Abstract
Classification of the structures of the complementarity determining regions (CDRs) of antibodies is critically important for antibody structure prediction and computational design. We have previously performed a clustering of antibody CDR conformations and defined a systematic nomenclature consisting of the CDR, length and an integer starting from the largest to the smallest cluster in the data set (e.g. L1-11-1). We present PyIgClassify (for Python-based immunoglobulin classification; available at http://dunbrack2.fccc.edu/pyigclassify/), a database and web server that provides access to assignments of all CDR structures in the PDB to our classification system. The database includes assignments to the IMGT germline V regions for heavy and light chains for several species. For humanized antibodies, the assignment of the frameworks is to human germlines and the CDRs to the germlines of mice or other species sources. The database can be searched by PDB entry, cluster identifier and IMGT germline group (e.g. human IGHV1). The entire database is downloadable so that users may filter the data as needed for antibody structure analysis, prediction and design.
INTRODUCTION
The vertebrate immune system produces a diverse set of antibody sequences and structures for the purpose of recognizing foreign antigens on the surfaces of microorganisms and bacteria as well as aberrant self-antigens. The sequences of antibody proteins are produced by immunoglobulin genes that have been rearranged by a process known as V(D)J recombination at distinct genetic loci that contain multiple copies of each segment of the final recombined gene, consisting of one choice each of the variable region (V), the diversity segment (D, found only in heavy chain genes), and the joining region (J), which is followed by the constant region (C) (1). Most mammalian, fish and avian antibodies consist of a heavy chain and a light chain, each of which is the product of V(D)J or VJ recombination, respectively. In each species, the light chain may be generated by one or more loci, generating additional diversity; for instance, in most mammals the kappa and lambda loci are used to generate light chain proteins.
Since the first antibody sequences and structures were determined in the 1960s and 1970s (2–4), attempts have been made to classify the complementarity determining regions or CDRs both by sequence and by structure. The earliest comprehensive attempts on structure were those of Chothia et al. (5,6), who coined the term ‘canonical structures’ for the antibody CDRs, indicating that each CDR (L1, L2, L3, H1, H2, H3) might only adopt a few common structures based on length and sequence. As more structures were determined, the early classifications were extended in the mid 1990s by Chothia et al. (7) and Thornton et al. (8). These classifications were updated periodically in the following decade (9), and other classifications have appeared of subsets of the current PDB (e.g. H3 CDRs or λ chains) (10–12). Nikoloudis et al. have recently presented a hierarchical clustering of antibody CDR structures, based on the PDB as of December 2011 (13), but not as a server or a database.
In 2011, we published a comprehensive quantitative classification of antibody CDR structures, based on a dihedral angle metric and an affinity-propagation clustering algorithm (14). By 2011, the number of unique antibody structures was more than 300 and it was possible to perform automatic clustering on a high-quality data set (i.e. removing structures with low resolution and/or high B-factors). In contrast to the Chothia system, we developed a systematic nomenclature for the antibody CDR clusters such that each cluster was named by CDR and length, followed by an integer starting with the largest cluster first, e.g. L1-11-1 was the largest cluster of CDR L1 length 11. Tentative associations of each cluster with gene locus (heavy, kappa and lambda) and species were provided. Recent databases of antibody CDR conformations have used our classification system (13,15) as a reference, and it has gained acceptance in the wider antibody literature (16,17) and in industry (18–20).
Classification of antibody structures and their correlation with locus, species and sequence leads to improved antibody structure prediction (21–23) and opportunities for antibody design (24,25). Because of this, we have implemented automatic assignments of CDR structures in the PDB to our CDR structure classification system (14), and in this paper, we present a comprehensive database and server of these assignments, PyIgClassify (for Python-based immunoglobulin classification), which will be updated periodically. PyIgClassify will also be updated with new clusters as the need arises. Even as of 2011, it is likely that all of the major clusters of conformations in human and mouse antibodies had already been observed and the only new conformations are either of lengths not previously observed due to somatic or engineered changes in CDR lengths from germline or from structures from new species not previously represented in the PDB.
Besides being up-to-date with the PDB, we have investigated the relationship between the CDR clusters and the germline V regions of the framework and CDR regions. Many of the antibodies in the PDB have undergone substantial maturation from germline sequences and in many cases have been heavily engineered. In some cases for therapeutic drugs, the CDRs are from one antibody and species, such as mouse, while the framework is primarily human in origin. Thus, assigning the correct germline V regions is a challenging problem. We have carefully determined the species and germline V region of each antibody in the PDB based on the IMGT nomenclature (26) and identified antibodies with grafts of CDRs from mouse or other species onto human frameworks. In many cases, the lengths of the CDR1 and CDR2 segments do not match the lengths of the same CDRs in the germline V region most similar to the framework sequence. These structures provide useful information on the possibility of grafting CDRs of different lengths onto commonly used, highly stable frameworks, such as the human IGHV3-66/IGKV1-39 framework, closely related to trastuzumab and other antibodies (27). We find that 9.5% of non-redundant antibodies in the PDB are mouse/human grafts and 16.6% contain mismatches between the CDR length and that of the framework germline, providing an ample data set to examine in terms of antibody computational design.
