Abstract
Plastids carry their own genetic material that encodes a variable set of genes that are limited in number but functionally important. Aside from orthology, the lineage-specific order and orientation of these genes are also relevant. Here, we develop a database, Plastid-LCGbase (http://lcgbase.big.ac.cn/plastid-LCGbase/), which focuses on organizational variability of plastid genes and genomes from diverse taxonomic groups. The current Plastid-LCGbase contains information from 470 plastid genomes and exhibits several unique features. First, through a genome-overview page generated from OrganellarGenomeDRAW, it displays general arrangement of all plastid genes (circular or linear). Second, it shows patterns and modes of all paired plastid genes and their physical distances across user-defined lineages, which are facilitated by a step-wise stratification of taxonomic groups. Third, it divides the paired genes into three categories (co-directionally-paired genes or CDPGs, convergently-paired genes or CPGs and divergently-paired genes or DPGs) and three patterns (separation, overlap and inclusion) and provides basic statistics for each species. Fourth, the gene pairing scheme is expandable, where neighboring genes can also be included in species-/lineage-specific comparisons. We hope that Plastid-LCGbase facilitates gene variation (insertion-deletion, translocation and rearrangement) and transcription-level studies of plastid genomes.
INTRODUCTION
Plastid is a vital organelle for the photosynthesis of eukaryotic species in broad taxa, including plants, alga and protists; it is also regarded as a favorable genetic material for transformation and manipulation for its extra-chromosomal status (1,2). The structure and number of plastids are different from those of mitochondrion and other subcellular organelles, as well as their phenotypes that are influenced by not only genetics but also environmental factors (3,4). According to the endosymbiotic theory, plastid originated from a cyanobacterium and experienced multiple evolutionary events, which had altered their primary, secondary and tertiary structures (5) to the extent that not every plastid contains a typical genome and not all plastid genes are involved in photosynthesis (6–8). The basic structure of plastid genomes, containing a number of essential protein-coding genes as well as rRNAs and tRNAs, is divided into four parts: a long single-copy, a small single-copy and two inverted repeated segments (9–13). The gene flow among plastid, mitochondrial and nuclear genomes starts at very early stages but may have heterogeneous rates (14–16). Plastid genomes produce considerable amount of essential and indispensable functional proteins for various functions, such as photosynthesis, respiration and translation (17), and the rest participants, proteins and RNAs, are contributed by nuclear genes, of which some are proposed to originate from cyanobacterium (18). The number and type of plastid genes vary among species and lineages but as a functional set still adequately maintains essential functions (17). In general, eukaryotic genomes are organized into gene clusters and these clustered genes often collaborate for function and appear not acting alone (19–22). For plastid genomes, it is known that there are several operon-like gene clusters that are co-regulated or co-expressed, composed of neighboring or consecutively ordered genes, and lead to improvement of transcription and translation efficiency (23).
Most of the current plastid relevant databases, such as GOBASE (24) and ChloroplastDB (25), emphasize gene structure and sequence annotation but pay less attention to genome organization. The only study related to plastid gene order included 32 species and merely displayed the text information (26), although there have been several databases built for visualizing the nuclear gene order in different evolutionary scales or in a limited scope of taxonomic grouping (27–32). As the rising popularity of high-throughput sequencing and the rapid accumulation of organellar genome sequences (33), the public data collection now has 470 plastid genomes. Here, based on conserved paired genes, our plastid-LCGbase provides general survey, visualization, comparative frameworks for plastid genomes in various user-defined phylogenetic grouping. We also define six patterns or modes and eight types of transcription start sites (TSS) distances for the plastid gene pairs. This database should become a useful repository for the study of plastid genome alignment and arrangement study as well as for the discovery of possible co-regulation of adjacent genes over evolutionary time scales.
