdhQ250 mice
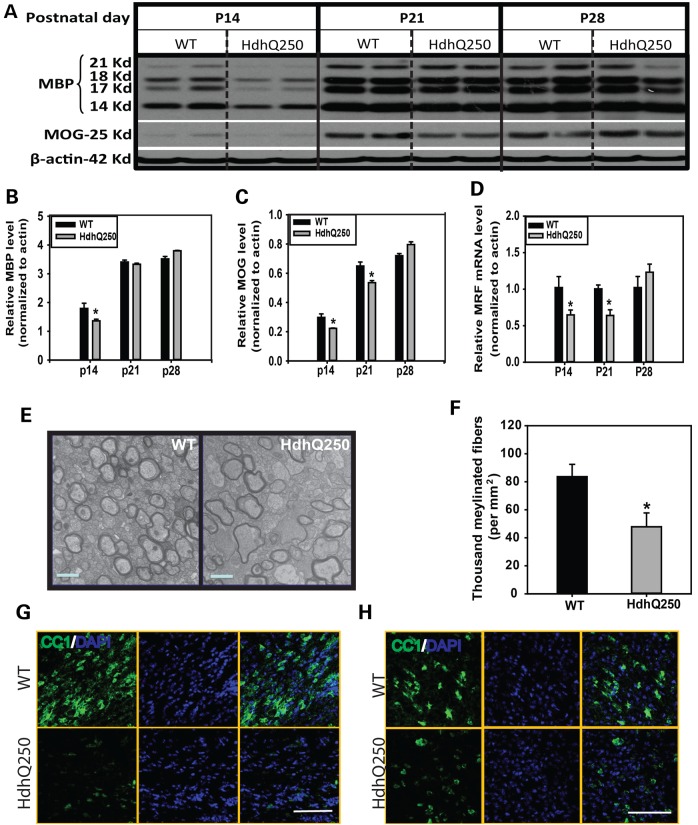
By behavioral, metabolic, pathological and biochemical characterization, our results suggest that HdhQ250 model exhibits accelerated phenotypes compared to all reported full-length KI mice and is suitable for preclinical therapeutic study. There is accumulating evidence that white matter abnormalities appear early, even before gray matter degeneration, in mutant HTT gene carriers (4,8,38–41). More important, there is a significant correlation between white matter changes and functional deficits in HD (42,43). We examined the white matter in HdhQ250 mice during a critical period in postnatal myelination, and found that the levels of all isoforms of myelin basic protein (MBP) and myelin oligodendrocyte glycoprotein (MOG) were significantly lower in the striatum of HdhQ250 mice than those in age-matched littermate WT controls at P14 (Fig. 5A–C). This abnormality of myelination is brain regional specific, as MBP levels in the cerebellum, a spared brain region in HD, are comparable between HdhQ250 mice and their littermate controls (Supplementary Material, Fig. S2). The decreased myelin-related proteins was consistent with decreased expression of a transcription factor, myelin regulatory factor (MRF) at P14 (Fig. 5D), which regulates expression of myelin-related proteins, and these abnormalities of myelination appear most significantly in the early postnatal myelination period (Supplementary Material, Fig. S3). Electron microscopy analysis of the ultrastructure of myelin indicated that HdhQ250 mice displayed significantly fewer myelinated axons in the corpus callosum compared with those in WT littermate controls at P14 (Fig. 5E and F). Our results indicated that mutant HTT interfered with myelination during the postantal myelination period and causes a delay of myelination. Moreover, the numbers of mature oligodendrocytes (CC1 positive cells) are dramatically less in the HdhQ250 brain than those in WT control mouse brain (Fig. 5G and H).
Figure 5.
Mutant Huntingtin leads to abnormal myelin development in HdhQ250 mice. (A) Western Blots of MBP (four different isoforms) in HdhQ250 mice (HdhQ250) or wild-type littermate (WT) controls at postnatal Day 14 (P14). (B and C) Quantification of densitometry of all isoforms of MBP and MOG in western blots. n = 3. (D) Expression of MRF in P14 brain in indicated genotypes. n = 3. *P < 0.05 compared with the values of WT mice by Student's t-tests. (E) Representative images were taken from the corpus callosum. Note: fewer myelinated axons in HdhQ250 mouse corpus callosum compared with those in wild-type (WT) mice. Scale bar = 1 µm. (F) Numbers of myelinated axons (fibers) in the corpus callosum of P14 mouse brain. n = 3. *P < 0.05 compared with the values of WT mice by Student's t-tests. (G and H) Post-mitotic oligodendrocytes are dramatically less in the corpus callosum (G) and striatum (H) of P14 HdhQ250 mouse brain than those in control mouse brain. Scale Bar = 100 µm.
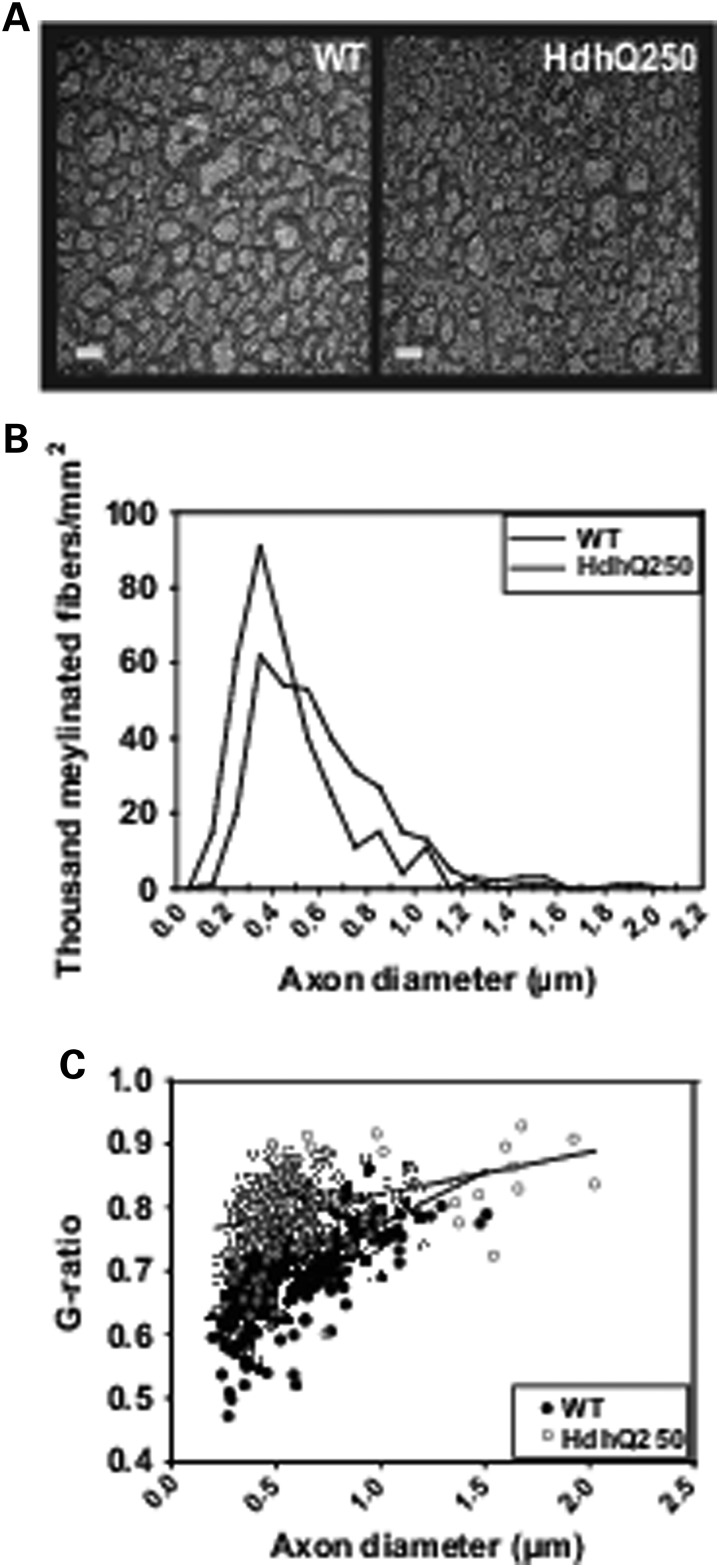
To determine whether the abnormalities of white matter were sustained in HdhQ250 mouse brain, we examined the ultrastructure of axons in the corpus callosum at 12 months of age. We found that HdhQ250 mice displayed numerous smaller axons that are hypomyelinated in the corpus callosum (Fig. 6A and B), and G-ratios (diameter of axon/outer diameter of the myelinated fiber) were significantly increased in the corpus callosum of HdhQ250 mice compared with those in WT littermate controls (Fig. 6C), indicating that the myelin sheaths were significantly thinner in adult HdhQ250 mice. Taken together, these results suggest that abnormalities in myelination are sustained in adult HdhQ250 mice.
Figure 6.
HdhQ250 mutation caused sustained dysmyelination in adult mice. (A) Electron microscopy reveals abnormalities in myelin in 12-month-old HdhQ250 mice (HD) compared with WT mice. Note the smaller caliber axons and hypomyelinated axons appear more in the corpus callosum of HdHQ250 brain. Scale bar = 1 µm. (B) Distribution of different diameters of myelinated axons in the corpus callosum of 12-month-old mice. (C) G-ratio (diameter of axon/outer diameter of the myelinated fiber) is significantly increased in the corpus callosum of HdhQ250 mice (red) compared with WT controls (blue). n = 3.