MATERIALS AND METHODS
The methods for determining which protein sequences in the PDB contain antibody VH and VL domains and for assigning IMGT V-region germlines to these sequences are described in the Supplemental Methods.
Determining antibody CDR cluster
For each PDB structure with an identified antibody VH or VL domain, we determine the CDR sequences and their lengths, which represents the first level of our classification system (e.g. L1-11, L2-8, etc.). For CDRs with complete backbone coordinates, we calculate the ω, ϕ and ψ dihedral angles of the residues in each CDR with in-house scripts.
The next level of classification is by the cis–trans pattern of the residues in the loop. Some CDR–length combinations commonly have cis-proline residues (e.g. L3-9 at position 7) while a surprising number of CDRs have cis-non-proline residues, probably due to low resolution and poor refinement of the structures (see the Results section). If the length was new (13 cases) or the cis–trans pattern was new (53 new cases), we labeled the loop with a generic cluster identifier (e.g. L3-5-* for CDR L3 of length 5 which did not appear in the curated 2011 data set; or L1-11-cis4-* for CDR L1 length 11 with a cis-residue at position 4). None of these clusters had more than seven non-redundant sequences (H2-11-*), and 47 of 66 (71%) had only one sequence. In the 2011 analysis, we excluded CDRs with cis-non-proline residues. The current database covers all antibody structures without a priorifiltering.
For each CDR length and cis–trans pattern with a cluster in our original analysis, we calculated the distance of the loop structure to each of the centroids of our clusters of the same length and cis–trans pattern for that CDR, using the same dihedral angle metric as in the 2011 work:
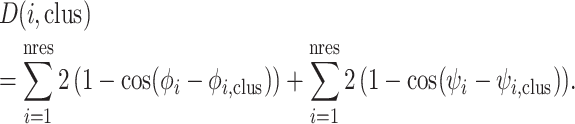 |
This is the proper distance between two angles used in directional statistics (28). The database and web server provide the distance from the centroid and we find that a cutoff mean dihedral angle distance of 40° and a backbone RMSD cutoff of 1.5 Å are reasonable to identify cluster members. CDRs that are more than 40° or 1.5 Å in RMSD from any existing cluster centroid are assigned to generic clusters of the form L1-11-*.
Databases and web site
The internal and ‘downloadable’ databases for PyIgClassify are SQlite (http://www.sqlite.org) relational databases due to its support and straightforward integration in a variety of computational languages and molecular modeling suites including R, Python, C++, BioPython and Rosetta (https://www.rosettacommons.org). Tab-delimited text versions of each database are also available. Each database contains at least four tables: cdr_data, SpeciesNames, GermlineAssignments and CdrClusterSum. The cdr_data table holds various pieces of information about the cluster, sequence, structure and germline for each CDR and framework of each identified antibody structure. The SpeciesNames table lists the species and their short names used in the databases and web site, while the GermlineAssignments table has the germline assignments for both CDRs and frameworks by comparing each antibody sequence to the IMGT (http://www.imgt.org/) germline sequences.
In each database, there is also a summary table (CDRClusterSum) for each CDR cluster. This table includes the number of unique sequences in a cluster and other useful summary information such as the median PDB, gene(s) and the identified PDB species where this cluster can be found. The average deviation of the dihedral angles from the cluster centroids (or medians) is calculated from the formula
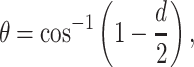 |
where d is the average of the normalized dihedral distances of each member in a cluster from the cluster median. In this file, PercentLoop is the number of structures in a particular cluster divided by the number of structures of that CDR (e.g. all L1). PercentUniqSeq is the number of unique sequences in a cluster divided by the number of unique sequences for that CDR in the database. Loop conformation is the conformation of the median loop in terms of the Ramachandran conformations, while ConsSeq is the consensus sequence for the sequences in the cluster (the most common residue at each position among the unique sequences in the cluster).
Databases are updated monthly to reflect the current state of the PDB. In addition, all antibodies identified are renumbered in the Honegger–Plückthun Numbering Scheme (29) and can be downloaded from the website.
RESULTS
Before showing examples of searches performed on the PyIgClassify server, we present some analysis of the current structural and germline coverage of antibody entries in the PDB (summarized in Supplementary Tables S1–S6).