MATERIALS AND METHODS
We collected genome information and sequences of protein-coding genes and non-coding RNAs of 470 plastids from NCBI Organelle Genome Resources (web and ftp server) based on careful selection of representative species. Taxonomic data were downloaded from the NCBI taxonomy ftp site and the keywords include names of kingdom, phylum, class, order, family, genus and species. The circular or linear maps for plastid genomes were drawn by using OrganellarGenomeDRAW (v1.1.1) (34) and other figures were plotted by using R software package. Comparisons between a reference genome and other genomes (99 in number) that are most similar to the reference were manipulated by using CGView (35). Similar determination was carried out by using Blastp (ncbi-blast-2.2.28+) with E-value = 1e-5 and max_target_seqs = 500 (36) for protein-coding genes and BlastR with E-value = 1e-5 for non-coding RNAs (37). Classification of gene families was based on Tribe-MCL (Markov Clustering) with I = 2 (38). The definition of conserved gene pairs was performed in a visual system. In details, once core data sets are imported into MYSQL database, an optimal index is created to make sure for fast user inquiry. PHP takes charge of the calculation modules, makes the reference chromosome fixed and searches along two opposite directions in other chromosomes in the process of comparisons. Once the computation is finished, a figure containing gene arrangement in both the reference and the searchable chromosomes is shown as an image, together with three types of textual formats. Colors and arrows are used to indicate homologous groups and transcriptional orientations, respectively; the colors are forced to distinguish different homologs. We recommend using ‘Google Chrome Browser’ to view this database since we have tested that this browser works compatible with different operating system for the full database functions.
RESULTS
We started by constructing phylogenetic trees for species involved using CVTree (39) based on proteomic data to provide a glance of evolutionary relationship among all the species for users (Figure 1A). In general, the database offers a genome map function to show an overview of gene distributions on the browse page (Figure 1G). At the same time, it provides graphic views for structural changes among plastid genomes at a global level, which include both DNAs and translated coding sequences (CDSs). For better display of structural features, we divided plastid genome into three gene groups: protein-coding, non-coding RNA and all genes (including the previous two groups). One of the functions for the database is to define paired genes into all three types and to discover conserved patterns of the gene pairs in different evolutionary lineages. In the search page, we provide eight different colors to distinguish the distance of neighboring genes (0–300 bp, 300–500 bp, 500–800 bp, 800–1000 bp, 1000–1200 bp, 1200–1500 bp, 1500–2000 bp and > 2000 bp) and multiple-checked boxes to determine the species of interest on the sorted display of taxa from kingdom all the way down to genus (Figure 1B). When a gene identifier is entered by a user, the resulting page produces a figure containing a list of conserved gene pairs (both homology-based and strand-specific) (Figure 1C). Since the distances between paired genes are color-coded, the dynamics of TSS of homologous gene pairs in different species can be visually compared. If query gene is unknown, the database provides two alternative choices since all featured data have been summarized in the species table (Figure 1D). One way is to browse the gene list in particular genomes to find their names in various nomenclature system (e.g. Gene Identifier, Protein ID, Gene ID and Product) and position information (e.g. Strand, Start and End) (Figure 1E). Another way is to view the gene pair list including their relationship and individual features (Figure 1F). We also calculated all conserved gene pairs in the 470 plastid genomes for browsing and downloading. Furthermore, we define operon-like structures as determined by concatenating highly-conserved gene pairs (at least conserved in 100 plastid genomes) in certain species. In addition, we classify gene pairs into nine categories based on whether they are co-directionally-paired genes (CDPGs), convergently-paired genes (CPGs) or divergently-paired genes (DPGs) and in ‘Separation’, ‘Overlap’ and ‘Inclusion’ as patterns. The former is an orientation parameter that defines gene clusters based on relative transcription direction of neighboring genes; the latter is a distance parameter that characterizes physical distance of neighboring genes (Figure 2). In addition, we plot densities of TSS distance in logarithmic scale for CDPGs, CPGs, DPGs (Figure 1H) and all paired genes, and show barplots of all nine paired gene types on the ‘Parameter’ page (Figure 1I). We offer processed gene pair data of all plastid genomes for free-download by users. Every figure in this database can be enlarged to display a high-resolution version. In order to establish connections between this database and external public databases, we linked many keywords to their NCBI definitions and annotation pages; for example, ‘Species’, ‘Protein GI’, ‘Locus’, ‘Protein Accession’ and ‘Gene ID’ are all appropriately linked.
Figure 1.