Enhanced proliferation of OPCs in HdhQ250 mice
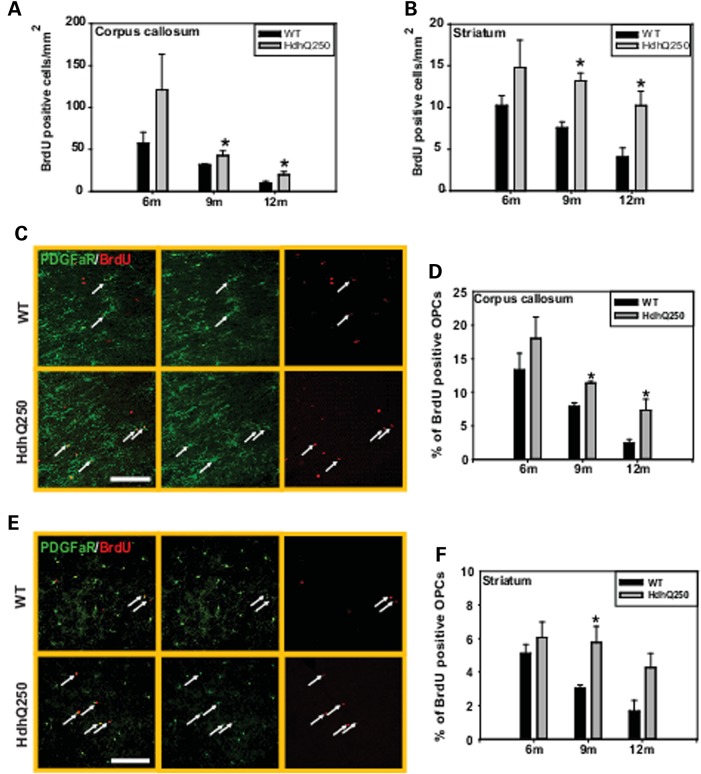
It has been reported that the degree of cell proliferation increased with pathological severity and increasing CAG repeat length, indicating a regenerative potential in human HD brain (44). The mammalian brain contains a widely distributed, abundant class of OPCs (also termed NG2+ cells), these cells co-express the alpha receptor for platelet-derived growth factor (PDGFαR) and NG2 (45,46), so that PDGFαR staining provides an excellent marker for labeling these OPCs. In regions of neurodegeneration, these OPCs exhibited increased proliferation (47). Similar to previous reports in mammalian brain (48,49), these OPCs are abundantly distributed among all the regions in the HdhQ250 brain (data not shown). The OPCs in the normal adult brain have a low level of proliferation that contributes to the regular replacement of oligodendrocytes (50). In response to injury, these OPCs often show enhanced proliferation that is crucial for myelin repair (51–54). To examine how OPCs respond to mutant HTT-induced white matter abnormalities, we assessed proliferation of OPCs in the corpus callosum and striatum of HdhQ250 mice. Mice were treated for 5 days with BrdU, a synthetic thymidine analog that is incorporated into the DNA during cell division; mice were then euthanized 48 h after the last BrdU injection, and examined by immunohistochemistry. The number of proliferating cells (BrdU+) increased significantly in both corpus callosum (Fig. 7A) and striatum (Fig. 7B) of 9- and 12-month-old HdhQ250 mice.
Figure 7.
HdhQ250 mutation causes enhanced proliferation of OPCs. (A and B) Density of proliferating cells in the corpus callosum (A) and striatum (B) in HdhQ250 mice versus control WT mice at indicated age. *P < 0.05 versus age-matched WT mice by standard Student t-tests, n = 3. (C) Representative imaging shows proliferating OPCs (PDGFαR+/BrdU+ cells, arrow pointed cells) in the corpus callosum of 9-month-old mice. Scale bar = 25 µm. (D) Percentage of proliferating OPCs in the corpus callosum of HdhQ250 mice or WT mice at indicated ages. n = 3, *P < 0.05 compared with the values of age-matched WT mice by standard Student's t-test. (E) Representative imaging shows proliferating OPCs in the striatum of 9-month-old mice. Scale bar = 100 µm. Note more proliferating OPCs in HdhQ250 mouse brain.(F) Percentage of proliferating OPCs in the striatum of HdhQ250 mice or WT mice at indicated ages. n = 3, *P < 0.05 compared with the values of age-matched WT mice by standard Student's t-test. At 12 month, the P-value was 0.051 between HdhQ250 group and WT.
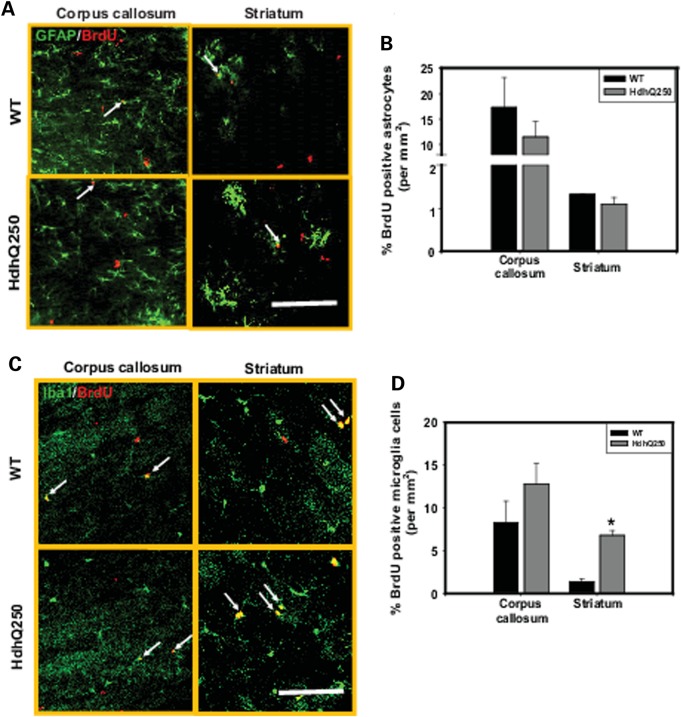
To determine whether the increased proliferation is a generic response of glia cells to mutant HTT or limited to OPCs in HdhQ250 mouse brain, we performed double-immunostaining using anti-BrdU and the OPC cell marker PDGFαR, the astrocyte marker GFAP and microglia marker Iba1. We observed that the percentage of BrdU+/PDGFαR+ (proliferating OPCs) increased significantly in both corpus callosum (Fig. 7C and D) and striatum (Fig. 7E and F) of HdhQ250 mice compare to age-matched controls. Interestingly, the total numbers of OPCs (PDGFαR+ cells) did not differ between HdhQ250 mice and WT controls (Supplementary Material, Fig. S4). In contrast, there is no difference in astrocytes proliferation between HdhQ250 mice and control mice (Fig. 8A and B), and increased proliferation of microglia only detected in the striatum of HdhQ250 mice (Fig. 8C and D). These results indicate that increased cell proliferation in the corpus callosum is restricted to the OPCs in HdhQ250 mouse brain, and increased cell proliferation in the striatum was contributed by both OPCs and microglia.
Figure 8.
None-oligodendrocytes glial cells proliferation in the corpus callosum and striatum of HdhQ250 mice. (A) Representative GFAP and BrdU co-immunostaining images in the corpus callosum and striatum of 9-month-old HdhQ250 mice and control wide-type (WT) mice. Scale bar = 100 um. (B) Percentage of proliferating astrocytes in the corpus callosum and striatum of HdhQ250 brain and wild-type (WT) mice. (C) Representative Iba1 and BrdU co-immunostaining images in the corpus callosum and striatum of 9-month-old HdhQ250 mice and control wide type (WT) mice. Scale bar = 100 um. (D) Percent of proliferating microglia in the corpus callosum and striatum of 9-month-old HdhQ250 mice and WT mice. *P < 0.05 versus control group by standard Student t-tests, n = 3.
In vivo proliferation of OPCs represents a response of these cells to neuronal/axonal degeneration induced by mutant HTT. To determine whether mutant HTT in OPCs has a direct effect on the proliferation ability of OPCs in the absence of influence from mutant HTT containing neurons, we cultured primary OPCs from P6 HdhQ250. First we confirmed the mutant HTT expression in the cultured OPCs from HdhQ250 mice (Fig. 9A). We then used S-phase length which was calculated with dual labeling method as described previously (55) to indicate the proliferation property. In this assay, cells were labeled with BrdU and nucleoside analog EdU (5-ethynyl-2′-deoxyuridine) (Fig. 9B), a longer S-phase would result in a greater number of dual labeled cells. We found that S-phase length was extended in the OPCs from HdhQ250 mice compared with OPCs from controls (11.4 ± 0.1 h in WT OPCs versus 14.1 ± 1.1 h in HdhQ250 OPCs, n = 3, P < 0.05 between two groups by Student's t-test) at 4 days in vitro (DIV 4) (Fig. 9C), indicating that mutant HTT may compromise the repair capacity of OPCs.
Figure 9.