Identifying antibody V regions
We identify antibody VH and VL regions using a set of eight hidden Markov models (HMMs) that cover the antibody VH, Vκ and Vλ regions (and one for the Vλ6 sequences that contain a framework insertion relative to other Vλ sequences) as well as the T-cell receptor α, β, γ and δ chains. While other immunoglobulin sequences are more distantly related, it is important to distinguish between antibody and T-cell receptor domains when clustering their CDR conformations. In Figure 1, we show a scatterplot of the highest scoring HMM (y-axis) versus the second highest scoring HMM (x-axis) for each positively scoring domain in the PDB. Empirically, the cutoff of a highest score of 90 across the four antibody HMMs is consistent with the annotations in the PDB for each sequence. The points are labeled by their assignments to heavy, Vkappa, Vlambda, TCR, nonAgR (for non-antigen-receptor) and constant domains (Cdom). The non-antigen receptors included shark Ig-NARs, CD8, the v-preB receptor and the human polio virus receptor. In total, we found 1897 PDB entries with one or more VH or VL domains of antibodies comprising 5711 chains and 17 260 CDRs. There were 240 entries with T-cell receptors.
Figure 1.
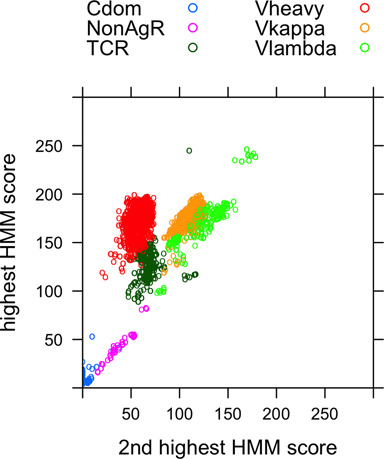
HMM scores of immunoglobulin domains in the PDB. For each sequence in the PDB with a V-set domain, the scores of each of the eight HMMs covering antibody VH and VL domains and TCR V domains were compared. The highest and second highest scores are plotted and the assignments that are consistent with the highest score are shown for those with score above 90, a threshold chosen such that the highest scoring HMM and annotations in the PDB were fully consistent. Domains whose highest score is below 90 were uniformly not antibody structures and were classified as either constant domains if they were in the same chains as antibody VH or VL domains or NonAgR for non-antigen receptors, including shark IgNARs, CD8, the poliovirus receptor and the preB-cell receptor.
Germline assignments
As described in the Supplemental Methods, we assigned germline V regions (but not D or J segments) to antibody sequences in the PDB based on the PDB's annotation of species and comparison of the full sequence, the framework sequence and the CDR sequences of the PDB antibodies with those in the IMGT germline repertoire for several species. From IMGT, we were able to obtain germline sequences for VH regions of human, mouse, rat, Danio rerio, macaque, llama, camel and rabbit; Vκ regions of human, mouse, rabbit, sheep, rat and pig; and Vλ regions of human, mouse, rabbit, rat and pig. A summary of these assignments to the current PDB is given in Supplementary Table S1 for all entries and for non-redundant entries (one for each unique concatenated sequence of the CDR sequences). For the 59 IMGT germline groups of human and mouse, only 11 are not present in the PDB currently: Hu_IGKV5, Hu_IGLV4, Hu_IGLV8, Mo_IGHV11, Mo_IGHV15, Mo_IGHV16, Mo_IGKV7, Mo_IGKV11, Mo_IGKV18, Mo_IGKV20 and Mo_IGLV2 (there are no Hu_IGLV9 or Mo_IGKV15 V-regions defined in IMGT).
For the human germline groups, the table also includes the number of structures that consist of mouse CDRs grafted onto these human (or humanized) frameworks (see the Supplemental Methods). The table shows the large number of antibodies based on the humanized 4D5 framework (27), which is closest to the human germline sequences IGHV3–66 and IGKV1–39 framework at ∼95% identity over the framework segments in most such antibodies. A total of 78 antibody structures with distinct CDR sequences use at least one of these frameworks and 36 different antibody structures use both. We note that the vast majority of humanized antibodies in the PDB (those with mouse CDRs but human-like frameworks) contain human κ light chain frameworks and all of these contain κ mouse CDRs. This is presumably because mice do not produce λ antibodies in substantial numbers (30) and grafts are almost always κ to κ, and not κ to λ or vice versa.
The table also shows the number of times the CDRs in the structures in each germline group are different from the length in the parent germline sequence. The distribution of length changes is shown in Figure 2. These may be due to engineered sequences or due to somatic mutation which can alter the lengths of CDRs by duplicating codons or eliminating a repeated codon (31). Most altered CDR lengths differ by only +1 or −1 amino acid from the germline CDR length. The V region covers the VH and VL domains through the first residue or two of CDR3, so data are only shown for CDR1 and CDR2 in Supplementary Table S1. The numbers are highly non-uniform due to differences in the variability of CDR lengths. For instance, nearly all mammalian L2 germline sequences are length 8 and so the IGKV and IGLV sequences for CDR2 do not show length mismatches, apparently because CDR L2 either does not undergo or does not tolerate somatic changes in CDR length. CDR H1 is length 13 in most mouse and human germ lengths, while CDR H2 shows a variety of lengths in the germline. It also shows more differences from framework germline in the human antibodies in the PDB.
