Abstract
MatrixDB (http://matrixdb.ibcp.fr) is a freely available database focused on interactions established by extracellular proteins and polysaccharides. It is an active member of the International Molecular Exchange (IMEx) consortium and has adopted the PSI-MI standards for annotating and exchanging interaction data, either at the MIMIx or IMEx level. MatrixDB content has been updated by curation and by importing extracellular interaction data from other IMEx databases. Other major changes include the creation of a new website and the development of a novel graphical navigator, iNavigator, to build and expand interaction networks. Filters may be applied to build sub-networks based on a list of biomolecules, a specified interaction detection method and/or an expression level by tissue, developmental stage, and health state (UniGene data). Any molecule of the network may be selected and its partners added to the network at any time. Networks may be exported under Cytoscape and tabular formats and as images, and may be saved for subsequent re-use.
INTRODUCTION
MatrixDB (http://matrixdb.ibcp.fr) is a freely available interaction database, first released in 2009, which focuses on interactions established by extracellular proteins and polysaccharides (1,2). MatrixDB is an active member of the International Molecular Exchange (IMEx) consortium, an international collaboration between a group of major public interaction data providers who have agreed to share curation effort and to capture full details of an interaction in a ‘deep’ curation model. The members of the IMEx consortium use a single set of curation rules when capturing interaction data from publications in peer-reviewed journals (3). MatrixDB is in charge since January 2009 of the curation of interaction data from articles published in Matrix Biology, a journal affiliated with the American Society for Matrix Biology and with the International Society for Matrix Biology. Other papers reporting interaction data for the extracellular matrix (ECM) full-length biomolecules and fragments (matricryptins) involved in angiogenesis, ECM maturation and neurodegenerative diseases are also curated. Previous curation performed by MatrixDB was at the MIMIx level (The minimum information required for reporting a molecular interaction experiment, (4)). MatrixDB now performs deep curation at the IMEx level and uses the web-based curation tool developed by IntAct, which is a common curation platform for 11 molecular interaction databases (5).
Other ECM-related databases collect data on the sequences or mutations of extracellular proteins or on a single extracellular protein family. For example, ElastoDB is a repository for elastin sequences (6), the laminin database stores information concerning laminins in health and disease (7), COLdb links mutations and genetic data to molecular function in fibrillar collagens (8) and the COL7A1 mutation database is focused on collagen VII (9). To the best of our knowledge MatrixDB is the only general-purpose database curating and storing interactions taking place in the ECM and at the cell surface.
MatrixDB is not restricted to protein–protein interactions like most other databases, but also focuses on protein–glycosaminoglycan interactions. Glycosaminoglycans are complex polysaccharides that are abundant in the ECM and at the cell surface, where they play key roles in adhesion and signaling (10). MatrixDB data have been used to build interaction networks of extracellular proteins (11,12), glycosaminoglycans (heparin and heparan sulfate, (13,14)), proteoglycans (15), basement membrane (a specialized ECM, (1)), the ECM (16) or physiological process (aging, (17)).
Several protein interaction network visualizers have been developed recently such as the Web-based Protein Interaction Network Visualizer, which allows interaction data to be explored interactively on line (18), or mentha, a resource for browsing and filtering integrated protein interaction networks (19). The updated version of MatrixDB features its own novel network navigator, iNavigator, which has been designed to build and query interaction networks in real time using several criteria.
MATRIXDB CONTENT
Sources of biomolecules
The biomolecules stored in MatrixDB are proteins, bioactive fragments of ECM biomolecules called matricryptins (20), glycosaminoglycans, synthetic peptides, ions, lipids and other inorganic compounds. Proteins are identified by their UniProtKB primary accession numbers (21), while custom MatrixDB identifiers are used for all other types of biomolecules: PFRAG_x_s for bioactive fragments (x being a number and s a species), GAG_x for glycosaminoglycans, SPEP_x for synthetic peptides, CAT_x for ions, LIP_x for lipids and INORG_x for other inorganic compounds. In addition, a number of ECM proteins and receptors (e.g. collagens, laminins, thrombospondins and integrins) are composed of several polypeptide chains and the native protein molecules are thus obligate complexes. They are termed MULT_x_s (for multimers, again x is a number and s a species). Protein data were imported from UniProtKB, and enriched by parsing additional annotations from Gene Ontology, InterPro, Pfam and UniGene (http://www.ncbi.nlm.nih.gov/unigene). The custom MatrixDB biomolecules are cross-referenced to relevant external databases when available. We reference the PRO feature of UniProtKB for bioactive fragments, the Chemical Entities of Biological Interest (CheBI, (22)) for glycosaminoglycans, ions, lipids and other inorganic compounds, the Lipid bank (http://www.lipidbank.jp/) for lipids and the complex portal (http://www.ebi.ac.uk/intact/complex/) for multimers.
