Abstract
The Gene Ontology Annotation (GOA) resource (http://www.ebi.ac.uk/GOA) provides evidence-based Gene Ontology (GO) annotations to proteins in the UniProt Knowledgebase (UniProtKB). Manual annotations provided by UniProt curators are supplemented by manual and automatic annotations from model organism databases and specialist annotation groups. GOA currently supplies 368 million GO annotations to almost 54 million proteins in more than 480 000 taxonomic groups. The resource now provides annotations to five times the number of proteins it did 4 years ago. As a member of the GO Consortium, we adhere to the most up-to-date Consortium-agreed annotation guidelines via the use of quality control checks that ensures that the GOA resource supplies high-quality functional information to proteins from a wide range of species. Annotations from GOA are freely available and are accessible through a powerful web browser as well as a variety of annotation file formats.
INTRODUCTION
GOA has been providing Gene Ontology (GO) annotations to proteins in the UniProt Knowledgebase for over 13 years. During that time, it has adapted to changing database technologies and GO annotation practice in order to provide users with the most current advances in GO curation and annotation file format. The year 2014 sees a significant change to the underlying structure of the GOA database. Until now, curators have only been able to assign annotations to proteins represented in UniProtKB, but a growing need for researchers to easily access the functional information of macromolecular complexes and non-coding RNAs has driven us to restructure the database and curation tools to allow annotation of these entities as well. These changes are currently underway and the ability to curate non-protein entities will be possible in the near future.
This article will report the developments of the database since its last description in the database issue in 2011 (1), as well as describe new sources of data that we have incorporated.
DATASET UPDATES IN THE GOA RESOURCE
The GOA resource consists of GO annotations from two different methods: automatic predictions and manual curation. Details of these two methods of GO annotation are published elsewhere (1,2). Curators contribute both to the manual curation of a wide range of species, by extracting the information from primary experimental literature, and to automatic predictions, by supplying keywords, subcellular locations and biochemical pathway information to UniProtKB entries. Curators also maintain the mapping files that are subsequently used in automatic annotation pipelines, namely UniProt keywords2GO, UniProt subcellular locations2GO and UniPathway2GO.
In addition to this, GOA supplements its annotation dataset with high-quality manual and automatic annotations from other annotation groups. The new sources of data that have been incorporated since our last update in 2011 are detailed in the next sections.
Automatic annotations
Automatic GO annotation predictions are an extremely valuable form of annotation because they provide a large number of high-quality functional annotations across a broad taxonomic range. For many species they are the only form of functional annotation available. Currently, in the GOA database there are over 52 million proteins from 483 000 taxonomic groups that are annotated using only automated methods (August 2014). Automatic annotations are created using algorithms based on sequence similarity, orthology or domain information or from pre-existing cross-references and keywords (1,3).
GOA now provides automatic annotations from nine sources, including InterPro2GO, UniProt keywords2GO and Enzyme Commission2GO. Since 2011 we have added two new sources: UniPathway2GO and EnsemblFungi. UniPathway is a manually curated resource for the representation and annotation of metabolic pathways (4). The UniPathway controlled vocabulary was mapped to GO terms by the GOA curators at the EBI to enable propagation of GO annotations to UniProtKB entries. UniPathway2GO is currently providing over 3.7 million annotations to 3.4 million proteins (August 2014). EnsemblFungi is an extension of the Ensembl Compara pipeline (5,6) to propagate GO annotations between orthologs. In this case, manual experimental annotations from Schizosaccharomyces pombe and Saccharomyces cerevisiae are propagated to over 20 other fungal species such as Neurospora crassa, Ashbya gossypii and Nectria haematococca. EnsemblFungi is currently providing around 27 000 annotations to almost 5800 fungal proteins (August 2014).
