Abstract
MyMpn (http://mympn.crg.eu) is an online resource devoted to studying the human pathogen Mycoplasma pneumoniae, a minimal bacterium causing lower respiratory tract infections. Due to its small size, its ability to grow in vitro, and the amount of data produced over the past decades, M. pneumoniae is an interesting model organisms for the development of systems biology approaches for unicellular organisms. Our database hosts a wealth of omics-scale datasets generated by hundreds of experimental and computational analyses. These include data obtained from gene expression profiling experiments, gene essentiality studies, protein abundance profiling, protein complex analysis, metabolic reactions and network modeling, cell growth experiments, comparative genomics and 3D tomography. In addition, the intuitive web interface provides access to several visualization and analysis tools as well as to different data search options. The availability and—even more relevant—the accessibility of properly structured and organized data are of up-most importance when aiming to understand the biology of an organism on a global scale. Therefore, MyMpn constitutes a unique and valuable new resource for the large systems biology and microbiology community.
INTRODUCTION
A huge amount of databases on the most diverse topics in biology is available nowadays via the world wide web. Some of those databases focus on general information about biological numbers (Bionumbers DB), enzymes (BRENDA) or proteins (UniProt), genes and pathways (KEGG) or about biological models (BioModels DB) (1–5). Others provide access to information related to a specific organism, such as EcoCyc covering genomic and metabolomic data on Escherichia coli K-12 (6) or SubtiWiki providing diverse information for Bacillus subtilis (7).
Mycoplasma pneumoniae, an obligate human parasite colonizing the lung epithelium, is involved in several diseases, amongst them walking pneumonia (8,9). It can be grown in vitro using rich and defined media (10,11), thus allowing to study its behavior under controlled laboratory conditions. Related to the reduced genome, the metabolic network of M. pneumoniae has been shown to be simple and linear in comparison to more complex organisms, such as E. coli (11–13). It is unable to produce the precursors of most cellular building blocks, for example amino acids, fatty acids or nucleic bases, on its own but depends on the supply of those nutrients by the host or the growth medium. On the other hand, the genome organization has been shown to be far more complex than anticipated including a large amount of ncRNAs and important regulatory, e.g. non-coding, genomic regions (14,15). As a result, M. pneumoniae is small enough to be analyzed on a global scale and at the same time complex enough to study general biological principles, rendering it one of the most promising model organisms for systems biology and toward understanding of an organism in its entirety. Accordingly, a large amount of genome-scale datasets has been produced during the past decades (11–25).
The MyMpn database, available at http://mympn.crg.eu, aims to provide access to properly structured and well organized data for M. pneumoniae. It includes a multitude of datasets on genomics, transcriptomics, proteomics and metabolomics resulting from experimental and computational analyses as well as information on general cellular properties. Experimental results include amongst others DNA, RNA and protein sequencing data, microarrays, mass spectrometry (MS) and tandem affinity purification (TAP) analyses, metabolic assays, growth curve (GC) measurements. Computational data for example has been obtained by determining the theoretical coding capabilities of the genome, by sequence comparison (internally and to other organisms), protein domain scans, and a metabolic model. The web interface is highly searchable, offering basic keyword queries, a BLAST (26) search mask and several menu browsing options, including an interactive genome browser (MyGBrowser: ‘Materials and Methods’) that is fully embedded in the logic of the website. In addition, it provides diverse visualization and analysis tools, e.g. an interactive tool for the comparison of growth curve measurements, a customizable genome browser (Gbrowse: (27)) to upload and visualize experimental data in the genomic context in a private section and a clickable metabolic map that links to diverse data on genes, proteins, metabolites and reactions. In order to provide the systems biology and microbiology community with most up-to-date information on M. pneumoniae, our database is regularly updated whenever new data are provided by the experimentalists to the MyMpn team.
MATERIALS AND METHODS
MyMpn architecture and implementation
MyMpn data is stored within a relational database management system, MySQL (http://www.mysql.com). The website was developed using the PHP language and the Apache web server. Our bioinformatics software is written in Perl and uses the Bioperl toolkit (28). Computational analyses were run on a Linux cluster running the Gentoo Linux distribution (http://www.gentoo.org) and the PBS scheduling system (http://www.openpbs.org).