Although MatrixDB is focused on Homo sapiens, relevant ECM interactions are sometimes experimentally demonstrated with biomolecules from other organisms. We therefore selected a few species of interest and imported all protein records from SwissProt (chicken, bovine, dog, guinea pig, human, mouse, pig, rat, sheep, zebrafish) and/or TrEMBL (bacteria, bovine, dictyostelium discoideum, human, pig, sheep, zebrafish) for these species.
Three lists of human proteins of particular interest, membrane proteins, secreted proteins and ECM proteins, were built based on selected UniProtKB keywords, Gene Ontology terms and the human matrisome data set (23). Membrane proteins were defined by the UniProtKB keyword 0472 and the GO term 0016020; secreted proteins by UniProtKB keyword 0964, GO terms 0005576 and 0005615 and the ‘matrisome-associated’ category from the matrisome data set (24); and ECM proteins were selected with UniProtKB keywords 0272 and 0084, GO terms 0005578, 0005201, 0005604, 0031012 and 0005605, and the ‘core-matrisome’ category (24). Furthermore, non-protein biomolecules were also annotated as belonging to one of these three lists when appropriate. The complete lists comprise 7144 membrane biomolecules, 2420 secreted biomolecules and 547 ECM biomolecules including 462 proteins and 85 other biomolecules (bioactive fragments, multimers and glycosaminoglycans).
Sources of interaction data
The content of MatrixDB has been updated with new interaction data manually curated by the MatrixDB team, and by importing interaction data from four interaction databases of the IMEx consortium via The Proteomics Standard Initiative Common QUery InterfaCe (PSICQUIC), a community standard for computational access to molecular-interaction data resources (25). The queried databases are IntAct (26), MINT (27), DIP (28) and InnateDB (29). All interactions involving at least one biomolecule from our ‘secreted’ or ‘ECM’ lists were imported, provided that the other partner originated from human, murine, bovine, chicken or zebrafish. Experimental details such as the participant features (e.g. binding sites, effect of mutations) and interaction parameters (e.g. affinity, kinetics) were imported when available.
In the previous version of MatrixDB, the use of homology to infer human interactions from interactions experimentally identified using non-human biomolecules was somewhat confusing. This has been clarified in the new version of the database, where a clear distinction is made between interactions supported by experimental evidence, tagged as ‘genuine’, and those inferred by homology, tagged as ‘inferred’ and linked back to the non-human interaction.
Data from the Human Protein Reference Database (HPRD) (30) were not included in the update because curated HPRD interaction data are limited to interactor names and experiment type, which is not reported with the controlled vocabulary used by the IMEx databases (Ontology Lookup Service, (31) whereas MatrixDB aims at providing a rich and detailed view of ECM interactions. Furthermore HPRD has not yet implemented the PSICQUIC service we use to retrieve extracellular interaction data in other databases. Interaction data from BioGRID (32) were not included because the BioGRID database captures interactions at the gene level. Although a mapping to the canonical protein is provided, interactions involving a specific protein isoform and/or a processed protein fragments (matricryptins), which play crucial roles in the ECM, cannot be represented.
In the current release MatrixDB contains 904 interactions supported by 1244 experiments, which have been manually curated from 237 publications, compared to 490 interactions supported by 847 experiments in the previous version of the database (2). This is the MatrixDB ‘core’ data set. Furthermore, 8947 ECM-related interactions supported by 13851 experiments have been imported, resulting in the ‘PSICQUIC-extended’ data set. Both data sets are freely available in MITAB 2.7 format on the ‘Download’ page. MatrixDB now contains 15095 experiments supporting 9851 interactions. These include 9262 protein–protein interactions, 139 protein–multimer interactions, 175 protein–glycosaminoglycan interactions, 22 protein–ion interactions, 5 protein–lipid interactions and 5 protein–synthetic peptide interactions compared to 2174 interactions available in the previous release.