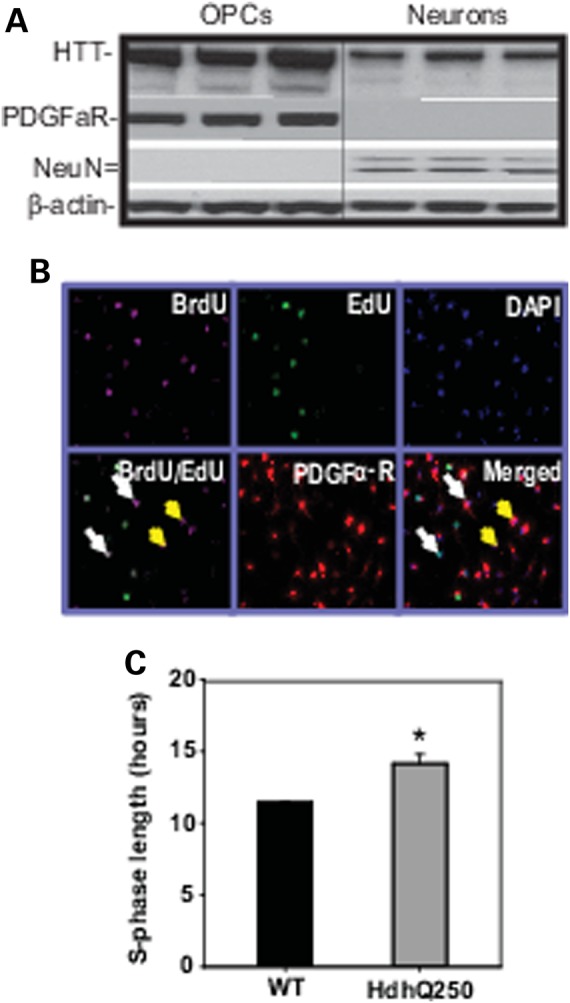
Effects of mutant HTT on OPC proliferation in vitro. (A) Huntingtin expression is detectable in cultured mouse OPCs and cortical neurons. Samples are from wild-type mouse cells. HTT was recognized by MAB2166 anti-HTT antibody. (B) Representative pictures of BrdU and EdU dual-labeling of cultured OPCs (PDGFαR positive cells). White arrows indicate the BrdU+/EdU+ cells; yellow arrows indicate BrdU+/EdU- cells, both are PDGFαR+ cells. (C) S-phase length in cultured OPCs from HdhQ250 mice and WT mice brain at Day 4 in vitro.*P < 0.05 versus control group by standard Student t-tests, n = 3.
Hdhq250 model is a robust preclinical full-length Hdh KI model
We next asked whether HdhQ250 line could be a robust mouse model for HD preclinical trials. To facilitate preclinical studies in HD, a mouse model should have a phenotype that not only recapitulates key features of the disease, but also has low phenotypic variability. Using the data obtained from the characterization of this line, we performed power analyses to estimate the number of mice that would be required to observe rescue of some of the disease-relevant characteristics identified in HdhQ250 mice. HdhQ250 mice had significant brain atrophy, reduced DARPP32 levels in the striatum and motor deficits. Power analyses indicated that if we used brain volume from 9-month-old mice as readout, nine mice per treatment group will have 95% power to observe a 20% rescue of the cortical and striatal volume. In addition, with a sample size feasible for preclinical studies, DARPP32 levels and motor functional assessment can be used to evaluate efficacy in this mode (Table 2). Thus, the power analyses suggest that HdhQ250 mice have robust pathological, molecular and behavioral features, establishing that HdhQ250 is a suitable full-length KI mouse model for HD preclinical studies.
Table 2.
Power analysis
| Phenotype | Age | Sample size |
||
|---|---|---|---|---|
| (month) | 20% rescue | 50% rescue | 80% rescue | |
| Cortical volume | 9 | 9 | 4 | 3 |
| Striatal volume | 9 | 9 | 4 | 3 |
| DARPP32 levels | 9 | 10 | 4 | 3 |
| Balance beam | 9 | 21 | 5 | 3 |
Power analysis indicated the number of experimental mice needed in order to have 95% power for observing a 20, 50 or 80% rescue of phenotypes at the significance level of P < 0.01 (α = 0.01, 95% power).
Discussion
We provide in vivo evidence of early and sustained white matter abnormalities and altered proliferation of OPCs in a novel mutant Hdh KI mouse model. The HdhQ250 mice have a much more robust phenotype than the original HdhQ150 mice (16) and its sister line HdhQ200 (21). Power analyses on the key behavioral and neuropathological outcomes provide strong evidence that the HdhQ250 line is a valuable model for preclinical studies. By replacing the short CAG repeat of the mouse Hdh with an expanded repeat, KI mice represent genetic replicas of human HD better than transgenic models in which mutant HTT is overexpressed. In term of gender difference, we found that female mice exhibited some features of HD, such as selective brain atrophy, motor deficits, altered brain metabolites (Supplementary Material, Fig. S5), in much slower progression comparing with male mice. We suggest using male mice for preclinical trials.
It is worth to mention that CAG repeat size of mutant HTT is much shorter in HD patients than that resulting in phenotypes in this mouse model. The perturbation induced by mutant Hdh in the mouse model may not equally recapitulate the human HD abnormality. Inherent differences between humans and mice must be considered in the search for efficacious treatments for HD. There is almost complete striatal degeneration in HD with 40–50 CAG repeat, but mice expressing full-length mutant HTT/Hdh with much longer CAG repeat (more than 100 CAG repeat in BACHD, YACHD, KI lines-Q140, 150, 175, 200) do not demonstrate such dramatic degeneration. Consistent with this observation, mutant HTT carriers have a smaller intracranial adult brain volume before the onset of the disease than controls, probably reflecting abnormal development (56), the brain volume did not show difference between HdhQ250 mice and littermate wild-type controls until 6 months of age, suggesting that HdhQ250 KI model may represent an adult-onset HD model in terms of phenotypes. In addition, protein levels, protein context, RNA structure are crucial determining factors of mutant HTT-induced pathogenesis. Nevertheless, HdhQ250 heterozygous mice demonstrated many features of human HD like symptoms, supporting that this model is a valuable full-length KI model for preclinical study.
We have noticed that the expression levels of full-length mutant Hdh are lower than those of WT Hdh in HdhQ250 mice. This phenomenon was also observed in another full-length KI mouse model (21). It has been reported that aberrant splicing of mutant HTT/Hdh in the early intron-1 region was detected in both human HD and KI mouse models, the aberrant splicing may result in fragment HTT/Hdh mRNA production and low levels of full-length mutant HTT/Hdh mRNA (57,58). Although the full-length mutant HTT/Hdh mRNA are reduced, the aberrant splicing form of fragment HTT/Hdh mRNA increases, the fragment mRNA itself can be more toxic and/or generate toxic fragment protein that leads to HD pathology (57). The current gene silencing-based therapeutic strategies by using antisense oligonucleotides, RNAi or small hairpin RNAs should also consider preventing the formation of these small fragment HTT/Hdh RNAs and/or their protein product.
HdhQ250 mice showed selective reductions in volume of the striatum and neocortex resembling neuropathology in human HD. These volumetric changes are detected by non-invasive structural MRI, which provides digitized data with full brain coverage free from distortions due to embedding and sectioning. Furthermore, such MRI-based volumetric determinations are routinely used in humans making preclinical mouse studies directly scalable to human clinical trials (59,60). Longitudinal in vivo imaging of HdhQ250 mice would allow a complete natural history of brain pathological changes to be developed during preclinical trials, and considerably increase the power to detect therapeutic efficacy compared with a single assessment. The current study of brain volume in the HdhQ250 mouse model suggests that this mouse model is suitable for preclinical trials.
The analyses of the HdhQ250 mice show several HD-like abnormalities that will be useful towards the goal of diminishing the pathological effects of the HD mutation. These abnormalities include selective striatal and cortical nuclear accumulation of aggregated mutant HTT as shown in other full-length HD mouse models (26–28,61). These studies formed the basis for the therapeutic strategy of blocking such aggregation which is based on the hypothesis that nuclear accumulation of mutant HTT aggregation may be a critical event required for selective neuropathogenesis in HD (30,62). The HdhQ250 mice also share a deficiency in BDNF, an important neurotrophic factor involved in regulating neuronal transmission (63), striatal neuronal survival (64) and HD pathogenesis (34,35). Thus the HdhQ250 model will be useful in establishing the potential role of cortical BDNF reduction in selective striatal pathogenesis in HD and determining the efficacy of BDNF replacement therapies.
Alterations in brain metabolism accompany HD. N-acetylaspartate (NAA) was reduced markedly in both presymptomatic and symptomatic HD patients, and reduced NAA correlated highly with the motor score of the Unified Huntington's Disease Rating Scale (65). Other studies also suggested disturbed brain metabolites in HD (66–68). These findings indicate that altered brain metabolite levels may serve as alternative biomarkers in clinical trials. We found that significant altered striatal metabolites were detected in 9- and 12-month-old HdhQ250 mice, these changes are reminiscent of some changes in human HD brain (69,70), such as decreased NAA and Glu levels and increased Taurine. The disturbed metabolites are not simply due to neuronal loss, as some metabolites (NAA, Glu) decreased, and the other (Taurine) increased in HD brain. Altered metabolites in the HdhQ250 mouse brain are also similar to the results in another full-length KI mouse model-zQ175 model (22), suggesting that brain metabolite changes may serve as alternative biomarkers in using these HD models. We do not know the reason why the changes of metabolites were only detectable in later ages of HdhQ250 mice, other studies in R6/2 mice and HdhQ111 KI mice suggest a potential compensatory response that may maintain energetic homeostasis from early ages through some manifest stages (71,72). Nevertheless, our longitudinal MRS data will instruct future preclinical trial design with this model, such as choosing appropriate ages and biomarkers. MRS data provide a non-invasive means of potentially determining the beneficial effects of a therapeutic for preclinical trials in mouse models that would be directly analogous to their use in similar clinical trials in HD patients. .
White matter atrophy has been reported in subjects with prodromal HD (56,73,74). In postmortem HD brain tissue, there are signs of myelin abnormalities, such as increased numbers of oligodendrocytes (13,75) and increased levels of ferritin iron (myelin breakdown product) (15). Nonetheless, it remains unclear whether these white matter changes represent a necessary step towards further neuropathology or are a response to neuronal damage. Recent MRI studies highlighted the possibility that white matter changes might not be due simply to the loss of neurons in cortical gray matter (76). Here, we detected deficient myelination during the critical postnatal myelination period and sustained white matter pathology in adulthood in HdhQ250 mice. The common theme underlying the myelination abnormalities we observed in the HD model is deregulation of the temporal profiles of myelination. These considerations presuppose that before the occurrence of HD-mediated striatal neuronal degeneration, abnormalities in supporting cells are present that reflect these putative pathological alterations in striatal function. Indeed, neuroimaging evidence is consistent with this notion (11). Therefore, despite the apparent normalization observed after the initial myelination delay, the early HD-associated aberrant myelination likely results in irrevocable changes in cellular homeostasis and stress responses, including processes known to lead to neurodegeneration. Thus, the disturbed myelination during postnatal period might be an important early pathogenic event in HD. Definitive examination of the role of these myelination abnormalities in HD pathogenesis will require substitution of the wild-type for the mutant HTT alleles in OPCs to assess their effects in differentially modifying the progression of the neurodegenerative phenotype.
