Abstract
Adverse drug reactions (ADRs) are noxious and unexpected effects during normal drug therapy. They have caused significant clinical burden and been responsible for a large portion of new drug development failure. Molecular understanding and in silico evaluation of drug (or candidate) safety in laboratory is thus so desired, and unfortunately has been largely hindered by misuse of ADR terms. The growing impact of bioinformatics and systems biology in toxicological research also requires a specialized ADR term system that works beyond a simple glossary. Adverse Drug Reaction Classification System (ADReCS; http://bioinf.xmu.edu.cn/ADReCS) is a comprehensive ADR ontology database that provides not only ADR standardization but also hierarchical classification of ADR terms. The ADR terms were pre-assigned with unique digital IDs and at the same time were well organized into a four-level ADR hierarchy tree for building an ADR–ADR relation. Currently, the database covers 6544 standard ADR terms and 34 796 synonyms. It also incorporates information of 1355 single active ingredient drugs and 134 022 drug–ADR pairs. In summary, ADReCS offers an opportunity for direct computation on ADR terms and also provides clues to mining common features underlying ADRs.
INTRODUCTION
As defined by the World Health Organization (WHO), adverse drug reaction (ADR) is ‘a response to a medicine which is noxious and unintended, and which occurs at doses normally used in man’ (1). ADRs, especially severe ADRs (SADRs), have caused significant clinical burden. In the USA alone, serious drug toxicities were estimated to cause over 100 000 deaths annually (2). Even in developing countries, national pharmacovigilance programme has been started for monitoring ADRs (3). Furthermore, SADRs were responsible for ∼28% of new drug development failures in clinical trials (4) and led to the eventual withdrawal of drugs (5,6). Hence, it is so desired that molecular investigation and in silico evaluation of drug (or candidate) safety could be demonstrated in advance in laboratory before the drug reaches the market. Unfortunately, the progress is largely hindered by some factors, one of which is the misuse of ADR terms. For example, ADR terms like ‘Aggressive reaction’, ‘Violent’, ‘Violent behavior’ and ‘Argumentativeness’ are actually synonyms of ‘Aggression’. In SIDER2, a database that is frequently used in computational toxicity studies, these terms are treated as independent terms. Besides, ambiguous word ‘GU pain’ is sometimes used, which ‘GU’ could be an abbreviation for ‘genito-urinary’ or ‘gastric ulcer’.
The WHO Adverse Reactions Terminology (WHO-ART) is the mostly recognized ADR terminology around the world. However, the most comprehensive and widely used ADR terminology for current pre-marketing development and post-marketing surveillance is the Medical Dictionary for Regulatory Activities (MedDRA) terminology (7). The MedDRA integrated several mainstream ADR dictionaries like WHO-ART, International Classification of Diseases (ICD-9), Coding Symbols for a Thesaurus of Adverse Reaction Terms (COSTART) and Japanese Adverse Reaction Terminology (J-ART). More than 95 000 terms are organized in a hierarchy of five levels: System Organ Class (SOC), High Level Group Term (HLGT), High Level Term (HLT), Preferred Term (PT) and Low Level Term (LLT). It should be noted that MedDRA is not specifically developed for ADR terminology. It also incorporates a number of medical terms for all phases of drug development, health effects and malfunction of devices. Unfortunately, MedDRA has not suggested a reliable method to aid in differentiating the ADR terms from others yet, which decreases its use in toxicity research.
With the growing impact of bioinformatics and systems biology in toxicological research, it requires a specialized ADR terminology system that works beyond a simple glossary (8–10). Contrary to current ADR-related repositories like MedDRA and SIDER2, the new ADR terminology system, or more precisely a prototype of ADR ontology system, should provide explicit and univocal descriptions of ADR terms so that they can be easily interpreted by the user or even the computer robot without misunderstanding (11). In practice, following problems need to be addressed at the first convenience: (i) many ADRs are ambiguous, especially between clinical and laboratory usage; (ii) the distance between ADRs is hard to be quantitatively measured; (iii) ADRs are inconvenient to be computed directly. Therefore, in this study, we constructed a comprehensive ADR ontology database, the Adverse Drug Reaction Classification System (ADReCS), to aid standardization and hierarchical classification of ADR terms.