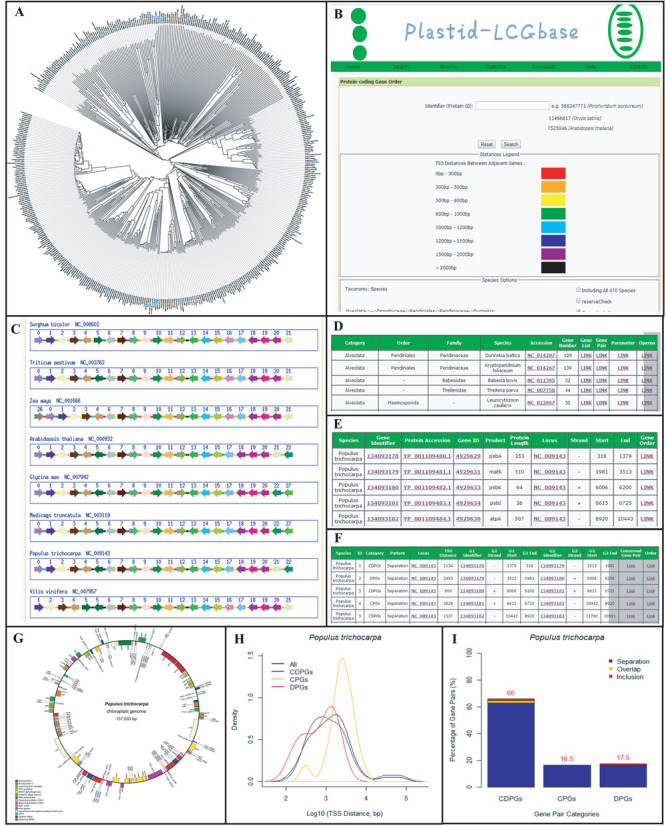
Screenshots for major functional modules of Plastid-LCGbase. (A) Phylogenetic trees for 470 species built from whole plastid proteomes. (B) The search page for determining conserved gene pairs and visualizing gene orders. (C) The result page from search pages. Arrows and orientation indicate genes and their transcription direction. (D) The browse page. (E) The gene list page. (F) The gene pair page. (G) The genome map page. (H) Distributions of TSS distances from the three types of gene pairs. (I) Barplots of the three types and three patterns of gene pairs.
Figure 2.
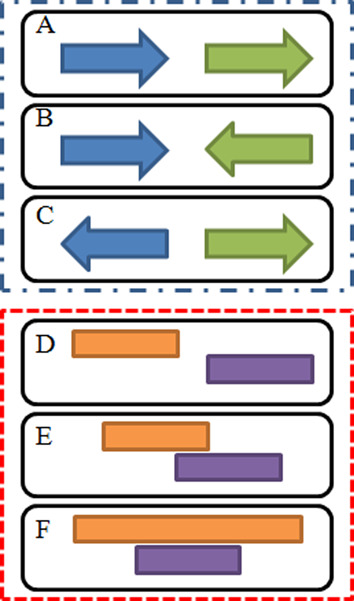
The definition of categories and patterns of gene pairs. On the basis of relative transcription direction, gene pairs are divided into three categories: (A) CDPG, gene transcribed in the same direction. (B) CPG, gene transcribed in the opposing direction but toward each other. (C) DPG, gene transcribed in the opposing transcription direction but toward distant directions. According to the relative positions of neighboring genes, gene pairs are classified into three patterns: (D) ‘Separation’, no overlapping regions. (E) ‘Overlap’, shared some regions but no one covers the other completely. (F) ‘Inclusion’, one gene covers all regions of the other gene.