BROWSING MATRIXDB
Tutorials, which are accessed by clicking on question marks, are available on the web site as videos.
Querying MatrixDB content
All MatrixDB queries are now performed using a novel search bar on the home page. Text typed by the user is searched for author and journal names, biomolecule common names and synonyms, titles and abstracts of publications, PubMed identifiers, UniProtKB, ChEBI, complex and MatrixDB identifiers, gene names and UniProtKB keywords. The results are presented as lists of relevant items in the following categories: biomolecules, UniProtKB keywords, publications and authors. The retrieved items of each category are ranked according to the number of interactions related to the item (e.g. the number of interactions involving a biomolecule or reported in a publication). If no match is found in a category for the query text, that category is not displayed. Retrieved biomolecules can be restricted to human-only thanks to a checkbox.
Biomolecule report page
Biomolecule data are displayed in several boxes on a single page (Figure 1A). A first box containing the MatrixDB identifier is always displayed, whereas the other boxes (biological function and location data, complex information, cross-references, domain annotation and 3D structure) are displayed only when the corresponding information is available (Figure 1A).
Figure 1.
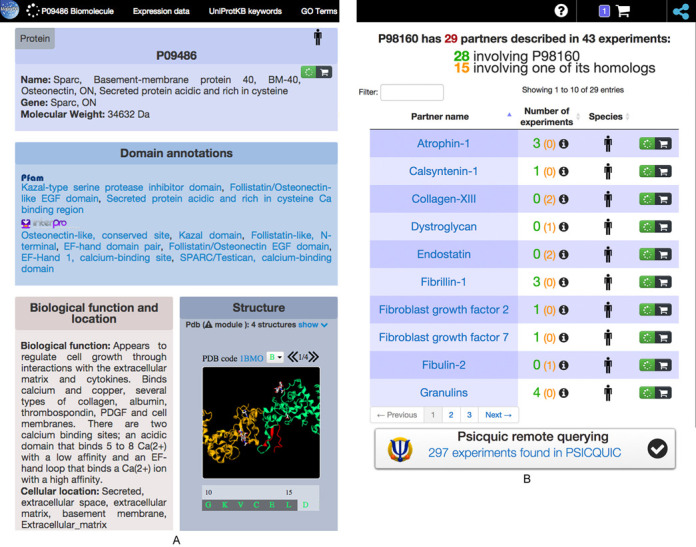
(A) The biomolecule report page: biomolecule data. The top bar allows to quickly scroll to the various subsections of the report page. Biomolecule-specific information is shown as boxes. When their 3D structures are available, they can be rotated. (B) The biomolecule report page: interaction data. The biomolecule's ECM partners stored in MatrixDB are listed, along with the number of individual experiments that support each interaction. Clicking on such a number pops up a window listing the experiments and linking to their report pages. The counts and experiments are colored green when involving the molecule itself, and orange when a homolog from another species was used in the experiment. Additional partners found in other cellular compartments can be identifed through the PSICQUIC remote-query widget. The number of items currently in the cart is displayed at the top, and provides a direct access to iNavigator (network icon).
The individual polypeptide chains comprising multimeric proteins referred to as multimers or complexes (e.g. collagens, laminins, thrombospondins) and their stoichiometry are listed in the ‘Complex composition’ box with a link to the polypeptide report page. Protein domains (Pfam, (33) and Interpro, (34)) are listed. The 3D structure of the protein, selected from the PDB entries selected by UniProtKB (35), is displayed and can be rotated. The cellular and tissue location of the biomolecule is annotated using UniProtKB keywords and GO ‘Cellular component’ terms.
The partners of the biomolecule, their identifier, their species and the number of experiments supporting their interaction with the biomolecule are listed (Figure 1B). They can be sorted alphabetically by their names, ranked by the number of experiments supporting the interaction and filtered according to their name or species. The partners can be added to a cart for later building of interaction networks as described below. The total number of additional partners available in all the PSICQUIC-enabled databases is also indicated, based on an on-the-fly query (Figure 1B). Expression data imported from UniGene are displayed as a bar chart for body sites (tissues), developmental stages and health states with a focus on tumors. UniProtKB keywords, which can be sorted alphabetically or by their identifiers, and GO terms annotating the biomolecule are listed in two separate tables with their definitions and external links.