Figure 2.
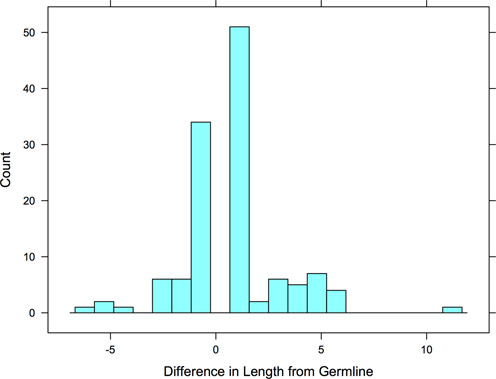
Histogram of CDR length changes relative to germline. For unique CDR sequences in the PDB, if the CDR length differs from that contained in the assigned framework germline sequence, the length change is counted in this histogram.
While the PDB is not a representative set of antibodies, it does contain information on the frequency with which VH and VL frameworks are associated with each other. A matrix of the most common associations of common human VH and VL domains is given in Supplementary Table S2. In parentheses, the ratio of observed versus expected counts is given for each pair. The IGKV1/IGHV3 combination represents the many structures based on the humanized 4D5 antibody (32). But other associations are noteworthy, including the tendency of IGHV4 regions to be associated with λ domains.
CDR conformational clusters
The largest clusters of CDR conformations for the light chains and heavy chains are given in Supplementary Tables S3 and S4, respectively. The total number of unique sequences in each cluster is given and the species and loci (κ or λ for L1, L2 and L3) present in each cluster. In cases, where only one or two germline V regions are present in the cluster (e.g. Mo_IGKV3 abbreviated to Mo_KV3 in the table) these are listed. The tables provide statistical information on the distribution of sequence lengths for each CDR and the distribution of clusters, both of which are highly uneven. For instance, 98.8% of L2 CDRs are of length 8 and 89.0% are in cluster L2-8-1. H1 is also very narrowly distributed with 91.8% of length 13 and 81.1% in cluster H1-13-1. L3 is the next CDR in terms of variable distribution with 83.0% of length 9 and 70.9% in cluster L3-8-cis7-1. The remaining CDRs, L1 and H2 are much more widely distributed in terms of lengths and clusters, especially L1.
A number of new CDR–length combinations are now present in the PDB which were not present in the PDB in 2011, as are some cis–trans configurations for some lengths. None of these had more than seven unique sequences. The thirteen new CDR–length combinations were: H1-9 (1 sequence), H1-11 (1), H1-18 (1), H1-20 (1), H1-24 (1), H2-11 (7), H2-14 (1), L1-7 (1), L1-8 (4), L1-9 (3), L3-5 (6), L3-6 (3) and L3-13 (1). There were 53 cis–trans patterns in the PDB not present in the 2011 analysis, although at that time we excluded CDRs with cis-non-proline residues. A total of 45 out of these 53 new cis–trans patterns are those with cis-non-proline residues and it is likely that a large majority of them are incorrectly refined structures. For instance, PDB entry 1OCW has 10 cis-residues (nine in VH and one in VL) that are not proline (33). At least it can be said that in most cases, the resolution of the structures does not support a structural feature that is very rare in the PDB (cis-non-proline residues) (34).
We were interested in the correlation between cluster and germline (in each direction) and so analyzed the prevalence of each germline V region group (e.g. Hu_IGHV1) for each cluster (e.g. H1-13-1) and vice versa. The results for the strongest correlations are shown in Supplementary Tables S5 and S6. Some of the largest clusters, such as H1-13-1, contain representatives from VH regions of mouse, human and other species and in fact 74% of unique H1 sequences belong to cluster H1-13-1. Other less common CDR lengths belong to only certain V regions of each species and further some light-chain clusters are locus specific (κ versus λ) or even species of specific germline group-specific. For example, L1-11-3, L1-14-1 and L1-14-2 contain only λ sequences. Further, it is interesting to note that the majority of mouse CDR grafts onto human frameworks belong to clusters consistent with the mouse CDR, in part because most such grafts have been added to human frameworks with similar CDR lengths and CDR clusters.
Supplementary Table S6 presents the predominant clusters for each major germline group. For some germline groups, the only CDR length of that group is also one that contains only one cluster. For instance, Hu_IGHV2 and Mo_IGHV8 germline sequences contain only CDR1s of length 15, and are all entirely in cluster H1-15-1. It is useful to note that several heavy-chain germlines sort either into H2-10-1 or H2-10-2, which may be useful in structure prediction. Hu_IGHV1, Hu_IGHV5, Mo_IGHV1, Mo_IGHV9 and Mo_IGHV15 are predominantly cluster H2-10-1, while Hu_IGHV3, Mo_IGHV4 and Mo_IGHV5 are predominantly in cluster H2-10-2. Hu_IGLV1 and Hu_IGLV6 neatly separate into clusters L1-13-1 and L1-13-2, respectively.