MyGBrowser
To assist researchers in the mining of genomic annotation stored in the MyMPN database, we have developed a dedicated genome browser, named MyGBrowser. This is a web-based tool (accessible from the MyMPN portal) that provides timely, convenient access to the high-quality and manually-curated genome sequence and annotation of M. pneumoniae. Given a coordinate range, this browser facilitates the searches and visualization of hosted genomic information. In particular, it allows to select several genome tracks which include operons, genes, non-coding RNA transcripts, transcriptional start sites, transcriptional termination sites, pribnow boxes and DNA and RNA hairpins. Images are generated on the fly using a Perl script that is based on sequence object methods available in BioPerl (http://www.bioperl.org (28)) and on image drawing functionalities from Bio::Graphics (available at the Comprehensive Perl Archive Network, http://www.cpan.org/modules/). In brief, this script retrieves all genomic features located in a given region from the MyMpn MySQL database, converts these features into Bio Perl sequence objects and finally generates an image (e.g. genomic view) by calculating the relative position of each of these sequences to the genomic reference. MyGBrowser web pages are written in PHP and are fully embedded in the logic of the MyMPN website. Images are clickable, and provide researches with an easy and visual entry point to the scientific content of the portal, including access to operon and gene centric pages.
Metabolic pathway map
To provide researchers with a quick and clear overview about the metabolic processes of M. pneumoniae, we implemented a web-based, zoomable and interactive pathway browser. Starting from a manually-curated model map in CellDesigner (http://www.celldesigner.org/ (29)), a GoogleMaps-based zoomable webpage was generated using the CellPublisher online interface (http://cellpublisher.gobics.de/ (30)) to produce different web files (HTML, CSS and JS). In order to better integrate the map with the MyMpn platform and other third party resources (such as the KEGG website (4)), we applied further modifications. For instance, we highlighted relevant metabolic pathways and classified existing entities into different groups (ions, simple molecules, proteins, complexes) by using Google Maps API (https://developers.google.com/maps/). Selecting those entities by clicking on their respective markers drawn in the metabolic pathway map opens drop down menus linking to additional information from the MyMpn database as well as to external websites.
MyMpn DATA
From the bench to the database
The mycoplasma project, an international collaboration between the ‘The Design of Biological Systems Group’ at the CRG Barcelona and different research groups at the EMBL Heidelberg, aims to understand M. pneumoniae in its entirety. To this end, a wealth of different ‘omics’ experiments have been conducted during the past years analyzing different aspects of the genome, transcriptome, the proteome and the metabolome of M. pneumoniae (11–15,18,20–25) and complementing earlier studies (10,31–35). However, the proper centralization of data produced in different laboratories is a much needed prerequisite for successful research. To this end, we designed a set of different spreadsheets (e.g. templates) that enable our lab collaborators to collect and properly curate their experimental data, metadata and results. Once ready, these templates are parsed using in-house Perl scripts that load the file content into a MySQL relational database. The use of pre-defined templates provides the lab scientists with a standard interface that can easily capture high diversity data as well as incorporate potential new data of different formats they might generate in the future. In its current version, our database hosts information relative to 1305 operons, 311 non-coding RNAs, 737 protein-coding genes, 306 reactions and 22 pathways. These genomic features are annotated with data obtained from hundreds of experiments and analysis, including gene expression profiling, gene essentiality studies, protein abundance profiling, protein complexes analysis, metabolic reactions, network modeling, cell growth experiments, comparative genomics and 3D tomography (Table 1).
Table 1. Omics datasets contained in the MyMpn database.
