Abstract
The iBeetle-Base (http://ibeetle-base.uni-goettingen.de) makes available annotations of RNAi phenotypes, which were gathered in a large scale RNAi screen in the red flour beetle Tribolium castaneum (iBeetle screen). In addition, it provides access to sequence information and links for all Tribolium castaneum genes. The iBeetle-Base contains the annotations of phenotypes of several thousands of genes knocked down during embryonic and metamorphic epidermis and muscle development in addition to phenotypes linked to oogenesis and stink gland biology. The phenotypes are described according to the EQM (entity, quality, modifier) system using controlled vocabularies and the Tribolium morphological ontology (TrOn). Furthermore, images linked to the respective annotations are provided. The data are searchable either for specific phenotypes using a complex ‘search for morphological defects’ or a ‘quick search’ for gene names and IDs. The red flour beetle Tribolium castaneum has become an important model system for insect functional genetics and is a representative of the most species rich taxon, the Coleoptera, which comprise several devastating pests. It is used for studying insect typical development, the evolution of development and for research on metabolism and pest control. Besides Drosophila, Tribolium is the first insect model organism where large scale unbiased screens have been performed.
INTRODUCTION
Next generation sequencing has been used for the identification of gene sequences in a plethora of insect species. Hence, our view on the gene complements present in insects is becoming ever more comprehensive. However, our knowledge on the function of these genes lags far behind because functional studies have long been restricted to organisms with a highly sophisticated genetic tool kit and a short generation time. These are the prerequisites for forward genetic screens, which have been used to identify gene function by large scale mutagenesis. So far, within insects, only the fruit fly Drosophila melanogaster has offered the possibility of saturated forward genetic screens and, hence, most of what we know about insect gene functions is derived from this species (1,2).
Recently, novel model species have become amenable to reverse genetic screens. Specifically, RNA interference (RNAi) has been widely used in a broad range of insects for knocking down gene function by injecting respective double stranded RNA (dsRNA) (3–6). So far, research has been focusing on genes, which were likely to be involved in a certain process based on Drosophila data like studies of gap gene (7–9) or pair rule gene orthologs (10) or based on vertebrates like studies on the Wnt pathway or head development (11–13). This candidate gene approach has been very fruitful but is biased towards highly conserved gene functions.
The iBeetle-Screen has been performed in order to overcome this bias and to identify unexpected gene functions. In this large scale RNAi screen, more than approximately one-third of the 16 505 genes were picked at random for knock-down allowing the detection of unexpected gene functions. The red flour beetle Tribolium castaneum has been selected for this endeavor because it has a strong and systemic RNAi response, which is transferred from the mother to its offspring (14). Further, in many respects Tribolium shows a more insect typical development than the fruit fly Drosophila. This includes axis formation, segmentation in an elongating germ band, head and leg development and oogenesis (15,16). Also metamorphosis reflects a more insect typical situation in that most larval cells are re-used for the adult epidermis instead of being replaced by imaginal cells (17). Furthermore, Tribolium is used as model for cuticle synthesis (18), hormonal control (19) and for as yet unstudied processes like stink gland biology (20).
The phenotypes listed in the iBeetle-Base will help scientists in the Tribolium community to identify novel genes involved in the process under study. For scientists from other fields, it will be a complementary source of information for putative functions of a given gene. While iBeetle-Base makes available gene-related information (like RNAi phenotypes, sequences and orthologs), the BeetleBase (hosted at Kansas State University, USA) (21) offers access to extensive genomic data with respect to Tribolium castaneum and related beetles. Together, these interlinked databases will foster research in Tribolium and other insects.
DATA SOURCE AND ANNOTATION
dsRNAs targeting several thousand genes were injected into female pupae and the resulting RNAi phenotypes were scored. This included phenotypes of the injected animal like morphological phenotypes arising during metamorphosis or sterility. The offspring of the injected animals was scored for defects in the muscles and the first instar larval cuticle. In parallel, the same dsRNAs were injected into 5th/6th instar larvae and resulting cuticle and muscle phenotypes arising during metamorphosis were annotated along with other aspects of metamorphic development.