DATABASE CONSTRUCTION
Data extraction
The drug-ADR information of ADReCS was mainly extracted from the drug labels in the DailyMed (http://dailymed.nlm.nih.gov/dailymed/about.cfm), a website managed by the U.S. National Library of Medicine (NLM) to provide comprehensive information about marketed drugs. ADReCS also derived the drug-ADR information from SIDER2 (12) and other reliable online sources like the U.S. Food and Drug Administration (US FDA). Finally, it yielded a total of 1355 single active ingredient drugs and 21 237 distinct ADR records. Of these data, ∼80% were derived from DailyMed, 16% were derived from SIDER2 and the remanding came from other sources. In addition, the frequency information for each drug–ADR pair was also collected, when available, in a form of either an exact frequency like ‘2.1%’ or a frequency range like ‘1%–3%’. The pharmacological and chemical information of these drugs, like drug description, indication, synonyms, structure and so on, was extracted from public medical repositories like Unified Medical Language System (UMLS) (13), DrugBank (14), PubChem (15) and KEGG (16) and Anatomical Therapeutic Chemical (ATC) Classification System. Then all the extracted data were first pre-formatted by removing those obvious typo errors before further data processing.
Standardization of ADR terms
The ADR records are normally indicated in a variety of conditions like physical findings, patient complaints, safety reports and laboratory results, which are often described in different strings (17). Therefore, it is hard to evaluate these medical terms straightforwardly before standardization. ADReCS adopted MedDRA and UMLS as major references for ADR term standardization. These two reference databases have made great efforts in medical term standardization, and their works are generally recognized. According to the principle of strict equivalence, the pre-formatted ADR records were first automatically mapped to MedDRA, which ∼1/3 of total ADR records can be then standardized by simple reference to MedDRA terms. The remaining 2/3 ADR records were then manually amended, with the help of UMLS. This was followed by second round or multiple rounds of MedDRA mapping. All the standard ADR terms were double-checked for validity. Even though, 27 ADR records cannot be properly linked with MedDRA terms so that they were assigned as new ADR terms (Supplementary Table S1). The detailed protocol of ADR standardization was also provided in the Supplementary Method.
Eventually, ADReCS achieved totally 4906 standard ADR terms out of 21 237 ADR records, and the remanding ADR records were taken as synonyms of standard terms after standardization. Furthermore, explicit and univocal definitions of the standard ADR terms were collected from the Medical Subject Heading vocabulary (MeSH), NCI Thesaurus (NCIt) and other public-available medical dictionaries, which give aid to reduce the risk of misinterpretation of the terms. Consensus definitions of the standard ADR terms also lead them to a meaningful data set of Common Data Elements (10).
Hierarchical classification of ADR terms
Hierarchy is an important mechanism for flexible data retrieval and clear presentation of data. The hierarchy provides degrees or levels of super ordination and subordination, and represents vertical links in the terminology. Most importantly, the hierarchy allows computation on different levels and supports logically linking ADRs to the underlying physiology.
ADReCS adopted similar hierarchy as MedDRA and WHO-ART. There are four levels in the ADR hierarchy tree of ADReCS: the SOC, the HLGT, the HLT and the PT. All ADR terms in ADReCS were assigned into the hierarchical tree. From SOC to PT, the ADR terms in ADReCS become more and more specific in describing medical concept. The SOC is the highest level, which normally describes the adverse effects in system organ level. On the contrary, the PT normally represents a specific, unique and unambiguous single ADR concept (Figure 1). Unlike MedDRA, the LLT is discarded in ADReCS as an independent level since LLTs are actually extensions of PTs in describing synonyms, lexical variants, quasi-synonyms or sub-elements of the same medical concepts. Hence, PTs are necessary and sufficient to characterize and differentiate single medical concepts of ADR terms.
Figure 1.
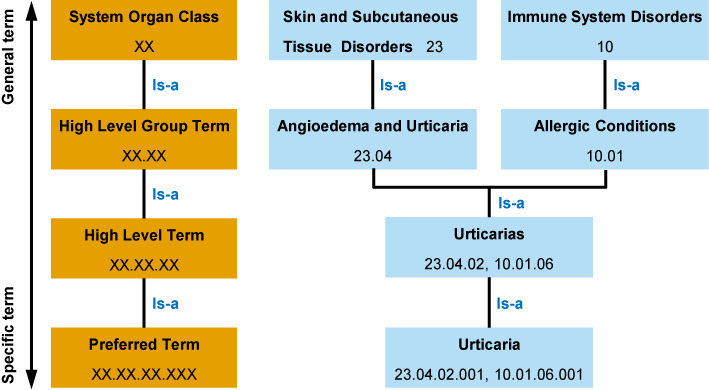
Hierarchy and digital IDs of ADReCS, using the ADR term ‘Urticaria’ as an example.