Manual annotations
The GOA resource continues to provide manual annotations based on experimental data in published literature. For several years, our strategy for choosing GOA annotation priorities has been based around a particular biological pathway or organelle. Our recent annotation projects have included curating proteins involved in kidney development and related processes (7), proteins located in the peroxisome (8) and those located in the exosome (http://www.ebi.ac.uk/GOA/exosome). We find these discrete projects very valuable in providing deep curation to a particular area of biology. UniProt curators at the EBI have also recently contributed manual annotation to assist with the second Critical Assessment of Functional Annotation challenge (9), which is designed to assess computational methods that predict protein function. Our provision of a set of high-quality manual annotations has enabled the organizers of the challenge to better validate the prediction methods that were submitted.
Curators at the EBI have been including additional contextual information in manual GO annotations since 2011. These so-called ‘Annotation Extensions’ can describe effector-target relationships such as the substrate acted upon by an enzyme or transcription factor targets, the subcellular location of an activity, the cell or tissue location of a gene product or activity, or the developmental or cell cycle phase during which a function or process occurs (10). The annotation extension is located in a separate field of the annotation file and consists of a Relation(Entity) expression, e.g. occurs_in(CL:1000606) (where CL:1000606 is the Cell Ontology (11) identifier for ‘kidney nerve cell’). This contextual information will increase the utility of functional annotation and support pathway analysis. Currently, the GOA database contains over 46 000 annotations that contain one or more annotation extension statements (August 2014).
The GOA dataset is supplemented with manual annotations from a variety of other annotation groups, both Model Organism Databases and specialist resources, most of which are also members of the GO Consortium. We incorporate annotations from 41 external groups and the level of contribution from each group can vary widely; since 2011 we have incorporated manual annotation datasets from the following groups: Aspergillus Genome Database (12), Pseudomonas Genome Database (13), the Community Assessment of Community Annotation with Ontologies Project (CACAO; http://gowiki.tamu.edu/wiki/index.php/Category:CACAO), Microbial Energy Processes Gene Ontology Project (MENGO; http://mengo.vbi.vt.edu/), the Syscilia Annotation Project (http://syscilia.org/), Parkinson's UK-University College London (http://www.ucl.ac.uk/functional-gene-annotation/neurological), the Alzheimer's Disease Annotation Project at the University of Toronto (http://wiki.geneontology.org/index.php/Alzheimer%27s_Disease_Annotation_Project), as well as annotations for two new species (Trypanosoma brucei and Leishmania major) from GeneDB (14).
GOA contributes to discussions about annotation guidelines and consistency within the GO Consortium and therefore adheres to the same guidelines and quality control checks that have been agreed by the Consortium. Additionally, to ensure that we are representing the biology correctly, we communicate with experts in the relevant field who, in collaboration with the GO Consortium ontology editors, provide input into the ontology structure, as well as advising on the proteins that should be curated with the GO terms. We have published several papers based on these collaborations that include the experts as co-authors (7,15,16). We have found this to be a very successful approach that we will continue with in future projects. GO Consortium collaborative projects can be found on the GO Consortium website (http://geneontology.org/collaborations). GO terms created as part of these collaborations are indicated in the QuickGO browser (17) with the logo of the appropriate funding agency, e.g. http://www.ebi.ac.uk/QuickGO-Beta/term/GO:1901207.
DEVELOPMENTS TO THE DATABASE AND CURATION TOOLS
We are a core member of the GO Consortium and as such contribute to the development of annotation guidelines, policies and new annotation file formats. These developments inevitably require our database and curation tools to adapt in order to keep pace and provide our users with annotations and tools that conform to the most current practices and technologies. The changes that have been necessary over the past 4 years are detailed below.
