Abstract
The ‘Human Immunodeficiency Virus Type 1 (HIV-1), Human Interaction Database’, available through the National Library of Medicine at http://www.ncbi.nlm.nih.gov/genome/viruses/retroviruses/hiv-1/interactions, serves the scientific community exploring the discovery of novel HIV vaccine candidates and therapeutic targets. Each HIV-1 human protein interaction can be retrieved without restriction by web-based downloads and ftp protocols and includes: Reference Sequence (RefSeq) protein accession numbers, National Center for Biotechnology Information Gene identification numbers, brief descriptions of the interactions, searchable keywords for interactions and PubMed identification numbers (PMIDs) of journal articles describing the interactions. In addition to specific HIV-1 protein–human protein interactions, included are interaction effects upon HIV-1 replication resulting when individual human gene expression is blocked using siRNA. A total of 3142 human genes are described participating in 12 786 protein–protein interactions, along with 1316 replication interactions described for each of 1250 human genes identified using small interfering RNA (siRNA). Together the data identifies 4006 human genes involved in 14 102 interactions. With the inclusion of siRNA interactions we introduce a redesigned web interface to enhance viewing, filtering and downloading of the combined data set.
INTRODUCTION
A large number of viral and cellular protein interactions are necessary for cellular immunity and competent viral infection. Knowledge of the protein–protein interactions that facilitate pathogenesis and disease has been critical to advancements in vaccine research, therapeutic drug discovery and cell biology. Many strides have been made in understanding the biology, pathogenesis and transmission of the Human Immunodeficiency Virus Type 1 (HIV-1) through the investigation of host immune responses, host and viral protein interactions and analysis of cell-type viral susceptibilities (1,2). These breakthroughs have been complemented or heralded by recent advances in biochemistry, bioengineering and bioinformatics. As a result, the number of publications related to HIV-1 research has grown steadily in number and complexity over the past three decades. In particular, the number of articles describing HIV-1 and human protein interactions has created an expanding universe of biological data from which scrutiny, analysis and interpretation is often difficult. Fortunately, the advent of electronic databases and tools is helping to mitigate these difficulties (3–6). Leveraging these advances, a collaboration between Southern Research Institute and the National Center for Biotechnology Information (NCBI) was established to catalog, in a searchable database, all the HIV-1 and human protein interactions published in peer-reviewed journals since 1984. Included in this database are brief descriptions of the respective interactions, National Library of Medicine (NLM) PubMed identification numbers (PMIDs) for the articles describing the interaction, NCBI Reference Sequence (RefSeq) protein accession numbers, NCBI Gene ID numbers and keywords that facilitate searches for the corresponding interaction.
GROWTH OF THE DATABASE
As previously described (3,4), journal abstracts and publications are identified and screened retrospectively (back to 1984) and prospectively for original (primary) research describing interactions between HIV-1 and human host proteins (3). Since the initial data set release in 2007, the number of protein interactions reported has grown to 12 786, an increase of nearly 150%. Supplementary Tables S1–S3 provide an updated summary of the protein interaction data set that was initially described (4). We recently added a new interaction type to the data set, ‘replication interactions’, which are gleaned from literature reports of effects upon HIV-1 replication that result when individual human gene expression is blocked using siRNA. In August 2014, 1316 replication interactions are reported for 1250 human gene knockdowns.
DATA ACCESS IN NCBI'S GENE DATABASE
Review of individual interactions is ongoing, and data are provided incrementally to NCBI as a set of comprehensive tab-delimited text files. To provide scientists in the field of HIV/AIDS research a concise, yet detailed, summary of all known interactions between HIV-1 and host cell proteins, the database tracks the following information for each protein–protein interaction identified in the literature:
NCBI Reference Sequence (RefSeq) protein accession numbers;
NCBI Gene ID numbers;
Brief descriptions of the protein–protein interactions;
Keywords to support searching for interactions;
NLM PMIDs for all journal articles describing the interactions.
Upon receipt the data files are parsed, identifiers are validated and data are loaded to the NCBI Gene database (7). Protein and replication interaction data are integrated into appropriate records in NCBI Gene, provided in a browseable interactions web site and made available for downloads.