Experiment report page
It comprises the interaction identifier, the name of the database which curated the interaction data, the interaction type, the interaction detection method, kinetics and affinity of the interactions when available and the PubMed reference of the publication(s) supporting the interaction (Figure 2). These information are provided for all interactions (i.e. those curated by MatrixDB and those imported from IMEx databases via PSICQUIC). Further information on participant features (e.g. biological role, experimental role, detection method, binding sites and effects of mutations) and on interactions (kinetics, affinity) are displayed when available.
Figure 2.
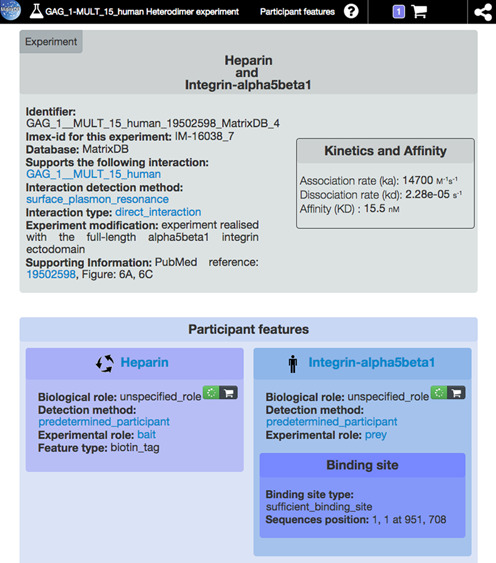
The experiment report page. As in the biomolecule report page, the top bar allows to scroll to subsections, displays the current cart size and provides direct access to iNavigator. The gray box displays information about the experiment (interaction type, detection method, external references and supporting information). When available, kinetics and affinity data are also shown. Specific information concerning each partner is presented below in blue boxes (e.g. binding sites, biochemical modifications and mutations).
BUILDING INTERACTION NETWORKS WITH iNAVIGATOR
When browsing MatrixDB, the user can select any publication, biomolecule or UniProtKB keyword and build the corresponding interaction network via iNavigator, a novel interactome network navigator we have developed (Figure 3). The selected items are collected in a cart, which is available on all MatrixDB web pages, and can be deleted from the cart at any time. Interaction networks may be modified either by changing the cart content or by clicking and dragging elements directly on the network. The network can be expanded either by adding the partners and interactions of biomolecules selected within the displayed network, or by querying MatrixDB via the search bar for further publications, biomolecules and keywords. New partners and interactions are added in real time. The process can be repeated iteratively to increase network coverage. Conversely, the user can delete biomolecules and interactions from the network.
Figure 3.
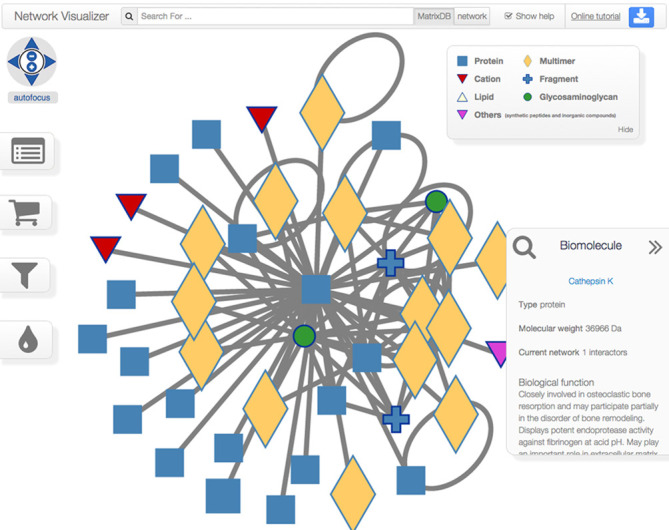
Visualization of a network in iNavigator. The navigation bar features a search box. The list of results can be toggled between hits found in MatrixDB or hits found in the displayed network. A button in the top-right corner is used to export the current network or reload a previously saved network. On the left-hand side, buttons provide access to the four main widgets: the tabular network widget, the cart, the network filter widget and a palette widget to change the color of selected nodes. The bottom-right ‘magnifying glass’ widget keeps track of recently visited network elements and allows for the quick inspection of biomolecules and interactions. Hovering the mouse over a node or edge in the network adds that node or edge to the ‘recently visited’ list.