Evidence types
The desire for the GO Consortium to capture more detailed types of supporting evidence within an annotation has seen the gradual adoption of Evidence Ontology (ECO) terms (18). ECO is a controlled vocabulary that describes types of scientific evidence and as such, provides more descriptive evidence terms than is possible with GO evidence codes. For example, the interaction methods ‘yeast 2-hybrid’ (ECO:0000068) and ‘co-immunoprecipitation’ (ECO:0000070) each have their own ECO term, whereas in a GO annotation we can represent both of these types of evidence only with ‘Inferred from Physical Interaction’ (IPI). All ECO terms used in GO annotation will map to an existing GO evidence code. UniProt has already adopted use of these terms within their protein entries and ECO codes became visible in UniProtKB on 1 October 2014. The GOA database also makes use of ECO terms, now all GO evidence codes are cross-referenced to ECO (e.g. Inferred from Direct Assay (IDA) is equivalent to ECO:0000314), therefore any use of a GO evidence in an annotation can be mapped to an ECO term (purl.obolibrary.org/obo/eco/gaf-eco-mapping.txt). The GOA curation tool (Protein2GO) now displays the equivalent ECO term codes alongside the GO evidence codes, and it is planned that curators will be able to choose any ECO term for their GO annotation in Protein2GO in the near future. Curators will be able to specify, for instance, that a Cellular Component annotation has supporting evidence from a green fluorescent protein transcript localization (ECO:0000296).
The GOA curation tool Protein2GO
Protein2GO is actively maintained and developed at the EBI and has served as the GO annotation tool for curators who wish to use it, both within and external to the EBI, for over 12 years and we welcome further contributors. All annotations are attributed to the contributing group and made available through the QuickGO browser (17) and annotation files. We describe here the annotation quality control mechanisms that are in place to ensure only high-quality annotations are added to our database, as a point of interest to the users of our annotations.
The most valuable feature of Protein2GO is its many inbuilt quality checks that prevent incorrectly formulated GO annotations from being added to the database. This is beneficial to both trainee and experienced curators as the rules for GO annotation become ever more complex. An example of one of these integral checks is shown in Figure 1. According to GO Consortium guidance, the evidence code ‘IC’ (Inferred by Curator) must refer to another GO term in the ‘With/From’ field of the annotation that has been used in a second annotation for the protein being curated. If no GO term is entered, or if a text string other than a GO term is entered, the curator is prevented from adding the annotation to the database. A warning is given detailing what the curator must rectify before the annotation is acceptable. A subsequent quality control check reports any IC-evidenced annotations that use a GO term in the ‘With/From’ field for which no experimentally evidenced annotations exist in that protein record.
Figure 1.
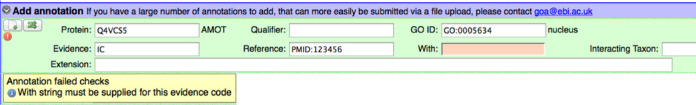
Annotation quality checks in the Protein2GO curation interface. If an annotation is incomplete or fails sanity checks, a warning is given and the curator is unable to add the annotation to the database. In the annotation pictured, an identifier is missing from the ‘With/From’ field.
Protein2GO has also been adapted to enable curators to add annotation extensions (see ‘Manual annotations’ section). There are complex rules on how annotation extensions must be used in curation; therefore these rules have been integrated into Protein2GO to assist curators in making extended annotations. For example, the annotation extension relation exists_during must only be used when the primary GO term used is from the Cellular Component ontology. Protein2GO will not allow the exists_during relationship to be used if the primary annotation uses a GO term from either the Molecular Function or Biological Process ontologies. The validation of annotation extensions relies on a service provided by QuickGO, which also provides services to allow Protein2GO to search for GO terms and gene product identifiers.
In the event that a curator notices an incorrect annotation, they are now able to raise a dispute or a query for that annotation. The curator who raises the dispute or query is prompted to provide a reason and then Protein2GO emails this information to the curator or group that supplied the annotation. We have found this a very efficient way of dealing with incorrect annotations, which previously would have to be dealt with by the curator identifying who made the annotation, finding their email address and then composing an email to explain the problem.
To assist those curators who do not use UniProtKB accessions as the primary identifiers for proteins from their particular species they may enter Model Organism Database (MOD) identifiers directly into Protein2GO. For example DictyBase curators may enter a DictyBase protein identifier into Protein2GO, which then accesses this identifier from cross-references in UniProtKB entries, via UniProtKB identifier mapping services, to find the corresponding UniProtKB accession. Protein2GO also has a lookup service that allows curators to search for model organism gene names and synonyms that are more familiar to the curator, to retrieve the appropriate UniProtKB accession.