Protein and replication interactions are reported in the HIV-1 Interactions section of the Gene report page (Figure 1); a link in the Gene report table of contents facilitates navigation to this section. Separate tables reporting one to many interaction descriptions are provided for each interaction type. For instance, the human gene for acetyl-CoA carboxylase beta (ACACB; GeneID 32) has a record with both replication and protein interaction data (Figure 1). The Gene database supports searching for HIV-1, human interaction data using properties (‘hiv1_protein_interactions[property]’ and ‘hiv1_replication_interactions[property]’). The data can also be queried, filtered and downloaded through the dedicated interactions web site described below. These interaction data are also reported in the Gene FTP site (ftp://ftp.ncbi.nlm.nih.gov/gene/GeneRIF/ in files hiv_interactions and hiv_siRNA_interactions).
Figure 1.
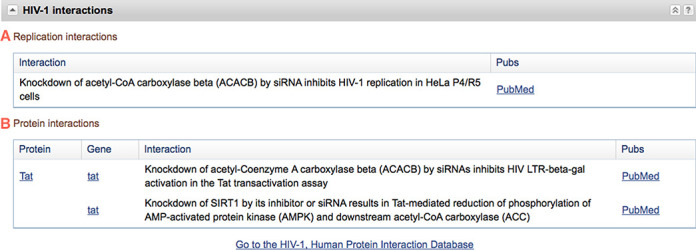
NCBI Gene report of HIV-1 Interactions. If involved in a protein–protein or siRNA established interaction the report page for such a human gene includes a section entitled ‘HIV-1 Interactions’ shown here. (A) Interactions described by siRNA (‘Replication Interactions’). (B) Protein–protein interactions. In both cases a description of the interaction is displayed as well as a link to the publication(s) supporting the described interaction. Included is a link to the main site for the HIV-1 Interaction Database.
HIV-1, HUMAN INTERACTION WEB SITE
HIV-1, human protein and replication interaction data can be browsed, filtered and downloaded from the HIV-1 Interactions web site (www.ncbi.nlm.nih.gov/genome/viruses/retroviruses/hiv-1/interactions/). The improved interface includes help documentation, supports structured queries against Gene and browsing of both the protein and replication interaction data sets. An updated retroviruses home page has also been provided. The interactions home page includes an interface to build a query against NCBI's Gene database. Options include restricting the query to protein or replication interaction data, by gene ontology terms (8) associated with Gene records, by a protein domain name annotated on a RefSeq protein record, or by Gene properties or keywords. These query options are documented on the Interactions Help page (www.ncbi.nlm.nih.gov/genome/viruses/retroviruses/hiv-1/help/). Submitting the search criteria returns results in the NCBI Gene resource where one may find additional gene-specific descriptions and links to related data. The home page also includes some predefined links to find related data in NCBI's Protein, Structure, Nucleotide, PopSets or PubMed databases. Links are provided at the top of the page to access a project description, help documentation, publications, database release history and to interactively browse the interaction data sets.