The results on white matter pathology emphasize the need to more closely look at pre-symptomatic HD carriers for white matter abnormalities to determine how closely the white matter abnormalities we see in HdhQ250 mice reflect the early pathology of HD in human. Our previous studies in a fragment HD model (R6/2) revealed white matter atrophy that correlated with findings in human HD (77). Further study of HdhQ250 mice might establish whether compromised white matter integrity can be detected by using diffusion tensor imaging technique, and determine whether neuroimaging changes in white matter can be detected in the early stage as shown in premanifest HD patients (4,39). Furthermore, it will be important to determine whether disturbed myelination during development contributes to neuronal dysfunction and subsequent neurodegeneration.
In human HD brain, increased cell proliferation in subependymal layer (12) and an increased density of oligodendrocytes is observed in the head of the caudate nucleus for the lower grades (grade 0 and 1) in postmortem HD brain, suggesting a possible compensatory increase due to deficient myelination in early stages of HD (13). Intriguingly, a recent study using carbon-14 dating approaches indicates that oligodendrocyte lineage cells have significantly lower turnover rates, and oligodendrocyte lineage cells are specifically depleted among all non-neuronal cells in the striatum of human HD grade 1–3 brains (14), indicating the vulnerability of these cells to mutant HTT. OPCs in the adult brain show increased rates of proliferation and often undergo dramatic reactive changes in response to acute injury or axonal degeneration (50). OPCs are essential not only for myelin assembly but also for the functional integrity and long-term survival of axons (78–81). In addition, oligodendrocytes provide metabolic and trophic support to axons independent of myelination (82). Specific degeneration of myelinated projection neurons with sparing of interneurons suggests that defective myelination may also be involved in HD pathogenesis (15). It is possible that oligodendrocytes have important bystander effects in HD that are not directly associated with demyelination. In the present study, we demonstrated, for the first time, enhanced proliferation of OPCs in HdhQ250 mouse brain and mutant Hdh directly altered the proliferation property of OPCs without the context of axons in cultures. The observation of increased proliferation of OPCs in HdhQ250 mouse brain could indicate a homeostatic response to oligodendrocyte degeneration, or a response to degenerating axons or neurons, which has been reported for another neurodegenerative disorder, ALS (47). It is not clear what factors signal the observed OPC proliferation, but it is clear that the enhanced OPC proliferation demonstrated in this study is insufficient to compensate for the progressive degeneration in HD. Although the percentage of proliferating OPCs increased in HdhQ250 brain, the total number of OPCs did not change and white matter pathology was sustained, suggesting the possibility that these proliferating OPCs may die quickly or not be able to differentiate into functional oligodendrocytes, which needs further investigation.
Taken in vivo and in vitro results together, increased proliferating OPCs in HdhQ250 mouse brain may reflect a response of these cells to degenerating white and/or gray matter, the mechanism underlying the increased proliferation of OPCs in vivo warrantees further investigation. The longer S-phase in mutant Hdh expressing OPCs suggests that mutant Hdh directly compromised the proliferation ability that might be needed for the repair process in response to damaged neurons and/or axons. These apparently controversial results suggest that mutant Hdh not only directly damage neurons, but also compromise the repair capacity of OPCs in response to neuronal damage in HdhQ250 mice.
Further analysis of the behavior of OPCs in HD in vivo may yield new insight into the factors that regulate their behavior, and the fate of these progenitors in HD. A recent study shows that oligodendrocyte lineage cells are specifically depleted among non-neuronal population in HD human brain (14), suggesting the high sensitivity of these lineage cells to mutant HTT. As oligodendrocytes are crucial for metabolic support of axons of neurons (83), altered OPCs may further accelerate neuronal degeneration. Our current work also leads to the hypothesis that neuronal dysfunction in HD might be due, in part, to the mutation affecting supporting oligodendrocytes which suggests therapeutic strategies aimed at supporting or replacing mutant oligodendrocytes. Such replacement would have a distinct advantage over proposed neuronal replacement therapies, in that neuronal connections already established might be preserved.
Taken together, the robust phenotype of HdhQ250 mice, resembling key features of HD, and the relatively small variability of several measures in this mouse line provides a novel model for identifying therapeutics. Most importantly, our study demonstrated white matter abnormalities, particularly disturbed myelin development and sustained white matter pathology in a novel KI mouse model of HD. These findings shed light on our understanding of HD pathogenesis and open a new avenue to develop therapeutic intervention for this devastating disease.
Materials and Methods
Generation of the HdhQ250 KI line, mouse colony maintenance and genotyping
HdhQ250 mice were generated by selective breeding that exploited the instability of germline repeat length in the HdhQ150 knock-in line described previously(84). Mice with large expansions were chosen for further breeding during backcrossing to C57BL/6J. After eight generations of backcrossing, this strategy yielded the mice used in this study with ∼250 CAG repeats (250 ± 7 CAGs). Unlike KI mouse with repeats 200 or shorter, it was not able to generate homozygous HdhQ250 mice. All experiments were performed with the HdhQ250 KI mice maintained on a C57BL/6J strain background. All in vivo data are from male mice. All mice were housed in cages grouped by gender and provided with food and water ad libitum. Animals were housed under specific pathogen-free conditions with a reversed 12-h light/dark cycle maintained at 23°C and provided with food and water ad libitum . All procedures were conducted in strict compliance with the Guide for the Animal Care and Use Committee of Johns Hopkins University.
In realizing the possibility of CAG instability among generations, our experimental mice have been genotyped and CAG size was determined for each individual mice. We selected the mice with 250 ± 5 CAG size as breeders and set up a large breeding colony to produce sufficient experimental mice in relatively short period. Of 200 meioses where the 250CAG allele was passed to the next generation, 95% mice produced offspring where the repeat size was within the 250 ± 7 range. This shows that the instability is minor enough to be easily overcome by setting up a few more breeding cages in this line. The CAG size from tail samples for our experimental mice is 250 ± 7. We also measured the CAG size in different brain regions in the experimental cohort, the CAG size is 241 ± 5 in the cerebral cortex, 230 ± 5 in the striatum and 255 ± 7 in the cerebellum. Our results also suggest that the instability is minor enough to be easily overcome by setting up a few more breeding cages in this line. Although the instability is a minor factor in this line, continuous monitoring of CAG size is necessary for comparing data among different labs.
Allele specific Hdh mRNA assay
Allele specific PCR was done by Mismatch Amplification Delay. Wild type mouse Hdh contains a CAG repeat that is interrupted by a single CAA (which also codes for glutamine). This interruption was removed during gene targeting the founder line creating a pure CAG repeat in all members of the allelic series (84). This sequence difference was used to develop an allele specific qRT–PCR assay which uses a reverse primer that binds to the first 12 bases of the repeat as well as upstream unique Hdh sequence (CTGCTGCTGCTGAAACGA) (85). Thus this primer perfectly binds the expanded repeat but contains a single mismatch when binding WT Hdh sequence. This primer was paired with the forward primer (GGCAGGAAGCCGTCAT) for Sybr green-based qRT–PCR. Using known quantities of mutant and wild-type DNAs, we empirically determined conditions where the CAA interruption in the WT CAG allele delayed amplification to threshold by 4.0 PCR cycles (300 nm each primer, 50°C 10 min, 95°C 60 s, then 40 cycles of 95°C 15 s, 61.5°C 60 s). These conditions also provided an average PCR efficiency of 1.94-fold amplification per cycle. Thus in a heterozygous sample, the wild-type cDNA is 14-fold under-represented versus mutant. A dilution series of known concentrations of WT and mutant Hdh PCR products that included amplicon were included in each qRT–PCR run to allow determination of copy number of cDNA present in each sample. Reverse transcription was performed replacing random primers from Applied Biosystems High Capacity Reverse transcription Kit with a single Hdh specific primer #4 on Supplementary Material, Figure S1b and c (GCG GCT GAG GGG GTT GA) at a final concentration of 1.25 ng/µl. Since extension of this primer crossed the CAG repeat before reaching the amplicon region, we measured the relative efficiency of reverse transcription across different sized alleles.
Transmission electron microscopy
To analyze the thickness of the myelin sheath, mice were anesthetized and perfused transcardially with PBS, followed by 4% paraformaldehyde (freshly prepared from EM grade), 2% glutaraldehyde, 3 mm CaCl2 in 0.1 m sodium cacodylate buffer, pH 7.2, for 30 min (perfusion speed at 2 ml/min). Samples were maintained in fixation solution (4% paraformaldehyde, 2% glutaraldehyde, 3 mm CaCl2, 1% sucrose in 0.1 m sodium cacodylate buffer (Caco), pH 7.2) at 4°C overnight, immersed in 8% sucrose in 0.1 m Na2HPO4 and NaH2PO4 buffer, and post-fixed in reduced 2% osmium tetroxide (2% osmium, 1.6% potassium ferrocyanide, 3 mm CaCl2, 0.1 m Caco) for 2 h on ice in the dark. Samples were rinsed in distilled water and treated with 1% aqueous uranyl acetate overnight, dehydrated in an ascending series of alcohol dilutions to 100%, followed by acetone. Samples were infiltrated with propylene oxide, embedded in Epon, and sectioned. Semi-thin sections (1 µm) were stained with toluidine blue for tissue identification. Ultrathin sections (90 nm) of corpus callosum from each mouse were cut and post-stained with uranyl acetate and lead citrate. Images were collected with a Hitachi 7600 transmission electron microscope (Hamamatsu City, Shizuoka, Japan). G-ratios (diameter of the axon/outer diameter of the myelinated fiber) of at least 300–500 myelinated axons per genotype and area were measured. Data are displayed as a scatter-plot against axon diameter. The number of axons with different diameters was also plotted against the axon diameter. To count the myelinated fibers in the brain of P14 HD mice, three brains from each genotype were processed and corpus callosum was sampled as described earlier. Twenty to thirty sections from each sample were randomly chosen and myelinated fibers were counted in each section. Data are expressed as mean numbers of myelinated fiber per square millimeter.