To increase our effort of engaging the scientific community, we have added a feature into Protein2GO that allows curators to send an email to the corresponding author of a paper they have comprehensively curated. The message provides a web link to the annotations in the QuickGO browser (17) for the author to view and so is intended to garner feedback and to make the author aware of the biocuration process. The emails are sent out coincident with our 4-weekly file release to ensure the annotations are visible to the author when they receive the email. The first set of author correspondence emails was sent in our September 2014 release.
The GO Consortium has seen an increasing need to use a common tool for curation to ensure consistent annotation among the member groups. As an initial step to enable this, the GO Consortium has adopted Protein2GO as a common tool for GO annotation of proteins. Groups including WormBase (19), the Saccharomyces Genome Database (20) and DictyBase (21) have already switched to using Protein2GO as their primary curation tool for protein GO annotation and more groups are preparing to switch in the near future. We continue to support smaller annotation groups by providing Protein2GO and incorporating their annotation directly into the GOA database. Since 2011, we have provided access to Protein2GO for curators from the following groups: the Alzheimer's Project at the University of Toronto (http://wiki.geneontology.org/index.php/Alzheimer%27s_Disease_Annotation_Project), the Community Assessment of Community Annotation with Ontologies project (CACAO; http://gowiki.tamu.edu/wiki/index.php/Category:CACAO), Parkinson's UK-University College London (http://www.ucl.ac.uk/functional-gene-annotation/neurological) and the Syscilia Annotation Project (http://syscilia.org/).
Curation of entities other than proteins
The GOA database and Protein2GO curation tool were developed to enable GO annotation of UniProtKB protein entries; however, there is an increasing need for the provision of GO annotations to entities such as RNAs and macromolecular complexes. For example, groups such as WormBase already supply annotations to RNAs, but currently are not able to curate these within Protein2GO. To enable this, we have begun to reconstruct the GOA database and Protein2GO to support annotation to macromolecular complexes from the IntAct Complex Portal (22) and RNA types from RNAcentral (23). For example, annotations may be made to ‘EBI-9008420’, which is the identifier for the human Hemoglobin HbA complex. The subunits/components of the complex are specified in the IntAct Complex Portal entry (http://www.ebi.ac.uk/intact/complex/details/EBI-9008420). Annotations made to non-protein identifiers will be provided in annotation files and made available alongside our current annotations in the QuickGO web browser (17).
The GOA web browser QuickGO
The GOA browser, QuickGO ((17); http://www.ebi.ac.uk/QuickGO), is currently undergoing improvements to its user interface in order to provide a more intuitive experience for users. QuickGO is a powerful, web-based tool for searching and viewing GO terms and annotations from the GOA database. However, it was not obvious how to use certain features and this was confirmed by user experience testing with QuickGO users and non-users. In order to provide more flexible searching, filtering and display, we have changed the underlying infrastructure of QuickGO to the Apache Solr™ search platform (http://lucene.apache.org/solr/). This has enabled us to include faceted filtering of the data (Figure 2) that is inline with the recent changes to the UniProt website (www.uniprot.org). A beta version of the improved QuickGO is available at http://www.ebi.ac.uk/QuickGO-Beta.
Figure 2.