The iBeetle Screen was designed as a first pass screen, i.e. the experiments were not repeated and the annotation policy was to avoid false negatives with the trade-off of elevated false positive annotations.
Phenotype annotation using the EQM system
For the annotation of phenotypic changes the EQM (entity, quality, modifier) system (22–24) was previously developed and implemented in several phenotypic databases (25,26). This system defines the use of controlled vocabularies for the affected entity E (e.g. leg), the quality of the change Q (e.g. number) and the nature of the change using a modifier M (e.g. increased). A larva in which more legs have developed after RNAi treatment was annotated with ‘larval_leg; number; increased’. Adhering to the EQM system has several advantages: First, it facilitates a systematic annotation of phenotypes. Second, it allows reading the descriptions of the phenotypes in phrases, which are well readable by both humans and machines. Third, the use of the same system in different phenotypic databases will facilitate data interconnection (23).
Structuring morphological information by the anatomical ontology TrOn
The Tribolium ontology (TrOn) (27) defines unique terms for the morphological entities of Tribolium castaneum (e.g. prothoracic_leg for the legs on the first thoracic segment). An ontology class comprises one such term together with its definition and is_a as well as part_of relations to other classes. TrOn covers all structures that were scored in the iBeetle screen. The annotation policy for the iBeetle screen was to annotate the most specifically affected (sub-) structure while the relations annotated in TrOn allow finding these phenotypes in searches using more general terms. For instance, an annotated defect in the specific structure ‘pretarsus’ (the most distal part of the larval leg) is found in a general search for ‘leg’ because TrOn stores the information that pretarsus along with coxa, tibia, tarsus, claw and other structures constitute a leg.
In TrOn all anatomical structures are modeled independently of a specific developmental stage (e.g. ‘leg’, which actually exists in different forms in embryos, larvae and adults). This category is called the generic subset and reflects the abstract anatomical concept of a certain structure. As such, it comprises many diverse realizations of that concept at different developmental stages (e.g. larva versus adult) or locations in different segments (e.g. fore- and hindwings) within one taxon or even in different species. A second category of ontology classes represents the visible occurrence of the respective structure at specific developmental stages (e.g. ‘larval_prothoracic_leg’ or ‘adult_leg’). The classes of this category, the concrete subset, reflect an anatomical entity at a specific developmental stage, i.e. a unique dissectible structure. The concrete subset comprises child terms of the generic classes. For example, the anatomical concept wing is expressed in TrOn with the generic class ‘wing’. Its children comprise concrete classes like ‘adult_wing’ or ‘pupal_wing’. Within the iBeetle-Base the annotated phenotypes are always linked to concrete classes while the generic classes can be used in the search.
The OBA service (ontology based answers) (28) was used to process the ontology and to integrate it in iBeetle-Base. The service was used to assign the ontology classes to the right subset and to transiently add the inverse relation has_part to the annotated part_of relation. During the search it is the task of the OBA service to retrieve the relevant downstream substructures (concrete classes) of the morphological structure the user selected (generic class).
THE WEB INTERFACE
Search for phenotypic data
In order to identify all genes required for the formation of a certain structure a search for morphological phenotypes is implemented on the start page of iBeetle-Base (Figure 1). Typically the user would specify the developmental stage and the morphological structure of interest. iBeetle-Base provides two functions to enable the user to pick the right morphological term. First, the input field has an autocomplete feature that suggests terms or synonyms represented in TrOn based on the typed characters (Figure 1A, arrow a). Second, a click on the three dots at the end of the input field (Figure 1A, arrow b) opens a dialog (Figure 1B) where all terms used in the screen are displayed and can be selected. The numbers following these terms indicate the number of annotations connected to these structures (independently of the selected developmental stage or other selections). An important additional search criterion is the penetrance with which a phenotype was detected during the screen. High penetrance phenotypes (>80%) are less likely to be false positives than low penetrance phenotypes (<30%). The number of data sets fulfilling the selected criteria are displayed right of the penetrance dropdown menu (Figure 1A, arrow c). In addition to this search for affected structures, also the nature of the phenotype can be specified by using the dropdown menus for ‘Temporal, local or logical specifications’, ‘Altered aspect’ and ‘Nature of change’. The content of these lists is reduced to those terms that result in at least one hit when combined with the previous selections (Figure 1C). For instance, after ‘number’ was selected as ‘Altered aspect’ the list of ‘Nature of change’ has been reduced from 37 terms to three. At the same time, the number of data sets that have been found has been updated from 205 (Figure 1A, arrow c) to 13 (Figure 1C).