| -omics | Table | Content description |
|---|---|---|
| Genomics | Genes | For annotated genes, identifiers, description, genomic localization, sequence, promoter and the encoded protein sequence are shown |
| Homology | Identification of orthologs using a reciprocal BLAST search against the UniProt (3) | |
| Gene essentiality | Essentiality of annotated genes and non-coding genomic sections based on combining theoretical coding capabilities and the experimental analysis of a comprehensive mutant library | |
| Operons | Estimation of operons based on combining information from microarray and tiling array experiments | |
| Transcriptomics | Microarrays | Gene expression along the growth curve determined by microarrays. A tool for the generation of gene expression plots is embedded |
| RNAseq | RNA sequencing results from different time points | |
| Proteomics | Proteins | For annotated proteins, internal and external identifiers, biochemical properties, structure predictions and functional features are displayed |
| Pfam domains | Determination of Pfam domains applying InterProScan to the protein sequences (E-value < 0.09) | |
| Complexes | Identification of protein complexes based on Tandem Affinity Purification-Mass Spectrometry (TAP-MS) and on molecular weight exclusion/MS | |
| Peptides | Quantitative mass spectrometry results for different growth conditions | |
| Metabolomics | Metabolic reactions | Metabolic reconstruction resulting from the combination of genomic and experimental data, literature mining, sequence analysis and metabolic modeling |
| Growth curves | Metabolite measurements along the growth curve under various conditions and medium compositions | |
| Metabolites | Metabolites identified by GC-MS, LC-MS or NMR |
The web interface
MyMpn data stored in the MySQL database is freely accessible through a web interface at the address: http://mympn.crg.eu. The web site has been designed to provide wet lab researchers with user-friendly tools to query and browse the annotation and experimental data available for M. pneumoniae. In brief, the interface allows several entry points for accessing the data:
searching by any key terms, such as gene symbols, accession numbers, gene ontology terms, protein domains, orthologs and IDs of major databases;
searching by sequence comparison against a local database of M. pneumoniae genome, gene and ncRNA sequences through the BLAST algorithm (26). Both nucleotide and amino acid sequences are allowed as query type. However, when using this search to compare other organisms to M. pneumoniae, an amino acid sequence comparison (pBLAST: (36)) is recommended since M. pneumoniae has a different codon usage as higher organisms (e.g. the TGA codon encodes tryptophan instead of indicating the end of a gene (16));
searching by genomic coordinates using the genomic search (available in DATA ACCESS) or the two genome browsers.
Each of these queries generates a list of genes that meet the search criteria. Information regarding the gene of interest is visualized in a specific page where a clickable genomic representation allows a close look at all the features present in the same genomic region (Figure 1). The web site also offers access to the different omics experiments conducted on the genome, the transcriptome, the proteome and the metabolome of M. pneumoniae. A list of the major omics tables is available in Table 1.
Figure 1.
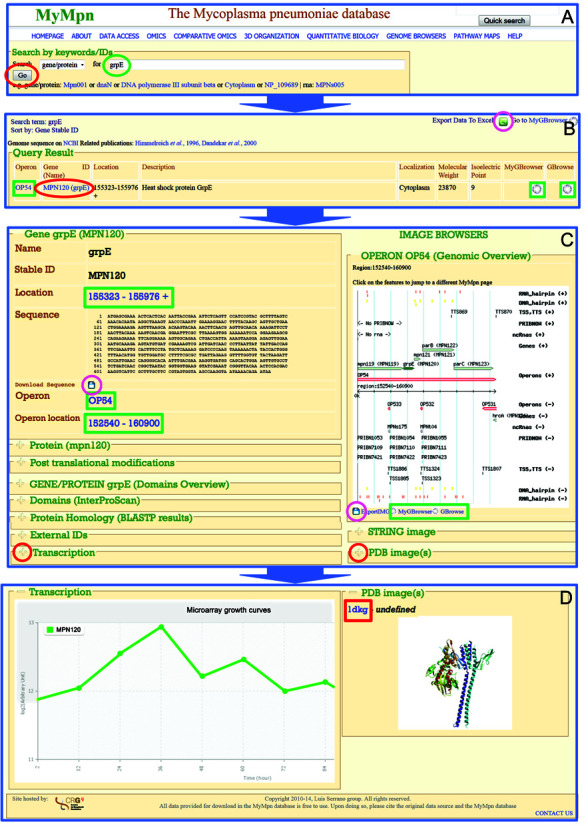
A keyword/ID example query of the MyMpn database. (A) To start a database request type a keyword (e.g. ‘grpE’) into the basic keyword/ID search mask and hit the ‘go’ button. (B) A list of DB entries matching to the search term is reported (e.g. for ‘grpE’ only one entry is found). Result entries of interest can be accessed by clicking on the ‘Gene ID (Name)’ (e.g. ‘MPN120 (grpE)’). (C) Information available for the selected DB entry is displayed: genetic information in detail by default, while additional information related to transcription, translation, domain prediction and function, protein structure, homology, network analysis as well as links to external databases is hidden in closed drop-down menus that can be expanded upon selection. (D) Expanding for example the ‘Transcription’ and the ‘PDB image(s)’ menu will show a graphic representation for gene expression data (microarray profiling experiments) and a PDB image with the link to the PDB (red box). Green circle—search term, red circles—buttons/links to click in order to be redirected (blue arrows) to the next sub-figure content, pink circles—download/data export buttons; green rectangles—links to other visualizations of the respective data internal to the MyMpn database.