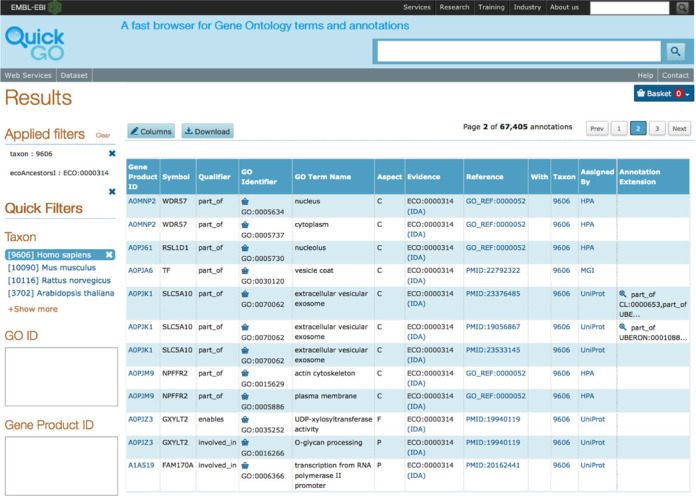
Faceted filtering of annotation data in QuickGO. Annotations in the GOA database can be filtered for particular subsets. Here the filters for human (taxon:9606) and direct assay evidence used in manual assertion (ECO identifier ECO:0000314) have been applied.
QUALITY CONTROL
As GO annotation guidelines continue to be refined, the GOA database and Protein2GO quality checks need to be reviewed and extended accordingly. The major additions to these checks since our last database update have previously been reported in (24), but a summary will be given below.
The introduction of subsets of GO terms that are deemed not suitable for direct annotation meant that it was necessary to report the annotations that use these terms in order for them to be corrected and to prevent curators from making new annotations to these terms. For example, the prohibition of cell cycle phase-type terms (e.g. ‘metaphase’ (GO:0051323)) in primary GO annotations led to a restriction being added to Protein2GO, which prevents curators from annotating directly to these GO terms, but curators are still able to use them in the annotation extension field. Any existing annotations using these terms had to be reviewed and either updated to a more appropriate term or deleted. Additionally, when importing data from other curation groups, these types of annotation are excluded from our database. When these GO terms are viewed in QuickGO, there is an alert informing that the term is not to be used for direct annotation (Figure 3).
Figure 3.
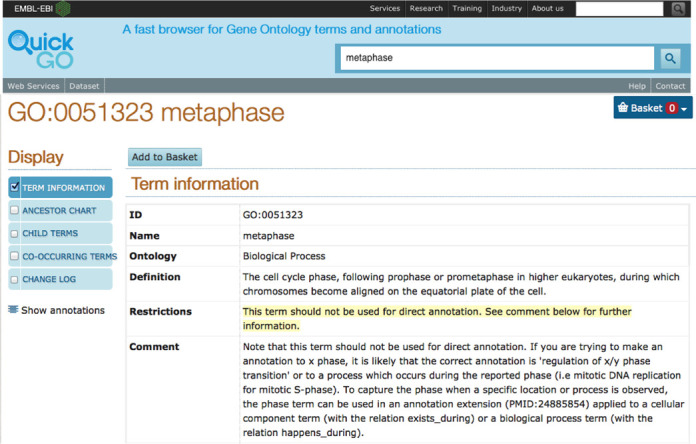
Usage restrictions for GO terms. Information about the usage of each term is included on the GO term page in QuickGO. In the case of ‘metaphase’ (GO:0051323), this term cannot be used in primary GO annotations.
We have improved the accuracy of automatic annotations by removing those annotations that violate taxon constraints. Some GO terms are applicable only to certain taxa and this is encoded in the GO taxon constraints (http://purl.obolibrary.org/obo/go/extensions/x-taxon-importer.owl). For example, if a GO term that is valid for use only with eukaryotes, e.g. ‘MAPK cascade’ (GO:0000165), is applied to a bacterial protein, the annotation would be incorrect and it would be deleted. This process has resulted in the deletion of approximately 106 000 incorrect automatic annotations.