OPC cell proliferation assay
BrdU (50 mg/kg) was injected intraperitoneally, twice a day for 1 week. Mice were deeply anesthetized with isoflurane and perfused transcardially with 4% paraformaldehyde (PFA in 0.1 m phosphate buffer, pH 7.4). Brains were post-fixed in 4% PFA overnight at 4°C, transferred to 30% sucrose solution (in PBS, pH 7.4) and stored at 4°C for more than 36 h. Tissue was sectioned (35 μm thick, unless stated otherwise) with a cryostat and immunofluorescence was examined on free-floating sections. Brain (coronal, bregma 0.5 to −1.9 mm) was first incubated in blocking solution (5% normal donkey serum, 0.3% Triton X-100 in PBS, pH 7.4) for 1 h at room temperature, then incubated overnight at 4°C with anti- PDGFαR antibody (1:250, a gift from Dr William Stallcup), or anti-GFAP (Millipore, 1:1000), anti-Iba 1 (Wako Chemicals,1:500), anti-CC1 (Calbiochem, 1:50). After washing with PBS, the sections were incubated with secondary antibody for 2 h at room temperature. Sections were then washed three times with PBS and Hoechst 33342 (Sigma, USA) was used for counterstaining to identify the nucleus. For BrdU staining, brain sections were denatured with 2 N HCl for 30 min at 37°C. Then sections were neutralized with 0.1 m borate buffer for 5 min twice, followed by the staining step described earlier. Mounted slides were imaged using an epifluorescence microscope (Zeiss Axio-imager M1), and Axiovision software (Zeiss), or a confocal laser scanning microscope (Zeiss LSM 510 Meta) by using appropriate excitation and emission filters. A total of 3–12 sections were examined per mouse, and three mice were analyzed per group. For all studies, areas were chosen randomly within the indicated brain regions. Data are presented as percent of double positive cells per mm2 brain area.
In vivo structural MRI and MRS acquisition and quantification
In vivo MRI and MRS were performed on a vertical 9.4 Tesla MR scanner (Bruker Biospin, Billerica, MA, USA) with a triple-axis gradient and a physiological monitoring system (EKG, respiration and body temperature). Mice were anesthetized with isoflurane (1%) mixed with oxygen and air at 1:3 ratios via a vaporizer and a facial mask and scanned longitudinally (the same mice were imaged repeatedly over 12 months period). We used a 20-mm diameter volume coil as the radiofrequency transmitter and receiver. Temperature was maintained by a heating block built into the gradient system. Respiration was monitored throughout the entire scan. High-resolution anatomical images were acquired using a three-dimensional (3D) T2-weighted fast spin echo sequence with the following parameters: echo time (TE)/repetition time (TR) = 40/700 ms, resolution = 0.1 mm × 0.1 mm × 0.25 mm, echo train length = 4, number of average = 2 and flip angle = 40°. Multi-slice T2-weighted images of the mouse brain was acquired using the RARE (Rapid Acquisition with Refocused Echoes) sequence with the following parameter [echo time (TE)/repetition time (TR) = 40 ms/1500 ms, RARE factor = 8, in-plane resolution = 0.125 mm × 0.125 mm, slice thickness = 1 mm, total imaging time <2 min] and used for the planning of the MRS voxel position and high-resolution anatomical imaging. The total imaging time was about 50 min per mouse. Mice recovered quickly once the anesthesia was turned off, and all mice survived the imaging sessions.
Structural MRI image analysis
Images were first rigidly aligned to a template image by using automated image registration software (http://bishopw.loni.ucla.edu/AIR5/, AIR). The template image was selected from one of the images acquired from age-matched littermate control mice (mouse had the medium brain volume among the control group), which had been manually adjusted to the orientation defined by the Paxinos atlas with an isotropic resolution of 0.1 mm × 0.1 mm × 0.1 mm per pixel. After rigid alignment, images had the same position and orientation as the template image, and image resolution was also adjusted to an isotropic resolution of 0.1 mm × 0.1 mm × 0.1 mm per pixel. Signals from non-brain tissue were removed manually (skull-stripping). Skull-stripped, rigidly aligned images were analyzed using Landmarker software (www.mristudio.org). Intensity values of the gray matter, white matter and cerebral spinal fluid were normalized to the values in the template images by using a piece-wise linear function. This procedure ensured that subject image and template image have similar intensity histograms. The intensity-normalized images were submitted by Landmarker software to a linux cluster, which runs Large Deformation Diffeomorphic Metric Mapping (LDDMM). The transformations were then used for quantitative measurement of changes in local tissue volume among different mouse brains, by computing the Jacobian values of the transformations generated by LDDMM.
MRS data analysis
Localized proton spectra were acquired using a Point Resolved Spectroscopy (PRESS) pulse sequence with the following parameters: TE/TR = 8.8 ms/3000 ms. A 3 × 3 × 3 mm3 voxel was placed in the frontal forebrain and covered the striatum. An unsuppressed water reference scan was acquired with two signal averages for quantification of the proton peak, and water suppressed signals were acquired with 1024 signal averages for quantification of other metabolites. Quantification of MRS spectra was performed using the LCModel (37,86) using unsuppressed water as internal reference. Specifically, pulse sequence parameters and molecular structure were used to generate basis sets for each metabolite. Then metabolites as well as water signals were fitted to simulated basis sets to acquire the resonance amplitudes. Then the metabolites concentrations were estimated by referencing to known water concentrations. Metabolite values with an LCModel fit of a Cramér–Rao lower bound (CRLB) above 20% were excluded. Water T1 and T2 values were chosen as 2200 and 25 ms, respectively, for 9.4 T magnetic field (87). Appropriate water attenuation correction was manually defined in the LCModel. The following equation was used to calculate the ‘real’ metabolite concentration [M]real:
In the above equation, CF is the correction factor; [M]LCModel is the concentration value calculated from LCModel; Nusup and Nsup are the number of signal averages in water-unsuppressed and -suppressed signal; Gainusup and Gainsup are the receiver gains used for acquiring unsuppressed water signals and suppressed signals. Both the signal averages and receiver gains were determined from the scanning protocol we used. ATTH2Odefault equals 0.7, a default attenuation correction factor assigned by LCModel. ATTH2Oreal is defined as
In our study, the calculated ATTH2Oreal was 0.52, and total time for MRS was 15 min.
Balance beam tests of motor function
Motor function was assessed on an 80-cm long and 5-mm wide square-shaped balance beam that was mounted on supports 50-cm in height. A bright light illuminated the start platform, and a darkened enclosed 1728 cm3 escape box (12 × 12 × 12 cm) was situated at the end of the beam. Disposable pads placed under the beam provided cushioning if an animal fell off the beam. Mice were trained to walk across the beam twice at least 1 h prior to testing. If a mouse stopped during training, the tail was gently pressed to encourage movement. After the training trial, mice were left undisturbed for at least an hour before testing. The time for each mouse to traverse the balance beam was recorded with a 60 s maximum cut-off, and falls were scored as 60 s.
Immunohistochemistry
Mice were anesthetized and perfused transcardially with phosphate-buffered saline (PBS) followed by 4% paraformaldehyde. Brains were post-fixed overnight followed by immersion into 30% sucrose for 24 h. Coronal brain sections (40 µm) were cut on a cryostat. Sections were stained with primary antibodies, including DARPP32 (1:500, Millipore, USA), NeuN (1:300, Millipore, USA), anti-huntingtin (N-18, 1:1000, Santa Cruz, USA) and PDGFαR (1:200, a gift from Dr William Stallcup), respectively. Briefly, the sections were washed three times with PBS for 10 min each time, then permeabilized by incubating with 0.3% Triton X-100 for 5 min, followed by incubation with blocking solution containing 5% donkey serum, 3% goat serum and 0.3% Triton X-100 for 1 h. Then the sections were incubated with primary antibody at 4°C overnight. After three washings with PBS, the sections were incubated with secondary antibody for 2 h at room temperature, followed by washing three times with PBS. DAPI (Sigma, USA) was used to counterstain the nucleus. Sections were mounted onto superfrost slides (Fisher Scientific, Pittsburgh, PA, USA) and dried and then covered with anti-fade mounting solution. Fluorescence images were acquired with a CCD camera attached to a fluorescence microscope (Zeiss).
For BrdU staining, brain sections were denatured with 2 N HCl for 30 min at 37°C. Then sections were neutralized with 0.1 M borate buffer twice for 5 min each time, followed by staining as described earlier. Images were taken with an Avixon 200 microscope, and BrdU-positive and PDGFαR-positive cells were counted in the striatum and corpus callosum from three brains in each group. Data are presented as percent of double positive cells per mm2 brain area.
Western blotting
Brain tissue samples were homogenized in a buffer containing 50 mm Tris–HCl, pH 8.0, 150 mm NaCl, 0.1% (w/v) SDS, 1.0% NP-40, 0.5% sodium deoxycholate and 1% (v/v) protease inhibitor mixture. For SDS–PAGE, 30–50 μg of proteins were separated in a 4–20% gradient gel and transferred to a nitrocellulose membrane. The membrane was blotted with the following primary antibodies: MW1 (1:5000), MAB2166 (1:1000), anti-NeuN (1:1000), anti-DARPP32 (1:1000), anti-MBP (1:500), anti-MOG (1:1000) and mouse anti-β-actin (Sigma, 1:5000). After incubation with HRP-conjugated secondary antibodies, the bound antibodies were visualized by chemiluminescence.