DATA OVERVIEW
In the database, we classified 470 plastids into 9 categories (Alveolata, Cryptophyta, Euglenozoa, Glaucocystophyceae, Haptophyceae, Rhizaria, Rhodophyta, Stramenopiles and Viridiplantae), 111 orders and 152 families, albeit some incomplete information for their order and family definitions. Most genomes are circular except 10 linear displays. We adopted four inflation parameters (I = 1.4, I = 2, I = 3 and I = 4) to deduce gene family classification for all the proteomes and found that the shape of their distributional curves are quite similar (data not shown). I = 1.4 generates more large gene families while I = 4 leads to more small gene families. We decided to choose a moderate one (I = 2) for the analysis. In details, the largest gene family contains 911 members for protein-coding genes and 857 members for non-coding RNAs; other measurements for gene family sizes are: 62 families for protein-coding genes and 25 families for non-coding RNAs > 400 members; 80 families and 36 families > = 100 members and 137 families and 47 families > = 30 members. We calculated some parameters for each genome to observe the complexity and sampled some representatives (Table 1). First, there are cases with extremely properties, such as the plastid of the parasitic Babesia bovis’ (category: Alveolata; family: Babesiidae); it is much smaller than the median value and has the strongest strand imbalance (i.e, all genes on the same strand). The plastid of Porphyra purpurea’ (category: Rhodophyta; family: Bangiaceae) is another interesting example, whose genome is much larger and has more DPGs. Second, we computed strand ratio to estimate the strand biased gene distribution and found that the median value is larger than 1, which indicates that the strand bias is common among plastid genomes. Third, CDPGs are the most popular patterns and the percentage of CPGs and DPGs are often comparable. The median percentages of ‘Separation’, ‘Overlap’ and ‘Inclusion’ are 95.7%, 3.4% and 1.2%, respectively (data not shown), which suggests that most of the gene pairs are not overlapping. Last, the median length of transcripts is smaller than the median distance between TSS, despite that their extents vary among different species. It is also noted that the current genomic data sets cover a large scope of robust data structures.
Table 1. A basic survey for protein-coding genes of 13 species as examples of Plastid-LCGbase.
| Species | Genome length (nt) | Gene number | Strand ratio | CDPGs% | CPGs% | DPGs% | Median transcript length | Median TSS distance |
|---|---|---|---|---|---|---|---|---|
| Durinskia baltica | 116470 | 129 | 1.67 | 79.7 | 10.2 | 10.2 | 419 | 533 |
| Babesia bovis | 35107 | 32 | 33.00 | 100.0 | 0.0 | 0.0 | 581 | 592 |
| Cryptomonas paramecium | 77717 | 82 | 2.11 | 75.3 | 12.3 | 12.3 | 486.5 | 608 |
| Emiliania huxleyi | 105309 | 119 | 1.95 | 73.7 | 13.6 | 12.7 | 416 | 587 |
| Porphyra purpurea | 191028 | 209 | 1.45 | 66.8 | 16.8 | 16.3 | 518 | 580 |
| Cuscuta exaltata | 125373 | 67 | 1.65 | 71.2 | 13.6 | 15.2 | 554 | 1405 |
| Colocasia esculenta | 162424 | 86 | 1.84 | 72.9 | 12.9 | 14.1 | 546.5 | 1230 |
| Acidosasa purpurea | 139697 | 82 | 1.21 | 79.0 | 9.9 | 11.1 | 510.5 | 1040 |
| Cathaya argyrophylla | 107122 | 70 | 1.32 | 73.9 | 13.0 | 13.0 | 416 | 1143 |
| Aethionema cordifolium | 154168 | 84 | 1.77 | 72.3 | 13.3 | 14.5 | 630.5 | 1009 |
| Cicer arietinum | 125319 | 75 | 1.41 | 74.3 | 12.2 | 13.5 | 605 | 1125 |
| Gossypium anomalum | 159507 | 86 | 1.84 | 75.3 | 11.8 | 12.9 | 579.5 | 1221 |
| Allosyncarpia ternata | 159593 | 85 | 1.72 | 70.2 | 14.3 | 15.5 | 605 | 1158.5 |
| Median of 470 genomes | 154425.5 | 85 | 1.69 | 74.7 | 12.0 | 13.1 | 554 | 1087.5 |
Note: Genome length, the length of whole genome; Gene number, the number of protein-coding genes; Strand ratio, (the number of genes in dominate strand +1)/(the number of genes in the other strand +1); CDPGs%, CPGs% and DPGs% indicate the percentages of CDPGs, CPGs and DPGs among all gene pairs. Median transcript length and median TSS distance indicate the median values of transcript length and the distance between neighboring transcription start sites.