Another quality control measure we have that is based on taxon constraints is the automated correction of annotations (24). This is necessary only for automatic annotations when it is not possible for the annotation group to update the annotations, for example when altering a mapping between a GO term and an InterPro domain would cause an unnecessarily large decrease in annotations. The GOA post-processing can make conservative changes to individual automatic annotations that fall into this category in order to correct the assigned GO term. An example of this is the phosphoribosylaminoimidazole synthetase, purM, from Prochlorococcus phage (UniProtKB:E3SNM7), which has a prediction to ‘cytoplasm’ (GO:0005737) from the InterPro annotation method. A slight change to this annotation prediction would lead to the correct term, ‘host cell cytoplasm’ (GO:0044165), being supplied in accordance with the GO taxon rules, which require the term ‘cytoplasm’ (GO:0005737) only be applied to cellular organisms (http://www.ebi.ac.uk/QuickGO-Beta/term/GO:0005737). All automatic annotations that are transformed by the GOA post-processing will use a ‘GO_REF’ reference that indicates to the user that such changes have occurred and which points users to the reference description: http://www.geneontology.org/cgi-bin/references.cgi. For example, UniProtKB: E3SNM7, as described in the example above would be displayed with the accompanying GO_REF:0000042.
During 2011 we implemented an ‘Annotation blacklist’, which specifies protein:GO term combinations that are not acceptable as annotations (24). For example, UniProtKB:B5X1G6, the Atlantic salmon AKT-interacting protein, belongs to the ubiquitin-conjugating enzyme family, but lacks the conserved Cys residue necessary for ubiquitin-conjugating enzyme E2 activity. It is therefore blacklisted for annotation with ‘ubiquitin-protein transferase activity’ (GO:0004842). The blacklist is used both by Protein2GO to prevent curators from making these annotations, and to stop these annotations from external groups from being imported into the GOA database. Additionally, automatic annotation providers apply the blacklist restrictions at source via access to a webservice (e.g. to retrieve all blacklist entries for taxon identifier 9031(chicken); http://www.ebi.ac.uk/QuickGO-Beta/ws/validate?type=taxon&taxon_id=9031&action=get_blacklist).
DATA ACCESS
QuickGO (http://www.ebi.ac.uk/QuickGO) (17) is the primary location for GOA data, where a full GO annotation set is made freely available to view, filter and download. Annotation data within QuickGO are updated on a weekly basis. In addition, users can also browse the GO hierarchies using QuickGO, which are updated daily.
Programmatic access to GOA data (annotations and GO terms) is available from QuickGO and described in http://www.ebi.ac.uk/QuickGO-Beta/webservices. We additionally provide webservices for the annotation blacklist, e.g. to retrieve all blacklist entries for taxon identifier 9606 (human); http://www.ebi.ac.uk/QuickGO-Beta/ws/validate?type=taxon&taxon_id=9606&action=get_blacklist, and taxon constraints; www.ebi.ac.uk/QuickGO-Beta/ws/validate?type=taxon&action=get_constraints.
As described in our previous NAR database article (1), we had begun to provide the entire set of GOA annotations in a new file format, GPAD (Gene Product Association Data) and its corresponding file GPI (Gene Product Information). Since then we have created GPAD/GPI versions for all of our annotation files, including the species-specific files such as human, dog, chicken and cow. In response to user feedback, we now also produce a set of Gene Association File (GAF), GPAD and GPI format files that are based on UniProt reference proteomes that provide one protein per gene. These are identified by the following file name formats:
gene_association.goa_ref_<species>
gp_association.goa_ref_<species>
gp_information.goa_ref_<species>
These files are located alongside all of our annotation files on the GOA ftp site:
ftp://ftp.ebi.ac.uk/pub/databases/GO/goa/
and on the GO Consortium ftp site (GAF format only):
ftp://ftp.geneontology.org/pub/go/gene-associations/
Annotation groups who wish to be considered for access to Protein2GO or who wish to provide annotations for inclusion into our database should contact us at goa@ebi.ac.uk.
Acknowledgments
GOA would like to thank curators from the UniProt Consortium and the GO Consortium, and all groups who contribute GO annotations for inclusion into our database.