ELISA analysis of BDNF protein levels
BDNF protein levels were quantified with a commercially available kit (Chemicon) as described previously (88). Briefly, samples were processed by acidification and subsequent neutralization. Wells of 96-well plates were coated with anti-BDNF antibody, incubated in the presence of ‘block and sample’ buffer, and washed in TBST (Tris-buffered saline with Tween-20). Samples (300 µg of protein) were added to triplicate wells, and serial dilutions of BDNF standard (0–500 pg/ml) were added to wells to generate a standard curve. Wells were washed five times with TBST, a hydrogen peroxide solution was added together with a peroxidase substrate, and plates were incubated for 10 min. Reactions were stopped by adding 100 μl of 1 m phosphoric acid, and absorbance was measured at 450 nm by using a plate reader. The concentrations of BDNF in each sample were determined in triplicate, and the average of the three values was used as the value for that mouse.
Quantitative RT-PCR
Total RNA was isolated from the HdhQ250 HD mouse brain by using Trizol reagent (Invitrogen) as the manufacturer recommended. First-strand cDNA was synthesized by using MuLV reverse transcriptase (ABI, USA) with hexamerrandom primer (ABI, USA). Quantitative RT–PCR (qRT–PCR) was carried out with the iTaq universal SYBR Green Supermix (Bio-Rad, USA) in ABI 7900HT Fast Real-Time PCR System. Sequences of the primers were: mouse Mrf forward primer 5′- catcaaagcagagccaaagg-3′, mouse Mrf reverse primer 5′-aactcccacgtggcatagag-3′. After 40 cycles of amplification, the melt curve was analyzed and relative expression of each gene was calculated after normalizing to β-actin expression by applying the comparative quantification method.
OPCs culture and dual labeling with BrdU and EdU to determine the S-phase length
Primary oligodendrocyte progenitor cell culture was performed as published (89). Cerebral cortices from Day 6 mouse pups were used. Sequentially exposing proliferating cells to BrdU and EdU allows differentiation between defined populations of cells, the relative sizes of these populations required direct determination of S-phase length (Ts) of the proliferating pool. OPC cells at DIV-4 were incubated sequentially with BrdU (10 mm) and EdU (20 mm). At the beginning of treatment, OPC cells were incubated with BrdU for 2 h and all cells in S-phase at the beginning of the experiment (T = 0) were labeled with BrdU. OPC cells were subsequently washed thoroughly with culture medium and cultured for another 4 h, and subsequently incubated with EdU for 2 h. Then OPC cells were washed with warmed PBS and fix with EM grade 4% PFA for 15 min at room temperature. Followed by the detection of EdU and BrdU incorporation as described previously (55).
Statistical analysis
Data are expressed as mean ± SEM. Repeated two-way (age and genotype) ANOVA was used for longitudinal body weight data and structural MRI data analysis. Student's t-test was used for other measures between WT and HdhQ250 groups at significance level set at P < 0.05.
Supplementary Material
Funding
The work was supported by the Cure Huntington's Disease Initiative (A3875 to W.D.), the National Institute of Health (NS072344, NS074196 and NS082338 to W.D., NS065306 to J.Z.).
Supplementary Material
Acknowledgements
We thank Dr Shin H. Kang for the expert guidance in the oligodendrocyte precursor cell proliferation study, Dr Bill Stallcup for providing PDGFαR and NG2 antibodies, Dr Alexey Shevelkin for sharing Iba1 antibody, Dr Nicolas Arbez for providing primary neurons and Dr Pamela Talalay for the editorial assistance.
Conflict of Interest statement. None declared.
References
- 1.The Huntington's Disease Collaborative Research Group. (1993) A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. Cell, 72, 971–983. [DOI] [PubMed] [Google Scholar]
- 2.Williams A.J., Paulson H.L. (2008) Polyglutamine neurodegeneration: protein misfolding revisited. Trends Neurosci., 31, 521–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Crotti A., Benner C., Kerman B.E., Gosselin D., Lagier-Tourenne C., Zuccato C., Cattaneo E., Gage F.H., Cleveland D.W., Glass C.K. (2014) Mutant Huntingtin promotes autonomous microglia activation via myeloid lineage-determining factors. Nat. Neurosci., 17, 513–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rosas H.D., Tuch D.S., Hevelone N.D., Zaleta A.K., Vangel M., Hersch S.M., Salat D.H. (2006) Diffusion tensor imaging in presymptomatic and early Huntington's disease: Selective white matter pathology and its relationship to clinical measures. Mov. Disord., 21, 1317–1325. [DOI] [PubMed] [Google Scholar]
- 5.Magnotta V.A., Kim J., Koscik T., Beglinger L.J., Espinso D., Langbehn D., Nopoulos P., Paulsen J.S. (2009) Diffusion tensor imaging in preclinical Huntington's disease. Brain Imaging Behav., 3, 77–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vandenberghe W., Demaerel P., Dom R., Maes F. (2009) Diffusion-weighted versus volumetric imaging of the striatum in early symptomatic Huntington disease. J. Neurol., 256, 109–114. [DOI] [PubMed] [Google Scholar]
- 7.Dumas E.M., van den Bogaard S.J., Ruber M.E., Reilman R.R., Stout J.C., Craufurd D., Hicks S.L., Kennard C., Tabrizi S.J., van Buchem M.A., et al. (2012) Early changes in white matter pathways of the sensorimotor cortex in premanifest Huntington's disease. Hum. Brain Mapp., 33, 203–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tabrizi S.J., Scahill R.I., Durr A., Roos R.A., Leavitt B.R., Jones R., Landwehrmeyer G.B., Fox N.C., Johnson H., Hicks S.L., et al. (2011) Biological and clinical changes in premanifest and early stage Huntington's disease in the TRACK-HD study: the 12-month longitudinal analysis. Lancet Neurol., 10, 31–42. [DOI] [PubMed] [Google Scholar]
- 9.Aylward E.H., Nopoulos P.C., Ross C.A., Langbehn D.R., Pierson R.K., Mills J.A., Johnson H.J., Magnotta V.A., Juhl A.R., Paulsen J.S., et al. (2011) Longitudinal change in regional brain volumes in prodromal Huntington disease. J. Neurol. Neurosurg. Psychiatry., 82, 405–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Poudel G., Stout J.C., Dominguez D.J., Salmon L., Churchyard A., Chua P., Georgiou-Karistianis N., Egan G.F. (2014) White matter connectivity reflects clinical and cognitive status in Huntington's disease. Neurobiol. Dis, 65, 180–187. [DOI] [PubMed] [Google Scholar]
- 11.Novak M.J., Seunarine K.K., Gibbard C.R., Hobbs N.Z., Scahill R.I., Clark C.A., Tabrizi S.J. (2014) White matter integrity in premanifest and early Huntington's disease is related to caudate loss and disease progression. Cortex., 52, 98–112. [DOI] [PubMed] [Google Scholar]
- 12.Curtis M.A., Penney E.B., Pearson A.G., van Roon-Mom W.M., Butterworth N.J., Dragunow M., Connor B., Faull R.L. (2003) Increased cell proliferation and neurogenesis in the adult human Huntington's disease brain. Proc. Natl Acad. Sci. USA, 100, 9023–9027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Myers R.H., Vonsattel J.P., Paskevich P.A., Kiely D.K., Stevens T.J., Cupples L.A., Richardson E.P., Jr., Bird E.D. (1991) Decreased neuronal and increased oligodendroglial densities in Huntington's disease caudate nucleus. J. Neuropathol. Exp. Neurol., 50, 729–742. [DOI] [PubMed] [Google Scholar]
- 14.Ernst A., Alkass K., Bernard S., Salehpour M., Perl S., Tisdale J., Possnert G., Druid H., Frisen J. (2014) Neurogenesis in the striatum of the adult human brain. Cell, 156, 1072–1083. [DOI] [PubMed] [Google Scholar]
- 15.Bartzokis G., Lu P.H., Tishler T.A., Fong S.M., Oluwadara B., Finn J.P., Huang D., Bordelon Y., Mintz J., Perlman S. (2007) Myelin breakdown and iron changes in Huntington's disease: pathogenesis and treatment implications. Neurochem. Res., 32, 1655–1664. [DOI] [PubMed] [Google Scholar]
- 16.Heng M.Y., Tallaksen-Greene S.J., Detloff P.J., Albin R.L. (2007) Longitudinal evaluation of the Hdh(CAG)150 knock-in murine model of Huntington's disease. J. Neurosci., 27, 8989–8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee C.Y., Cantle J.P., Yang X.W. (2013) Genetic manipulations of mutant huntingtin in mice: new insights into Huntington's disease pathogenesis. FEBS J., 280, 4382–4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Menalled L.B., Sison J.D., Dragatsis I., Zeitlin S., Chesselet M.F. (2003) Time course of early motor and neuropathological anomalies in a knock-in mouse model of Huntington's disease with 140 CAG repeats. J. Comp. Neurol., 465, 11–26. [DOI] [PubMed] [Google Scholar]
- 19.Ferrante R.J. (2009) Mouse models of Huntington's disease and methodological considerations for therapeutic trials. Biochim. Biophys. Acta, 1792, 506–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heng M.Y., Detloff P.J., Albin R.L. (2008) Rodent genetic models of Huntington disease. Neurobiol. Dis., 32, 1–9. [DOI] [PubMed] [Google Scholar]