Searching PyIgClassify web site
There are four types of searches that can be done on the PyIgClassify website: (i) a PDB ID or a PDB ID with chain specified; (ii) a CDR cluster selected from the list boxes (e.g. L1-11-1); (iii) a CDR or CDR–length combination selected from the list boxes (e.g. L1 or L1-11); (iv) an IMGT germline group (e.g. Hu_IGHV1). Figures 3 and 4 show the results of a cluster search and a germline search of PyIgClassify, respectively.
Figure 3.
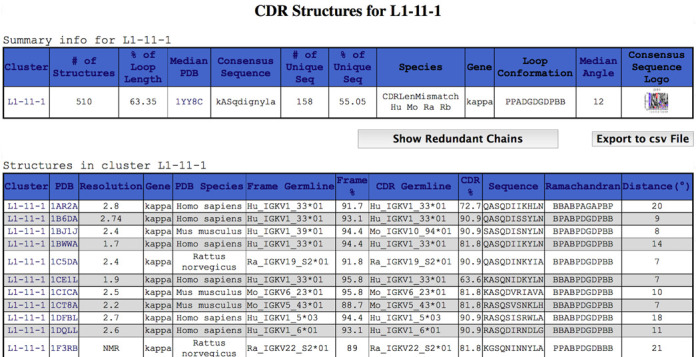
Fragment of a result of a cluster search on PyIgClassify. A screenshot of results for the L1-11-1 cluster is shown. This page can be accessed directly from the search page or by clicking on the result for a particular PDB entry that contains a member of the cluster (e.g. PDB entry 1N8Z). Note that chain 1BJ1J is a humanized antibody with a primarily human germline framework (Hu_IGKV1_39*01 at 94.4% sequence identity over the human framework germline sequence) and a mouse CDR (Mo_IGKV10_94*01 at 90.9% sequence identity over 11 residues of the mouse CDR germline sequence). Information on the entire cluster is shown at the top of the figure, including a link to a sequence logo for the 158 unique sequences in the cluster.
Figure 4.
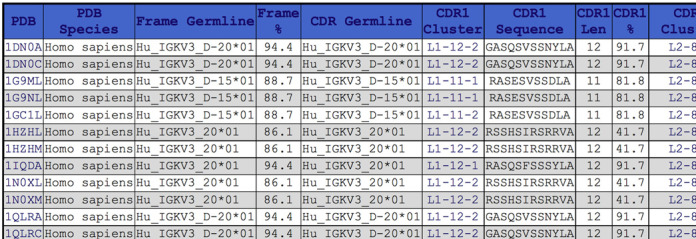
Fragment of a result of an IMGT germline group search on PyIgClassify. A screenshot of the results for IMGT germline group Hu_IGKV3 is shown. Only a horizontal fragment showing the CDR1 information is shown, while the other CDR sequences and clusters would be shown further to the right in the snapshot. Even within the same germline family (Hu_IGKV3), different germline V-regions may have different length CDRs (11 or 12 in this case).
A PDB ID query, such as 1N8Z (32), will return a list of CDRs and CDR clusters in the input structure. 1N8Z is a humanized mouse antibody (hum4D5 or trastuzumab) which has frameworks that are 94% identical to human IMGT germline sequences (Hu_IGKV1_39*01 and Hu_IGHV3_66*02). The closest germline V-region for the light chain CDRs is Mo_IGKV6_17*01 and for the heavy chain CDRs Mo_IGHV14_3*02. The table also contains the sequence length, cluster ID, distance from the cluster median (°), sequence and Ramachandran conformation.
From the results of a PDB search (by clicking on a cluster identifier) or from a direct search for CDR clusters, a user can obtain all of the structures that exist for that particular cluster, as shown in Figure 3 for cluster L1-11-1. The sequence logo icon in the upper right can be clicked to show a larger image. Clicking the ‘Show Non-Redundant Chains Only’ button will display only the representative sequences (the highest resolution structure for each sequence). The ‘Export to csv File’ can export any PyIgClassify query result page to a ‘comma-separated-value’ formatted text file, which can be easily parsed or imported into a variety of programs including Microsoft Excel. The majority of H3 loops occur in clusters labeled with an asterisk because they do not cluster well, e.g. H3-24-*. These pages can be used to view the sequences of each length and the framework germlines they occur in.
There are three options for searching by germlines. The list box contains all germlines identified in the current antibody sequences from IMGT. The user can search for structures with a framework in that germline group, with CDRs in that germline group, or both. An example is shown in Figure 4 for human IGKV3 sequences (Hu_IGKV3). Only the CDR1 portion of the table is shown. The germlines and sequence identities of the frameworks and CDRs to those germline are shown as are the clusters for each CDR in the chain.