Viewing information in the network
Information associated with biomolecules and interactions are displayed in the ‘Tabular network widget’, accessed by clicking on its icon (left-hand side of the screen). This widget provides additional information on biomolecules (identifier, common name with a link to the ‘Biomolecule report page’ and number of interactions) and interactions (genuine or inferred to human, identifier of both participants, number of experiments supporting the interaction). It also allows modifying the cart content and the displayed network. Biomolecules can be selected either individually or collectively. By clicking on the button ‘Toggle node labels’ one can display the biomolecule name, species, the gene symbol and the number of partners.
Querying displayed interaction networks
The user can search for free text, biomolecule names or identifiers (UniProtKB, CheBI and MatrixDB, EBI complexes) using the search bar displayed at the top of the network visualizer. Matching biomolecules are highlighted in red and yellow. The iNavigator can also search for biomolecules based on the existence - or absence - of partners after selection and filter applications of the initial network. The iNavigator uses graphical layers to project information on the network. In particular, tissue and/or health state specific sub-networks can be built thanks to the ‘Filter network widget’ found below the ‘Tabular network widget’ and ‘Cart’ icons (left-hand side of the screen). The user can select one or more health states and tissues of interest and specify an expression threshold in transcripts per million for each selected tissue (Figure 4). Biomolecules expressed above the threshold in all selected tissues are displayed in green, while others are displayed in red. Biomolecules lacking expression data may be included or not in the interaction network thanks to the MDE (Missing Data Elements) checkbox. Using the ‘Filter network widget’, one can also build a network comprised of interactions identified by one or several experimental methods. Interactions detected by at least one of the selected methods are colored in green, whereas those detected by other methods are colored in red. Red biomolecules and/or interactions are masked when clicking ‘Apply’.
Figure 4.
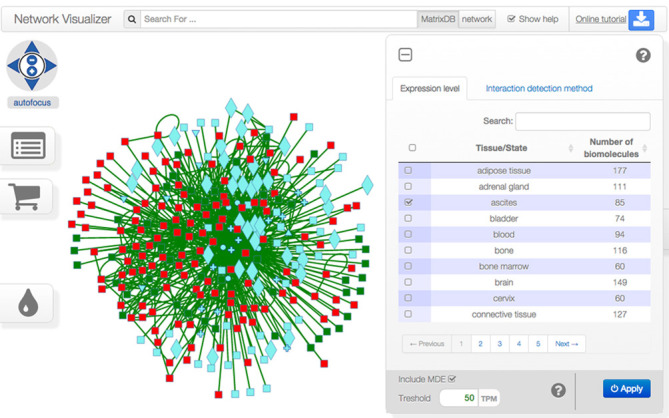
The network filter widget. The widget displays two filters: ‘expression level’ (in transcripts per million) for biomolecules, and ‘detection method’ for interactions. Checking several boxes imposes all criteria to be met (i.e. logical AND). An expression level (TPM) threshold can be specified, and applies to all selected tissues/health state/developmental stages. The widget uses a color code to preview the effect of a filter on network elements: green elements meet the criteria while red ones do not. Biomolecules that lack expression data are colored in cyan and called Missing Data Elements (MDE). They can be excluded from the network by unchecking the MDE box, hence appearing in red. Clicking on the apply button will hide all the red elements.
Export of interaction networks generated with the iNavigator
Interaction networks can be exported under three different formats: a tabular format (MITAB 2.7) for data exchange, the .sif format for visualizing and analysing interaction networks in Cytoscape (36) and the .png format for saving graphical displays of the networks. In addition, networks built with iNavigator can be saved directly on the MatrixDB server at any time, and subsequently retrieved for one month using a unique private key.
Implementation
iNavigator has been developed using two well-established JavaScript libraries, namely D3 (37) and jQuery (https://jquery.com/). The tables of the report pages use the DataTables plugin (http://www.datatables.net/) and the visualization component for 3D structures of biomolecules relies on GLmol (http://sourceforge.jp/projects/webglmol/). iNavigator visualization widgets were built as individual reusable components, following the BioJs open-source standard for biological data visualisation (38). Additionally, iNavigator strives to minimize response time. In particular, all data queries are performed server-side against our local MatrixDB database, rather than querying PSICQUIC in real time.
We applied the Ajax methodology to decouple the network's construction and the retrieval of data associated with biomolecules and interactions. This ensures that networks can be displayed and modified via iNavigator while data are still being retrieved in the background. All modifications (e.g. filters) made on the networks are reversible. Indeed, biomolecules and/or interactions deleted from the networks are in fact only masked and remain accessible. More importantly, the interactions involving masked biomolecules are integrated when new biomolecules are added to the network.