CASE STUDIES
The first case
A pair of CDPG, atpG (568247771) and atpF (568247772), responsible for ATP synthesis, from Porphyridium purpureum (Category: Rhodophyta; Order: Porphyridiales; Family: Porphyridiaceae) and their counterparts in other species appear in several lower plant species (Alveolata, Cryptophyta, Glaucocystophyceae, Haptophyceae, Rhodophyta and Stramenopiles) but they are absent in all Viridiplantae. This observation indicates that this gene pair has an ancient origin but suffered from gene loss when species are evolving. The TSS distances between the gene pair range 500–800 bp in most species with an exception of Porphyridium purpureum, whose TSS distance is slightly larger: around 1000–1200 bp. Furthermore, a conservation pattern appears expanded to their four neighboring genes, forming a cluster of atpI-atpH-atpG-atpF-atpD-atpA in all species but not in Cyanophora paradoxa, due to the loss of atpI, reflecting the unique feature of Glaucocystophyceae (Supplementary Figure S1).
The second case
A pair DPG involving psbN (11466817) and psbH (11466818), are part of the photosystem II in Oryza sativa (Category: Viridiplantae; Order: Poales; Family: Poaceae), which is shared among 432 species. Their TSS distances are very small (0–300 bp) in most species but become larger in Nephroselmis olivacea (300–500 bp), Chlamydomonas reinhardtii (500–800 bp) and Pleodorina starrii (500–800 bp). The short TSS is ancient pattern since it exists in both Viridiplantae and non-Viridiplantae species. When looking at the Family Fabaceae, the gene clusters containing the pair becomes separated in different species, showing subtle differences. For example, a cluster of 20 consecutive genes concerning the pair (petL-petG-psaJ-rpl33-rps18-rpl20-rps12-clpP-psbB-psbT-psbN-psbH-petB-petD-rpoA-rps11-rpl36-rps8-rpl14-rpl16-rps3-rps19) are conserved not only in nine Glycine subspecies (Glycine canescens, Glycine cyrtoloba, Glycine dolichocarpa, Glycine falcata, Glycine max, Glycine soja, Glycine stenophita, Glycine syndetika and Glycine tomentella) but also in other related family members such as Lotus japonicus, Lupinus luteus, Medicago truncatula, Millettia pinnata and Castanea mollissima. However, subtle changes are found in other Fabaceae species. When comparing Lathyrus sativus with Lotus japonicus, we observed gene inversion and insertion-deletion: the left of psbB, a unit of eight genes (petL-petG-psaJ-rpl33-rps18-rpl20-rps12-clpP), was inversed and then rps12 was deleted between clpP and rpl20 in Lathyrus sativus (Supplementary Figure S2).
The third case
A pair of CPGs, petA (7525046) and psbJ (7525047), of Arabidopsis thaliana (Category: Viridiplantae; Order: Brassicales; Family: Brassicaceae), is part of the cytochrome complex and photosystem II reaction center protein, respectively. Their orthologs have been identified only in Viridiplantae, especially in Amborella trichopoda, which separates from other flowering plants in the very early stage of evolution (40). In particular, most of the TSS distances of such a pair are larger than 1500 bp. Together with the observations in various species, we speculate that this cluster (ndhJ-ndhK-ndhC-atpE-atpB-rbcL-accD-psaI-ycf4-cemA-petA-psbJ-psbL-psbF-psbE-petL-petG-psaJ-rpl33-rps18-rpl20-rps12) is ancestral among Angiosperms. However, there are still modifications of the cluster, which are found in different branches of plant taxa. For instance, an insertion of a hypothetical protein (134093208) between rbcL and accD in Populus trichocarpa and a deletion of accD between rbcL and psaI in several species, such as Brachypodium distachyon and Triticum aestivum, have been found (Supplementary Figure S3).
DISCUSSION AND CONCLUSION
Genomes and their genes, large or small, are always organized in order and orientation. The variation and conservation of such organizations in the context of lineages and closely related taxa and under mutation and selection over time are considered as an important part of genomic signatures. Information on plastid genomes is therefore of importance and worthy of a dedicated database. We started with analysis of paired genes to provide a window for gene co-regulation. The dynamics of neighboring gene pairs can be defined as loss of genes or loss of relationship, and it is useful in recognizing important evolutionary events and common ancestors. In fact, whole plastome has been used to construct phylogenetic trees for plants and to delineate the timing of speciation based on both sequence feature and gene order (41–43). We also believe that visualization of comparative genomics data helps the discovery of rules and patterns in gene orders and orientations. In addition, the precise measure of TSS distances between paired genes and the display of these distances are all useful in defining gene co-regulations, and the dynamic process of gene losses in plastid genomes and plastid-associated nuclear genes are all relevant in defining the functional network of plastid genes (44). We anticipate that plastid-LCGbase will be developed to become a principle bioinformatic resource for plastid study.