A user can also submit a sequence or a PDB-formatted structure to our web site. The server identifies the CDRs for the input sequence, or CDRs and clusters for the submitted structure, and allows the user to download the resulting Honegger–Plückthun-renumbered PDB coordinate file (29).
The entire database is available for download by clicking the Download button on the main PyIgClassify page, http://dunbrack2.fccc.edu/pyigclassify.
DISCUSSION
Many antibody servers and databases have been published in recent years with the dramatic rise in the number of available antibody structures in the PDB as well as the ability to quickly sequence an individual's antibody repertoire. Many of these efforts, such as NEP (35) and Paratome (36) have focused on the identification of antigen epitopes and paratopes, respectively. Servers such as IgBLAST (37) and DigIt (38) have introduced tools for the sequence analysis of antibody variable domains and their associated CDR regions.
The SAbDab server (15), like PyIgClassify, provides a clustering of the CDR conformations of antibodies in the PDB. SAbDab is based on hierarchical clustering with an RMSD metric and allows the user to create clusters at any input RMSD cutoff value. Our cluster designations as well as those of Chothia are provided for each of the output clusters, if at least one PDB in the SAbDab cluster was present in our 2011 paper or in Chothia's papers. As such, the output of SAbDab differs from PyIgClassify that directly recompiles clusters of CDR structures based on a fixed nomenclature and clustering scheme. The on-the-fly clustering has its advantages but so too does a stable set of clusters for the most common conformations in the PDB. PyIgClassify provides a dihedral angle distance to the cluster centroids, which readily identifies potential outliers or members of the cluster that deviate too far from the centroid to be considered true members.
SAbDab provides IMGT subgroups (e.g. IGHV1), but it does not provide the full IMGT designation (e.g. IGHV1-69*01) nor does it analyze the framework and CDR sequence separately or provide sequence identity to germline. It only provides the species information given by the PDB, which is unfortunately inaccurate in many cases. At least 150 antibody chains in the PDB are labeled mouse or human when the VH and VL domains are entirely human or mouse, respectively. In some of these cases, the species designation may belong to the constant domains and not the V regions. SAbDab does not specify the species of the IMGT germline subgroup. This is problematic because human and mouse (and other species) germline subgroups are not numbered in the same way. For instance, human IGKV1 is closest to mouse IGKV16 and IGKV10 and is quite distantly related to mouse IGKV1. Thus, PyIgClassify provides complete and accurate information on the association of CDR clusters and IMGT germline information.
Finally, our aim in developing the PyIgClassify database is to provide information suitable for the prediction of antibody structures and more importantly antibody computational design. We believe that the sequence variation in large clusters provides ample information that can be used to guide design programs such as Rosetta (39) to sample amino acid types that are compatible with well-represented structural clusters in the PDB, a principle that has been used for other protein families (40). Further, accurate germline assignments enable an examination of both sequence and structure variation on a given germline framework and its CDRs which can be utilized in making sequence changes on a particular starting antibody with the same germline or germline group. To enable these types of projects, all data are available for download from the PyIgClassify website.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
We thank Greg Adams and Matthew Robinson for useful discussions.
FUNDING
National Institutes of Health (NIH) [R01 GM084453 to R.L.D.]. Funding for open access charge: NIH [R01 GM084453].
Conflict of interest statement. None declared.
REFERENCES
- 1.Tonegawa S. Somatic generation of antibody diversity. Nature. 1983;302:575–581. doi: 10.1038/302575a0. [DOI] [PubMed] [Google Scholar]