MatrixDB literature-curated interactions are provided to the community as downloadable PSI-MI TAB files as well as through a PSICQUIC web service. In this release of MatrixDB, these files and the web service have been upgraded to the MITAB 2.7 format.
Prospects include the development of the PSI-MI XML 3.0 format to export MatrixDB data. Further improvements include the visualization of binding sites and/or mutations interfering with interactions on the 3D structure of biomolecules displayed on the Biomolecule report page. Further criteria (affinity, kinetic parameters) will be added to the filter network widget to prioritize interactions in the networks.
Acknowledgments
We thank Christophe Blanchet and Alexis Michon (FR 3302, Institut de Biologie et Chimie des Protéines, Lyon, France) for helpful discussions and technical assistance, Sylvain D. Vallet for his contribution to the curation process, Marine Dumousseau (European Bioinformatics Institute, Hinxton, UK) for her help regarding data exchange and Sandra Orchard (European Bioinformatics Institute, Hinxton, UK) for her expert assistance with curation issues.
FUNDING
Centre National de la Recherche Scientifique [PEPS BMI R13M to G.L., N.T.M. and S.R.B.]; European Union FP7 (PSIMEx) [FP7-HEALTH-2007-223411 to S.R.B.]. Funding for open access charge: Centre National de la Recherche Scientifique [PEPS BMI R13M].
Conflict of interest statement. None declared.
REFERENCES
- 1.Chautard E., Ballut L., Thierry-Mieg N., Ricard-Blum S. MatrixDB, a database focused on extracellular protein-protein and protein-carbohydrate interactions. Bioinforma. Oxf. Engl. 2009;25:690–691. doi: 10.1093/bioinformatics/btp025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chautard E., Fatoux-Ardore M., Ballut L., Thierry-Mieg N., Ricard-Blum S. MatrixDB, the extracellular matrix interaction database. Nucleic Acids Res. 2011;39:D235–D240. doi: 10.1093/nar/gkq830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Orchard S., Kerrien S., Abbani S., Aranda B., Bhate J., Bidwell S., Bridge A., Briganti L., Brinkman F., Cesareni G., et al. Protein interaction data curation: the International Molecular Exchange (IMEx) consortium. Nat. Methods. 2012;9:345–350. doi: 10.1038/nmeth.1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Orchard S., Salwinski L., Kerrien S., Montecchi-Palazzi L., Oesterheld M., Stümpflen V., Ceol A., Chatr-aryamontri A., Armstrong J., Woollard P., et al. The minimum information required for reporting a molecular interaction experiment (MIMIx) Nat. Biotechnol. 2007;25:894–898. doi: 10.1038/nbt1324. [DOI] [PubMed] [Google Scholar]
- 5.Orchard S., Ammari M., Aranda B., Breuza L., Briganti L., Broackes-Carter F., Campbell N.H., Chavali G., Chen C., del-Toro N., et al. The MIntAct project–IntAct as a common curation platform for 11 molecular interaction databases. Nucleic Acids Res. 2014;42:D358–D363. doi: 10.1093/nar/gkt1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.He D., Chung M., Chan E., Alleyne T., Ha K.C.H., Miao M., Stahl R.J., Keeley F.W., Parkinson J. Comparative genomics of elastin: Sequence analysis of a highly repetitive protein. Matrix Biol. J. Int. Soc. Matrix Biol. 2007;26:524–540. doi: 10.1016/j.matbio.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 7.Golbert D.C.F., Santana-van-Vliet E., Mundstein A.S., Calfo V., Savino W., de Vasconcelos A.T.R. Laminin-database v.2.0: an update on laminins in health and neuromuscular disorders. Nucleic Acids Res. 2014;42:D426–D429. doi: 10.1093/nar/gkt901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bodian D.L., Klein T.E. COLdb, a database linking genetic data to molecular function in fibrillar collagens. Hum. Mutat. 2009;30:946–951. doi: 10.1002/humu.20978. [DOI] [PubMed] [Google Scholar]
- 9.Wertheim-Tysarowska K., Sobczyńska-Tomaszewska A., Kowalewski C., Skroński M., Swięćkowski G., Kutkowska-Kaźmierczak A., Woźniak K., Bal J. The COL7A1 mutation database. Hum. Mutat. 2012;33:327–331. doi: 10.1002/humu.21651. [DOI] [PubMed] [Google Scholar]
- 10.Sarrazin S., Lamanna W.C., Esko J.D. Heparan sulfate proteoglycans. Cold Spring Harb. Perspect. Biol. 2011;3 doi: 10.1101/cshperspect.a004952. doi:10.1101/cshperspect.a004952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Faye C., Chautard E., Olsen B.R., Ricard-Blum S. The first draft of the endostatin interaction network. J. Biol. Chem. 2009;284:22041–22047. doi: 10.1074/jbc.M109.002964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Salza R., Peysselon F., Chautard E., Faye C., Moschcovich L., Weiss T., Perrin-Cocon L., Lotteau V., Kessler E., Ricard-Blum S. Extended interaction network of procollagen C-proteinase enhancer-1 in the extracellular matrix. Biochem. J. 2014;457:137–149. doi: 10.1042/BJ20130295. [DOI] [PubMed] [Google Scholar]
- 13.Ori A., Wilkinson M.C., Fernig D.G. A systems biology approach for the investigation of the heparin/heparan sulfate interactome. J. Biol. Chem. 2011;286:19892–19904. doi: 10.1074/jbc.M111.228114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peysselon F., Ricard-Blum S. Heparin-protein interactions: from affinity and kinetics to biological roles. Application to an interaction network regulating angiogenesis. Matrix Biol. J. Int. Soc. Matrix Biol. 2014;35:73–81. doi: 10.1016/j.matbio.2013.11.001. [DOI] [PubMed] [Google Scholar]
- 15.Peysselon F., Fatoux-Ardore M., Ricard-Blum S. From binary interactions of glycosaminoglycan and proteoglycans to interaction networks. In: Balazs EA, editor. Structure and Function of Biomatrix: Control of Cell Behavior and Gene Expression . Vol. 5. Edgewater, NJ: Matrix Biology Institute; 2012. pp. 139–174. [Google Scholar]
- 16.Cromar G.L., Xiong X., Chautard E., Ricard-Blum S., Parkinson J. Toward a systems level view of the ECM and related proteins: a framework for the systematic definition and analysis of biological systems. Proteins. 2012;80:1522–1544. doi: 10.1002/prot.24036. [DOI] [PubMed] [Google Scholar]
- 17.Chautard E., Thierry-Mieg N., Ricard-Blum S. Interaction networks as a tool to investigate the mechanisms of aging. Biogerontology. 2010;11:463–473. doi: 10.1007/s10522-010-9268-5. [DOI] [PubMed] [Google Scholar]
- 18.Salazar G.A., Meintjes A., Mazandu G.K., Rapanoël H.A., Akinola R.O., Mulder N.J. A web-based protein interaction network visualizer. BMC Bioinformatics. 2014;15 doi: 10.1186/1471-2105-15-129. doi:10.1186/1471-2105-15-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Calderone A., Castagnoli L., Cesareni G. mentha: a resource for browsing integrated protein-interaction networks. Nat. Methods. 2013;10:690–691. doi: 10.1038/nmeth.2561. [DOI] [PubMed] [Google Scholar]
- 20.Ricard-Blum S., Salza R. Matricryptins and matrikines: biologically active fragments of the extracellular matrix. Exp. Dermatol. 2014;23:457–463. doi: 10.1111/exd.12435. [DOI] [PubMed] [Google Scholar]
- 21.UniProt Consortium. Activities at the Universal Protein Resource (UniProt) Nucleic Acids Res. 2014;42:D191–D198. doi: 10.1093/nar/gkt1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hastings J., de Matos P., Dekker A., Ennis M., Harsha B., Kale N., Muthukrishnan V., Owen G., Turner S., Williams M., et al. The ChEBI reference database and ontology for biologically relevant chemistry: enhancements for 2013. Nucleic Acids Res. 2013;41:D456–D463. doi: 10.1093/nar/gks1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Naba A., Hoersch S., Hynes R.O. Towards definition of an ECM parts list: an advance on GO categories. Matrix Biol. J. Int. Soc. Matrix Biol. 2012;31:371–372. doi: 10.1016/j.matbio.2012.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Naba A., Clauser K.R., Hoersch S., Liu H., Carr S.A., Hynes R.O. The matrisome: in silico definition and in vivo characterization by proteomics of normal and tumor extracellular matrices. Mol. Cell. Proteomics. 