The default Browse page loads all interactions and supports downloading the complete data sets or finding (and downloading) subsets of the data by filtering for interaction type, specific protein and/or interaction terms as shown in (Figure 2). For instance, to browse the siRNA-based replication interactions that enhance HIV-1 virus replication simply select ‘enhanced by knockdown of human gene’ from the Replication interaction menu. To browse the HIV-1 protein interaction data, start by either selecting an interaction type then the HIV-1 protein (or all proteins) to browse data for a specific type of interaction, or by first selecting the HIV-1 protein product of interest. Upon selecting an individual HIV-1 protein option, such as the Pol protein, the interaction type filter menu automatically refreshes to display only those interaction types that are stored in the database for Pol. The results table refreshes on-the-fly as different filter combinations are selected. The filtered results are presented in a 20 row paginated table with interactions anchored on the human gene record; additional details of the interaction are presented upon clicking on the ‘more’ link (Figure 2). The page also supports navigating to view the gene set in NCBI's Gene database by clicking on the ‘View NCBI Gene records’ button. The tabular results presented on the web page can also be downloaded as a comma separated text file. The download file includes more columns than are displayed, such as the NCBI GeneID, RefSeq protein accession, protein name and interaction data modification date. Note that a human gene can be listed more than once if there are multiple distinct interaction descriptions. For example, there are three rows returned for the human APOBEC3F gene when selecting HIV protein ‘vif, Vif’ and interaction ‘disrupts’. Each row is associated with a distinct interaction description with Pubmed ID numbers:
‘Protein: NP_660341.2, GeneRIF: HIV-1 Vif W21A, S32A, W38A, Y69A, E76A, W79A, H108A, C114S, C133S and H139A mutants have no viral ability to neutralize APOBEC3F. Pubmed ID: 20299747’
‘Protein: NP_660341.2, GeneRIF: All residues except N175 in the (171)EDRWN(175) domain of Vif are equally important for regulation of A3F neutralization. Pubmed ID: 20335268’
‘Protein: NP_660341.2, GeneRIF: The T(Q/D/E)x(5)ADx(2)(I/L) motif, located at residues 96 to 107 in HIV-1 Vif, plays a critical role in neutralizing activity toward A3F. This motif regulates Vif interaction with Cul5. Pubmed ID: 20592083’
Figure 2.
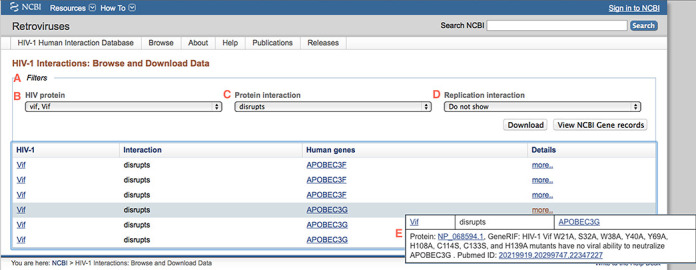
Filtering results while browsing. The ‘Filters’ section (A) allows users to choose criteria from three menus: (B) ‘HIV protein’, (C) ‘Protein interaction’ and (D) ‘Replication interaction’. In this example, the filtering results in presenting only protein interactions where the HIV interactant is ‘vif’ and the interaction with a product of each indicated human gene ‘disrupts’ an activity. Clicking on ‘more…’ on any listed interaction yields a popup menu (E) with details including the human protein product RefSeq accession, the full text interaction description and the PubMed ID(s). Also visible (middle right) are the ‘Download’ and ‘View NCBI Gene records’ selection buttons, described in the text.
VALUE OF THE DATABASE TO THE HIV/AIDS RESEARCH COMMUNITY
The database provides a unique and streamlined approach for HIV/AIDS investigators to quickly aggregate and parse data relevant to their field of interest. To date over 90 research articles and reviews have cited the database, particularly as a source that supports their observations and/or predictions (9–11). Likewise, some investigators have used the database as a primary data source for their experimental analyses (12–14) while others have utilized the database for developing tools that analyze and visualize the HIV-1-host protein–protein interaction network (15–17). Interestingly, the database has also served in a predictive capacity for some investigators (18–20). Highlighting the increasing utilization of genome-wide analyses, various groups have also used the database for the characterization of HIV-1 infection and replication mechanisms (21,22). And, results from genome-wide transcriptomes of primary monocytes from HIV-positive patients on highly active antiretroviral therapy were compared to the database for gene set enrichment analysis (23). As would be expected, the database has also provided useful information for identifying novel anti-HIV drugs (24). Finally, the database has to some extent been used to create protein-level analysis for other pathogen–host interactions (25,26). Overall, the HIV, human interaction database has been utilized not only by the HIV/AIDS research community, but also by investigators in other fields of infectious disease. As such, it has the potential to not only elucidate critical HIV-1-human protein interactions but also serve as a tool for understanding a broad array of other host–pathogen interactions.
UPDATED HIV-1 GENOME ANNOTATION
Annotation of the RefSeq HIV-1 genome record (accession NC_001802.1) was updated in June 2014 with revisions that add experimental evidence supporting the annotated proteins. In addition, gene and coding sequence (CDS) annotation was added for a highly conserved minus-strand encoded protein (antisense protein) of unknown function (GeneID 19424028; protein accession YP_009028572.1) with marked similarity to the HTLV-1 encoded protein, HBZ.