FUNDING
National Human Genome Research Institute of the National Institutes of Health [U41HG006104, U41HG007822 to UniProt and U41HG002273 to GO Consortium]; National Institutes of Health; British Heart Foundation [RG/13/5/30112]; Parkinson's UK [G-1307]; European Molecular Biology Laboratory (EMBL) core funds. Funding for open access charge: EMBL core funds.
Conflict of interest statement. None declared.
REFERENCES
- 1.Dimmer E.C., Huntley R.P., Alam-Faruque Y., Sawford T., O'Donovan C., Martin M.J., Bely B., Browne P., Mun Chan W., Eberhardt R., et al. The UniProt-GO Annotation database in 2011. Nucleic Acids Res. 2011;40:D565–D570. doi: 10.1093/nar/gkr1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balakrishnan R., Harris M.A., Huntley R., Van Auken K., Cherry J.M. A guide to best practices for Gene Ontology (GO) manual annotation. Database. 2013;2013:doi:10.1093/database/bat054. doi: 10.1093/database/bat054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burge S., Kelly E., Lonsdale D., Mutowo-Muellenet P., McAnulla C., Mitchell A., Sangrador-Vegas A., Yong S.Y., Mulder N., Hunter S. Manual GO annotation of predictive protein signatures: the InterPro approach to GO curation. Database. 2012;2012:doi:10.1093/database/bar068. doi: 10.1093/database/bar068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morgat A., Coissac E., Coudert E., Axelsen K.B., Keller G., Bairoch A., Bridge A., Bougueleret L., Xenarios I., Viari A. UniPathway: a resource for the exploration and annotation of metabolic pathways. Nucleic Acids Res. 2012;40:D761–D769. doi: 10.1093/nar/gkr1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barrell D., Dimmer E., Huntley R.P., Binns D., O'Donovan C., Apweiler R. The GOA database in 2009—an integrated Gene Ontology Annotation resource. Nucleic Acids Res. 2009;37:D396–D403. doi: 10.1093/nar/gkn803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vilella A.J., Severin J., Ureta-Vidal A., Heng L., Durbin R., Birney E. EnsemblCompara GeneTrees: complete, duplication-aware phylogenetic trees in vertebrates. Genome Res. 2009;19:327–335. doi: 10.1101/gr.073585.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alam-Faruque Y., Hill D.P., Dimmer E.C., Harris M.A., Foulger R.E., Tweedie S., Attrill H., Howe D.G., Thomas S.R., Davidson D., et al. Representing kidney development using the gene ontology. PLoS One. 2014;9:e99864. doi: 10.1371/journal.pone.0099864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mutowo-Meullenet P., Huntley R.P., Dimmer E.C., Alam-Faruque Y., Sawford T., Jesus Martin M., O'Donovan C., Apweiler R. Use of Gene Ontology Annotation to understand the peroxisome proteome in humans. Database. 2013;2013:doi: 10.1093/database/bas062. doi: 10.1093/database/bas062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wass M.N., Mooney S.D., Linial M., Radivojac P., Friedberg I. The automated function prediction SIG looks back at 2013 and prepares for 2014. Bioinformatics. 2014;30:2091–2092. doi: 10.1093/bioinformatics/btu117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huntley R.P., Harris M.A., Alam-Faruque Y., Blake J.A., Carbon S., Dietze H., Dimmer E.C., Foulger R.E., Hill D.P., Khodiyar V.K., et al. A method for increasing expressivity of Gene Ontology annotations using a compositional approach. BMC Bioinformatics. 2014;15:155. doi: 10.1186/1471-2105-15-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meehan T.F., Masci A.M., Abdulla A., Cowell L.G., Blake J.A., Mungall C.J., Diehl A.D. Logical development of the cell ontology. BMC Bioinformatics. 