FUTURE PLANS
We have plans in mind to improve the current status of the database, including both the content and technique. First, we will incorporate gene family information to differentiate paralogs into different subcategories by estimating the timing of speciation and duplication. Second, we would like to develop intelligent modules to identify specific events for gene orientation and sequence changes to cope with user demands. Third, we also plan to tag evolutionarily conserved gene sets to their functional roles in terms of metabolic pathways and networks for studying mechanisms of co-regulation. Fourth, we will attempt to improve visual effects and make better gene alignment by introducing the concept of ‘gaps’, adding user-friendly operational options. Last, we will continue to update the database with newly acquired genomes and annotations and build automatic protocols for processing data and generating results at lesser key strikes.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China [31301030]; CAS Youth Innovation Promotion Association [to D.W.]. Funding for open access charge: National Natural Science Foundation of China [31301030] [to D.W.].
Conflict of interest statement. None declared.
REFERENCES
- 1.Elghabi Z., Ruf S., Bock R. Biolistic co-transformation of the nuclear and plastid genomes. Plant J. 2011;67:941–948. doi: 10.1111/j.1365-313X.2011.04631.x. [DOI] [PubMed] [Google Scholar]
- 2.Svab Z., Hajdukiewicz P., Maliga P. Stable transformation of plastids in higher plants. Proc. Natl Acad. Sci. U.S.A. 1990;87:8526–8530. doi: 10.1073/pnas.87.21.8526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gould S.B., Waller R.F., McFadden G.I. Plastid evolution. Annu. Rev. Plant Biol. 2008;59:491–517. doi: 10.1146/annurev.arplant.59.032607.092915. [DOI] [PubMed] [Google Scholar]
- 4.Bendich A.J. Why do chloroplasts and mitochondria contain so many copies of their genome. BioEssays. 1987;6:279–282. doi: 10.1002/bies.950060608. [DOI] [PubMed] [Google Scholar]
- 5.Archibald J.M. The puzzle of plastid evolution. Curr. Biol. 2009;19:R81–R88. doi: 10.1016/j.cub.2008.11.067. [DOI] [PubMed] [Google Scholar]
- 6.Smith D.R., Lee R.W. A plastid without a genome: evidence from the nonphotosynthetic green algal genus Polytomella. Plant Physiol. 2014;164:1812–1819. doi: 10.1104/pp.113.233718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Molina J., Hazzouri K.M., Nickrent D., Geisler M., Meyer R.S., Pentony M.M., Flowers J.M., Pelser P., Barcelona J., Inovejas S.A., et al. Possible loss of the chloroplast genome in the parasitic flowering plant Rafflesia lagascae (Rafflesiaceae) Mol. Biol. Evol. 2014;31:793–803. doi: 10.1093/molbev/msu051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barbrook A.C., Howe C.J., Purton S. Why are plastid genomes retained in non-photosynthetic organisms. Trends Plant Sci. 2006;11:101–108. doi: 10.1016/j.tplants.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 9.Oudot-Le Secq M.P., Grimwood J., Shapiro H., Armbrust E.V., Bowler C., Green B.R. Chloroplast genomes of the diatoms Phaeodactylum tricornutum and Thalassiosira pseudonana: comparison with other plastid genomes of the red lineage. Molecular Genet. Genomics. 2007;277:427–439. doi: 10.1007/s00438-006-0199-4. [DOI] [PubMed] [Google Scholar]