- 21.Heng M.Y., Duong D.K., Albin R.L., Tallaksen-Greene S.J., Hunter J.M., Lesort M.J., Osmand A., Paulson H.L., Detloff P.J. (2010) Early autophagic response in a novel knock-in model of Huntington disease. Hum. Mol. Genet., 19, 3702–3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heikkinen T., Lehtimaki K., Vartiainen N., Puolivali J., Hendricks S.J., Glaser J.R., Bradaia A., Wadel K., Touller C., Kontkanen O., et al. (2012) Characterization of neurophysiological and behavioral changes, MRI brain volumetry and 1H MRS in zQ175 knock-in mouse model of Huntington's disease. PLoS One, 7, e50717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Menalled L.B., Kudwa A.E., Miller S., Fitzpatrick J., Watson-Johnson J., Keating N., Ruiz M., Mushlin R., Alosio W., McConnell K., et al. (2012) Comprehensive behavioral and molecular characterization of a new knock-in mouse model of Huntington's disease: zQ175. PLoS One, 7, e49838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saleh N., Moutereau S., Durr A., Krystkowiak P., Azulay J.P., Tranchant C., Broussolle E., Morin F., Bachoud-Levi A.C., Maison P. (2009) Neuroendocrine disturbances in Huntington's disease. PLoS One, 4, e4962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hodgson J.G., Agopyan N., Gutekunst C.A., Leavitt B.R., LePiane F., Singaraja R., Smith D.J., Bissada N., McCutcheon K., Nasir J., et al. (1999) A YAC mouse model for Huntington's disease with full-length mutant huntingtin, cytoplasmic toxicity, and selective striatal neurodegeneration. Neuron, 23, 181–192. [DOI] [PubMed] [Google Scholar]
- 26.Li H., Li S.H., Johnston H., Shelbourne P.F., Li X.J. (2000) Amino-terminal fragments of mutant huntingtin show selective accumulation in striatal neurons and synaptic toxicity. Nat. Genet., 25, 385–389. [DOI] [PubMed] [Google Scholar]
- 27.Wheeler V.C., White J.K., Gutekunst C.A., Vrbanac V., Weaver M., Li X.J., Li S.H., Yi H., Vonsattel J.P., Gusella J.F., et al. (2000) Long glutamine tracts cause nuclear localization of a novel form of huntingtin in medium spiny striatal neurons in HdhQ92 and HdhQ111 knock-in mice. Hum. Mol. Genet., 9, 503–513. [DOI] [PubMed] [Google Scholar]
- 28.Menalled L.B., Sison J.D., Wu Y., Olivieri M., Li X.J., Li H., Zeitlin S., Chesselet M.F. (2002) Early motor dysfunction and striosomal distribution of huntingtin microaggregates in Huntington's disease knock-in mice. J. Neurosci., 22, 8266–8276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Slow E.J., van Raamsdonk J., Rogers D., Coleman S.H., Graham R.K., Deng Y., Oh R., Bissada N., Hossain S.M., Yang Y.Z., et al. (2003) Selective striatal neuronal loss in a YAC128 mouse model of Huntington disease. Hum. Mol. Genet., 12, 1555–1567. [DOI] [PubMed] [Google Scholar]
- 30.Van Raamsdonk J.M., Murphy Z., Slow E.J., Leavitt B.R., Hayden M.R. (2005) Selective degeneration and nuclear localization of mutant huntingtin in the YAC128 mouse model of Huntington disease. Hum. Mol. Genet., 14, 3823–3835. [DOI] [PubMed] [Google Scholar]
- 31.DiFiglia M., Sapp E., Chase K.O., Davies S.W., Bates G.P., Vonsattel J.P., Aronin N. (1997) Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science, 277, 1990–1993. [DOI] [PubMed] [Google Scholar]
- 32.Gutekunst C.A., Li S.H., Yi H., Mulroy J.S., Kuemmerle S., Jones R., Rye D., Ferrante R.J., Hersch S.M., Li X.J. (1999) Nuclear and neuropil aggregates in Huntington's disease: relationship to neuropathology. J. Neurosci., 19, 2522–2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuemmerle S., Gutekunst C.A., Klein A.M., Li X.J., Li S.H., Beal M.F., Hersch S.M., Ferrante R.J. (1999) Huntington aggregates may not predict neuronal death in Huntington's disease. Ann. Neurol., 46, 842–849. [PubMed] [Google Scholar]
- 34.Zuccato C., Ciammola A., Rigamonti D., Leavitt B.R., Goffredo D., Conti L., MacDonald M.E., Friedlander R.M., Silani V., Hayden M.R., et al. (2001) Loss of huntingtin-mediated BDNF gene transcription in Huntington's disease. Science, 293, 493–498. [DOI] [PubMed] [Google Scholar]
- 35.Gauthier L.R., Charrin B.C., Borrell-Pages M., Dompierre J.P., Rangone H., Cordelieres F.P., De Mey J., MacDonald M.E., Lessmann V., Humbert S., et al. (2004) Huntingtin controls neurotrophic support and survival of neurons by enhancing BDNF vesicular transport along microtubules. Cell, 118, 127–138. [DOI] [PubMed] [Google Scholar]
- 36.Powers W.J., Videen T.O., Markham J., McGee-Minnich L., Antenor-Dorsey J.V., Hershey T., Perlmutter J.S. (2007) Selective defect of in vivo glycolysis in early Huntington's disease striatum. Proc. Natl Acad. Sci. USA, 104, 2945–2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Provencher S.W. (1993) Estimation of metabolite concentrations from localized in vivo proton NMR spectra. Magn. Reson. Med., 30, 672–679. [DOI] [PubMed] [Google Scholar]
- 38.Paulsen J.S., Nopoulos P.C., Aylward E., Ross C.A., Johnson H., Magnotta V.A., Juhl A., Pierson R.K., Mills J., Langbehn D., et al. (2010) Striatal and white matter predictors of estimated diagnosis for Huntington disease. Brain Res. Bull., 82, 201–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rosas H.D., Lee S.Y., Bender A.C., Zaleta A.K., Vangel M., Yu P., Fischl B., Pappu V., Onorato C., Cha J.H., et al. (2010) Altered white matter microstructure in the corpus callosum in Huntington's disease: implications for cortical "disconnection". Neuroimage, 49, 2995–3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weaver K.E., Richards T.L., Liang O., Laurino M.Y., Samii A., Aylward E.H. (2009) Longitudinal diffusion tensor imaging in Huntington's Disease. Exp. Neurol., 216, 525–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kloppel S., Draganski B., Golding C.V., Chu C., Nagy Z., Cook P.A., Hicks S.L., Kennard C., Alexander D.C., Parker G.J., et al. (2008) White matter connections reflect changes in voluntary-guided saccades in pre-symptomatic Huntington's disease. Brain, 131, 196–204. [DOI] [PubMed] [Google Scholar]
- 42.Nucifora P.G., Verma R., Lee S.K., Melhem E.R. (2007) Diffusion-tensor MR imaging and tractography: exploring brain microstructure and connectivity. Radiology, 245, 367–384. [DOI] [PubMed] [Google Scholar]
- 43.Delmaire C., Dumas E.M., Sharman M.A., van den Bogaard S.J., Valabregue R., Jauffret C., Justo D., Reilmann R., Stout J.C., Craufurd D., et al. (2013) The structural correlates of functional deficits in early huntington's disease. Hum. Brain Mapp, 34, 2141–2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Curtis M.A., Connor B., Faull R.L. (2003) Neurogenesis in the diseased adult human brain--new therapeutic strategies for neurodegenerative diseases. Cell Cycle, 2, 428–430. [PubMed] [Google Scholar]
- 45.Nishiyama A., Lin X.H., Giese N., Heldin C.H., Stallcup W.B. (1996) Co-localization of NG2 proteoglycan and PDGF alpha-receptor on O2A progenitor cells in the developing rat brain. J. Neurosci. Res., 43, 299–314. [DOI] [PubMed] [Google Scholar]
- 46.Rivers L.E., Young K.M., Rizzi M., Jamen F., Psachoulia K., Wade A., Kessaris N., Richardson W.D. (2008) PDGFRA/NG2 glia generate myelinating oligodendrocytes and piriform projection neurons in adult mice. Nat. Neurosci., 11, 1392–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kang S.H., Fukaya M., Yang J.K., Rothstein J.D., Bergles D.E. (2010) NG2+ CNS glial progenitors remain committed to the oligodendrocyte lineage in postnatal life and following neurodegeneration. Neuron, 68, 668–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang Q.K., Xiong J.X., Yao Z.X. (2013) Neuron-NG2 Cell Synapses: Novel Functions for Regulating NG2 Cell Proliferation and Differentiation. Biomed. Res. Int., 2013, 402843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Richardson W.D., Young K.M., Tripathi R.B., McKenzie I. (2011) NG2-glia as multipotent neural stem cells: fact or fantasy? Neuron, 70, 661–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Levine J.M., Reynolds R., Fawcett J.W. (2001) The oligodendrocyte precursor cell in health and disease. Trends Neurosci., 24, 39–47. [DOI] [PubMed] [Google Scholar]
- 51.Gensert J.M., Goldman J.E. (1997) Endogenous progenitors remyelinate demyelinated axons in the adult CNS. Neuron, 19, 197–203. [DOI] [PubMed] [Google Scholar]
- 52.Keirstead H.S., Levine J.M., Blakemore W.F. (1998) Response of the oligodendrocyte progenitor cell population (defined by NG2 labelling) to demyelination of the adult spinal cord. Glia, 22, 161–170. [PubMed] [Google Scholar]
- 53.Dawson M.R., Levine J.M., Reynolds R. (2000) NG2-expressing cells in the central nervous system: are they oligodendroglial progenitors? J. Neurosci. Res., 61, 471–479. [DOI] [PubMed] [Google Scholar]
- 54.Di Bello I.C., Dawson M.R., Levine J.M., Reynolds R. (1999) Generation of oligodendroglial progenitors in acute inflammatory demyelinating lesions of the rat brain stem is associated with demyelination rather than inflammation. J. Neurocytol., 28, 365–381. [DOI] [PubMed] [Google Scholar]
- 55.Martynoga B., Morrison H., Price D.J., Mason J.O. (2005) Foxg1 is required for specification of ventral telencephalon and region-specific regulation of dorsal telencephalic precursor proliferation and apoptosis. Dev. Biol., 283, 113–127. [DOI] [PubMed] [Google Scholar]
- 56.Nopoulos P.C., Aylward E.H., Ross C.A., Mills J.A., Langbehn D.R., Johnson H.J., Magnotta V.A., Pierson R.K., Beglinger L.J., Nance M.A., et al. (2011) Smaller intracranial volume in prodromal Huntington's disease: evidence for abnormal neurodevelopment. Brain, 134, 137–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sathasivam K., Neueder A., Gipson T.A., Landles C., Benjamin A.C., Bondulich M.K., Smith D.L., Faull R.L., Roos R.A., Howland D., et al. (2013) Aberrant splicing of HTT generates the pathogenic exon 1 protein in Huntington disease. Proc. Natl Acad. Sci. USA, 110, 2366–2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gipson T.A., Neueder A., Wexler N.S., Bates G.P., Housman D. (2013) Aberrantly spliced HTT, a new player in Huntington's disease pathogenesis. RNA Biol., 10, 1647–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rosas H.D., Goodman J., Chen Y.I., Jenkins B.G., Kennedy D.N., Makris N., Patti M., Seidman L.J., Beal M.F., Koroshetz W.J. (2001) Striatal volume loss in HD as measured by MRI and the influence of CAG repeat. Neurology, 57, 1025–1028. [DOI] [PubMed] [Google Scholar]
- 60.Rosas H.D., Doros G., Gevorkian S., Malarick K., Reuter M., Coutu J.P., Triggs T.D., Wilkens P.J., Matson W., Salat D.H., et al. (2014) PRECREST: a phase II prevention and biomarker trial of creatine in at-risk Huntington disease. Neurology, 82, 850–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Glodzik-Sobanska L., Slowik A., Borratynska A., Szczudlik A. (2003) Depressive symptoms following ischemic stroke. Neurol. Neurochir. Pol., 37, 17–25. [PubMed] [Google Scholar]
- 62.Ross C.A. (2002) Polyglutamine pathogenesis: emergence of unifying mechanisms for Huntington's disease and related disorders. Neuron, 35, 819–822. [DOI] [PubMed] [Google Scholar]
- 63.Lu B. (2003) BDNF and activity-dependent synaptic modulation. Learn. Mem., 10, 86–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Baquet Z.C., Gorski J.A., Jones K.R. (2004) Early striatal dendrite deficits followed by neuron loss with advanced age in the absence of anterograde cortical brain-derived neurotrophic factor. J. Neurosci., 24, 4250–4258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sanchez-Pernaute R., Garcia-Segura J.M., del Barrio Alba A., Viano J., de Yebenes J.G. (1999) Clinical correlation of striatal 1H MRS changes in Huntington's disease. Neurology, 53, 806–812. [DOI] [PubMed] [Google Scholar]
- 66.Reynolds N.C., Jr., Prost R.W., Mark L.P. (2005) Heterogeneity in 1H-MRS profiles of presymptomatic and early manifest Huntington's disease. Brain Res., 1031, 82–89. [DOI] [PubMed] [Google Scholar]
- 67.Sturrock A., Laule C., Decolongon J., Dar Santos R., Coleman A.J., Creighton S., Bechtel N., Reilmann R., Hayden M.R., Tabrizi S.J., et al. (2010) Magnetic resonance spectroscopy biomarkers in premanifest and early Huntington disease. Neurology, 75, 1702–1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.van den Bogaard S.J., Dumas E.M., Teeuwisse W.M., Kan H.E., Webb A., Roos R.A., van der Grond J. (2011) Exploratory 7-Tesla magnetic resonance spectroscopy in Huntington's disease provides in vivo evidence for impaired energy metabolism. J. Neurol., 258, 2230–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Unschuld P.G., Edden R.A., Carass A., Liu X., Shanahan M., Wang X., Oishi K., Brandt J., Bassett S.S., Redgrave G.W., et al. (2012) Brain metabolite alterations and cognitive dysfunction in early Huntington's disease. Mov. Disord., 27, 895–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gramsbergen J.B., Veenma-Van der Duin L., Venema K., Korf J. (1986) Cerebral cation shifts and amino acids in Huntington's disease. Arch. Neurol., 43, 1276–1281. [DOI] [PubMed] [Google Scholar]
- 71.Zacharoff L., Tkac I., Song Q., Tang C., Bolan P.J., Mangia S., Henry P.G., Li T., Dubinsky J.M. (2012) Cortical metabolites as biomarkers in the R6/2 model of Huntington's disease. J. Cereb. Blood Flow. Metab., 32, 502–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tkac I., Henry P.G., Zacharoff L., Wedel M., Gong W., Deelchand D.K., Li T., Dubinsky J.M. (2012) Homeostatic adaptations in brain energy metabolism in mouse models of Huntington disease. J. Cereb. Blood Flow. Metab., 32, 1977–1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Paulsen J.S., Langbehn D.R., Stout J.C., Aylward E., Ross C.A., Nance M., Guttman M., Johnson S., MacDonald M., Beglinger L.J., et al. (2008) Detection of Huntington's disease decades before diagnosis: the Predict-HD study. J. Neurol. Neurosurg. Psychiatry, 79, 874–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tabrizi S.J., Langbehn D.R., Leavitt B.R., Roos R.A., Durr A., Craufurd D., Kennard C., Hicks S.L., Fox N.C., Scahill R.I., et al. (2009) Biological and clinical manifestations of Huntington's disease in the longitudinal TRACK-HD study: cross-sectional analysis of baseline data. Lancet Neurol., 8, 791–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gomez-Tortosa E., MacDonald M.E., Friend J.C., Taylor S.A., Weiler L.J., Cupples L.A., Srinidhi J., Gusella J.F., Bird E.D., Vonsattel J.P., et al. (2001) Quantitative neuropathological changes in presymptomatic Huntington's disease. Ann. Neurol., 49, 29–34. [PubMed] [Google Scholar]
- 76.Stoffers D., Sheldon S., Kuperman J.M., Goldstein J., Corey-Bloom J., Aron A.R. (2010) Contrasting gray and white matter changes in preclinical Huntington disease: an MRI study. Neurology, 74, 1208–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Xiang Z., Valenza M., Cui L., Leoni V., Jeong H.K., Brilli E., Zhang J., Peng Q., Duan W., Reeves S.A., et al. (2011) Peroxisome-proliferator-activated receptor gamma coactivator 1 alpha contributes to dysmyelination in experimental models of Huntington's disease. J. Neurosci., 31, 9544–9553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Griffiths I., Klugmann M., Anderson T., Yool D., Thomson C., Schwab M.H., Schneider A., Zimmermann F., McCulloch M., Nadon N., et al. (1998) Axonal swellings and degeneration in mice lacking the major proteolipid of myelin. Science, 280, 1610–1613. [DOI] [PubMed] [Google Scholar]
- 79.Lappe-Siefke C., Goebbels S., Gravel M., Nicksch E., Lee J., Braun P.E., Griffiths I.R., Nave K.A. (2003) Disruption of Cnp1 uncouples oligodendroglial functions in axonal support and myelination. Nat. Genet., 33, 366–374. [DOI] [PubMed] [Google Scholar]
- 80.Kassmann C.M., Lappe-Siefke C., Baes M., Brugger B., Mildner A., Werner H.B., Natt O., Michaelis T., Prinz M., Frahm J., et al. (2007) Axonal loss and neuroinflammation caused by peroxisome-deficient oligodendrocytes. Nat. Genet., 39, 969–976. [DOI] [PubMed] [Google Scholar]
- 81.Yin X., Crawford T.O., Griffin J.W., Tu P., Lee V.M., Li C., Roder J., Trapp B.D. (1998) Myelin-associated glycoprotein is a myelin signal that modulates the caliber of myelinated axons. J. Neurosci., 18, 1953–1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nave K.A. (2010) Myelination and the trophic support of long axons. Nat. Rev. Neurosci., 11, 275–283. [DOI] [PubMed] [Google Scholar]
- 83.Nave K.A. (2010) Oligodendrocytes and the "micro brake" of progenitor cell proliferation. Neuron, 65, 577–579. [DOI] [PubMed] [Google Scholar]
- 84.Lin C.H., Tallaksen-Greene S., Chien W.M., Cearley J.A., Jackson W.S., Crouse A.B., Ren S., Li X.J., Albin R.L., Detloff P.J. (2001) Neurological abnormalities in a knock-in mouse model of Huntington's disease. Hum. Mol. Genet., 10, 137–144. [DOI] [PubMed] [Google Scholar]
- 85.Dixon K.T., Cearley J.A., Hunter J.M., Detloff P.J. (2004) Mouse Huntington's disease homolog mRNA levels: variation and allele effects. Gene Expr., 11, 221–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Provencher S.W. (2001) Automatic quantitation of localized in vivo 1H spectra with LCModel. NMR Biomed., 14, 260–264. [DOI] [PubMed] [Google Scholar]
- 87.Kuo Y.T., Herlihy A.H., So P.W., Bhakoo K.K., Bell J.D. (2005) In vivo measurements of T1 relaxation times in mouse brain associated with different modes of systemic administration of manganese chloride. J. Magn. Reson. Imaging, 21, 334–339. [DOI] [PubMed] [Google Scholar]
- 88.Duan W., Peng Q., Masuda N., Ford E., Tryggestad E., Ladenheim B., Zhao M., Cadet J.L., Wong J., Ross C.A. (2008) Sertraline slows disease progression and increases neurogenesis in N171-82Q mouse model of Huntington's disease. Neurobiol. Dis., 30, 312–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Dugas J.C., Emery B. (2013) Purification of oligodendrocyte precursor cells from rat cortices by immunopanning. Cold Spring Harb. Protoc., 2013, 745–758. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.