FUTURE DEVELOPMENTS
Non-coding RNAs (ncRNAs) are a diverse family of untranslated transcripts that play crucial roles in different kinds of cellular function, including repression of gene expression by microRNA (miRNA), regulation of mRNA splicing by small nuclear RNA, modulation of ribosomal activity by small nucleolar RNA and infrastructure of transcriptional and translational functions by tRNA and rRNA (27–30). The HIV-1 life cycle involves utilization of the cellular machinery and evading the immune system. A profound challenge for developing effective HIV-1 treatments is viral latency and regulation of gene expression. Recent developments provide some insights into ncRNA-mediated regulation of viral latency and gene expression (31,32). miRNA expression profile changes were also reported in recent studies in HIV-1-transfected human cells and in HIV-1 seropositive individuals (33,34). Moreover, 7SK small nuclear RNA demonstrated a significant role for controlling HIV-1 transcription by inhibiting the CDK9/cyclin T1 kinase (35). Therefore, plans are being developed to expand the database to include information on the involvement of ncRNAs in HIV replication as additional research is published in this area. Future updates to the database are planned to be released on a quarterly to bi-annual basis.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
We thank additional staff from Southern Research Institute including Mr. Michael Inskeep and Ms. Erika Maynor for their efforts in reviewing and editing interaction descriptions, and Dr. Marintha Heil for her support as Principal Investigator of NIAID contract HHSN272200700042C.
Footnotes
Present address: Kim Pruitt, National Institute of Biotechnology Information, National Library of Medicine, National Institutes of Health, 8600 Rockville Pike, Bethesda, MD 20894, USA.
FUNDING
Intramural Research Program of the National Institutes of Health, National Library of Medicine. Division of AIDS (DAIDS), National Institute of Allergy and Infectious Diseases, National Institutes of Health [HHSN272200700042C] entitled ‘In Vitro Testing Resources for AIDS Therapeutic Development, PART B: Specialized In Vitro Virological Assays for HIV Therapeutics and Topical Microbicides’ under the direction of Dr. Roger Miller (DHHS, NIH, NIAID, DAIDS, BSP). Funding for open access charge: The Intramural Research Program of the National Institutes of Health, National Library of Medicine.
Conflict of interest statement. None declared.
REFERENCES
- 1.Barre-Sinoussi F., Chermann J.C., Rey F., Nugeyre M.T., Chamaret S., Gruest J., Dauguet C., Axler-Blin C., Vezinet-Brun F., Rouzioux C., et al. Isolation of a T-lymphotropic retrovirus from a patient at risk for acquired immune deficiency syndrome (AIDS) Science. 1983;220:868–871. doi: 10.1126/science.6189183. [DOI] [PubMed] [Google Scholar]
- 2.Gallo R.C., Salahuddin S.Z., Popovic M., Shearer G.M., Kaplan M., Haynes B.F., Palker T.J., Redfield R., Oleske J., Safai B., et al. Frequent detection and isolation of cytopathic retroviruses (HTLV-III) from patients with AIDS and at risk for AIDS. Science. 1984;224:500–503. doi: 10.1126/science.6200936. [DOI] [PubMed] [Google Scholar]
- 3.Fu W., Sanders-Beer B.E., Katz K.S., Maglott D.R., Pruitt K.D., Ptak R.G. Human immunodeficiency virus type 1, human protein interaction database at NCBI. Nucleic Acids Res. 2009;37:D417–D422. doi: 10.1093/nar/gkn708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ptak R.G., Fu W., Sanders-Beer B.E., Dickerson J.E., Pinney J.W., Robertson D.L., Rozanov M.N., Katz K.S., Maglott D.R., Pruitt K.D., et al. Cataloguing the HIV type 1 human protein interaction network. AIDS Res. Hum. Retroviruses. 2008;24:1497–1502. doi: 10.1089/aid.2008.0113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.NCBI resource coordinators. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2013;41:D8–D20. doi: 10.1093/nar/gks1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Oliveira T., Deforche K., Cassol S., Salminen M., Paraskevis D., Seebregts C., Snoeck J., van Rensburg E.J., Wensing A.M., van de Vijver D.A., et al. An automated genotyping system for analysis of HIV-1 and other microbial sequences. Bioinformatics. 2005;21:3797–3800. doi: 10.1093/bioinformatics/bti607. [DOI] [PubMed] [Google Scholar]