- 10.Turmel M., Otis C., Lemieux C. The complete chloroplast DNA sequence of the green alga Nephroselmis olivacea: insights into the architecture of ancestral chloroplast genomes. Proc. Natl Acad. Sci. U.S.A. 1999;96:10248–10253. doi: 10.1073/pnas.96.18.10248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Funk H.T., Berg S., Krupinska K., Maier U.G., Krause K. Complete DNA sequences of the plastid genomes of two parasitic flowering plant species, Cuscuta reflexa and Cuscuta gronovii. BMC Plant Biol. 2007;7:45. doi: 10.1186/1471-2229-7-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Koning A.P., Keeling P.J. The complete plastid genome sequence of the parasitic green alga Helicosporidium sp. is highly reduced and structured. BMC Biol. 2006;4:12. doi: 10.1186/1741-7007-4-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cai Z., Penaflor C., Kuehl J.V., Leebens-Mack J., Carlson J.E., dePamphilis C.W., Boore J.L., Jansen R.K. Complete plastid genome sequences of Drimys, Liriodendron, and Piper: implications for the phylogenetic relationships of magnoliids. BMC Evol. Biol. 2006;6:77. doi: 10.1186/1471-2148-6-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Janouskovec J., Liu S.L., Martone P.T., Carre W., Leblanc C., Collen J., Keeling P.J. Evolution of red algal plastid genomes: ancient architectures, introns, horizontal gene transfer, and taxonomic utility of plastid markers. PloS One. 2013;8:e59001. doi: 10.1371/journal.pone.0059001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deusch O., Landan G., Roettger M., Gruenheit N., Kowallik K.V., Allen J.F., Martin W., Dagan T. Genes of cyanobacterial origin in plant nuclear genomes point to a heterocyst-forming plastid ancestor. Mol. Biol. Evol. 2008;25:748–761. doi: 10.1093/molbev/msn022. [DOI] [PubMed] [Google Scholar]
- 16.Wang D., Wu Y.W., Shih A.C., Wu C.S., Wang Y.N., Chaw S.M. Transfer of chloroplast genomic DNA to mitochondrial genome occurred at least 300 MYA. Mol. Biol. Evol. 2007;24:2040–2048. doi: 10.1093/molbev/msm133. [DOI] [PubMed] [Google Scholar]
- 17.Race H.L., Herrmann R.G., Martin W. Why have organelles retained genomes. Trends Genet. 1999;15:364–370. doi: 10.1016/s0168-9525(99)01766-7. [DOI] [PubMed] [Google Scholar]
- 18.Raven J.A., Allen J.F. Genomics and chloroplast evolution: what did cyanobacteria do for plants. Genome Biol. 2003;4:209. doi: 10.1186/gb-2003-4-3-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davila Lopez M., Martinez Guerra J.J., Samuelsson T. Analysis of gene order conservation in eukaryotes identifies transcriptionally and functionally linked genes. PloS One. 2010;5:e10654. doi: 10.1371/journal.pone.0010654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ballouz S., Francis A.R., Lan R., Tanaka M.M. Conditions for the evolution of gene clusters in bacterial genomes. PLoS Comput. Biol. 2010;6:e1000672. doi: 10.1371/journal.pcbi.1000672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu X., Han B. Evolutionary conservation of neighbouring gene pairs in plants. Gene. 2009;437:71–79. doi: 10.1016/j.gene.2009.02.012. [DOI] [PubMed] [Google Scholar]
- 22.Xie B., Wang D., Duan Y., Yu J., Lei H. Functional networking of human divergently paired genes (DPGs) PloS One. 2013;8:e78896. doi: 10.1371/journal.pone.0078896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stoebe B., Kowallik K.V. Gene-cluster analysis in chloroplast genomics. Trends Genet. 1999;15:344–347. doi: 10.1016/s0168-9525(99)01815-6. [DOI] [PubMed] [Google Scholar]
- 24.O'Brien E.A., Zhang Y., Wang E., Marie V., Badejoko W., Lang B.F., Burger G. GOBASE: an organelle genome database. Nucleic Acids Res. 2009;37:D946–D950. doi: 10.1093/nar/gkn819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cui L., Veeraraghavan N., Richter A., Wall K., Jansen R.K., Leebens-Mack J., Makalowska I., dePamphilis C.W. ChloroplastDB: the chloroplast genome database. Nucleic Acids Res. 2006;34:D692–D696. doi: 10.1093/nar/gkj055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kurihara K., Kunisawa T. A gene order database of plastid genomes. Data Sci. J. 2004;3:60–79. [Google Scholar]
- 27.Maguire S.L., OhEigeartaigh S.S., Byrne K.P., Schroder M.S., O'Gaora P., Wolfe K.H., Butler G. Comparative genome analysis and gene finding in Candida species using CGOB. Mol. Biol. Evol. 2013;30:1281–1291. doi: 10.1093/molbev/mst042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lopez M.D., Samuelsson T. eGOB: eukaryotic gene order browser. Bioinformatics. 2011;27:1150–1151. doi: 10.1093/bioinformatics/btr075. [DOI] [PubMed] [Google Scholar]