Furthermore, the web interface provides several data analysis and visualization tools, including (i) an interactive tool for the comparison of growth curve measurements, (ii) a customizable genome browser (27) where users can create personal tracks to upload and compare their datasets to the M. pneumoniae genome annotation and (iii) a clickable metabolic map to explore the metabolic network designed with the CellDesigner (29) and CellPublisher (30) tools and linked to diverse data on genes, enzymes, metabolites and reactions (see ‘Materials and Methods’ section). Sequences and data stored in our database are also provided as downloadable files in the download section, along with a general description of the dataset and the original data source. The ‘About MyMpn database’ page contains more detailed information about the different sections of the web interface. Information about how to properly use the different tools of the web interface as well as answers to frequently asked questions (FAQs) are given in the ‘Help’ section.
CONCLUDING REMARKS
In summary, MyMpn constitutes a unique and comprehensive resource for the study of the human pathogen M. pneumoniae. We want to provide researchers and other interested users around the world access to the multitude of information about this minimal bacterium and, thus, invite others to participate in the challenge of understanding an organism in its entirety. The portal allows to quickly find genomic features of interest, to access a vast collection of diverse experimental and computational datasets, and to ultimately compare those to other internal data, to external data and to annotations. This analysis potential and the amount of available data, in combination with the facts that M. pneumoniae has clinical relevance and is one of the most promising model organisms in systems biology, render the MyMpn database interesting to the large community of microbiology and systems biology researchers.
Acknowledgments
We thank Lope Flórez for help on implementing the interactive metabolic map, Bernhard Petzold for providing us his light-microscopy pictures of M. pneumoniae for the illustration of the MyMpn homepage and the members of the Serrano group for using the database during the testing phase and providing us their feedback for improvements. In addition, we would like to thank all the core facilities of the CRG as well as the ITC department for their support.
Disclaimer: This article reflects only the authors views and the European Community is not liable for any use that may be made of the information contained.
Footnotes
The authors wish it to be known that, in their opinion, the first two authors should be regarded as Joint First Authors.
FUNDING
European Union through the European Research Council (ERC) [‘Celldoctor’ GA 232913]; Spanish Ministerio de Economía y Competitividad Plan Nacional [BIO2007-61762]; Fundación Marcelino Botin [to L.S.]. laCaixa-CRG Fellowship, Fundació laCaixa [to J.W.]; Spanish Ministry of Economy and Competitiveness No [PTA2010-4446-I to A.H.; PTA2011-6729-I to L.C.]. Funding for open access charge: ERC [to L.S.].
Conflict of interest statement. None declared.
REFERENCES
- 1.Milo R., Jorgensen P., Moran U., Weber G., Springer M. BioNumbers–the database of key numbers in molecular and cell biology. Nucleic Acids Res. 2010;38:D750–D753. doi: 10.1093/nar/gkp889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schomburg I., Chang A., Placzek S., Söhngen C., Rother M., Lang M., Munaretto C., Ulas S., Stelzer M., Grote A., et al. BRENDA in 2013: integrated reactions, kinetic data, enzyme function data, improved disease classification: new options and contents in BRENDA. Nucleic Acids Res. 2013;41:D764–D772. doi: 10.1093/nar/gks1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.The Uniprot Consortium. Activities at the Universal Protein Resource (UniProt) Nucleic Acids Res. 2014;42:D191–D198. doi: 10.1093/nar/gkt1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kanehisa M., Goto S., Sato Y., Kawashima M., Furumichi M., Tanabe M. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res. 2014;42:D199–D205. doi: 10.1093/nar/gkt1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li C., Donizelli M., Rodriguez N., Dharuri H., Endler L., Chelliah V., Li L., He E., Henry A., Stefan M.I., et al. BioModels Database: an enhanced, curated and annotated resource for published quantitative kinetic models. BMC Sys. Bio. 2010;4:92. doi: 10.1186/1752-0509-4-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Keseler I.M., Mackie A., Peralta-Gil M., Santos-Zavaleta A., Gama-Castro S., Bonavides-Martínez C., Fulcher C., Huerta A.M., Kothari A., Krummenacker M., et al. EcoCyc: fusing model organism databases with systems biology. Nucleic Acids Res. 2013;41:D605–D612. doi: 10.1093/nar/gks1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Michna R.H., Commichau F.M., Tödter D., Zschiedrich C.P., Stülke J. SubtiWiki-a database for the model organism Bacillus subtilis that links pathway, interaction and expression information. Nucleic Acids Res. 2014;42:D692–D698. doi: 10.1093/nar/gkt1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chiner E., Signes-Costa J., Andreu A.L., Andreu L. Mycoplasma pneumoniae pneumonia: an uncommon cause of adult respiratory distress syndrome. An. Med. Int. 2003;20:597–598. [PubMed] [Google Scholar]
- 9.Waites K.B., Talkington D.F. Mycoplasma pneumoniae and its role as a human pathogen. Clin. Microbiol. Rev. 2004;17:697–728. doi: 10.1128/CMR.17.4.697-728.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chanock R.M., Hayflick L., Barile M.F. Growth on artificial medium of an agent associated with atypical pneumonia and its identification as a PPLO. PNAS. 1962;48:41–49. doi: 10.1073/pnas.48.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yus E., Maier T., Michalodimitrakis K., van Noort V., Yamada T., Chen W.-H., Wodke J.A.H., Güell M., Martínez S., Bourgeois R., et al. Impact of genome reduction on bacterial metabolism and its regulation. Science. 2009;326:1263–1268. doi: 10.1126/science.1177263. [DOI] [PubMed] [Google Scholar]
- 12.Wodke J.A.H., Puchalka J., Lluch-Senar M., Marcos J., Yus E., Godinho M., Gutiérrez-Gallego R., Martins dos Santos V.A., Serrano L., Klipp E. Dissecting the energy metabolism in Mycoplasma pneumoniae through genome-scale metabolic modeling. Mol. Syst. Biol. 2013;9:653. doi: 10.1038/msb.2013.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maier T., Marcos J., Wodke J.A.H., Paetzold B., Liebeke M., Gutiérrez-Gallego R., Serrano L. Large-scale metabolome analysis and quantitative integration with genomics and proteomics data in Mycoplasma pneumoniae. Mol. Biosyst. 2013;9:1743–1755. doi: 10.1039/c3mb70113a. [DOI] [PubMed] [Google Scholar]
- 14.Yus E., Güell M., Vivancos A.P., Chen W.-H., Lluch-Senar M., Delgado J., Gavin A.-C., Bork P., Serrano L. Transcription start site associated RNAs in bacteria. Mol. Syst. Biol. 2012;8:585. doi: 10.1038/msb.2012.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lluch-Senar M., Luong K., Lloréns-Rico V., Delgado J., Fang G., Spittle K., Clark T.A., Schadt E., Turner S.W., Korlach J., et al. Comprehensive methylome characterization of Mycoplasma genitalium and Mycoplasma pneumoniae at single-base resolution. PLoS Genet. 2013;9:e1003191. doi: 10.1371/journal.pgen.1003191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Himmelreich R., Hilbert H., Plagens H., Pirkl E., Li B.C., Herrmann R. Complete sequence analysis of the genome of the bacterium Mycoplasma pneumoniae. Nucleic Acids Res. 1996;24:4420–4449. doi: 10.1093/nar/24.22.4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dandekar T., Huynen M., Regula J.T., Ueberle B., Zimmermann C.U., Andrade M.A., Doerks T., Sánchez-Pulido L., Snel B., Suyama M., et al. Re-annotating the Mycoplasma pneumoniae genome sequence: adding value, function and reading frames. Nucleic Acids Res. 2000;28:3278–3288. doi: 10.1093/nar/28.17.3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seybert A., Herrmann R., Frangakis A.S. Structural analysis of Mycoplasma pneumoniae by cryo-electron tomography. J. Struct. Biol. 2006;156:342–354. doi: 10.1016/j.jsb.2006.04.010. [DOI] [PubMed] [Google Scholar]