- 2.Wu T.T., Kabat E.A. An analysis of the sequences of the variable regions of Bence Jones proteins and myeloma light chains and their implications for antibody complementarity. J. Exp. Med. 1970;132:211–250. doi: 10.1084/jem.132.2.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Poljak R.J., Amzel L.M., Avey H.P., Chen B.L., Phizackerley R.P., Saul F. Three-dimensional structure of the Fab’ fragment of a human immunoglobulin at 2,8-A resolution. Proc. Natl Acad. Sci. U.S.A. 1973;70:3305–3310. doi: 10.1073/pnas.70.12.3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schiffer M., Girling R.L., Ely K.R., Edmundson A.B. Structure of a λ-type Bence-Jones protein at 3.5-Å resolution. Biochemistry. 1973;12:4620–4631. doi: 10.1021/bi00747a013. [DOI] [PubMed] [Google Scholar]
- 5.Chothia C., Lesk A.M. Canonical structures for the hypervariable regions of immunoglobulins. J. Mol. Biol. 1987;196:901–917. doi: 10.1016/0022-2836(87)90412-8. [DOI] [PubMed] [Google Scholar]
- 6.Chothia C., Lesk A.M., Tramontano A., Levitt M., Smith-Gill S.J., Air G., Sheriff S., Padlan E.A., Davies D., Tulip W.R., et al. Conformations of immunoglobulin hypervariable regions. Nature. 1989;342:877–883. doi: 10.1038/342877a0. [DOI] [PubMed] [Google Scholar]
- 7.Al-Lazikani B., Lesk A.M., Chothia C. Standard conformations for the canonical structures of immunoglobulins. J. Mol. Biol. 1997;273:927–948. doi: 10.1006/jmbi.1997.1354. [DOI] [PubMed] [Google Scholar]
- 8.Martin A.C.R., Thornton J.M. Structural families in loops of homologous proteins: automatic classification, modeling, and application to antibodies. J. Mol. Biol. 1996;263:800–815. doi: 10.1006/jmbi.1996.0617. [DOI] [PubMed] [Google Scholar]
- 9.Whitelegg N., Rees A.R. Antibody Engineering. Totowa, NJ: Springer; 2004. pp. 51–91. [Google Scholar]
- 10.Shirai H., Kidera A., Nakamura N. H3-rules: identification of CDR-H3 structures in antibodies. FEBS Lett. 1999;455:188–197. doi: 10.1016/s0014-5793(99)00821-2. [DOI] [PubMed] [Google Scholar]
- 11.Oliva B., Bates P.A., Querol E., Aviles F.X., Sternberg M.J. Automated classification of antibody complementarity determining region 3 of the heavy chain (H3) loops into canonical forms and its application to protein structure prediction. J. Mol. Biol. 1998;279:1193–1210. doi: 10.1006/jmbi.1998.1847. [DOI] [PubMed] [Google Scholar]
- 12.Chailyan A., Marcatili P., Cirillo D., Tramontano A. Structural repertoire of immunoglobulin lambda light chains. Proteins. 2011;79:1513–1524. doi: 10.1002/prot.22979. [DOI] [PubMed] [Google Scholar]
- 13.Nikoloudis D., Pitts J.E., Saldanha J.W. A complete, multi-level conformational clustering of antibody complementarity-determining regions. PeerJ. 2014;2:e456. doi: 10.7717/peerj.456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.North B., Lehmann A., Dunbrack R.L. Jr. A new clustering of antibody CDR loop conformations. J. Mol. Biol. 2011;406:228–256. doi: 10.1016/j.jmb.2010.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dunbar J., Krawczyk K., Leem J., Baker T., Fuchs A., Georges G., Shi J., Deane C.M. SAbDab: the structural antibody database. Nucleic Acids Res. 2014;42:D1140–D1146. doi: 10.1093/nar/gkt1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rynkiewicz M.J., Lu Z., Hui J.H., Sharon J., Seaton B.A. Structural analysis of a protective epitope of the Francisella tularensis O-polysaccharide. Biochemistry. 2012;51:5684–5694. doi: 10.1021/bi201711m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Robles V.M., Maréchal J.-D., Bahloul A., Sari M.-A., Mahy J.-P., Golinelli-Pimpaneau B. Crystal structure of two anti-porphyrin antibodies with peroxidase activity. PloS One. 2012;7:e51128. doi: 10.1371/journal.pone.0051128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nilvebrant J., Dunlop D.C., Sircar A., Wurch T., Falkowska E., Reichert J.M., Helguera G., Piccione E.C., Brack S., Berger S. IBC's 22nd Annual Antibody Engineering and 9th Annual Antibody Therapeutics International Conferences and the 2011 Annual Meeting of The Antibody Society. mAbs. 2012;4:153–181. doi: 10.4161/mabs.4.2.19495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Almagro J.C., Gilliland G.L., Scott J., Larrick J.W., Plückthun A., Veldman T., Adams G.P., Parren P.W., Chester K.A., Bradbury A., et al. Antibody Engineering and Therapeutics Conference: The Annual Meeting of the Antibody Society. mAbs. 2013;5:817–825. doi: 10.4161/19420862.2014.971627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ultsch M., Bevers J., Nakamura G., Vandlen R., Kelley R.F., Wu L.C., Eigenbrot C. Structural basis of signaling blockade by anti-IL-13 antibody lebrikizumab. J. Mol. Biol. 2013;425:1330–1339. doi: 10.1016/j.jmb.2013.01.024. [DOI] [PubMed] [Google Scholar]
- 21.Marcatili P., Rosi A., Tramontano A. PIGS: automatic prediction of antibody structures. Bioinformatics. 2008;24:1953–1954. doi: 10.1093/bioinformatics/btn341. [DOI] [PubMed] [Google Scholar]
