Abstract
The Universal PBM Resource for Oligonucleotide Binding Evaluation (UniPROBE) serves as a convenient source of information on published data generated using universal protein-binding microarray (PBM) technology, which provides in vitro data about the relative DNA-binding preferences of transcription factors for all possible sequence variants of a length k (‘k-mers’). The database displays important information about the proteins and displays their DNA-binding specificity data in terms of k-mers, position weight matrices and graphical sequence logos. This update to the database documents the growth of UniPROBE since the last update 4 years ago, and introduces a variety of new features and tools, including a new streamlined pipeline that facilitates data deposition by universal PBM data generators in the research community, a tool that generates putative nonbinding (i.e. negative control) DNA sequences for one or more proteins and novel motifs obtained by analyzing the PBM data using the BEEML-PBM algorithm for motif inference. The UniPROBE database is available at http://uniprobe.org.
INTRODUCTION
Characterizing and predicting transcription factor (TF) DNA-binding specificities are crucial tasks for understanding the functioning of cellular regulatory networks. The particular binding affinities of a TF govern its set of target genes and thus play an important role in cellular functions and differentiation. The development of universal protein-binding microarray (PBM) technology (1) has allowed for comprehensive high-resolution profiling of the DNA-binding specificity of a given TF by evaluating its binding affinity for all possible k-mer DNA sequences. The Universal PBM Resource for Oligonucleotide Binding Evaluation (UniPROBE) database (2) was created to provide appropriate curation, easy searching and an informative display interface for universal PBM data.
An update to UniPROBE was published in 2011 (3). Since that time, many new features have been added to the web interface. In addition, numerous data sets have been deposited into UniPROBE. Here, we discuss these data and features, which include a new data deposition pipeline, a negative control sequence generation tool and motifs derived using BEEML-PBM (4).
DATABASE ADDITIONS
Table 1 describes 12 new publications whose PBM data sets have been introduced into UniPROBE since the last update (5–16). The 96 TFs from these publications come from 19 highly diverse species, many of which are new to the database. At the time of this manuscript's preparation, UniPROBE hosts 515 non-redundant proteins and complexes. A number of additional data depositions are planned for the near future: e.g. Nowak-Lovato et al., 2012 (17); Weirauch et al., 2013 (18); Siggers et al., 2014 (19); Lindemose et al., 2014 (20); Oberstaller et al., 2014 (21). We anticipate that most future depositions will likely be performed by the authors themselves using our new data deposition pipeline.
Table 1. New PBM data sets added into UniPROBE.
| Reference | Number of proteins or complexes | Species |
|---|---|---|
| Alibés et al. (5) | 2 | Homo sapiens, Saccharomyces cerevisiae |
| Campbell et al. (6) | 19 | Plasmodium falciparum |
| Gordân et al. (7) | 27 | S. cerevisiae |
| Del Bianco et al. (8) | 9 | H. sapiens |
| Cheatle Jarvela et al. (9) | 2 | Patiria miniata, Strongylocentrotus purpuratus |
| Busser et al. (Development) (10) | 10 | Drosophila melanogaster |
| Nakagawa et al. (11) | 20 | Acanthamoeba castellanii, Allomyces macrogynus, Ashbya gossypii, Aspergillus nidulans, D. melanogaster, H. sapiens, Kluyveromyces lactis, Monosiga brevicollis, Mus musculus, Mycosphaerella graminicola, Nematostella vectensis, S. purpuratus, Trichoplax adhaerens, Tuber melanosporum |
| Soruco et al. (12) | 1 | D. melanogaster |
| Busser et al. (PNAS) (13) | 1 | D. melanogaster |
| Peterson et al. (14) | 3 | M. musculus |
| De Masi et al. (15) | 1 | Caenorhabditis elegans |
| Helfer et al. (16) | 1 | Arabidopsis thaliana |
| Total number of new proteins/complexes: | 96 | |
| Total, last described (3): | 404 | |
| Total number of non-redundant proteins/complexes in UniPROBE: | 515 | |
DATA DEPOSITION PIPELINE
Among the most significant features recently added to UniPROBE is a web-based pipeline for deposition of new PBM data sets. The link for this tool is found conveniently in a header near the top of the front page or by accessing it directly by URL at http://thebrain.bwh.harvard.edu/pbms/webworks_pub_dev/admin.php. Previously, uploading data manually into the MySQL database was inefficient and error-prone; therefore, we designed several linked scripts to automate the process.
Figure 1A shows the main page for this pipeline, which also outlines the control flow of the deposition for users. In the first five steps, the user can input information into the database concerning the proteins involved in their study. While the most convenient way to do this is by preparing an appropriately formatted spreadsheet file (for steps 2, 4 and 5; see Figure 1B), alternatively the input can be done one entry at a time using an HTML form if a user prefers that method. Currently, the user must prepare a folder with all of the data files they wish to make public. Instructions for data file preparation are given (and are also provided in Supplementary Text 1), and several helpful scripts are available for download to aid the process. The user then uploads the folder to the UniPROBE server as a zip file. The remaining steps fully integrate the data files into the web interface, including constructing sequence logos for each protein and making all the data easily searchable and available for download. The UniPROBE administrator will then finalize the deposition by ensuring proper insertion and moving the new data into the public version of the web site. Data depositors may contact the UniPROBE administrator to specify a release date for prepublication data submissions.
Figure 1.
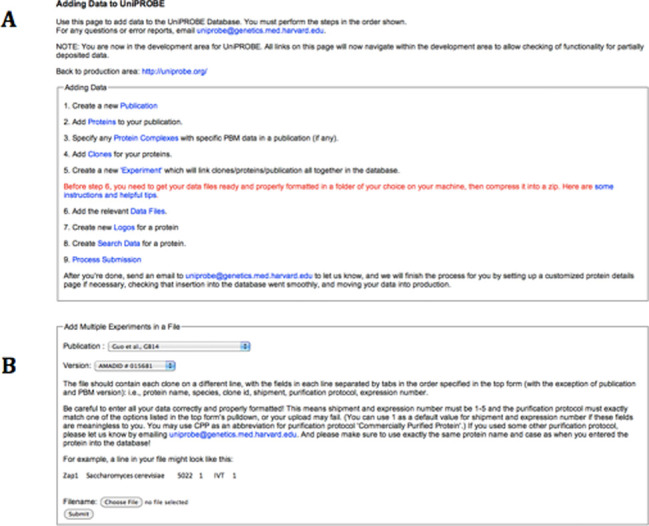
Data deposition pipeline. (A) The main page for the UniPROBE data deposition pipeline provides an outline of the data deposition procedure. The user successively clicks each link and follows the instructions in each step. Some steps require only the click of a button, whereas others require either submission of an input file or some extra actions on the command line. (B) The instructions for file-based input in step 5. Steps 2 and 4 have similar instructions. File-based input makes it easy for the user to simultaneously provide all the relevant information to add to the database, and has formatting, error checking and rollback functionality built in.
INCORPORATION OF BEEML-PBM MOTIFS
All of the raw PBM data posted in UniPROBE until recently have been handled in the same manner: the Seed-and-Wobble algorithm, introduced jointly with universal PBM technology (1,22), is used to generate a position weight matrix (PWM) (23,24), which in turn is used to generate sequence logos (25) that are displayed on the protein's Details page (e.g. see Figure 2A). Since the development of universal PBM technology, other algorithms have been developed to derive PWMs from the PBM data. BEEML-PBM employs a maximum likelihood approach, using a weighted nonlinear least-squares regression to infer free energy parameters for TF–DNA interactions (4). BEEML-PBM was one of the top two algorithms in the DREAM5 challenge (18) and provided PWMs with better performance than Seed-and-Wobble for the majority of TFs. We have generated PWMs using BEEML-PBM for the PBM data from all publications whose data have been incorporated into UniPROBE, including those mentioned in this paper (1,5–16,26–32). The free energy parameters derived from BEEML-PBM were converted into PWM frequencies by applying a Boltzmann distribution probability mass function to each matrix column. Figure 2 shows an example of Seed-and-Wobble and BEEML-PBM logos in UniPROBE. All of the new logos are currently viewable on the appropriate protein pages and the PWMs are available for download either individually on these pages or in bulk on the Downloads page.
Figure 2.
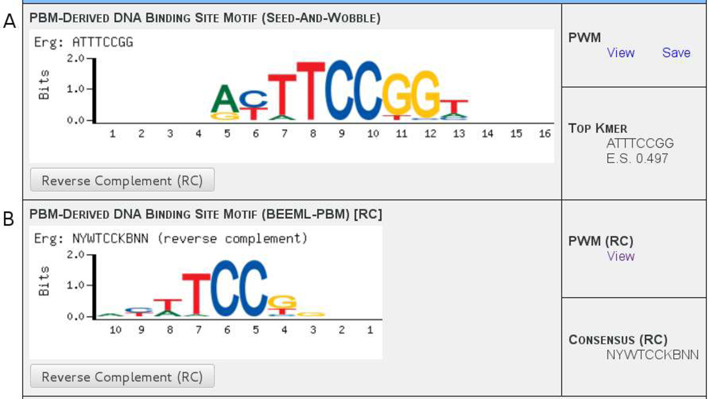
Seed-and-Wobble and BEEML-PBM motif displays. Examples of displays for data generated using the (A) Seed-and-Wobble and (B) BEEML-PBM algorithms for the Erg protein, from Wei et al., 2010 (31). (A) The Seed-and-Wobble data displays a sequence logo, links for downloading the PWM data and the top-scoring k-mer along with its PBM enrichment score. (B) The BEEML-PBM data display format is essentially the same, but because k-mers and enrichment scores are not utilized in this algorithm, an IUPAC consensus sequence derived from the PWM is instead displayed above the motif. The reverse complement sequence orientation can be displayed for either data set individually by clicking the appropriate button; this changes the logo, the PWM file link and the displayed sequence. Assignment of ‘forward’ versus ‘reverse complement’ orientation is arbitrary for each PWM—here, the BEEML-PBM data have been switched to ‘reverse complement’ mode in order to display a more obvious comparison between the logos, since its ‘forward’ orientation happens to correspond more closely to the Seed-and-Wobble data's ‘reverse complement’ orientation.
NEGATIVE CONTROL SEQUENCE GENERATOR
UniPROBE's main ‘toolbox’, found on the front and Browse pages, includes: a basic text search with different options; a tool that finds proteins with a sufficiently close match to a query DNA motif; a tool that scans a DNA sequence for putative TF-binding sites (2); and a blastp search tool for matching protein sequences (3). In addition to predicting specific protein–DNA interactions, it is sometimes desirable to find a sequence that is predicted not to be bound by a given protein(s); e.g. when designing negative controls for in vivo reporter experiments or nonspecific competitor DNA for in vitro assays. An important new addition to this toolbox is a negative control (nonbinding) sequence generator for such purposes; the search interface for this tool is displayed in Figure 3A. This tool takes a list of proteins stored in UniPROBE as input along with a few parameters (PBM k-mer enrichment score threshold for TF binding and minimum and maximum length cutoffs) for the desired sequence to be generated. The output is a DNA sequence which is predicted to have little to no specific binding by any of the proteins selected as input based on the PBM data available for that protein in UniPROBE.
Figure 3.
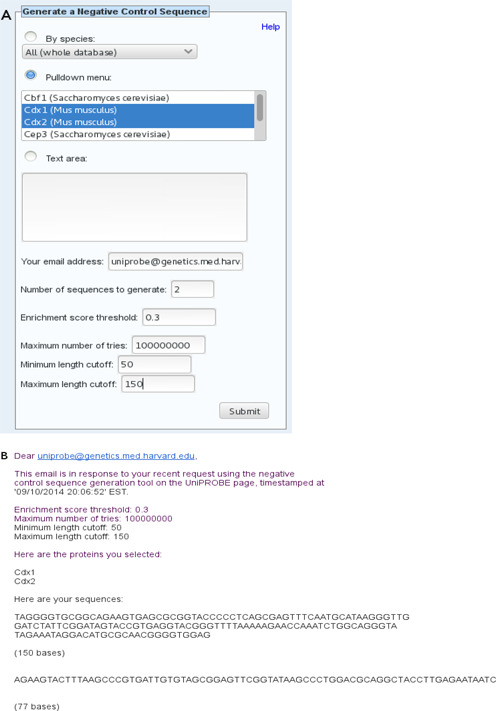
Examples of input and output from the Negative Control Sequence Generator tool. (A) Form for the Negative Control Sequence Generator tool. In this example, the user has selected two proteins using the pulldown menu, but alternatively, the user can select all proteins in the database from a given species or enter the proteins he/she wants into the text area. The user has requested two sequences between 50 and 150 bp in length. The enrichment score threshold and ‘maximum number of tries’ parameter values used here are the defaults. Clicking on the ‘Help’ link in this box on the web page provides more information about the various parameters. (B) The text of an email reply containing the results from the Negative Control Sequence Generator tool for the input shown in (A).
Briefly, the algorithm works as follows. First, it assembles a list of all contiguous 8-mers such that every selected protein has scored below the enrichment score threshold for binding to that 8-mer in every PBM data set for that protein. Then, it generates putative nonbinding DNA sequences by randomly concatenating suitable k-mers such that no disallowed 8-mer—i.e. no 8-mer not in the input list—will appear at any point in the sequence. This is ensured by the construction and use of a mapping in which every 7-mer corresponds to a list of the bases allowed to directly follow it in the next sequential nucleotide. During each addition to the sequence, the next nucleotide added is selected from this list to ensure that no disallowed 8-mer is created. Note that since the addition of k-mers is performed randomly, this algorithm is non-deterministic; thus, the user can also specify the number of sequences to be generated. The results are emailed to the user once the computation has finished; an example is provided in Figure 3B.
OTHER NEW FEATURES
The blastp search feature introduced in the last published update (3) has been further improved by adding a visualization of the alignment between the query and result sequences within the search results.
Links to the TFBSshape database (33) have been included in the Details pages of proteins with available TFBSshape data. TFBSshape describes the structural features of DNA at TF binding sites, and has entries for proteins corresponding to entries in JASPAR (34,35) and UniPROBE. Figure 4 shows an example of a link and its corresponding TFBSshape web page. Publications with data in UniPROBE whose protein pages currently link to TFBSshape (and vice versa) are: Berger et al., 2006 (1); Berger et al., 2008 (26); Zhu et al., 2009 (27); Badis et al., 2009 (28); Lesch et al., 2009 (30); Scharer et al., 2009 (32). We will continue to correspond with the TFBSshape administrators and provide links for additional publications as they become available in the TFBSshape database.
Figure 4.
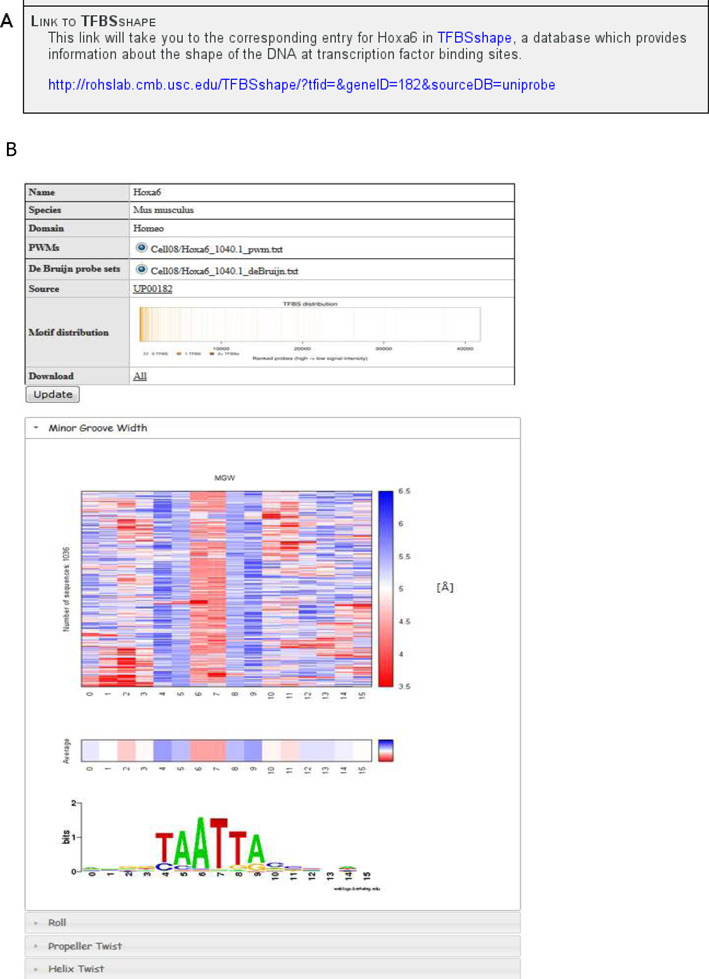
TFBSshape links. (A) An example of a link to the TFBSshape database from the Protein Details page for Hoxa6, from Berger et al., 2008 (26). (B) The TFBSshape page for Hoxa6, to which the link in (A) leads.
Finally, migration to a new, faster server has been completed, and we expect a concomitant speedup in web operation times.
DISCUSSION
There are many opportunities for further improvements to UniPROBE in the near future. To start, there are additional published PBM data sets still awaiting deposition into the database. To expedite data deposition, we encourage authors of such studies to submit their data themselves into UniPROBE using our new data deposition pipeline.
Additional improvements could be made to the data deposition pipeline. Currently, the pipeline does not account explicitly for variations in the structure of the data files available for download from each publication; for example each protein from a publication may have different sets of PBM data reflecting distinct binding activity for different clones, protein complexes of which a particular protein is a component or data from replicate PBM experiments. In some cases, data are available from experiments using different PBM array versions (which may themselves have multiple replicates).
Similarly, the structure of the Protein Details page must properly match the file structure in order to optimally display the data. The default template for the Details page has not yet been configured to handle the amount of potential variability in the file structure, and currently a new template page must be generated by the database administrator for any publication newly deposited into UniPROBE that does not have strictly one set of files per protein, without any complexes. In the future, we hope to automate this process by creating one or more pre-written page templates that can handle variation in data file structure. Users should still be able to request customization of their publication's Details pages if necessary.
Further planned improvements to the deposition process include the ability to request specific UniPROBE accession numbers for proteins (see Robasky and Bulyk, 2011 (3) for a description of protein accession numbers in UniPROBE). We also plan to generate accession numbers to publications for reference and to allow users to specify particular publication data sets for searches.
PWM data derived using other motif finding algorithms in addition to Seed-and-Wobble and BEEML-PBM will also be added. Among those on which we may choose to focus initially are FeatureREDUCE (manuscript in preparation) and MatrixREDUCE (36), which also performed well in the DREAM5 challenge (18). BEEML-PBM data will also be generated for the remaining publications that have been deposited in UniPROBE.
Finally, users will also soon be able to do a bulk download as a FASTA file of the protein sequences of all the TF clones used in the PBM experiments.
We welcome feedback and suggestions for further improvements from our users. A new UniPROBE administrative email account can now be reached with any questions, comments or suggestions at uniprobe@genetics.med.harvard.edu.
AVAILABILITY
As before, the data in UniPROBE are freely available at the database web site (http://uniprobe.org), and the sequences of the 60-mer DNA probes on the custom-designed oligonucleotide arrays are available under the terms of an academic research use license available at http://thebrain.bwh.harvard.edu/uniprobe/academic-license.php.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
We thank Ivan Adzhubey for technical assistance, Chirag Parmar for work on documentation and Kimberly Robasky for helpful discussions.
FUNDING
National Institutes of Health [R01 HG003985 to M.L.B.]; National Science Foundation Graduate Research Fellowship [to L.A.B.]. Funding for open access charge: National Institutes of Health [R01 HG003985 to M.L.B.].
Conflict of interest statement. None declared.
REFERENCES
- 1.Berger M.F., Philippakis A.A., Qureshi A.M., He F.S., Estep P.W., 3rd, Bulyk M.L. Compact, universal DNA microarrays to comprehensively determine transcription-factor binding site specificities. Nat. Biotechnol. 2006;24:1429–1435. doi: 10.1038/nbt1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Newburger D.E., Bulyk M.L. UniPROBE: an online database of protein binding microarray data on protein–DNA interactions. Nucleic Acids Res. 2009;37:D77–D82. doi: 10.1093/nar/gkn660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Robasky K., Bulyk M.L. UniPROBE, update 2011: expanded content and search tools in the online database of protein-binding microarray data on protein–DNA interactions. Nucleic Acids Res. 2011;39:D124–D128. doi: 10.1093/nar/gkq992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhao Y., Stormo G.D. Quantitative analysis demonstrates most transcription factors require only simple models of specificity. Nat. Biotechnol. 2011;29:480–483. doi: 10.1038/nbt.1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alibés A., Nadra A.D., De Masi F., Bulyk M.L., Serrano L., Stricher F. Using protein design algorithms to understand the molecular basis of disease caused by protein–DNA interactions: the Pax6 example. Nucleic Acids Res. 2010;38:7422–7431. doi: 10.1093/nar/gkq683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campbell T.L., De Silva E.K., Olszewski K.L., Elemento O., Llinás M. Identification and genome-wide prediction of DNA binding specificities for the ApiAP2 family of regulators from the malaria parasite. PLoS Pathog. 2010;6:e1001165. doi: 10.1371/journal.ppat.1001165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gordân R., Murphy K.F., McCord R.P., Zhu C., Vedenko A., Bulyk M.L. Curated collection of yeast transcription factor DNA binding specificity data reveals novel structural and gene regulatory insights. Genome Biol. 2011;12:R125. doi: 10.1186/gb-2011-12-12-r125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Del Bianco C., Vedenko A., Choi S.H., Berger M.F., Shokri L., Bulyk M.L., Blacklow S.C. Notch and MAML-1 complexation do not detectably alter the DNA binding specificity of the transcription factor CSL. PLoS One. 2010;5:e15034. doi: 10.1371/journal.pone.0015034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheatle Jarvela A.M., Brubaker L., Vedenko A., Gupta A., Armitage B.A., Bulyk M.L., Hinman V.F. Modular evolution of DNA-binding preference of a Tbrain transcription factor provides a mechanism for modifying gene regulatory networks. Mol. Biol. Evol. 2014;31:2672–2688. doi: 10.1093/molbev/msu213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Busser B.W., Shokri L., Jaeger S.A., Gisselbrecht S.S., Singhania A., Berger M.F., Zhou B., Bulyk M.L., Michelson A.M. Molecular mechanism underlying the regulatory specificity of a Drosophila homeodomain protein that specifies myoblast identity. Development. 2012;139:1164–1174. doi: 10.1242/dev.077362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakagawa S., Gisselbrecht S.S., Rogers J.M., Hartl D.L., Bulyk M.L. DNA-binding specificity changes in the evolution of forkhead transcription factors. Proc. Natl. Acad. Sci. U.S.A. 2013;110:12349–12354. doi: 10.1073/pnas.1310430110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Soruco M.M., Chery J., Bishop E.P., Siggers T., Tolstorukov M.Y., Leydon A.R., Sugden A.U., Goebel K., Feng J., Xia P., et al. The CLAMP protein links the MSL complex to the X chromosome during Drosophila dosage compensation. Genes Dev. 2013;27:1551–1556. doi: 10.1101/gad.214585.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Busser B.W., Huang D., Rogacki K.R., Lane E.A., Shokri L., Ni T., Gamble C.E., Gisselbrecht S.S., Zhu J., Bulyk M.L., et al. Integrative analysis of the zinc finger transcription factor Lame duck in the Drosophila myogenic gene regulatory network. Proc. Natl. Acad. Sci. U.S.A. 2012;109:20768–20773. doi: 10.1073/pnas.1210415109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peterson K.A., Nishi Y., Ma W., Vedenko A., Shokri L., Zhang X., McFarlane M., Baizabal J.M., Junker J.P., van Oudenaarden A., et al. Neural-specific Sox2 input and differential Gli-binding affinity provide context and positional information in Shh-directed neural patterning. Genes Dev. 2012;26:2802–2816. doi: 10.1101/gad.207142.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Masi F., Grove C.A., Vedenko A., Alibés A., Gisselbrecht S.S., Serrano L., Bulyk M.L., Walhout A.J. Using a structural and logics systems approach to infer bHLH-DNA binding specificity determinants. Nucleic Acids Res. 2011;39:4553–4563. doi: 10.1093/nar/gkr070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Helfer A., Nusinow D.A., Chow B.Y., Gehrke A.R., Bulyk M.L., Kay S.A. LUX ARRHYTHMO encodes a nighttime repressor of circadian gene expression in the Arabidopsis core clock. Curr. Biol. 2011;21:126–133. doi: 10.1016/j.cub.2010.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nowak-Lovato K.L., Hickmott A.J., Maity T.S., Bulyk M.L., Dunbar J., Hong-Geller E. DNA binding site analysis of Burkholderia thailandensis response regulators. J. Microbiol. Methods. 2012;90:46–52. doi: 10.1016/j.mimet.2012.03.019. [DOI] [PubMed] [Google Scholar]
- 18.Weirauch M.T., Cote A., Norel R., Annala M., Zhao Y., Riley T.R., Saez-Rodriguez J., Cokelaer T., Vedenko A., Talukder S., et al. Evaluation of methods for modeling transcription factor sequence specificity. Nat. Biotechnol. 2013;31:126–134. doi: 10.1038/nbt.2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Siggers T., Reddy J., Barron B., Bulyk M.L. Diversification of transcription factor paralogs via noncanonical modularity in C2H2 zinc finger DNA binding. Mol. Cell. 2014;55:640–648. doi: 10.1016/j.molcel.2014.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lindemose S., Jensen M.K., de Velde J.V., O'Shea C., Heyndrickx K.S., Workman C.T., Vandepoele K., Skriver K., De Masi F. A DNA-binding-site landscape and regulatory network analysis for NAC transcription factors in Arabidopsis thaliana. Nucleic Acids Res. 2014;42:7681–7693. doi: 10.1093/nar/gku502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oberstaller J., Pumpalova Y., Schieler A., Llinás M., Kissinger J.C. The Cryptosporidium parvum ApiAP2 gene family: insights into the evolution of apicomplexan AP2 regulatory systems. Nucleic Acids Res. 2014;42:8271–8284. doi: 10.1093/nar/gku500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berger M.F., Bulyk M.L. Universal protein-binding microarrays for the comprehensive characterization of the DNA-binding specificities of transcription factors. Nat. Protoc. 2009;4:393–411. doi: 10.1038/nprot.2008.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stormo G.D., Schneider T.D., Gold L., Ehrenfeucht A. Use of the ‘Perceptron’ algorithm to distinguish translational initiation sites in E. coli. Nucleic Acids Res. 1982;10:2997–3011. doi: 10.1093/nar/10.9.2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stormo G.D. DNA binding sites: representation and discovery. Bioinformatics. 2000;16:16–23. doi: 10.1093/bioinformatics/16.1.16. [DOI] [PubMed] [Google Scholar]
- 25.Schneider T.D., Stephens R.M. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berger M.F., Badis G., Gehrke A.R., Talukder S., Philippakis A.A., Pena-Castillo L., Alleyne T.M., Mnaimneh S., Botvinnik O.B., Chan E.T., et al. Variation in homeodomain DNA binding revealed by high-resolution analysis of sequence preferences. Cell. 2008;133:1266–1276. doi: 10.1016/j.cell.2008.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu C., Byers K.J., McCord R.P., Shi Z., Berger M.F., Newburger D.E., Saulrieta K., Smith Z., Shah M.V., Radhakrishnan M., et al. High-resolution DNA-binding specificity analysis of yeast transcription factors. Genome Res. 2009;19:556–566. doi: 10.1101/gr.090233.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Badis G., Berger M.F., Philippakis A.A., Talukder S., Gehrke A.R., Jaeger S.A., Chan E.T., Metzler G., Vedenko A., Chen X., et al. Diversity and complexity in DNA recognition by transcription factors. Science. 2009;324:1720–1723. doi: 10.1126/science.1162327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grove C.A., De Masi F., Barrasa M.I., Newburger D.E., Alkema M.J., Bulyk M.L., Walhout A.J. A multiparameter network reveals extensive divergence between C. elegans bHLH transcription factors. Cell. 2009;138:314–327. doi: 10.1016/j.cell.2009.04.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lesch B.J., Gehrke A.R., Bulyk M.L., Bargmann C.I. Transcriptional regulation and stabilization of left–right neuronal identity in C. elegans. Genes Dev. 2009;23:345–358. doi: 10.1101/gad.1763509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wei G.H., Badis G., Berger M.F., Kivioja T., Palin K., Enge M., Bonke M., Jolma A., Varjosalo M., Gehrke A.R., et al. Genome-wide analysis of ETS-family DNA-binding in vitro and in vivo. EMBO J. 2010;29:2147–2160. doi: 10.1038/emboj.2010.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scharer C.D., McCabe C.D., Ali-Seyed M., Berger M.F., Bulyk M.L., Moreno C.S. Genome-wide promoter analysis of the SOX4 transcriptional network in prostate cancer cells. Cancer Res. 2009;69:709–717. doi: 10.1158/0008-5472.CAN-08-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang L., Zhou T., Dror I., Mathelier A., Wasserman W.W., Gordân R., Rohs R. TFBSshape: a motif database for DNA shape features of transcription factor binding sites. Nucleic Acids Res. 2014;42:D148–D155. doi: 10.1093/nar/gkt1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sandelin A., Alkema W., Engstrom P., Wasserman W.W., Lenhard B. JASPAR: an open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res. 2004;32:D91–D94. doi: 10.1093/nar/gkh012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mathelier A., Zhao X., Zhang A.W., Parcy F., Worsley-Hunt R., Arenillas D.J., Buchman S., Chen C.Y., Chou A., Ienasescu H., et al. JASPAR 2014: an extensively expanded and updated open-access database of transcription factor binding profiles. Nucleic Acids Res. 2014;42:D142–D147. doi: 10.1093/nar/gkt997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Foat B.C., Morozov A.V., Bussemaker H.J. Statistical mechanical modeling of genome-wide transcription factor occupancy data by MatrixREDUCE. Bioinformatics. 2006;22:e141–e149. doi: 10.1093/bioinformatics/btl223. [DOI] [PubMed] [Google Scholar]