- 7.Brown G.R., Hem V., Katz K.S., Ovetsky M., Wallin C., Ermolaeva O., Tolstoy I., Tatusova T., Pruitt K.D., Maglott D.R., et al. Gene: a gene-centered information resource atNCBI. Nucleic Acids Res. 2014:doi:10.1093/nar/gku1055. doi: 10.1093/nar/gku1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harris M.A., Clark J., Ireland A., Lomax J., Ashburner M., Foulger R., Eilbeck K., Lewis S., Marshall B., Mungall C., et al. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004;32:D258–D261. doi: 10.1093/nar/gkh036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Linde M.E., Colquhoun D.R., Ubaida Mohien C., Kole T., Aquino V., Cotter R., Edwards N., Hildreth J.E., Graham D.R. The conserved set of host proteins incorporated into HIV-1 virions suggests a common egress pathway in multiple cell types. J. Proteome Res. 2013;12:2045–2054. doi: 10.1021/pr300918r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chan E.Y., Sutton J.N., Jacobs J.M., Bondarenko A., Smith R.D., Katze M.G. Dynamic host energetics and cytoskeletal proteomes in human immunodeficiency virus type 1-infected human primary CD4 cells: analysis by multiplexed label-free mass spectrometry. J. Virol. 2009;83:9283–9295. doi: 10.1128/JVI.00814-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Navare A.T., Sova P., Purdy D.E., Weiss J.M., Wolf-Yadlin A., Korth M.J., Chang S.T., Proll S.C., Jahan T.A., Krasnoselsky A.L., et al. Quantitative proteomic analysis of HIV-1 infected CD4+ T cells reveals an early host response in important biological pathways: protein synthesis, cell proliferation, and T-cell activation. Virology. 2012;429:37–46. doi: 10.1016/j.virol.2012.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bozek K., Lengauer T. Positive selection of HIV host factors and the evolution of lentivirus genes. BMC Evol. Biol. 2010;10:186. doi: 10.1186/1471-2148-10-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chinn L.W., Tang M., Kessing B.D., Lautenberger J.A., Troyer J.L., Malasky M.J., McIntosh C., Kirk G.D., Wolinsky S.M., Buchbinder S.P., et al. Genetic associations of variants in genes encoding HIV-dependency factors required for HIV-1 infection. J. Infect. Dis. 2010;202:1836–1845. doi: 10.1086/657322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qian X., Yoon B.J. Comparative analysis of protein interaction networks reveals that conserved pathways are susceptible to HIV-1 interception. BMC Bioinformat. 2011;12(Suppl. 1):S19. doi: 10.1186/1471-2105-12-S1-S19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Macpherson J.I., Pinney J.W., Robertson D.L. JNets: exploring networks by integrating annotation. BMC Bioinformat. 2009;10:95. doi: 10.1186/1471-2105-10-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sargeant D., Deverasetty S., Luo Y., Baleta A., Zobrist S., Rathnayake V., Russo J.C., Vyas J., Muesing M.A., Schiller M.R. HIVToolbox, an integrated web application for investigating HIV. PLoS ONE. 2011;6:e20122. doi: 10.1371/journal.pone.0020122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fahey M.E., Bennett M.J., Mahon C., Jager S., Pache L., Kumar D., Shapiro A., Rao K., Chanda S.K., Craik C.S., et al. GPS-Prot: a web-based visualization platform for integrating host-pathogen interaction data. BMC Bioinformat. 2011;12:298. doi: 10.1186/1471-2105-12-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qi Y., Tastan O., Carbonell J.G., Klein-Seetharaman J., Weston J. Semi-supervised multi-task learning for predicting interactions between HIV-1 and human proteins. Bioinformatics. 