In building relationship between ADR terms, ADReCS always used direct taxonomic relation (is-a) to avoid partonomic relation (part-of) whenever possible. Each ADR term has at least one complete route from the PT to the SOC according to clinical relevance, and an ADR may evolve via multiple routes. The multi-axiality characteristics of ADR hierarchy are the results of many factors like different lesions, various causes, or complex medical conditions, hinting different molecular mechanisms underlying ADRs. For example, the PT ‘Urticaria’ is linked to two SOCs, ‘Skin and Subcutaneous Tissue Disorders’ (manifestation site) and ‘Immune System Disorders’ (aetiology) (Figure 1).
Digital ID of ADR terms
Digital ID was designed for each ADR term to aid standardization and hierarchical classification. The digital ID is a combination of four fixed length digital strings like xx.xx.xx.xxx, where x stands for a numeric character from 0 to 9. The digital strings, from left to right, represent four levels of ADR hierarchy: SOC, HLGT, HLT and PT, respectively. Currently, the most left digital string (representing the SOC) ranges from 01 to 26, standing for ADRs in 26 organ systems. The digital ID shows the evolution routes of an ADR term and the relationship between the ADR terms of different levels. As shown in Figure 1, the PT ‘Urticaria’ has two IDs, 23.04.02.001 and 10.01.06.001, which suggests two routes to the SOCs ‘Skin and Subcutaneous Tissue Disorders’ and ‘Immune System Disorders’, respectively. Theoretically, the digital ID supports ADReCS to expand maximum up to 2.6 × 108 PTs, which is far beyond our estimation of ∼104 distinct ADR PTs in practice.
DATA ACCESS
Data retrieval
The ADReCS database can be accessed through a simple and user-friendly interface at: http://bioinf.xmu.edu.cn/ADReCS. This interface allows users to view or retrieve ADR ontology in different methods. These data retrieval methods were summarized and illustrated in a schematic view (Figure 2). The most convenient and direct way for acquiring ADR ontology information is via the ADR hierarchy tree provided in the database BROWSE page (Figure 2C), where the standard ADR terms were pre-arranged in an expansible tree according to their clinical relevance (or their digital IDs). In addition, ADReCS also provides ‘Browse By Drugs’ method to facilitate reviewing drug–ADR associations in alphabetical order of drug names. Clicking on an ADR term in the ADR hierarchy tree or a drug name in the drug tables will redirect to the page of ADR Ontology or Drug information, respectively.
Figure 2.
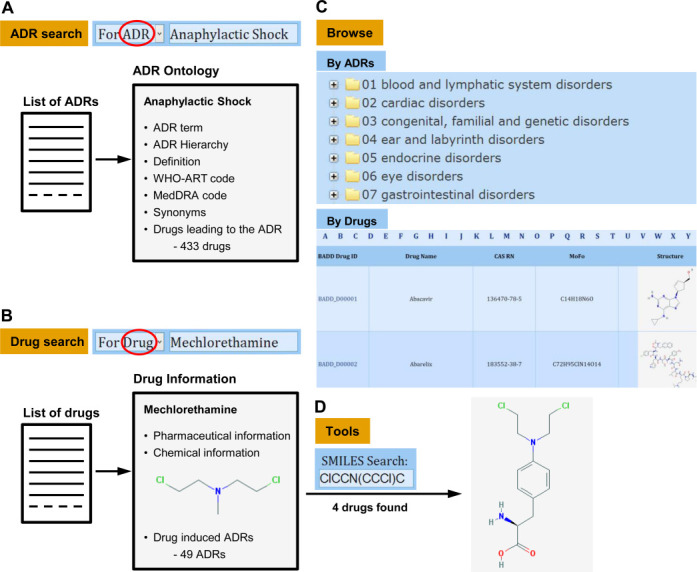
Schematic view of ADRs/drugs search and navigation supported in ADReCS: (A) ADR search, (B) Drug search, (C) Browse by ADRs or drugs and (D) Tools for structure search.