- 19.Halbedel S., Stülke J. Tools for the genetic analysis of Mycoplasma. Int. J. Med. Microbiol. 2007;297:37–44. doi: 10.1016/j.ijmm.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 20.Güell M., van Noort V., Yus E., Chen W.-H., Leigh-Bell J., Michalodimitrakis K., Yamada T., Arumugam M., Doerks T., Kühner S., et al. Transcriptome complexity in a genome-reduced bacterium. Science. 2009;326:1268–1271. doi: 10.1126/science.1176951. [DOI] [PubMed] [Google Scholar]
- 21.Kühner S., van Noort V., Betts M.J., Leo-Macias A., Batisse C., Rode M., Yamada T., Maier T., Bader S., Beltran-Alvarez P., et al. Proteome organization in a genome-reduced bacterium. Science. 2009;326:1235–1240. doi: 10.1126/science.1176343. [DOI] [PubMed] [Google Scholar]
- 22.Schmidl S.R., Gronau K., Pietack N., Hecker M., Becher D., Stülke J. The phosphoproteome of the minimal bacterium Mycoplasma pneumoniae: analysis of the complete known Ser/Thr kinome suggests the existence of novel kinases. Mol. Cell Proteomics. 2010;9:1228–1242. doi: 10.1074/mcp.M900267-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maier T., Schmidt A., Güell M., Kühner S., Gavin A.-C., Aebersold R., Serrano L. Quantification of mRNA and protein and integration with protein turnover in a bacterium. Mol. Syst. Biol. 2011;7:511. doi: 10.1038/msb.2011.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Güell M., Yus E., Lluch-Senar M., Serrano L. Bacterial transcriptomics: what is beyond the RNA horiz-ome. Nat. Rev. Microbiol. 2011;9:658–669. doi: 10.1038/nrmicro2620. [DOI] [PubMed] [Google Scholar]
- 25.van Noort V., Seebacher J., Bader S., Mohammed S., Vonkova I., Betts M.J., Kühner S., Kumar R., Maier T., O'Flaherty M., et al. Cross-talk between phosphorylation and lysine acetylation in a genome-reduced bacterium. Mol. Syst. Biol. 2012;8:571. doi: 10.1038/msb.2012.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 27.Donlin M.J. Using the generic genome browser. In: Bateman A, Pearson WR, Stein LD, Stormo GD, Yates JR III, editors. Current Protocols in Bioinformatics. New York: John Wiley & Sons, Inc.; 2009. [Google Scholar]
- 28.Stajich J.E., Block D., Boulez K., Brenner S.E., Chervitz S.A., Dagdigian C., Fuellen G., Gilbert J.G.R., Korf I., Lapp H., et al. The Bioperl toolkit: Perl modules for the life sciences. Genome Res. 2002;12:1611–1618. doi: 10.1101/gr.361602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kitano H., Funahashi A., Matsuoka Y., Oda K. Using process diagrams for the graphical representation of biological networks. Nat. Biotechnol. 2005;23:961–966. doi: 10.1038/nbt1111. [DOI] [PubMed] [Google Scholar]
- 30.Flórez L.A., Lammers C.R., Michna R., Stülke J. CellPublisher: a web platform for the intuitive visualization and sharing of metabolic, signalling and regulatory pathways. Bioinformatics. 2010;26:2997–2999. doi: 10.1093/bioinformatics/btq585. [DOI] [PubMed] [Google Scholar]
- 31.Chanock R.M., James W.D., Fox H.H., Turner H.C., Mufson M.A., Hayflick L. Growth of Eaton PPLO in broth and preparation of complement fixing antigen. Proc. Soc. Exp. Bio. Med. 1962;110:884–889. doi: 10.3181/00379727-110-27681. [DOI] [PubMed] [Google Scholar]
- 32.Razin S., Aragaman M., Avigan J. Chemical composition of Mycoplasma cells and membranes. J. Gen. Microbiol. 1963;33:477–487. doi: 10.1099/00221287-33-3-477. [DOI] [PubMed] [Google Scholar]
- 33.Pollack J.D., Somerson N.L., Senterfit L. Chemical composition and serology of Mycoplasma pneumoniae lipids. J. Infect. Dis. 1973;127:S32–S35. doi: 10.1093/infdis/127.supplement_1.s32. [DOI] [PubMed] [Google Scholar]
- 34.Manolukas J.T., Barile M.F., Chandler D.K., Pollack J.D. Presence of anaplerotic reactions and transamination, and the absence of the tricarboxylic acid cycle in mollicutes. J. Gen. Microbiol. 1988;134:791–800. doi: 10.1099/00221287-134-3-791. [DOI] [PubMed] [Google Scholar]
- 35.Razin S., Yogev D., Naot Y. Molecular biology and pathogenicity of mycoplasmas. Microbiol. Mol. Biol. Rev. 1998;62:1094–1156. doi: 10.1128/mmbr.62.4.1094-1156.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Altschul S.F., Madden T.L., Schäffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]