2012;11 doi: 10.1074/mcp.M111.014647. doi: 10.1074/mcp.M111.014647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aranda B., Blankenburg H., Kerrien S., Brinkman F.S.L., Ceol A., Chautard E., Dana J.M., De Las Rivas J., Dumousseau M., Galeota E., et al. PSICQUIC and PSISCORE: accessing and scoring molecular interactions. Nat. Methods. 2011;8:528–529. doi: 10.1038/nmeth.1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kerrien S., Aranda B., Breuza L., Bridge A., Broackes-Carter F., Chen C., Duesbury M., Dumousseau M., Feuermann M., Hinz U., et al. The IntAct molecular interaction database in 2012. Nucleic Acids Res. 2012;40:D841–D846. doi: 10.1093/nar/gkr1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Licata L., Briganti L., Peluso D., Perfetto L., Iannuccelli M., Galeota E., Sacco F., Palma A., Nardozza A.P., Santonico E., et al. MINT, the molecular interaction database: 2012 update. Nucleic Acids Res. 2012;40:D857–D861. doi: 10.1093/nar/gkr930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Salwinski L., Miller C.S., Smith A.J., Pettit F.K., Bowie J.U., Eisenberg D. The Database of Interacting Proteins: 2004 update. Nucleic Acids Res. 2004;32:D449–D451. doi: 10.1093/nar/gkh086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Breuer K., Foroushani A.K., Laird M.R., Chen C., Sribnaia A., Lo R., Winsor G.L., Hancock R.E.W., Brinkman F.S.L., Lynn D.J. InnateDB: systems biology of innate immunity and beyond–recent updates and continuing curation. Nucleic Acids Res. 2013;41:D1228–D1233. doi: 10.1093/nar/gks1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Keshava Prasad T.S., Goel R., Kandasamy K., Keerthikumar S., Kumar S., Mathivanan S., Telikicherla D., Raju R., Shafreen B., Venugopal A., et al. Human Protein Reference Database–2009 update. Nucleic Acids Res. 2009;37:D767–D772. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Côté R., Reisinger F., Martens L., Barsnes H., Vizcaino J.A., Hermjakob H. The Ontology Lookup Service: bigger and better. Nucleic Acids Res. 2010;38:W155–W160. doi: 10.1093/nar/gkq331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chatr-Aryamontri A., Breitkreutz B.-J., Heinicke S., Boucher L., Winter A., Stark C., Nixon J., Ramage L., Kolas N., O'Donnell L., et al. The BioGRID interaction database: 2013 update. Nucleic Acids Res. 2013;41:D816–D823. doi: 10.1093/nar/gks1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Finn R.D., Bateman A., Clements J., Coggill P., Eberhardt R.Y., Eddy S.R., Heger A., Hetherington K., Holm L., Mistry J., et al. Pfam: the protein families database. Nucleic Acids Res. 2014;42:D222–D230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hunter S., Jones P., Mitchell A., Apweiler R., Attwood T.K., Bateman A., Bernard T., Binns D., Bork P., Burge S., et al. InterPro in 2011: new developments in the family and domain prediction database. Nucleic Acids Res. 2012;40:D306–D312. doi: 10.1093/nar/gkr948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martin A.C.R. Mapping PDB chains to UniProtKB entries. Bioinformatics. 2005;21:4297–4301. doi: 10.1093/bioinformatics/bti694. [DOI] [PubMed] [Google Scholar]
- 36.Lotia S., Montojo J., Dong Y., Bader G.D., Pico A.R. Cytoscape app store. Bioinformatics. 2013;29:1350–1351. doi: 10.1093/bioinformatics/btt138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bostock M., Ogievetsky V., Heer J. D3: Data-Driven Documents. IEEE Trans. Vis. Comput. Graph. 2011;17:2301–2309. doi: 10.1109/TVCG.2011.185. [DOI] [PubMed] [Google Scholar]
- 38.Gómez J., García L.J., Salazar G.A., Villaveces J., Gore S., García A., Martín M.J., Launay G., Alcántara R., Del-Toro N., et al. BioJS: an open source JavaScript framework for biological data visualization. Bioinformatics. 2013;29:1103–1104. doi: 10.1093/bioinformatics/btt100. [DOI] [PMC free article] [PubMed] [Google Scholar]