Besides, ADReCS offers a conventional search method for customized data retrieval. To start a query, the user is asked to choose either ‘For ADR’ or ‘For Drug’ option in the quick search form. Meanwhile, complete or partial keyword is required to trigger the search. Empty input is not allowed. The search form supports both accurate search (for complete keyword) and fuzzy search (for partial keyword). Optionally, the search can be further constrained by choosing one of 12 keyword types like ADR term, Drug name, ATC code, CAS number, DrugBank ID, WHO-ART code, MedDRA code and so on. When search ‘For ADR’ (Figure 2A), ADR records that meet the query criteria are responded in the ascending alphabet order of ADR term, along with MedDRA code and WHO-ART code. Clicking on the ADR term will lead to the page of ADR Ontology, where ADR information is listed in order when available, including ADR hierarchy (and its digital ID), definition, synonyms, MedDRA code, WHO-ART code and drugs leading to the ADR. When search ‘For Drug’ (Figure 2B), drugs that meet the query criteria are responded in the ascending order of drug names, along with ADReCS drug ID, CAS RN and structure. Clicking on the drug name will redirect to the detailed information page of drug. In this page, drug information is arranged in three sections: the pharmaceutical information, the chemical information and the drug-induced ADRs. Cross-links to some useful resources such as DrugBank, PubChem and KEGG via ID mapping are also provided when available. The cross-links to these public databases allow ADReCS to expand by continuous integration of heterogeneous data; vice versa, the cross-links enable ADReCS be easily integrated.
Additional tools for structure search
In addition to the search and browse method, ADReCS implements a structural search tool for database access. The users are allowed to conduct drug search by providing a complete or partial SMILES string of chemical (Figure 2D). Furthermore, a structure drawing board JSDraw is embedded, where a chemical structure can be drawn or uploaded (for example, *.mol format) for automatically converting into SMILES string. The structural similarity search of ADReCS uses regular string comparison method upon SMILES strings. To start a search, the query SMILES string is first transformed into its canonical SMILES string using an underlying java tool openbabel.jar. This transformed SMILES string is then compared to the canonical SMILES strings of drugs deposited in the database. For fuzzy search, it requires partial string match; while for accurate search, it requires exact string match. This tool allows a researcher to retrieve drugs with similar sub-structures in order to facilitate the structure–ADR relationship analysis.
Data download
ADReCS provides two ways for data distribution. Users can freely download the full ADReCS data at the DOWNLOAD page (http://bioinf.xmu.edu.cn/ADReCS/download.jsp). Login is required to proceed the downloading; however, registration is completely free for academic users. The data are distributed in XML format. Besides, users can customize the data downloading via the search form. The records that meet the query criteria are downloadable in XML format as well.
DATA STATISTICS
Currently, the ADReCS covers total 6544 standard ADR terms and 34 796 synonyms. The standard ADR terms are classified into 26 SOCs, 322 HLGTs, 1290 HLTs and 4906 PTs. Majority (4879 out of 4906, or 99.4%) of ADReCS PTs are overlapped with MedDRA PTs except 27 new terms (Supplementary Table S1). Besides, 21 468 ADR terms (including synonyms) can be mapped into the UMLS. Of 6544 standard ADR terms, 2078 were given with explicit and univocal definitions. Moreover, ADReCS also includes 1355 single active ingredient drugs and 134 022 non-redundant drug–ADR pairs. A preliminary statistic of the database was demonstrated, showing that altogether, ∼60% of drugs can induce ∼10 to 100 different ADRs (Figure 3A). About 70% of ADRs occur to less than 10 drugs and only 7% of all ADRs occur to more than 100 drugs (Figure 3B). It seems that most drugs can induce multiple ADRs and most ADRs are rare ADRs occurring to few drugs. These results are roughly agreed with the statistics of SIDER2 (12). In addition, 50 490 (or 38%) of all drug–ADR pairs have frequency information, including 17 171 (or 34%) pairs with exact frequency and 33 319 (or 66%) pairs with frequency range. Statistics of drug–ADR pairs by the ADR frequency (Figure 3C and D) suggests that majority of ADRs (∼80%) occur in small frequency (<5%). The median frequency of ‘frequent’ ADRs (occurring in >1% of patients) is ∼4%.
Figure 3.
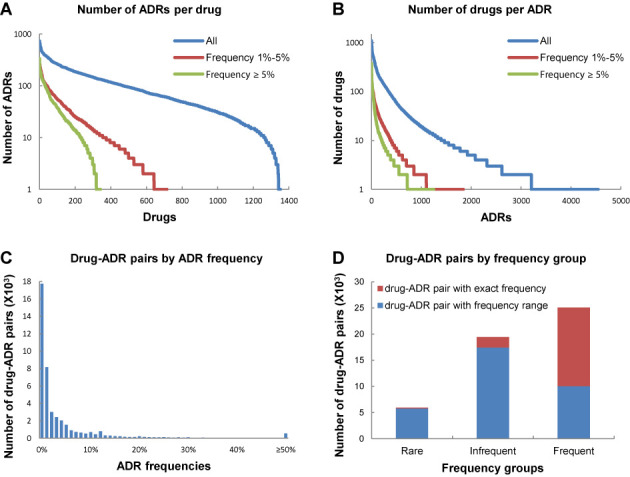
Statistics of ADReCS. (A) The number of ADRs counted by drug with frequency thresholds of 1%–5% (red, 717 drugs), above 5% (green, 344 drugs) and all (blue, 1355 drugs), respectively. (B) The number of drugs counted by ADR with frequency thresholds of 1%–5% (red, 1842 ADRs), above 5% (green, 1250 ADRs) and all (blue, 4545 ADRs), respectively. (C) Statistics of drug–ADR pairs by ADR frequency. (D) Statistics of drug–ADR pairs by ADR frequency group. The drug–ADR pairs were classified into three groups: Rare (frequency thresholds of <0.1%), Infrequent (frequency thresholds of 0.1%–1%) and Frequent (frequency thresholds of >1%).
POTENTIAL APPLICATIONS
ADReCS can be easily applied in various toxicological applications. As an ontology database, ADReCS facilitates ADR mechanism study. The users can freely select an ADR (PT) or ADR group (HLT or HLGT) from the ADR hierarchy tree, and retrieve the corresponding list of drugs that could induce the selected ADR. By seeking the commonality of these drugs such as their physiochemical properties or protein target bindings, it has chance to reveal the molecular mechanisms underlying the particular ADR or ADR groups so as to aid future rational drug design. Moreover, by comparing different drug profiles, it helps to detect the distinct characteristics between sibling ADR terms.
The digital ID is one of the key features of ADReCS. Each standard ADR term in ADReCS is represented by a unique digital ID, indicating its position in the ADR hierarchy tree and its connection with other ADR terms. Using the digital IDs and the hierarchy tree, the pharmacovigilance and the corresponding association analyses can be clearly and efficiently carried out upon any selected ADR or ADR group. All ADR terms under the selected ADR group as well as their synonyms are involved in the analyses, which make the vigilance more complete and robust. For example, vigilance can be flexibly demonstrated upon either ADR group ‘Visual color distortions’ (HLT, ADReCS_ID: 06.02.05, as well as all PTs under this group) or any of its subordinate ADR PTs with IDs 06.02.05.xxx like Cyanopsia (ADReCS_ID: 06.02.05.003) without missing any subordinate ADR or falsely including ADRs outside this group. In another example, ‘Abdominal discomfort’ cannot be found in WHO-ART but can be found in MedDRA (MedDRA_ID: 10000059) and ADReCS (ADReCS_ID: 07.01.06.001). Synonyms of ‘Abdominal discomfort’ in ADReCS like ‘Abdo. discomfort’ (MedDRA_ID: 10000042), ‘Distress abdominal’ (MedDRA_ID: 10013488), ‘Gastrointestinal upset’ (MedDRA_ID: 10018028), ‘GI upset’ (MedDRA_ID: 10018249) and so on are independent terms (LLTs) in MedDRA. Simple keyword search of ‘Abdominal discomfort’ (MedDRA_ID: 10000059) in PubMed literature database retrieves 3847 related articles; however, combinational keyword search like ‘abdominal discomfort OR distress abdominal OR gastrointestinal upset’ (ADReCS_ID: 07.01.06.001) hits 6226 related articles. Hence, pharmacovigilance of ‘Abdominal discomfort’ based on WHO-ART or MedDRA terminology may completely or partially lose signals if the LLTs are not explicitly involved. Moreover, ADReCS allows direct and reliable computation on ADRs, for example, in drug safety evaluation and prediction. Conventionally, the drug safety predictions were demonstrated on ADR terms themselves that derived from databases like SIDER2 and MedDRA (18,19). In these applications, few parent–child or sibling relations between ADRs were considered. The ADR hierarchy and the digital IDs of ADReCS may help to achieve better performance by providing clear ADR connections.
CONCLUSION AND COMPARED WITH OTHER RESOURCES
ADReCS is so far the first ADR ontology database that provides both standardization and hierarchical classification of ADR terms. In current ADR terminology databases, WHO-ART is the most authoritative source and usually serves as a reference for other ADR terminologies. However, WHO-ART is limited in ADR terms. MedDRA integrates several ADR dictionaries so that it contains the largest ADR-related vocabulary. Unfortunately, MedDRA also incorporates a large number of medical terms other than ADRs. Besides the ADR terminology databases, there are some ADR-specialized information databases like SIDER2 and PROTECT. These two databases provide information of drug–ADR relations but are lack of a hierarchy to address ADR–ADR relations. Compared to current ADR terminology or information databases, ADReCS has its advantages: (i) ADReCS is specialized in ADR; (ii) ADReCS has clear ADR hierarchy for ADR–ADR relation; (iii) ADReCS provides hierarchical digital IDs for direct and reliable ADR computation. The database comparison was briefly summarized in Table 1.
Table 1. Comparison of ADReCS to some typical ADR databases.
| Resources | ADReCS v1.2 | MedDRA 17.0 | WHO-ART (Jan 2014) | SIDER2 | PROTECT ADR database |
|---|---|---|---|---|---|
| Hierarchical structure | Four levels | Five levels | Four levels | N/A | N/A |
| ID of ADR terma | 10.01.03.002 | 10001738b | 1058 001c | N/A | N/A |
| Definition of ADR | Yes | N/A | N/A | Yes | N/A |
| Number of PTs | 4906 | 20 559 | 2178 | 4192 | 5676 |
| Number of synonyms/LLTs | 34 796 | 72 637 | 5813 | >6000 | N/A |
| Number of drugs | 1353 | N/A | N/A | 996 | 677 |
| Number of drug–ADR pairs | 134 022 | N/A | N/A | 99 423 | 46 687 |
N/A = Not available
aUsing ‘Allergy’ as an example.
bMedDRA codes are assigned in alphabetical order starting with 10000001.
cWHO-ART codes are assigned by combining the high-level term ID (e.g. 1058) with a sequential number of ADR term (starting from 001).
Further improvements are expected to keep ADReCS useful, up-to-date and relevant for the community of scientists working on both laboratory toxicity and clinical ADRs. Although best efforts have been made, many more still need to be done continuously and gradually to make ADReCS a valuable source for different research fields, especially toxicology and pharmacology. Generally, ADReCS will be updated continuously: small improvements will be made once they are ready. New data are scheduled to be incorporated in batches once half a year. Major updates like frame change and new function implement will be undertaken annually when available. We believe, with the advances in systems toxicology and in silico drug safety evaluation, ADReCS will serve as an informative ontology source that provides framework for linking ADRs with underlying biological mechanisms, establishes molecular correlations between ADRs and allows ADR computation in a high-throughput manner in early drug discovery stage.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
The authors are grateful to former colleagues at Bioinformatics-Aided Drug Discovery Group, Xiamen University, for their contributions to the database.
FUNDING
National Natural Science Foundation of China [30873159, 31271405]. Funding for open access charge: National Natural Science Foundation of China [31271405].
Conflict of interest statement. None declared.