Figure 1.
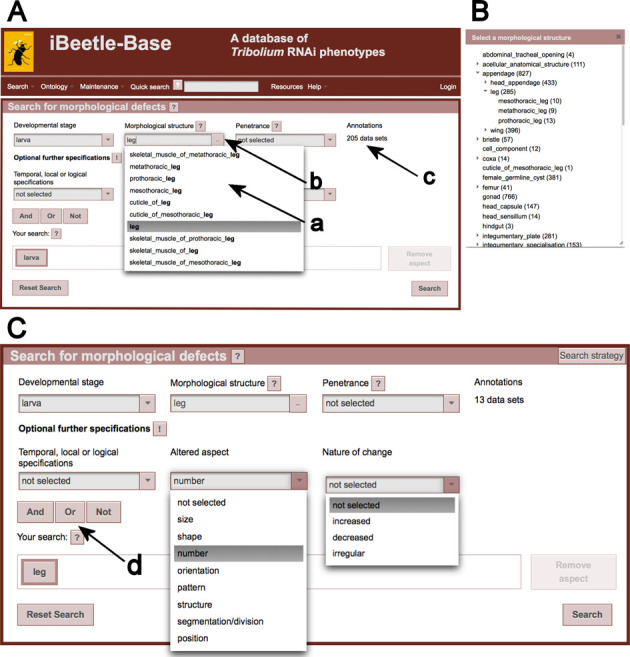
The start page of iBeetle Base. (A) An overview of the start page, where the user can either perform a quick search for a gene, ID's or a morphological structure of interest. A search for data sets affecting the leg is shown. To facilitate the search the input field suggests terms or synonyms based on the typed characters (black arrow a). (B) Clicks on the button at the end of the input field (black arrow b) show all terms used in the screen providing an alternative way to find terms. (C) A detailed view of different drop-down menus is shown (‘Temporal, local or logical specifications’, ‘Altered aspect’ and ‘Nature of change’), which further specify the nature of the phenotype. The respective lists are reduced to the terms that are used or combined in the screen. Arrow d points to the buttons, which enables the user to combine two or more search aspects.
Complex queries can be assembled by combining several search aspects with logical operators. After the search criterion for the first aspect is entered in the search form, a new aspect can be added by choosing one of the buttons ‘And’, ‘Or’, ‘Not’ (Figure 1, arrow d). In the ‘Your search’ field all aspects are shown and can be selected for editing the search form.
To identify most relevant phenotypes with low number of false positives, it is recommended to start with a rather broad search for an affected anatomical entity at a certain stage and a penetrance >50%. Any further restriction will make the hits more specific but will also lead to an increase of false negatives.
Clicking the search button will retrieve an overview of the search results (Figure 2). In the resulting table all annotations fitting the search criteria are listed along with the EQM annotations (Figure 2, arrow a) and respective penetrance (Figure 2, arrow b). Previews of the images documenting the respective annotations can be displayed in different sizes. Further information on the gene and additional phenotypes can be found upon click on the details page (see below). Based on these pieces of information the user can deselect data sets (Figure 2, arrow c) and a consolidated list is provided upon clicking on ‘Refresh’. The resulting list can be exported or saved (Figure 2, arrow d).
Figure 2.