2010;26:i645–i652. doi: 10.1093/bioinformatics/btq394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bol S.M., Moerland P.D., Limou S., van Remmerden Y., Coulonges C., van Manen D., Herbeck J.T., Fellay J., Sieberer M., Sietzema J.G., et al. Genome-wide association study identifies single nucleotide polymorphism in DYRK1A associated with replication of HIV-1 in monocyte-derived macrophages. PLoS ONE. 2011;6:e17190. doi: 10.1371/journal.pone.0017190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mukhopadhyay A., Ray S., Maulik U. Incorporating the type and direction information in predicting novel regulatory interactions between HIV-1 and human proteins using a biclustering approach. BMC Bioinformat. 2014;15:26. doi: 10.1186/1471-2105-15-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bushman F.D., Malani N., Fernandes J., D'Orso I., Cagney G., Diamond T.L., Zhou H., Hazuda D.J., Espeseth A.S., Konig R., et al. Host cell factors in HIV replication: meta-analysis of genome-wide studies. PLoS Pathog. 2009;5:e1000437. doi: 10.1371/journal.ppat.1000437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Troyer J.L., Nelson G.W., Lautenberger J.A., Chinn L., McIntosh C., Johnson R.C., Sezgin E., Kessing B., Malasky M., Hendrickson S.L., et al. Genome-wide association study implicates PARD3B-based AIDS restriction. J. Infect. Dis. 2011;203:1491–1502. doi: 10.1093/infdis/jir046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu J.Q., Sasse T.R., Saksena M.M., Saksena N.K. Transcriptome analysis of primary monocytes from HIV-positive patients with differential responses to antiretroviral therapy. Virol. J. 2013;10:361. doi: 10.1186/1743-422X-10-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li B.Q., Niu B., Chen L., Wei Z.J., Huang T., Jiang M., Lu J., Zheng M.Y., Kong X.Y., Cai Y.D. Identifying chemicals with potential therapy of HIV based on protein-protein and protein-chemical interaction network. PLoS ONE. 2013;8:e65207. doi: 10.1371/journal.pone.0065207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mairiang D., Zhang H., Sodja A., Murali T., Suriyaphol P., Malasit P., Limjindaporn T., Finley R.L., Jr Identification of new protein interactions between dengue fever virus and its hosts, human and mosquito. PLoS ONE. 2013;8:e53535. doi: 10.1371/journal.pone.0053535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou H., Gao S., Nguyen N.N., Fan M., Jin J., Liu B., Zhao L., Xiong G., Tan M., Li S., et al. Stringent homology-based prediction of H. sapiens-M. tuberculosis H37Rv protein-protein interactions. Biol. Direct. 2014;9:5. doi: 10.1186/1745-6150-9-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mattick J.S. Non-coding RNAs: the architects of eukaryotic complexity. EMBO Rep. 2001;2:986–991. doi: 10.1093/embo-reports/kve230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kwek K.Y., Murphy S., Furger A., Thomas B., O'Gorman W., Kimura H., Proudfoot N.J., Akoulitchev A. U1 snRNA associates with TFIIH and regulates transcriptional initiation. Nat. Struct. Biol. 2002;9:800–805. doi: 10.1038/nsb862. [DOI] [PubMed] [Google Scholar]
- 29.Bachellerie J.P., Cavaille J., Huttenhofer A. The expanding snoRNA world. Biochimie. 2002;84:775–790. doi: 10.1016/s0300-9084(02)01402-5. [DOI] [PubMed] [Google Scholar]
- 30.Fabian M.R., Sonenberg N., Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu. Rev. Biochem. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- 31.Eilebrecht S., Schwartz C., Rohr O. Non-coding RNAs: novel players in chromatin-regulation during viral latency. Curr. Opin. Virol. 2013;3:387–393. doi: 10.1016/j.coviro.2013.04.001. [DOI] [PubMed] [Google Scholar]
- 32.Groen J.N., Morris K.V. Chromatin, non-coding RNAs, and the expression of HIV. Viruses. 2013;5:1633–1645. doi: 10.3390/v5071633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yeung M.L., