Abstract
Over the past decade, increasing evidence has implied a significant connection between caspase-6 activity and the pathogenesis of Huntington's disease (HD). Consequently, inhibiting caspase-6 activity was suggested as a promising therapeutic strategy to reduce mutant Huntingtin toxicity, and to provide protection from mutant Huntingtin-induced motor and behavioral deficits. Here, we describe a novel caspase-6 inhibitor peptide based on the huntingtin caspase-6 cleavage site, fused with a cell-penetrating sequence. The peptide reduces mutant Huntingtin proteolysis by caspase-6, and protects cells from mutant Huntingtin toxicity. Continuous subcutaneous administration of the peptide protected pre-symptomatic BACHD mice from motor deficits and behavioral abnormalities. Moreover, administration of the peptide in an advanced disease state resulted in the partial recovery of motor performance, and an alleviation of depression-related behavior and cognitive deficits. Our findings reveal the potential of substrate-based caspase inhibition as a therapeutic strategy, and present a promising agent for the treatment of HD.
Introduction
Huntington's disease (HD) is a fatal neurodegenerative disease with autosomal dominant inheritance. It is characterized by progressive motor, cognitive and psychiatric symptoms. The average age of HD onset is 40, and death occurs after ∼10–15 years. Currently, no disease-modifying treatment is available (1).
The protein responsible for the pathogenesis of HD is Huntingtin (Htt), and the mutated form of Htt carries >35 glutamine repeats in its N-terminus. Htt is a substrate for caspase cleavage and can be cleaved by caspase-3 at Asp513 and Asp552, caspase-2 at Asp552 and caspase-6 at Asp586 (2,3). Caspase cleavage of mHtt precedes neurodegeneration and generates N-terminal fragments that aggregate in the cytoplasm and nucleus of neurons (3). Caspase-6 in particular serves an important role in HD pathogenesis: its activation is an early pathogenic event in HD mutation carriers, and the level of activation is directly correlated with CAG repeat length and inversely correlated with age of onset (4). In addition, activated caspase-6 and cleaved fragments of Htt are found in the nuclei of striatal neurons after the initiation of cellular stress, which correlates with increased toxicity (5). Recent evidence shows that caspase-6 activity induced by mHtt is not restricted to the central nervous system (CNS), but can be found in muscle tissue from HD patients and in HD mouse models (6), suggesting that caspase-6 might be involved in central as well as peripheral features of HD.
The importance of mHtt proteolysis at Asp586 was revealed in a mouse model genetically engineered to express Caspase-6 Resistant (C6R) mHtt. Mice expressing C6R mHtt do not develop striatal neurodegeneration and are protected from motor deficits and the depressive phenotype seen in HD mouse models. Furthermore, these mice are protected from neurotoxicity induced by multiple stressors including NMDA, quinolinic acid and staurosporine, demonstrating the protective effects of C6R mHtt expression (7,8). In a complementary study, transgenic mice expressing the 586 amino acid N-terminal fragment of mHtt demonstrated severe neurologic abnormalities and aggregate formation (9). These observations imply that inhibition of mHtt proteolysis by caspase-6 would both reduce the load of toxic N-terminal fragments and maintain the neuroprotective function of wild-type (WT) Htt. Therefore, specific inhibition of caspase-6 was suggested as a disease-modifying therapeutic strategy.
Here, we describe a novel peptide inhibitor based on the Htt caspase-6 cleavage site (amino acids 550–560). This inhibitor is expected to compete with Htt for the caspase-6 active site, and thus reduce Htt cleavage. To facilitate cell penetration, we utilized the HIV-1 TAT-derived peptide. TAT (amino acids 48–60) is known to enable efficient cellular and nuclear penetration, as well as transportation through the blood–brain barrier (BBB) without causing apparent toxic effects (10–12). These considerations led to the peptide sequence GRKKRRQRRRPPQSSEIVLDGTDN (Supplementary material, Fig. S1), designated as ED11.
Results
ED11 inhibits caspase-6 activity
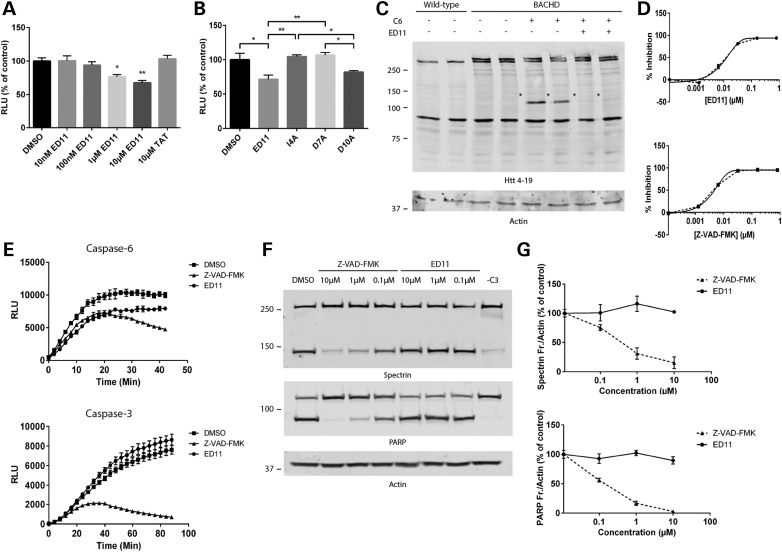
To assess the ability of ED11 to directly inhibit caspase-6, cleavage of Z-VEID-aminoluciferin was monitored. We found that Z-VEID-aminoluciferin cleavage by caspase-6 was reduced by ED11 in a dose-dependent matter, while a control peptide containing the TAT (48–60) sequence alone did not affect the reaction (Fig. 1A). Replacement of amino acids at positions Ile4 or Asp7 within the caspase recognition site of ED11 eliminated the ability of ED11 to inhibit caspase-6 (Fig. 1B), indicating that these amino acids are crucial for ED11's effectiveness.
Figure 1.
ED11 potently and selectively inhibits caspase-6 proteolytic activity. (A) Cleavage of Z-VEID-aminoluciferin by caspase-6 was quantified by light emission (n = 3). (B) Influence of alanine substitution of the indicated amino acids within ED11 on the inhibition of caspase-6 was evaluated. The peptides were administrated at a concentration of 10 µM each. (C) Western blots of BACHD striatal lysates incubated with caspase-6 and labeled with mab2166 antibody (*caspase-6-generated fragments). (D) FRET assay to monitor Htt cleavage by caspase-6 for the determination of ED11 and Z-VAD-FMK IC50 values. (E) Effect of ED11 and Z-VAD-FMK on the cleavage of Z-VEID-aminoluciferin by caspase-6 or caspase-3 (n = 3). (F) Western blots of HEK293 cell lysates incubated with caspase-3 in the presence of ED11 or Z-VAD-FMK and stained with anti-Spectrin and anti-PARP antibodies. (G) Quantification of caspase-3 generated Spectrin and PARP fragments as a function of ED11 and Z-VAD-FMK concentrations (n = 2). *P < 0.05, **P < 0.01 one-way ANOVA followed by Tukey's post hoc test. All data are expressed as mean ± SEM.
To test the ability of ED11 to interfere with caspase-6 cleavage of mHtt, striatal extracts from BACHD mice were incubated with caspase-6 in the presence of ED11. In this experiment, the addition of 10 µM ED11 abolished proteolysis of the human mHtt transgene by caspase-6 (Fig. 1C). To determine IC50 values for the inhibition of caspase-6 by ED11, a FRET-based Htt cleavage assay was used. This assay measures Htt cleavage using FRET between an N-terminal Htt antibody and a neo-epitope antibody directed against the 586 cleavage site (5). The IC50 value determined for ED11 was 12.12 nM, whereas the IC50 for the commercial pan-caspase inhibitor Z-VAD-FMK was determined as 4.62 nM (Fig. 1D).
Due to the structural similarity of active sites among the caspase family, small molecule caspase-6 inhibitors frequently demonstrate cross-reactivity with other caspases. This is evident especially with caspase-3, which is the most abundant caspase in adult rodent brain (13–15). Therefore, we examined the specificity of ED11 to caspase-6 over caspase-3, using Z-VEID-aminoluciferin as a substrate and the synthetic pan-caspase inhibitor Z-VAD-FMK as a control. When monitoring the influence on caspase-6 activity, we observed that both ED11 and Z-VAD-FMK inhibited the proteolytic process to a similar extent. However, when caspase-3 was used in the experiment, no influence of ED11 on Z-VEID-aminoluciferin cleavage was recorded, whereas Z-VAD-FMK still strongly inhibited the luminescent signal (Fig. 1E). To confirm these findings, caspase-3 was incubated with HEK293 cell lysate, and the cleavage of PARP and Spectrin, two endogenous caspase-3 targets, was quantified by western blot. We observed that Z-VAD-FMK inhibited caspase-3-mediated cleavage of PARP and Spectrin, as expected, whereas ED11 did not influence the reaction, indicating that its influence on caspase-3 is insignificant (Fig. 1F and G). To test the inhibitory effect of ED11 on a wider range of caspases, we tested Htt protein cleavage by caspases 1–10 and the effect of ED11 on fragment generation. We have demonstrated recently that these caspases can cleave Htt at different sites (16). After incubation of the different caspases with Huntingtin, western blot staining by BKP1 antibody demonstrated that the inhibitory effect of ED11 was most pronounced for the generation of the 586 fragment by caspase-6. A lesser effect of the inhibitor was found on caspase-1 and -10-mediated generation of the 513 fragment and caspase-2-mediated generation of the 552 fragment (Supplementary material, Fig. S2).
ED11 protects cells from 145Q-mHtt-induced toxicity
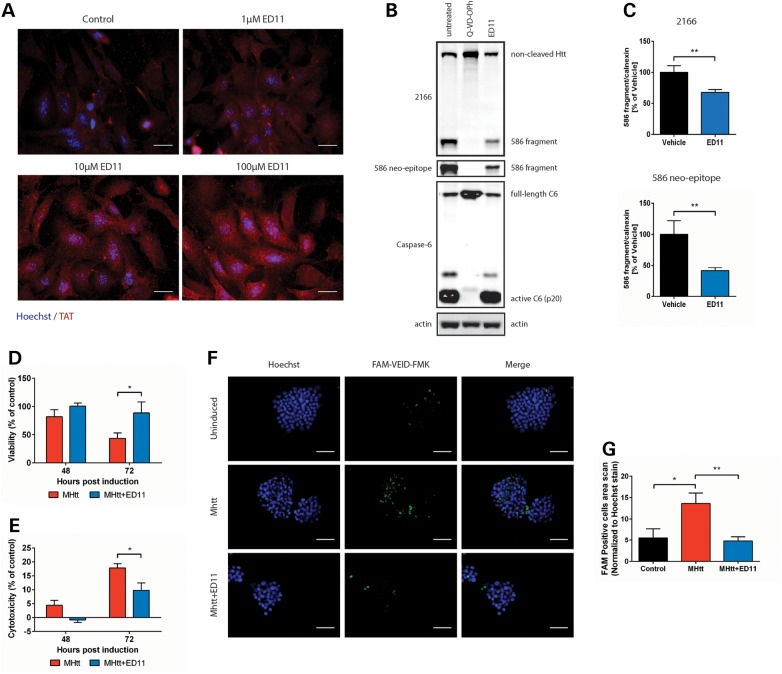
To confirm the ability of ED11 to penetrate cellular membranes, different concentrations of ED11 were incubated with mouse embryonic fibroblast (MEF) cells. Staining with an anti-TAT antibody demonstrated the dose-dependent accumulation of ED11 inside the cells (Fig. 2A). We furthermore confirmed that ED11 does not affect cell viability, proliferation or cell cycle status in a basal state (Supplementary material, Fig. S3).
Figure 2.
ED11 penetrates the cell membrane, inhibits caspase-6 activity intracellularly and protects cells from mHtt-induced toxicity. (A) Representative images of MEF cells incubated with the indicated concentrations of ED11 and labeled with anti-TAT antibody. (B) Western blots of HEK293 cells co-transfected with caspase-6 and Htt, and stained with mab2166 and 586 neo-epitope antibodies. (C) Quantification of the Htt fragment generated by caspase-6 that was detected by mab2166 and 586 neo-epitope antibodies (n = 4–8). (D) Viability measurement of 145Q-mHtt-expressing, serum-deprived PC12 cells by Alamar blue viability assay (n = 5). (E) Cell death measurement of 145Q-mHtt-expressing, serum-deprived PC12 cells by LDH release assay (n = 5). (F) Caspase activity measurement of 145Q-mHtt-expressing, serum-deprived PC12 cells by incubation with FAM-VEID-FMK. (G) Quantification of caspase activity-dependent FAM-VEID-FMK fluorescence signal normalized to Hoechst nuclear staining (n = 7). Scale bars indicate 50 µM. *P < 0.05, **P < 0.01 one-way ANOVA followed by Tukey's post hoc test. All data are expressed as mean ± SEM.
To evaluate the ability of ED11 to reduce intracellular Htt cleavage by caspase-6, we co-transfected HEK293 cells with caspase-6 and the N-terminal 1212 amino acids of 15Q-Htt and added ED11 to the medium. Htt 586 fragment levels were quantified 24 h after transfection using anti-Htt (mab2166) and anti-Htt586 neo-epitope antibodies. Treatment with ED11 resulted in a significant reduction of the 586 fragment resulting from caspase-6 cleavage (Fig. 2B and C), whereas no effect on the auto-activation of caspase-6 was observed. The cell-permeable pan-caspase inhibitor Q-VD-OPh, however, inhibited both caspase-6 auto-activation and Htt cleavage (Fig. 2B). These findings suggest that ED11 is able to block the intracellular cleavage of Htt by caspase-6.
To evaluate ED11's ability to protect cells from mHtt toxicity, PC12 cells harboring an inducible 145Q-mHtt-expressing vector were placed under chronic serum deprivation stress and induced to express mHtt for 72 h. Cell viability measurements showed that while 145Q-mHtt expression gradually decreases viability, treatment with ED11 resulted in the preservation of cell viability (Fig. 2D). Measuring LDH release revealed that ED11 treatment attenuated cell death that was evident after 72 h of 145Q-mHtt expression in untreated cells (Fig. 2E). Next, we evaluated caspase activity levels in 145Q-mHtt-expressing cells with a FAM-VEID-FMK caspase-6 activity reporter. Whereas the induction of 145Q-mHtt significantly increased caspase activity in vehicle-treated cells, ED11-treated cells maintained baseline levels of caspase activation (Fig. 2F and G), confirming that the peptide is able to inhibit caspase-6 intracellularly.
Pre-symptomatic ED11 treatment protects BACHD mice from motor and behavioral deficits
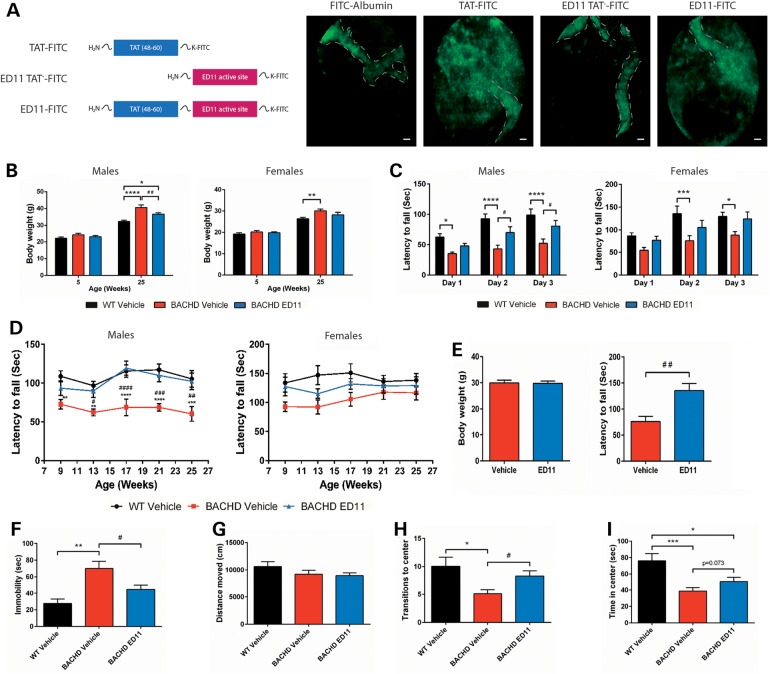
To evaluate the ability of ED11 to penetrate the BBB, we labeled ED11 with a fluorescein isothiocyanate (FITC) tag at the C-terminus. Then, we monitored the extravasation of labeled ED11 into the brain parenchyma. While FITC-conjugated albumin and ED11 lacking the cell-penetrating sequence TAT (48–60) remained inside the blood vessels, FITC-conjugated TAT and ED11 can be seen in the brain parenchyma (Fig. 3A).
Figure 3.
ED11 penetrates the CNS and protects BACHD mice from motor and behavioral deficits. (A) In vivo fluorescence tracking of the indicated peptides, 40 min after IV injection. The estimated blood vessel pattern is marked with a dashed line. (B–F) BACHD mice were treated with ED11 starting from the age of 5 weeks and the indicated parameters were tested. (B) Body weight measurements of pre- and post-treatment animals (n = 10–11 per sex). (C) Rotarod training performance was measured by latency to fall from an accelerating rod 3 weeks after treatment commencement for 3 consecutive days, average of three trials per day. (D) Long-term rotarod performance was measured by latency to fall from an accelerating rod every 4 weeks. (E) Body weight matching for the ED11 and vehicle-treated mice and their corresponding rotarod performance (n = 6). (F) Depressive-like behavior as measured by time spent immobile in the FST (n = 20–22). (G–I) Distance moved, time spent in center and transitions to center in the open field test (n = 20–22). Scale bars indicate 20 µM. Asterisk indicates statistical difference from the WT-vehicle group, whereas hash indicates statistical difference from the BACHD ED11 group. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. #P < 0.05, ##P < 0.01, ###P < 0.001, ####P < 0.0001. (B–D) Two-way ANOVA followed by Tukey's post hoc test. (E) Two-tailed unpaired Student's t-test. (F–I) One-way ANOVA followed by Tukey's post hoc test. All data are expressed as mean ± SEM.
To evaluate the ability of ED11 to protect from mHtt toxicity in vivo, the full-length 97Q-mHtt transgenic BACHD mouse model was used. ED11 treatment was commenced at the age of 5 weeks with a subcutaneously implanted Alzet mini-pump, which infused the peptide continuously for 28 days at a dose of 4 mg/kg/day. The pump was replaced every 28 days for the duration of the experiment. Since mHtt expression causes a metabolic imbalance manifested by an excessive body weight gain in BACHD mice (17), animals were weighed regularly throughout the study. Notably, treatment with ED11 attenuated the excess body weight gain in BACHD mice (Fig. 3B).
Mice were tested for motor performance 4 weeks after treatment initiation by measuring the latency to fall on an accelerating rotarod. During the training session, BACHD mice treated with ED11 demonstrated higher motor learning capabilities than the vehicle-treated BACHD mice (Fig. 3C). Long-term motor performance was monitored starting at the age of 9 weeks, and was evaluated monthly. Male BACHD mice treated with ED11 showed preserved motor performance compared with WT mice. The same trend was detected in the female group, which did not show a significant motor impairment in our cohort (Fig. 3D). Since body weight differences were observed, it was important to verify that the effect of ED11 on motor performance was not weight-dependent. To this end, we matched male subjects with the same body weight and compared their rotarod performance. The results showed that the beneficial effect of ED11 cannot be attributed to the lower body weight of the ED11-treated mice (Fig. 3E).
To evaluate the influence of ED11 on the depressive-like behavior of the BACHD mice, the forced swim test (FST) was conducted at 5 months of age. ED11-treated mice did not exhibit the increased immobility observed in the vehicle-treated BACHD mice (Fig. 3F), indicating that ED11 protects against this depression-related phenotype. No correlation was found between motor performance in the rotarod test and time spent immobile in the FST (Pearson correlation coefficient r = 0.057, P-value = 0.83, two-tailed Student's t-test), confirming that the protective effect of ED11 was not directly related to motor ability.
To assess the effect of ED11 on basal locomotor activity, exploratory activity and anxiety-related behavior the open field test were examined at 22 weeks of age. In this test, no significant differences in total distance traveled were observed between the treatment groups, indicating that basal locomotor activity was unchanged (Fig. 3G). Exploratory activity and anxiety-related behavior were measured by quantification of the time spent in the center and number of transitions to the center. Whereas BACHD mice show decreased exploratory and increased anxiety-related behavior, the ED11-treated mice showed more transitions to the center and a trend toward increased time spent in the center compared with vehicle-treated BACHD mice, indicating lower anxiety levels and improved exploratory behavior (Fig. 3H and I).
In agreement with published data, where mHtt-586 fragments were only detectable in 13- to 15-month-old BACHD animals (18), the levels of this fragment were below detection in our 6-month-old cohorts (data not shown). Similarly, mHtt aggregation cannot be detected at this age in BACHD mice (19,20), precluding an assessment of ED11-mediated effects on this readout.
Post-symptomatic ED11 treatment partially restores motor ability in BACHD mice and protects from behavioral and cognitive deficits
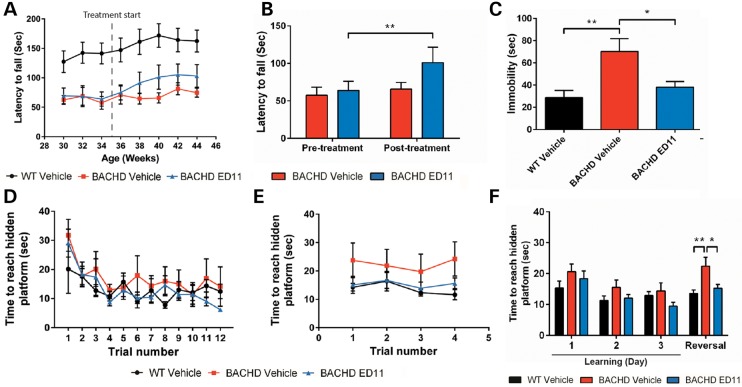
To evaluate whether treatment with ED11 is also effective in a more advanced disease state, we treated BACHD mice at the age of 36 weeks, when HD-related phenotypes are already more pronounced. For motor evaluation, BACHD mice were trained on the accelerating rotarod at the age of 30 weeks and assigned to treatment groups according to a similar baseline motor performance. Treatment with ED11 or vehicle was started at 36 weeks of age. While the performance of vehicle-treated BACHD mice remained at the same level throughout the testing period, ED11-treated mice demonstrated an increase in rotarod performance, indicating that ED11 was able to exert a restorative effect in these mice (Fig. 4A). This effect was statistically significant in relation to the pretreatment state of BACHD mice (Fig. 4B).
Figure 4.
ED11 treatment partially restores motor ability and reverses behavioral and cognitive deficits in progressive disease stage BACHD mice. (A) Determination of rotarod performance in an advanced disease state with the rotarod test. The time point of treatment initiation is indicated with a dashed line. (B) Motor performance in the rotarod test was measured at different time points. The pretreatment state was recorded at the age of 34 weeks, and post-treatment values were recorded at the age of 40 weeks, 5 weeks after treatment initiation. (C) The effect of ED11 on the depressive-like behavior as measured by time spent immobile in the FST. (D) Time to reach the hidden platform in the learning phase of the swimming T-maze test for 3 consecutive days, four trials per day. (E) Time to reach hidden platform in the strategy shifting phase of the swimming T-maze test, four trials a day following the learning phase. (F) Daily averages of the time to reach the hidden platform. *P < 0.05, **P < 0.01. (B) Paired samples Student's t-test. (C and F) One-way ANOVA followed by Tukey's post hoc test. All data are expressed as mean ± SEM.
To evaluate the effect of ED11 on the depression-related phenotype in symptomatic BACHD mice, mice were tested using the FST at 11 months of age. While vehicle-treated BACHD mice were found to be more immobile than their WT littermates, the immobility of ED11-treated mice was similar to WT animals, indicating a reversal of the depressive-like phenotype (Fig. 4C). The effect of ED11 on the mobility state in the FST is not related to motor ability, as no correlation between motor performance in the rotarod test and time spent immobile in the FST was detected (Pearson correlation coefficient r = 0.12, P-value = 0.73, two-tailed Student's t-test).
To address the possible effect of ED11 treatment on cognitive deficits observed in the BACHD mice, we used the swimming T-maze test. During the 3-day learning period, all experimental groups learned to swim directly to the platform, as measured by a progressive decrease in the time to reach the target (Fig. 4D and F). On the fourth day, the strategy shifting ability was tested by relocating the hidden platform to the opposite arm. We found impairment in the strategy shifting ability of vehicle-treated BACHD mice, as their time to reach the hidden platform was longer than for WT mice. In contrast, the performance of ED11-treated BACHD mice was comparable to their WT littermates (Fig. 4E and F), indicating a reversal of cognitive rigidity by ED11.
MRI volumetric measurements were performed at the age of 12 months to examine the possible effect of ED11 treatment on striatal, cortical and hippocampal atrophy. However, we did not detect significant reduction in the BACHD mice compared with WT mice (Supplementary material, Fig. S4), precluding any evaluation of ED11-mediated effects on brain atrophy.
Discussion
In our study, we used a peptide encompassing the human Htt sequence around amino acid Asp586 to compete for caspase-6 activity and inhibit the proteolysis of Htt. We show that the peptide inhibits Htt cleavage, rescues a cell culture model from mHtt toxicity and significantly reduces the motor deficits and behavioral abnormalities in BACHD mice. Moreover, treatment in an advanced disease state resulted in partial recovery of motor performance, an alleviation of the depressive-like behavior and cognitive deficits.
Subsequent to the discovery of Htt as a substrate for caspase cleavage, it was shown that caspase-6 activation plays a major pathogenic role in human HD patients and in HD models. However, the importance of caspase-6 in mHtt pathogenesis has recently been challenged in studies that crossed caspase-6-deficient mice and HD mouse models (18,21). These reports have demonstrated the continued presence of 586 amino acid mHtt fragments in the absence of caspase-6, suggesting that additional proteases are involved in mHtt fragmentation at this site. Nonetheless, caspase-6 deficiency provided significant beneficial effects including a decrease of full-length mHtt and mHtt fragments and a significant reduction in aggregate formation. Furthermore, body weight levels and motor deficits were attenuated.
In contrast to the genetic ablation of caspase-6, compound-based inhibition of caspase-6 activity in adulthood may be more efficient in providing protection from mHtt toxicity in vivo. Caspase-6 knockout mice demonstrate alterations in striatum and cortex (22), regions that are specifically affected in HD. Furthermore, caspase-6 is important in neurodevelopment, as it is profoundly involved in axonal pruning during development (23). Its absence may lead to a compensatory upregulation of other members of the caspase family, a scenario that has been described for caspase-3−/− mice (24). Our approach to use an Htt-derived peptide as a sink for excessive caspase-6 activity, on the other hand, should also trap other proteases that can recognize and cleave at amino acid 586. It is a target-independent approach to prevent mHtt proteolysis and thus has significant advantages over both caspase-6 ablation and classical caspase inhibitors, which are nonspecific, lack sufficient cell and brain permeability and could cause nonspecific toxic effects when added at concentrations required to inhibit intracellular caspases (25–27). Interestingly, our in vitro Htt cleavage experiment demonstrates that ED11 may profoundly alter caspase cleavage patterns of this protein. We recently found that Htt can be cleaved by caspases 1–10 in vitro, and we now show that ED11 inhibits caspase-1, -2 and -10 as well as increases caspase-5-mediated Htt cleavage. For most of these caspases, the in vivo relevance of their Htt cleavage processes as well as their contribution to HD pathogenesis remains unknown. However, the inhibition of caspase-2-mediated Htt cleavage by ED11 may contribute to the preservation and improvement of the in vivo behavioral function, since the ablation of this enzyme has been associated with therapeutic benefits in YAC128 mice in a previous study (28).
The ability of ED11 to reverse HD-like phenotypes in post-symptomatic BACHD mice is likely due to the restoration of neuronal function prior to the occurrence of neuronal death. We did not observe any significant changes by MRI in vehicle-treated BACHD mice compared with WT animals at 12 months of age, suggesting that in these mice, a window for therapeutic intervention exists between the onset of HD-like behavioral phenotypes and irreversible brain atrophy. In patients, treatment should similarly be most effective prior to neuronal loss and with the availability of an accurate genetic prediction could even be envisaged pre-symptomatically.
Due to the absence of striatal, cortical or hippocampal atrophy in our BACHD cohort, we were however unable to evaluate the effects of ED11 on the prevention of brain atrophy. These neuroanatomical endpoints will have to be analyzed in a more long-term study, which will furthermore determine whether ED11 treatment is likely to provide the long-lasting therapeutic benefits that are necessary to successfully treat a chronic illness like HD.
Our study demonstrates that the use of substrate-based peptides is a feasible concept for inhibition of caspase-mediated protein cleavage for therapeutic purposes. In view of its potency and selectivity, we suggest ED11 as a treatment for HD. Further studies should be performed to translate our findings from animal models to HD patients. In addition, our findings might be applicable in other neurodegenerative or neurological diseases where caspase-6 activity plays a part in the pathogenic process.
Materials and Methods
Peptide synthesis and dissolvent
ED11 (GRKKRRQRRRPPQSSEIVLDGTDN), TAT-only (GRKKRRQRRRPPQ), ED11-FITC (GRKKRRQRRRPPQSSEIVLDGTDNK-FITC), ED11-No TAT-FITC (SSEIVLDGTDNK-FITC), TAT-FITC (YGRKKRRK-FITC), I4A (GRKKRRQRRRPPQSSEAVLDGTDN), D7A (GRKKRRQRRRPPQSSEIVLAGTDN) and D10A (GRKKRRQRRRPPQSSEIVLDGTAN) were synthesized by China-Peptides Ltd (Shanghai, China) with a purification level at over 95%. For the purified caspase activity assays, peptides were dissolved in DMSO. For the cell culture involved experiments, peptides were initially dissolved in cell culture grade H2O and further diluted by phosphate-buffered saline (PBS) to the desired concentrations. For in vivo studies, peptides were initially dissolved in cell culture grade H2O and further diluted by 0.9% saline.
Luminescent caspase inhibition assays
To determine caspase inhibition by ED11, the Caspase-Glo® 6 assay (Promega, Fitchburg, WI, USA) was carried out according to the manufacturer's instructions. Briefly, 0.1 U/ml of purified caspase-6 (Enzo Life Sciences, Farmingdale, NY, USA) or caspase-3 (Abcam, Cambridge, UK) was incubated with 5 µM Z-VEID-aminoluciferin in the presence of 5 µM ED11 or DMSO as a vehicle control, and the luminescent signal of released aminoluciferin was detected using a Synergy HT multi-mode micro-plate reader (Bio-Tek, Winooski, VT, USA).
Protein caspase cleavage assays
For brain lysate preparation, FVB/N BACHD mice and their WT littermates were euthanized by decapitation. Striata were dissected, suspended in a lysis buffer (200 mm HEPES, 150 mm NaCl, 1 mm Na3VO4, 5 mm EDTA, 1% NP-40, 0.5% DOC and 50 mm NaF) with protease inhibitors (Roche), homogenized with a dounce homogenizer and placed for 1 h on ice. Tissue debris was removed by centrifugation at 20 000g for 15 min at 4°C. Protein concentration was determined by the BCA method (Pierce). About 50 µg of lysate proteins were exposed to 100 U/ml of purified active human caspase-6 for 45 min at 37°C in the presence of 10 µm ED11 or DMSO as a vehicle control. Lysates were subsequently loaded on 7.5% SDS–PAGE gels and transferred to 0.45 µm nitrocellulose membranes. The membranes were probed with rabbit anti-Htt 4–19 (1 : 1000, CHDI) and mouse anti-β-actin (1 : 10 000, Sigma). The secondary antibodies used were IRDye 800CW Goat anti-rabbit IgG and IRDye 680RD Goat anti-Mouse IgG (Licor Ltd), respectively. The fluorescent signal was read using the Odyssey imaging system (Li-Cor Biosciences, Nebraska, NE, USA).
FRET assay for caspase-6 activity
The N-terminal 1212 amino acids of Htt with 15Q and the 4c mutations (D513A, D552A, D530A and D589A) (5) were transiently transfected and overexpressed in COS-7 cells. Cells were lysed in SDP buffer (50 mm Tris pH 8, 150 mm NaCl, 1% Igepal and 1× Roche complete protease inhibitor) and cleared lysates were used as a substrate for the activity assay. Caspase-6 enzyme (BioMol GmbH, Hamburg, Germany) was diluted to 0.125 U/µl in FRET buffer (10 mm HEPES pH 7.4, 100 mm NaCl, 0.05% gelatin, 0.1% CHAPS and 2 mm DTT) and mixed with different concentrations of ED11 at a final volume of 22 µl in a white 384-well plate. Samples were incubated for 1 h at room temperature, and then 28 µl of a mixture of COS-7 lysate containing Htt protein (90 ng/µl), Tb-labeled BKP1 antibody (36 ng/ml) and D2-labeled monoclonal 586 neo-epitope antibody (360 ng/ml) (5) in FRET buffer were added. The plate was incubated at 37°C for 6 h, and then stored for an additional 18 h at 4°C. Plates were measured with a Xenon lamp Victor Plate Reader (Perkin Elmer, Waltham, MA, USA) after excitation at 340 nm (time delay 50 ms and window 200 ms). The signal measured at 615 nm resulted from the emission of the Terbium-labeled antibody and was used for normalization of potential signal artifacts. The cleaved Htt-specific signal at 665 nm resulted from the emission of the D2-labeled neo-epitope antibody after time-delayed excitation by the Terbium. Caspase-6 activity was measured by the amount of cleaved Htt generated and represented by the 665/615 nm ratio.
PARP cleavage by caspase-3
HEK293 cells were lysed in MCB buffer (50 mm HEPES pH 7.4, 100 mm NaCl, 1% Igepal, 1 mm EDTA, 10% glycerol, Roche complete protease inhibitors), and DTT was added to 10 mm after the protein concentration was measured in cleared lysates. Caspase-3 enzyme (50 U) was mixed with ED11 or Z-VAD-FMK in MCB buffer supplemented with 10 mm DTT at a total volume of 5 µl and incubated at room temperature for 1 h. Then, 40 µg of HEK lysate (adjusted to 2 µg/µl) were added and the samples were incubated at 37°C for 1 h. SDS–PAGE loading dye was added, samples were denatured and run on a 3–8% Tris-acetate SDS–PAGE gel (Invitrogen, Carlsbad, CA, USA). After transfer to PVDF membranes, the blots were probed with Spectrin (Enzo), PARP (Cell Signaling Technology, Beverly MA, USA) and actin (Millipore, Billerica, MA, USA) antibodies in the Li-Cor system.
Cleavage of Htt by caspases 1–10
COS7 cells were transiently transfected with the 3949-15Q-HTT construct (5) using the Xtreme gene 9 transfection reagent (Roche Applied Science, Quebec, Canada), according to the manufacturer's protocol.
Twenty-four hours post-transfection, cells were harvested and lysed. Fifty units of purified active human caspase-1 through 10 each (Enzo Life Sciences) were mixed with 10 µM ED11 in caspase buffer (100 mm HEPES pH 7.4, 200 mm NaCl, 0.2% CHAPS, 2 mm EDTA, 20% glycerol and 10 mm DTT) and incubated for 30 min at room temperature. About 40 µg of total protein from cell lysates were added, and the reaction was incubated for 1 h at 37°C. A HTT 15Q fragment truncated at amino acid 586 was overexpressed in COS7 cells and used as a size control on the western blot. SDS–PAGE loading dye was added, and samples were denatured and run on a 10% Tris-Glycine SDS–PAGE gel. After transfer to PVDF membranes, the blots were probed with the polyclonal antibody BKP1(29) directed against amino acids 1–17 of Htt. Membranes were scanned and quantified using the Li-Cor system.
TAT antibody staining in MEFs
MEFs derived from FVB/N WT mice (30) were treated with different amounts of ED11 in the media for 16 h. Cells were washed with PBS, fixed with 4% paraformaldehyde in PBS for 30 min at room temperature and processed for immunofluorescence with an anti-TAT antibody (Abcam), Alexa-488-labeled secondary antibody (Invitrogen) and Hoechst nuclear counterstaining.
Htt–caspase-6 co-transfection
COS-7 cells were transiently co-transfected with equal amounts of FLAG-tagged human caspase-6 and the N-terminal 1212 amino acids of Htt with 15Q (5) using the X-treme Gene reagent (Roche, Basel, Switzerland) according to manufacturer's instructions. One hour after transfection, ED11 peptide was added to the growth medium. Cells were harvested in the medium 24 h after transfection and the presence of cleaved Htt was assessed by western blotting using the Htt antibody 2166 (Millipore) or the neo-epitope antibody against Htt cleaved at amino acid 586 (5). Signals were normalized to calnexin as a loading control.
Cell culture
Human neuroblastoma cells, SH-SY5Y cells (ATCC), were grown on tissue culture plates (Greiner, Frickenhausen, Germany) in Dulbecco's Modified Eagle's Medium (DMEM), supplemented with 10% fetal calf serum (FCS), 1% l-glutamine and 1% SPN antibiotics (Biological Industries, Beit Haemek, Israel). Inducible 145Q-MHtt-expressing PC12 cells were obtained from CHDI (by Coriell Institute). Cells were grown in suspension in 75 cm3 culture flasks (Corning, Corning, NY, USA) in DMEM, supplemented with 15% Horse serum, 2.5% FCS, 0.1 mg/ml of G418 (Gibco, Paisley, UK), 0.1 mg/ml of Zeocin (Invitrogen), 1% l-glutamine and 1% SPN antibiotics (Biological Industries). Both cell types were incubated at 37°C in a humidified atmosphere with 5% CO2, and passaged twice a week. For cell proliferation and cell cycle analysis, SH-SY5Y cells were pre-incubated with 25 µM ED11 for 1h. Then, 10 µM 5-bromo-2-deoxyuridine (BrdU) was added for 2 h. The medium was discarded and cells were fixed with 70% ethanol. DNA denaturation was conducted with 1.5 M HCl exposure for 30 min. FITC-conjugated anti-BrdU antibody was used to mark incorporated BrdU, and DNA staining was performed using propidium iodide. A total of 15 000 cells per sample were read using a FACS-Calibur flow cytometer, and cell cycle analysis was conducted after doublet discrimination.
Mutant Huntingtin-induced toxicity
To induce mHtt expression, inducible 145Q-mHtt-expressing PC12 cells were incubated with 25 µM ponasterone A (PA, Invitrogen) for the indicated time periods, in the presence of 25 µM ED11 or vehicle as a control. For viability assessment, the medium was depleted and washed twice with PBS, and Alamar blue dye (AbD serotec, Kidlington, UK, 1/10 in culture medium) was added to the cells, as instructed by the manufacturer. Fluorescence was monitored with a 530–560 nm excitation and a 590 nm emission wavelength, and viability was calculated as a percentage of the untreated control. LDH release was measured with the LDH Cytotoxicity Detection Kit (Clontech, Mountain View, CA, USA), following the manufacturer's instructions. Briefly, PC12 cells were grown on six-well plate at a density of 7 × 105 cells/ml, and samples were taken at the indicated time points. Subsequently, samples were centrifuged, transferred to a 96-well plate and incubated with the reaction mixture for 30 min in the dark. The plate was read at 492 and 690 nm as a reference in the Synergy HT multi-mode micro-plate reader, and calculations were done with reference to a non-induced control. For FLICA caspase-6 assay (ICT, USA), cells were adjusted to 1.5 × 106 cells/ml and were incubated with FAM-VEID-FMK for 1 h at 37°C as described. Hoechst stain was used as nucleus counterstaining. The images were recorded with a fluorescence microscope (Olympus, Tokyo, Japan) and image analysis was performed using the Image-J software.
Animal studies
All animal-related procedures were approved by the Tel-Aviv University Committee of Animal Use for Research and Education, under protocols M-13-024, M-13-061 and M-13-110. BACHD mice were obtained from Jackson Laboratory (Bar Harbor, ME, USA). The mice were placed under 12 h light/dark conditions and housed in individually ventilated cages with ad libitum access to food and water.
Assessment of BBB penetration
To assess ED11's ability to penetrate the BBB and enter the brain parenchyma, a modified in vivo brain delivery detection method was used. A previously reported two photon-based brain delivery detection method (31) was adjusted to an endoscopic microscopy probe-based method (32) to enable deep brain detection. FVB/N mice (n = 2) were injected IV with one of the following compounds: FITC-conjugated albumin, a marker of intact blood vessels (10 mg in 200 µl of saline), FITC-conjugated TAT, as a positive control for extravasation from the blood vessels, FITC-conjugated ED11 lacking the TAT cell-penetrating sequence (50 mg/kg) and FITC-conjugated ED11 (50 mg/kg). Twenty minutes after compound injection, the mice were anesthetized with a mixture of ketamine (100 mg/kg) and xylazine (8 mg/kg). The mice were then placed in a stereotaxic device, and a 0.3 mm fluorescence detection probe was inserted into the central caudate–putamen. Coordinates with respect to the bregma used were: anterior-posterior +0.5, medial-lateral +2, dorsal-ventral −2.7. Forty minutes after compound injection, fluorescence was monitored using a 488 nm excitation laser.
ED11 in vivo efficacy trials
To evaluate ED11's ability to provide protection from mHtt toxicity in vivo, the HD mouse model BACHD (19) and their FVB/N WT littermates were used. Mice were assigned to different groups according to littermates to minimize inherent variability. Mice were housed in mixed genotype and treatment cages, and the researcher was blinded with regard to mouse genotype and treatment throughout all of the behavioral tests. The tested compounds were Vehicle, which was consisted of sterile water (20%) and normal saline (80%) only, or ED11 dissolved in an equal vehicle solution. Delivery of the tested compound was done by a subcutaneously implanted mini-pump alzet model 1004 (DURECT, Cupertino, CA, USA) according to the manufacturer's instructions. The pumps infused continuously for 28 days, at a dose of 4 mg/kg/day. Replacement with freshly prepared pumps was conducted every 28 days until the completion of the experiment.
Rotarod test
Motor coordination and strength were assessed using the accelerating rotarod according to an adapted, previously reported, protocol (19). During the training period, mice were placed on an accelerating rod (0–21 rpm in 4 min), and latency to fall from the rod was recorded. The mice were tested three times per day with a 2-h intertrial rest for 3 consecutive days. For the rotarod test, mice were placed on the accelerating rod (0–21 rpm in 4 min) for three consecutive trials with a 5-min intertrial rest, and average score was taken for analysis.
For emotional and cognitive evaluation, mice were tested in the following behavioral tasks, separated by a minimum of 1-week interval between each task.
Forced swim test
The FST was conducted as previously described (8) with minor modifications. The mice were placed in a 15 cm diameter 40 cm height cylinder, filled with water (23–25°C) to a depth of 20 cm. Animal behavior was recorded by a side camera for 6 min. For mobility analysis, the animal mobility state in the last 4 min was evaluated using the Noldus Ethovision (Wageningen, The Netherlands) video-tracking software
Open field test
For the open field test, an unfamiliar open field (50 × 50 cm) was used. Mice were placed in one corner of the arena and their behavior was recorded for 20 min and scored with the Ethovision video-tracking system. Total distance traveled, total time spent in the center and the number of entries to the center were measured.
Light dark choice test
The light dark choice test was conducted as previously described (33). The arena consisted of two compartments: a dark compartment (14 × 27 × 26 cm) and a compartment illuminated by 1050 lux (30 × 27 × 26 cm), connected by a small passage. Mice were placed in the light compartment to initiate a 5-min test session. The time spent in the dark compartment, the number of entries to the dark compartment and the latency of entering the dark zone were measured. The indices collected in these tests were quantified using the Ethovision video-tracking system.
Swimming T-maze strategy shift test
The swimming T-maze strategy shifting test was conducted as previously described (34). The apparatus consisted of three arms (9 × 50 cm each) filled with water to a depth of 15 cm. On the first 3 days, the mice were subjected to a learning period. In the learning period, a transparent platform was hidden at the end of the right arm. The mouse was placed at the stern of the T-maze four times a day with a 45-min interval between trials. On the fourth day, the strategy shift paradigm was initiated, as the platform was relocated to the left arm, and the mice were placed at the stern four times with a 45-min interval between trials. The time taken for the mice to reach hidden platform was recorded and analyzed using the Ethovision video-tracking system.
MRI protocols and analysis
MRI scanning and analysis was performed by BioImage (Haifa, Israel). MRI was performed with a 7 T MRI scanner (Bruker, Billerica, MA, USA) with a 30 cm bore and a gradient strength of up to 400 mT/m, using a quadrature head coil. The mice were anesthetized with 1–2% isoflurane and oxygen, and were maintained at 37°C. Animal breathing was monitored with a breathing sensor. T2-weighted imaging was performed with the following parameters: multi-slice multi-echo sequence, repetition time 2000 ms, 10 different echo times (ms):10, 20, 30, 40, 50, 60, 70, 80, 90, 100 and spatial resolution: 0.07 × 0.07 × 0.8 mm3. For image analysis, T2-relaxation maps were calculated using BioImage software written in Matlab (Mathworks©), using least-squares algorithm. For volumetric analysis, the images were rotated and cropped, and the regions of interest were outlined manually. A trained operator manually segmented the caudate–putamen, cortex, frontal cortex, motor cortex and hippocampus to evaluate volume changes.
Statistical analysis
Statistical analysis was performed using GraphPad prism version 6.0. Statistical significance for differences between two groups was evaluated by Student's unpaired t-test. When three groups were addressed, statistical evaluation was made by one-way ANOVA followed by Tukey's multiple comparison post hoc test. When addressing time point-dependent alterations, two-way ANOVA with Tukey's multiple comparison post hoc test was used. Data are presented as mean ± SEM, and the level of P < 0.05 was accepted as statistically significant.
Supplementary Material
Funding
This work was supported by the Israel Ministry of Science and Technology, the Colton foundation and Teva Pharmaceutical Industries Ltd under the Israeli National Network of Excellence in Neuroscience (NNE) established by Teva. M.R.H. is a Killam Professor, holds a Canada Research Chair in Human Genetics and received funding from the Canadian Institutes of Health Research (grant MOP 84438).
Supplementary Material
Acknowledgements
We thank Yael Zilberstein from the Sackler Cellular and Molecular Imaging Center at Tel-Aviv University for her dedicated help with the in vivo fluorescence imaging.
Conflict of Interest statement. D.O. and I.A. are registered as co-inventors in the patent filed for the caspase-6 peptide inhibitor ED11. M.R.H. is an employee of Teva Pharmaceuticals. Teva did not have any influence on the design of the study or the analysis and interpretation of data.
References
- 1.Ross C.A., Tabrizi S.J. (2011) Huntington's disease: from molecular pathogenesis to clinical treatment. Lancet Neurol., 10, 83–98. [DOI] [PubMed] [Google Scholar]
- 2.Goldberg Y.P., Nicholson D.W., Rasper D.M., Kalchman M.A., Koide H.B., Graham R.K., Bromm M., Kazemi-Esfarjani P., Thornberry N.A., Vaillancourt J.P., et al. (1996) Cleavage of huntingtin by apopain, a proapoptotic cysteine protease, is modulated by the polyglutamine tract. Nat. Genet., 13, 442–449. [DOI] [PubMed] [Google Scholar]
- 3.Wellington C.L., Ellerby L.M., Gutekunst C.A., Rogers D., Warby S., Graham R.K., Loubser O., van Raamsdonk J., Singaraja R., Yang Y.Z. (2002) Caspase cleavage of mutant huntingtin precedes neurodegeneration in Huntington's disease. J. Neurosci., 22, 7862–7872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Graham R.K., Deng Y., Carroll J., Vaid K., Cowan C., Pouladi M.A., Metzler M., Bissada N., Wang L., Faull R.L.M., et al. (2010) Cleavage at the 586 amino acid caspase-6 site in mutant huntingtin influences caspase-6 activation in vivo. J. Neurosci., 30, 15019–15029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Warby S.C., Doty C.N., Graham R.K., Carroll J.B., Yang Y., Singaraja R.R., Overall C.M., Hayden M.R. (2008) Activated caspase-6 and caspase-6-cleaved fragments of huntingtin specifically colocalize in the nucleus. Hum. Mol. Genet., 17, 2390–2404. [DOI] [PubMed] [Google Scholar]
- 6.Ehrnhoefer D.E., Skotte N.H., Ladha S., Nguyen Y.T.N., Qiu X., Deng Y., Huynh K.T., Engemann S., Nielsen S.M., Becanovic K., et al. (2014) p53 increases caspase-6 expression and activation in muscle tissue expressing mutant huntingtin. Hum. Mol. Genet., 23, 717–729. [DOI] [PubMed] [Google Scholar]
- 7.Graham R.K., Deng Y., Slow E.J., Haigh B., Bissada N., Lu G., Pearson J., Shehadeh J., Bertram L., Murphy Z., et al. (2006) Cleavage at the caspase-6 site is required for neuronal dysfunction and degeneration due to mutant huntingtin. Cell, 125, 1179–1191. [DOI] [PubMed] [Google Scholar]
- 8.Pouladi M.A., Graham R.K., Karasinska J.M., Xie Y., Santos R.D., Petersén Å, Hayden M.R. (2009) Prevention of depressive behaviour in the YAC128 mouse model of Huntington disease by mutation at residue 586 of huntingtin. Brain, 132, 919–932. [DOI] [PubMed] [Google Scholar]
- 9.Tebbenkamp A.T.N., Green C., Xu G., Denovan-Wright E.M., Rising A.C., Fromholt S.E., Brown H.H., Swing D., Mandel R.J., Tessarollo L., et al. (2011) Transgenic mice expressing caspase-6-derived N-terminal fragments of mutant huntingtin develop neurologic abnormalities with predominant cytoplasmic inclusion pathology composed largely of a smaller proteolytic derivative. Hum. Mol. Genet., 20, 2770–2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schwarze S.R., Ho A., Vocero-Akbani A., Dowdy S.F. (1999) In vivo protein transduction: delivery of a biologically active protein into the mouse. Science, 285, 1569–1572. [DOI] [PubMed] [Google Scholar]
- 11.Hällbrink M., Florén A., Elmquist A., Pooga M., Bartfai T., Langel Ü. (2001) Cargo delivery kinetics of cell-penetrating peptides. Biochim. Biophys. Acta, 1515, 101–109. [DOI] [PubMed] [Google Scholar]
- 12.Thorén P.E.G., Persson D., Isakson P., Goksör M., Önfelt A., Nordén B. (2003) Uptake of analogs of penetratin, TAT(48–60) and oligoarginine in live cells. Biochem. Biophys. Res. Commun., 307, 100–107. [DOI] [PubMed] [Google Scholar]
- 13.Chéreau D., Kodandapani L., Tomaselli K.J., Spada A.P., Wu J.C. (2003) Structural and functional analysis of caspase active sites. Biochemistry, 42, 4151–4160. [DOI] [PubMed] [Google Scholar]
- 14.Leyva M.J., DeGiacomo F., Kaltenbach L.S., Holcomb J., Zhang N., Gafni J., Park H., Lo D.C., Salvesen G.S., Ellerby L.M., et al. (2010) Identification and evaluation of small molecule pan-caspase inhibitors in Huntington's disease models. Chem. Biol., 17, 1189–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Le D.A., Wu Y., Huang Z., Matsushita K., Plesnila N., Augustinack J.C., Hyman B.T., Yuan J., Kuida K., Flavell R.A., et al. (2002) Caspase activation and neuroprotection in caspase-3- deficient mice after in vivo cerebral ischemia and in vitro oxygen glucose deprivation. Proc. Natl. Acad. Sci. USA, 99, 15188–15193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wong B.K.Y., Ehrnhoefer D.E., Graham R.K., Martin D.D.O., Ladha S., Uribe V., Stanek L.M., Franciosi S., Qiu X., Deng Y., et al. Partial rescue of some features of Huntington disease in the genetic absence of caspase-6 in YAC128 mice. Neurobiol. Dis., doi:10.1016/j.nbd.2014.12.030. [DOI] [PubMed] [Google Scholar]
- 17.Hult S., Soylu R., Björklund T., Belgardt B., Mauer J., Brüning J., Kirik D., Petersén Å. (2011) Mutant huntingtin causes metabolic imbalance by disruption of hypothalamic neurocircuits. Cell Metab., 13, 428–439. [DOI] [PubMed] [Google Scholar]
- 18.Gafni J., Papanikolaou T., DeGiacomo F., Holcomb J., Chen S., Menalled L., Kudwa A., Fitzpatrick J., Miller S., Ramboz S. (2012) Caspase-6 activity in a BACHD mouse modulates steady-state levels of mutant huntingtin protein but is not necessary for production of a 586 amino acid proteolytic fragment. J. Neurosci., 32, 7454–7465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gray M., Shirasaki D.I., Cepeda C., André V.M., Wilburn B., Lu X., Tao J., Yamazaki I., Li S., Sun Y.E., et al. (2008) Full-length human mutant huntingtin with a stable polyglutamine repeat can elicit progressive and selective neuropathogenesis in BACHD mice. J. Neurosci., 28, 6182–6195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pouladi M.A., Stanek L.M., Xie Y., Franciosi S., Southwell A.L., Deng Y., Butland S., Zhang W., Cheng S.H., Shihabuddin L.S., et al. (2012) Marked differences in neurochemistry and aggregates despite similar behavioural and neuropathological features of Huntington disease in the full-length BACHD and YAC128 mice. Hum. Mol. Genet., 21, 2219–2232. [DOI] [PubMed] [Google Scholar]
- 21.Landles C., Weiss A., Franklin S., Howland D., Bates G. (2012) Caspase-6 does not contribute to the proteolysis of mutant huntingtin in the HdhQ150 knock-in mouse model of Huntington's disease. PLoS Curr., doi:10.1371/4fd085bfc9973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Uribe V., Wong B.K.Y., Graham R.K., Cusack C.L., Skotte N.H., Pouladi M.A., Xie Y., Feinberg K., Ou Y., Ouyang Y., et al. (2012) Rescue from excitotoxicity and axonal degeneration accompanied by age-dependent behavioral and neuroanatomical alterations in caspase-6-deficient mice. Hum. Mol. Genet., 21, 1954–1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nikolaev A., McLaughlin T., O'Leary D.D.M., Tessier-Lavigne M. (2009) APP binds DR6 to trigger axon pruning and neuron death via distinct caspases. Nature, 457, 981–989. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 24.Houde C., Banks K.G., Coulombe N., Rasper D., Grimm E., Roy S., Simpson E.M., Nicholson D.W. (2004) Caspase-7 expanded function and intrinsic expression level underlies strain-specific brain phenotype of caspase-3-null mice. J. Neurosci., 24, 9977–9984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Berger A.B., Sexton K.B., Bogyo M. (2006) Commonly used caspase inhibitors designed based on substrate specificity profiles lack selectivity. Cell Res., 16, 961–963. [DOI] [PubMed] [Google Scholar]
- 26.Chauvier D., Ankri S., Charriaut-Marlangue C., Casimir R., Jacotot E. (2006) Broad-spectrum caspase inhibitors: from myth to reality? Cell Death Differ., 14, 387–391. [DOI] [PubMed] [Google Scholar]
- 27.Pereira N.A., Song Z. (2008) Some commonly used caspase substrates and inhibitors lack the specificity required to monitor individual caspase activity. Biochem. Biophys. Res. Commun., 377, 873–877. [DOI] [PubMed] [Google Scholar]
- 28.Carroll J., Southwell A., Graham R., Lerch J., Ehrnhoefer D., Cao L., Zhang W., Deng Y., Bissada N., Henkelman R., et al. (2011) Mice lacking caspase-2 are protected from behavioral changes, but not pathology, in the YAC128 model of Huntington disease. Mol. Neurodegener., 6, 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kalchman M.A., Koide H.B., McCutcheon K., Graham R.K., Nichol K., Nishiyama K., Kazemi-Esfarjani P., Lynn F.C., Wellington C., Metzler M., et al. (1997) HIP1, a human homologue of S. cerevisiae Sla2p, interacts with membrane-associated huntingtin in the brain. Nat. Genet., 16, 44–53. [DOI] [PubMed] [Google Scholar]
- 30.Ehrnhoefer D.E., Skotte N.H., Savill J., Nguyen Y.T., Ladha S., Cao L.P., Dullaghan E., Hayden M.R. (2011) A quantitative method for the specific assessment of caspase-6 activity in cell culture. PLoS ONE, 6, e27680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koffie R.M., Farrar C.T., Saidi L., William C.M., Hyman B.T., Spires-Jones T.L. (2011) Nanoparticles enhance brain delivery of blood–brain barrier-impermeable probes for in vivo optical and magnetic resonance imaging. Proc. Natl. Acad. Sci. USA, 108, 18837–18842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jung J.C., Mehta A.D., Aksay E., Stepnoski R., Schnitzer M.J. (2004) In vivo mammalian brain imaging using one- and two-photon fluorescence microendoscopy. J. Neurophysiol., 92, 3121–3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bourin M., Hascoët M. (2003) The mouse light/dark box test. Eur. J. Pharmacol., 463, 55–65. [DOI] [PubMed] [Google Scholar]
- 34.Marco S., Giralt A., Petrovic M.M., Pouladi M.A., Martinez-Turrillas R., Martinez-Hernandez J., Kaltenbach L.S., Torres-Peraza J., Graham R.K., Watanabe M., et al. (2013) Suppressing aberrant GluN3A expression rescues synaptic and behavioral impairments in Huntington's disease models. Nat. Med., 19, 1030–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.