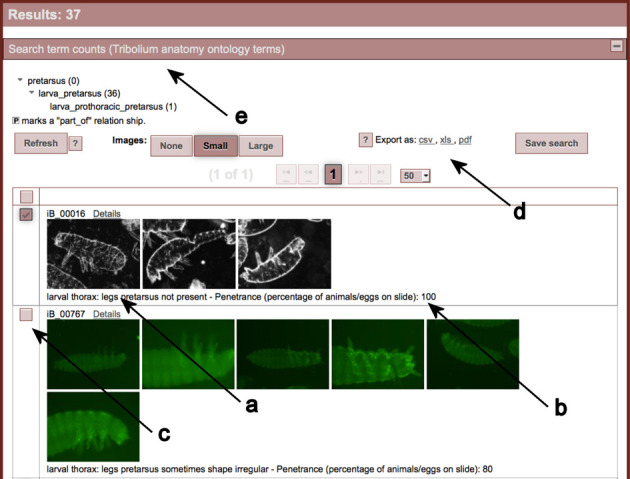
Partial view of the table with the search results. All research results are shown, which fulfil the selected criteria. In the shown example, the search for data sets affecting the pretarus leads to 37 results (not all data sets are shown). The results are listed including the EQM annotations (arrow a) and the respective penetrance (arrow b). Images can be displayed in different sizes. Arrow c points to the button, which allows the user to deselect specific data sets. The corresponding results list can be exported or saved (arrow d). Arrow e points to the search term counts, which includes all terms used for the semantic search. The numbers indicate how many annotations were retrieved for a specific morphological term.
For further information or optimization of the search strategy, the number of annotations retrieved for a specific morphological term is displayed by expanding the panel ‘Search terms count’ (Figure 2, arrow e).
Quick search for genes
A complementary way of using iBeetle-Base is a search for the functions of a given gene. To this end, gene identifier (i.e. TC number) or Drosophila ortholog (e.g. CG number or name) are entered in the ‘Quick search’ field in the menu bar (Figure 1A). If the search term is non-ambiguous the user is redirected to the corresponding details page (Supplementary Material, Figure S1). If several data sets fit the search term, a list is displayed where the user is able to choose the preferred ones.
The quick search can also be used to directly jump to specific data sets using the iBeetle identifier (e.g. iB_01757). For searches in the quick search, the prefix ‘iB_’ can be omitted.
Details pages for comprehensive gene and phenotypic information
The details pages of iBeetle-Base display information about a given gene and the iBeetle RNAi phenotypes associated with it (Supplementary Figure S1). The top section (Gene information) (Supplementary Figure S1A) provides access to gene and protein sequence data of the respective Tribolium gene. Furthermore, it provides links to the BeetleBase (21) genome browser where the genomic structure and location are displayed. A link to OrthoDB (29) allows the identification of orthologs of the respective gene. Where appropriate, information about the Drosophila ortholog is provided by a link to FlyBase (30).
If a certain gene was included in the iBeetle screen, the sequence of the respective dsRNA fragment is shown below of the gene information (Supplementary Figure S1B) (‘iBeetle fragment’) along with the RNAi phenotypes elicited with this iBeetle fragment. The observed phenotypes are presented with the annotations in the EQM scheme, pictures, free text comments and technical remarks. The results of the two parts of the iBeetle screen (pupal injection and larval injection screens) are shown in separate sections (Figure 2 C and D).
The dsRNA sequences of the iBeetle screen were mapped to the current gene models after a major re-annotation effort (Stanke, personal communication). Due to the changes in the gene predictions the association between an iB ID and a gene may change. For example, if two genes covered with iB fragments were merged in the new annotation, there are now two iB IDs assigned to the same gene. In such cases, both sets of results are displayed below the same TC number (Supplementary Figure S1E) and the former TC number is given in the section ‘Gene information’ (Supplementary Figure S1A). An example is the gene TC032760 (also used for Supplementary Figure S1).
Links to morphological definitions provided by TrOn and additional resources
The Tribolium Ontology TrOn is not only utilized in the background to improve the search algorithm but can also be searched and browsed on the web interface. This allows the user to access the definitions of morphological structures and their relations, which will foster the use of common terms in the community. The classes of the ontology are displayed as a tree which shows the hierarchy of the is_a relations (Supplementary Material, Figure S2, left column). Alternatively, a term can specifically be searched for (‘Search & control’). For a selected node in the tree additional information is shown like synonyms, the definition (including links to the respective source of information), or relations to other ontology classes from TrOn (Supplementary Figure S2, arrow a). In addition, each morphological term is linked to those details pages, which mention this term (Supplementary Figure S2, arrow b). Further, the number of links of this class and all of its downstream classes is given (Supplementary Figure S2, arrow c).
Statistics
Basic information about all the 16 505 Tribolium genes of the current gene set is available in iBeetle-Base. 5180 different dsRNAs have been injected leading to 39 156 EQM annotations, documented with 14 487 pictures. 1007 morphological terms and definitions provided by TrOn can be browsed in the iBeetle-Base. Two-hundred-and-seven of them are used in the screen to describe morphological structures.
Linking to and from other resources
All data are available without restriction. Only for saving individual searches, a login is required. iBeetle-Base is intended to become the main gene-centered information hub for Tribolium research. Therefore, it provides links to additional resources like blast, genome browser, a community listserver and a link collection to some Tribolium labs.
Besides the possibility to search and browse the data of iBeetle-Base, it is welcomed that other projects cross link to this data resource. To link to a gene, iBeetle ID or TrOn ID stable URL patterns are administered. In detail these are:
iBeetle IDs: http://ibeetle-base.uni-goettingen.de/details/iB_00000
TC numbers: http://ibeetle-base.uni-goettingen.de/details/TC000000
Ontology terms: http://ibeetle-base.uni-goettingen.de/ontology/tron/TrOn_0000000
FUTURE DIRECTIONS
iBeetle-Base is the first and currently only resource giving access to RNAi phenotypes gathered in an hypothesis independent way in any insect outside Drosophila. It is intended as a central resource for the increasing community working on aspects of the emerging model organism Tribolium castaneum. Beyond the Tribolium community, it will be used by scientists working on other insects in order to gather additional information on genes they are working on. To this end we plan to include links from FlyBase to iBeetle-Base. As part of the continuation of the iBeetle screen, additional data are currently being produced and will be added to the iBeetle-Base in the future. Finally, iBeetle-Base will be extended in a way to allow the community to contribute data. These data will be: complementary information to the already described phenotypes (e.g. confirmation), additional RNAi phenotypes (published or unpublished) and additional data, like gene expression data or literature.
iBeetle-Base is hosted at the computing center of the University of Göttingen (GWDG). The University of Göttingen and the GWDG will provide long term hosting and maintenance of iBeetle-Base as part of the eResearch initiative of the university.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
We thank the screeners of the first phase of the iBeetle project for their accurate and laborious work: Van Anh Dao, Daniela Grossmann, Upalparna Majumdar, Christian Schmitt-Engel, Dorothea Schultheis, Nadi Stroehlein, Jonas Schwirz, Nicole Troelenberg and Tobias Richter. Further, we thank Dr Mario Stanke for providing the new gene set prior to publication and Marita Büscher for her linguistic advice.
FUNDING
German Research Foundation (DFG) [BU1443/6-1]. Funding for open access charge: German Research Foundation; Open Access Publication Funds of the Göttingen University.
Conflict of interest statement. None declared.
REFERENCES
- 1.Nüsslein-Volhard C., Wieschaus E. Mutations affecting segment number and polarity in Drosophila. Nature. 1980;287:795–801. doi: 10.1038/287795a0. [DOI] [PubMed] [Google Scholar]
- 2.St Johnston D. The art and design of genetic screens: Drosophila melanogaster. Nat. Rev. Genet. 2002;3:176–188. doi: 10.1038/nrg751. [DOI] [PubMed] [Google Scholar]
- 3.Brown S.J., Mahaffey J.P., Lorenzen M.D., Denell R.E., Mahaffey J.W. Using RNAi to investigate orthologous homeotic gene function during development of distantly related insects. Evol. Dev. 1999;1:11–15. doi: 10.1046/j.1525-142x.1999.99013.x. [DOI] [PubMed] [Google Scholar]
- 4.Lynch J.A., Desplan C. A method for parental RNA interference in the wasp Nasonia vitripennis. Nat. Protoc. 2006;1:486–494. doi: 10.1038/nprot.2006.70. [DOI] [PubMed] [Google Scholar]
- 5.Hughes C.L., Kaufman T.C. RNAi analysis of Deformed, proboscipedia and Sex combs reduced in the milkweed bug Oncopeltus fasciatus: novel roles for Hox genes in the hemipteran head. Development. 2000;127:3683–3694. doi: 10.1242/dev.127.17.3683. [DOI] [PubMed] [Google Scholar]
- 6.Miyawaki K., Mito T., Sarashina I., Zhang H., Shinmyo Y., Ohuchi H., Noji S. Involvement of Wingless/Armadillo signaling in the posterior sequential segmentation in the cricket, Gryllus bimaculatus (Orthoptera), as revealed by RNAi analysis. Mech. Dev. 2004;121:119–130. doi: 10.1016/j.mod.2004.01.002. [DOI] [PubMed] [Google Scholar]
- 7.Marques-Souza H., Aranda M., Tautz D. Delimiting the conserved features of hunchback function for the trunk organization of insects. Dev. Camb. Engl. 2008;135:881–888. doi: 10.1242/dev.018317. [DOI] [PubMed] [Google Scholar]
- 8.Cerny A.C., Grossmann D., Bucher G., Klingler M. The Tribolium ortholog of knirps and knirps-related is crucial for head segmentation but plays a minor role during abdominal patterning. Dev. Biol. 2008;321:284–294. doi: 10.1016/j.ydbio.2008.05.527. [DOI] [PubMed] [Google Scholar]
- 9.Bucher G., Klingler M. Divergent segmentation mechanism in the short germ insect Tribolium revealed by giant expression and function. Dev. Camb. Engl. 2004;131:1729–1740. doi: 10.1242/dev.01073. [DOI] [PubMed] [Google Scholar]
- 10.Choe C.P., Miller S.C., Brown S.J. A pair-rule gene circuit defines segments sequentially in the short-germ insect Tribolium castaneum. Proc. Natl. Acad. Sci. U.S.A. 2006;103:6560–6564. doi: 10.1073/pnas.0510440103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bolognesi R., Farzana L., Fischer T.D., Brown S.J. Multiple Wnt genes are required for segmentation in the short-germ embryo of Tribolium castaneum. Curr. Biol. CB. 2008;18:1624–1629. doi: 10.1016/j.cub.2008.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fu J., Posnien N., Bolognesi R., Fischer T.D., Rayl P., Oberhofer G., Kitzmann P., Brown S.J., Bucher G. Asymmetrically expressed axin required for anterior development in Tribolium. Proc. Natl. Acad. Sci. U.S.A. 2012;109:7782–7786. doi: 10.1073/pnas.1116641109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Posnien N., Koniszewski N.D.B., Hein H.J., Bucher G. Candidate gene screen in the red flour beetle Tribolium reveals six3 as ancient regulator of anterior median head and central complex development. PLoS Genet. 2011;7:e1002418. doi: 10.1371/journal.pgen.1002416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bucher G., Scholten J., Klingler M. Parental RNAi in Tribolium (Coleoptera) Curr. Biol. 2002;12:R85–R86. doi: 10.1016/s0960-9822(02)00666-8. [DOI] [PubMed] [Google Scholar]
- 15.Klingler M. Tribolium. Curr. Biol. 2004;14:R639–R640. doi: 10.1016/j.cub.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 16.Schröder R., Beermann A., Wittkopp N., Lutz R. From development to biodiversity—Triboliumcastaneum, an insect model organism for short germband development. Dev. Genes Evol. 2008;218:119–126. doi: 10.1007/s00427-008-0214-3. [DOI] [PubMed] [Google Scholar]
- 17.Snodgrass R. Insect Metamorphosis: Smithsonian Miscellaneous Collections. Washington, DC: V122, No. 9 Literary Licensing; 1954. [Google Scholar]
- 18.Chaudhari S.S., Arakane Y., Specht C.A., Moussian B., Boyle D.L., Park Y., Kramer K.J., Beeman R.W., Muthukrishnan S. Knickkopf protein protects and organizes chitin in the newly synthesized insect exoskeleton. Proc. Natl. Acad. Sci. U.S.A. 2011;108:17028–17033. doi: 10.1073/pnas.1112288108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Konopova B., Jindra M. Juvenile hormone resistance gene Methoprene-tolerant controls entry into metamorphosis in the beetle Triboliumcastaneum. Proc. Natl. Acad. Sci. U.S.A. 2007;104:10488–10493. doi: 10.1073/pnas.0703719104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li J., Lehmann S., Weißbecker B., Naharros I., Schütz S., Joop G., Wimmer E.A. Odoriferous Defensive Stink Gland Transcriptome to Identify Novel Genes Necessary for Quinone Synthesis in the Red Flour Beetle, Tribolium castaneum. PLoS Genet. 2013;9:e1003596. doi: 10.1371/journal.pgen.1003596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim H.S., Murphy T., Xia J., Caragea D., Park Y., Beeman R.W., Lorenzen M.D., Butcher S., Manak J.R., Brown S.J. BeetleBase in 2010: revisions to provide comprehensive genomic information for Triboliumcastaneum. Nucleic Acids Res. 2010;38:D437–D442. doi: 10.1093/nar/gkp807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mungall C.J., Gkoutos G.V., Smith C.L., Haendel M.A., Lewis S.E., Ashburner M. Integrating phenotype ontologies across multiple species. Genome Biol. 2010;11:R2. doi: 10.1186/gb-2010-11-1-r2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Washington N.L., Haendel M.A., Mungall C.J., Ashburner M., Westerfield M., Lewis S.E. Linking human diseases to animal models using ontology-based phenotype annotation. PLoS Biol. 2009;7:e1000247. doi: 10.1371/journal.pbio.1000247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beck T., Morgan H., Blake A., Wells S., Hancock J.M., Mallon A.-M. Practical application of ontologies to annotate and analyse large scale raw mouse phenotype data. BMC Bioinformatics. 2009;10:S2. doi: 10.1186/1471-2105-10-S5-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sprague J., Bayraktaroglu L., Bradford Y., Conlin T., Dunn N., Fashena D., Frazer K., Haendel M., Howe D.G., Knight J., et al. The Zebrafish Information Network: the zebrafish model organism database provides expanded support for genotypes and phenotypes. Nucleic Acids Res. 2008;36:D768–D772. doi: 10.1093/nar/gkm956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grumbling G., Strelets V. FlyBase: anatomical data, images and queries. Nucleic Acids Res. 2006;34:D484–D488. doi: 10.1093/nar/gkj068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dönitz J., Grossmann D., Schild I., Schmitt-Engel C., Bradler S., Prpic N.-M., Bucher G. TrOn: an anatomical ontology for the beetle Triboliumcastaneum. PLoS One. 2013;8:e70695. doi: 10.1371/journal.pone.0070695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dönitz J., Wingender E. The ontology-based answers (OBA) service: a connector for embedded usage of ontologies in applications. Bioinforma. Comput. Biol. 2012;3:197. doi: 10.3389/fgene.2012.00197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Waterhouse R.M., Tegenfeldt F., Li J., Zdobnov E.M., Kriventseva E.V. OrthoDB: a hierarchical catalog of animal, fungal and bacterial orthologs. Nucleic Acids Res. 2013;41:D358–D365. doi: 10.1093/nar/gks1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McQuilton P., St Pierre S.E., Thurmond J., FlyBase Consortium FlyBase 101–the basics of navigating FlyBase. Nucleic Acids Res. 2012;40:D706–D714. doi: 10.1093/nar/gkr1030. [DOI] [PMC free article] [PubMed] [Google Scholar]