2011;12:6. doi: 10.1186/1471-2105-12-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cerqueira G.C., Arnaud M.B., Inglis D.O., Skrzypek M.S., Binkley G., Simison M., Miyasato S.R., Binkley J., Orvis J., Shah P., et al. The Aspergillus Genome Database: multispecies curation and incorporation of RNA-Seq data to improve structural gene annotations. Nucleic Acids Res. 2014;42:D705–D710. doi: 10.1093/nar/gkt1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Winsor G.L., Lam D.K.W., Fleming L., Lo R., Whiteside M.D., Yu N.Y., Hancock R.E.W., Brinkman F.S.L. Pseudomonas Genome Database: improved comparative analysis and population genomics capability for Pseudomonas genomes. Nucleic Acids Res. 2011;39:D596–D600. doi: 10.1093/nar/gkq869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Logan-Klumpler F.J., De Silva N., Boehme U., Rogers M.B., Velarde G., McQuillan J.A., Carver T., Aslett M., Olsen C., Subramanian S., et al. GeneDB—an annotation database for pathogens. Nucleic Acids Res. 2012;40:D98–D108. doi: 10.1093/nar/gkr1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alam-Faruque Y., Huntley R.P., Khodiyar V.K., Camon E.B., Dimmer E.C., Sawford T., Martin M.J., O'Donovan C., Talmud P.J., Scambler P., et al. The impact of focused Gene Ontology curation of specific mammalian systems. PLoS One. 2011;6:e27541. doi: 10.1371/journal.pone.0027541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lovering R.C., Dimmer E., Khodiyar V.K., Barrell D.G., Scambler P., Hubank M., Apweiler R., Talmud P.J. Cardiovascular GO annotation initiative year 1 report: why cardiovascular GO. Proteomics. 2008;8:1950–1953. doi: 10.1002/pmic.200800078. [DOI] [PubMed] [Google Scholar]
- 17.Binns D., Dimmer E., Huntley R., Barrell D., O'Donovan C., Apweiler R. QuickGO: a web-based tool for Gene Ontology searching. Bioinformatics. 2009;25:3045–3046. doi: 10.1093/bioinformatics/btp536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chibucos M.C., Mungall C.J., Balakrishnan R., Christie K.R., Huntley R.P., White O., Blake J.A., Lewis S.E., Giglio M. Standardized description of scientific evidence using the Evidence Ontology (ECO) Database. 2014;2014 doi: 10.1093/database/bau075. doi:10.1093/database/bau075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harris T.W., Baran J., Bieri T., Cabunoc A., Chan J., Chen W.J., Davis P., Done J., Grove C., Howe K., et al. WormBase 2014: new views of curated biology. Nucleic Acids Res. 2014;42:D789–D793. doi: 10.1093/nar/gkt1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Costanzo M.C., Engel S.R., Wong E.D., Lloyd P., Karra K., Chan E.T., Weng S., Paskov K.M., Roe G.R., Binkley G., et al. Saccharomyces genome database provides new regulation data. Nucleic Acids Res. 2014;42:D717–D725. doi: 10.1093/nar/gkt1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Basu S., Fey P., Pandit Y., Dodson R., Kibbe W.A., Chisholm R.L. DictyBase 2013: integrating multiple Dictyostelid species. Nucleic Acids Res. 2013;41:D676–D683. doi: 10.1093/nar/gks1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meldal B.H.M., Forner-Martinez O., Costanzo M.C., Dana J., Demeter J., Dumousseau M., Dwight S.S., Gaulton A., Licata L., Melidoni A.N., et al. The complex portal—an encyclopaedia of macromolecular complexes. Nucleic Acids Res. 2014. doi:10.1093/nar/gku975. [DOI] [PMC free article] [PubMed]
- 23.Bateman A., Agrawal S., Birney E., Bruford E.A., Bujnicki J.M., Cochrane G., Cole J.R., Dinger M.E., Enright A.J., Gardner P.P., et al. RNAcentral: a vision for an international database of RNA sequences. RNA. 2011;17:1941–1946. doi: 10.1261/rna.2750811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huntley R.P., Sawford T., Martin M.J., O'Donovan C. Understanding how and why the Gene Ontology and its annotations evolve: the GO within UniProt. Gigascience. 2014;3:4. doi: 10.1186/2047-217X-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]