- 29.Louis A., Muffato M., Roest Crollius H. Genomicus: five genome browsers for comparative genomics in eukaryota. Nucleic Acids Res. 2013;41:D700–D705. doi: 10.1093/nar/gks1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang D., Zhang Y., Fan Z., Liu G., Yu J. LCGbase: a comprehensive database for lineage-based co-regulated genes. Evol. Bioinform. Online. 2012;8:39–46. doi: 10.4137/EBO.S8540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Proost S., Van Bel M., Sterck L., Billiau K., Van Parys T., Van de Peer Y., Vandepoele K. PLAZA: a comparative genomics resource to study gene and genome evolution in plants. Plant Cell. 2009;21:3718–3731. doi: 10.1105/tpc.109.071506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kandimalla E.R., Bhagat L., Wang D., Yu D., Sullivan T., La Monica N., Agrawal S. Design, synthesis and biological evaluation of novel antagonist compounds of Toll-like receptors 7, 8 and 9. Nucleic Acids Res. 2013;41:3947–3961. doi: 10.1093/nar/gkt078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burger G., Lavrov D.V., Forget L., Lang B.F. Sequencing complete mitochondrial and plastid genomes. Nat. Protoc. 2007;2:603–614. doi: 10.1038/nprot.2007.59. [DOI] [PubMed] [Google Scholar]
- 34.Lohse M., Drechsel O., Kahlau S., Bock R. OrganellarGenomeDRAW–a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 2013;41:W575–W581. doi: 10.1093/nar/gkt289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grant J.R., Arantes A.S., Stothard P. Comparing thousands of circular genomes using the CGView Comparison Tool. BMC Genomics. 2012;13:202. doi: 10.1186/1471-2164-13-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., Madden T.L. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bussotti G., Raineri E., Erb I., Zytnicki M., Wilm A., Beaudoing E., Bucher P., Notredame C. BlastR–fast and accurate database searches for non-coding RNAs. Nucleic Acids Res. 2011;39:6886–6895. doi: 10.1093/nar/gkr335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Enright A.J., Van Dongen S., Ouzounis C.A. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 2002;30:1575–1584. doi: 10.1093/nar/30.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu Z., Hao B. CVTree update: a newly designed phylogenetic study platform using composition vectors and whole genomes. Nucleic Acids Res. 2009;37:W174–W178. doi: 10.1093/nar/gkp278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Soltis D.E., Soltis P.S. Amborella not a ‘basal angiosperm’? Not so fast. Am. J. Bot. 2004;91:997–1001. doi: 10.3732/ajb.91.6.997. [DOI] [PubMed] [Google Scholar]
- 41.Jansen R.K., Cai Z., Raubeson L.A., Daniell H., Depamphilis C.W., Leebens-Mack J., Muller K.F., Guisinger-Bellian M., Haberle R.C., Hansen A.K., et al. Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc. Natl Acad. Sci. U.S.A. 2007;104:19369–19374. doi: 10.1073/pnas.0709121104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.De Las Rivas J., Lozano J.J., Ortiz A.R. Comparative analysis of chloroplast genomes: functional annotation, genome-based phylogeny, and deduced evolutionary patterns. Genome Res. 2002;12:567–583. doi: 10.1101/gr.209402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yoon H.S., Hackett J.D., Ciniglia C., Pinto G., Bhattacharya D. A molecular timeline for the origin of photosynthetic eukaryotes. Mol. Biol. Evol. 2004;21:809–818. doi: 10.1093/molbev/msh075. [DOI] [PubMed] [Google Scholar]
- 44.Yagi Y., Shiina T. Recent advances in the study of chloroplast gene expression and its evolution. Front. Plant Sci. 2014;5:61. doi: 10.3389/fpls.2014.00061. [DOI] [PMC free article] [PubMed] [Google Scholar]