REFERENCES
- 1.WHO. Safety of Medicines: A guide to detecting and reporting adverse drug reactions. World Health Organization. 2002 WHO/EDM/QSM/2002.2. [Google Scholar]
- 2.Giacomini K.M., Krauss R.M., Roden D.M., Eichelbaum M., Hayden M.R., Nakamura Y. When good drugs go bad. Nature. 2007;446:975–977. doi: 10.1038/446975a. [DOI] [PubMed] [Google Scholar]
- 3.Kavitha D. Adverse drug reaction (ADR) monitoring and pharmacovigilance. Asian J. Pharm. Res. Health Care. 2010;2:127–134. [Google Scholar]
- 4.Arrowsmith J., Miller P. Trial watch: phase II and phase III attrition rates 2011–2012. Nat. Rev. Drug Discov. 2013;12:569. doi: 10.1038/nrd4090. [DOI] [PubMed] [Google Scholar]
- 5.Paludetto M.N., Olivier-Abbal P., Montastruc J.L. Is spontaneous reporting always the most important information supporting drug withdrawals for pharmacovigilance reasons in France. Pharmacoepidemiol. Drug Saf. 2012;21:1289–1294. doi: 10.1002/pds.3333. [DOI] [PubMed] [Google Scholar]
- 6.Clarke A., Deeks J.J., Shakir S.A. An assessment of the publicly disseminated evidence of safety used in decisions to withdraw medicinal products from the UK and US markets. Drug Saf. 2006;29:175–181. doi: 10.2165/00002018-200629020-00008. [DOI] [PubMed] [Google Scholar]
- 7.Brown E.G., Wood L., Wood S. The medical dictionary for regulatory activities (MedDRA) Drug Saf. 1999;20:109–117. doi: 10.2165/00002018-199920020-00002. [DOI] [PubMed] [Google Scholar]
- 8.Waters M.D., Fostel J.M. Toxicogenomics and systems toxicology: aims and prospects. Nat. Rev. Genet. 2004;5:936–948. doi: 10.1038/nrg1493. [DOI] [PubMed] [Google Scholar]
- 9.Berger S.I., Iyengar R. Role of systems pharmacology in understanding drug adverse events. Wiley Interdiscip. Rev. Syst. Biol. Med. 2011;3:129–135. doi: 10.1002/wsbm.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhichkin P.E., Athey B.D., Avigan M.I., Abernethy D.R. Needs for an expanded ontology-based classification of adverse drug reactions and related mechanisms. Clin. Pharmacol. Ther. 2012;91:963–965. doi: 10.1038/clpt.2012.41. [DOI] [PubMed] [Google Scholar]
- 11.Bousquet C., Sadou E., Souvignet J., Jaulent M.C., Declerck G. Formalizing MedDRA to support semantic reasoning on adverse drug reaction terms. J. Biomed. Inform. 2014;49:282–291. doi: 10.1016/j.jbi.2014.03.012. [DOI] [PubMed] [Google Scholar]
- 12.Kuhn M., Campillos M., Letunic I., Jensen L.J., Bork P. A side effect resource to capture phenotypic effects of drugs. Mol. Syst. Biol. 2010;6:343. doi: 10.1038/msb.2009.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bodenreider O. The Unified Medical Language System (UMLS): integrating biomedical terminology. Nucleic Acids Res. 2004;32:D267–D270. doi: 10.1093/nar/gkh061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Law V., Knox C., Djoumbou Y., Jewison T., Guo A.C., Liu Y., Maciejewski A., Arndt D., Wilson M., Neveu V., et al. DrugBank 4.0: shedding new light on drug metabolism. Nucleic Acids Res. 2014;42:D1091–D1097. doi: 10.1093/nar/gkt1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Y., Suzek T., Zhang J., Wang J., He S., Cheng T., Shoemaker B.A., Gindulyte A., Bryant S.H. PubChem BioAssay: 2014 update. Nucleic Acids Res. 2014;42:D1075–D1082. doi: 10.1093/nar/gkt978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kanehisa M., Goto S., Sato Y., Kawashima M., Furumichi M., Tanabe M. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res. 2014;42:D199–D205. doi: 10.1093/nar/gkt1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Richesson R.L., Fung K.W., Krischer J.P. Heterogeneous but “standard” coding systems for adverse events: issues in achieving interoperability between apples and oranges. Contemp. Clin. Trials. 2008;29:635–645. doi: 10.1016/j.cct.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu M., Wu Y., Chen Y., Sun J., Zhao Z., Chen X.W., Matheny M.E., Xu H. Large-scale prediction of adverse drug reactions using chemical, biological, and phenotypic properties of drugs. J. Am. Med. Inform. Assoc. 2012;19:e28–e35. doi: 10.1136/amiajnl-2011-000699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lounkine E., Keiser M.J., Whitebread S., Mikhailov D., Hamon J., Jenkins J.L., Lavan P., Weber E., Doak A.K., Cote S., et al. Large-scale prediction and testing of drug activity on side-effect targets. Nature. 2012;486:361–367. doi: 10.1038/nature11159. [DOI] [PMC free article] [PubMed] [Google Scholar]