- 22.Sircar A., Kim E.T., Gray J.J. RosettaAntibody: antibody variable region homology modeling server. Nucleic Acids Res. 2009;37:W474–W479. doi: 10.1093/nar/gkp387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Almagro J., Teplyakov A., Luo J., Sweet R., Kodangattil S., Hernandez-Guzman F., Gilliland G. Second antibody modeling assessment (AMA-II) Proteins. 2014;82:1553–1562. doi: 10.1002/prot.24567. [DOI] [PubMed] [Google Scholar]
- 24.Rees A.R., Staunton D., Webster D.M., Searle S.J., Henry A.H., Pedersen J.T. Antibody design: beyond the natural limits. Trends Biotechnol. 1994;12:199–206. doi: 10.1016/0167-7799(94)90083-3. [DOI] [PubMed] [Google Scholar]
- 25.Kuroda D., Shirai H., Jacobson M.P., Nakamura H. Computer-aided antibody design. Protein Eng. Des. Sel. 2012;25:507–521. doi: 10.1093/protein/gzs024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lefranc M.P., Giudicelli V., Ginestoux C., Jabado-Michaloud J., Folch G., Bellahcene F., Wu Y., Gemrot E., Brochet X., Lane J., et al. IMGT, the international ImMunoGeneTics information system. Nucleic Acids Res. 2009;37:D1006–D1012. doi: 10.1093/nar/gkn838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carter P., Presta L., Gorman C.M., Ridgway J., Henner D., Wong W., Rowland A.M., Kotts C., Carver M.E., Shepard H.M. Humanization of an anti-p185HER2 antibody for human cancer therapy. Proc. Natl Acad. Sci. U.S.A. 1992;89:4285–4289. doi: 10.1073/pnas.89.10.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mardia K.V., Jupp P.E. Directional Statistics. London: Wiley; 2000. [Google Scholar]
- 29.Honegger A., Pluckthun A. Yet another numbering scheme for immunoglobulin variable domains: an automatic modeling and analysis tool. J. Mol. Biol. 2001;309:657–670. doi: 10.1006/jmbi.2001.4662. [DOI] [PubMed] [Google Scholar]
- 30.Ramsden D.A., Wu G.E. Mouse kappa light-chain recombination signal sequences mediate recombination more frequently than do those of lambda light chain. Proc. Natl Acad. Sci. U.S.A. 1991;88:10721–10725. doi: 10.1073/pnas.88.23.10721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Wildt R.M., van Venrooij W.J., Winter G., Hoet R., Tomlinson I.M. Somatic insertions and deletions shape the human antibody repertoire. J. Mol. Biol. 1999;294:701–710. doi: 10.1006/jmbi.1999.3289. [DOI] [PubMed] [Google Scholar]
- 32.Cho H.-S., Mason K., Ramyar K.X., Stanley A.M., Gabelli S.B., Denney D.W., Leahy D.J. Structure of the extracellular region of HER2 alone and in complex with the Herceptin Fab. Nature. 2003;421:756–760. doi: 10.1038/nature01392. [DOI] [PubMed] [Google Scholar]
- 33.James L.C., Roversi P., Tawfik D.S. Antibody multispecificity mediated by conformational diversity. Science. 2003;299:1362–1367. doi: 10.1126/science.1079731. [DOI] [PubMed] [Google Scholar]
- 34.Jabs A., Weiss M.S., Hilgenfeld R. Non-proline Cis peptide bonds in proteins. J. Mol. Biol. 1999;286:291–304. doi: 10.1006/jmbi.1998.2459. [DOI] [PubMed] [Google Scholar]
- 35.Chuang G.-Y., Liou D., Kwong P.D., Georgiev I.S. NEP: web server for epitope prediction based on antibody neutralization of viral strains with diverse sequences. Nucleic Acids Res. 2014;42:W64–W71. doi: 10.1093/nar/gku318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kunik V., Ashkenazi S., Ofran Y. Paratome: an online tool for systematic identification of antigen-binding regions in antibodies based on sequence or structure. Nucleic Acids Res. 2012;40:W521–W524. doi: 10.1093/nar/gks480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ye J., Ma N., Madden T.L., Ostell J.M. IgBLAST: an immunoglobulin variable domain sequence analysis tool. Nucleic Acids Res. 2013;41:W34–W40. doi: 10.1093/nar/gkt382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chailyan A., Tramontano A., Marcatili P. A database of immunoglobulins with integrated tools: DIGIT. Nucleic Acids Res. 2012;40:D1230–D1234. doi: 10.1093/nar/gkr806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leaver-Fay A., Tyka M., Lewis S.M., Lange O.F., Thompson J., Jacak R., Kaufman K., Renfrew P.D., Smith C.A., Sheffler W., et al. Rosetta3 an object-oriented software suite for the simulation and design of macromolecules. Methods Enzymol. 2011;487:545–574. doi: 10.1016/B978-0-12-381270-4.00019-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dai L., Yang Y., Kim H.R., Zhou Y. Improving computational protein design by using structure-derived sequence profile. Proteins. 2010;78:2338–2348. doi: 10.1002/prot.22746. [DOI] [PMC free article] [PubMed] [Google Scholar]


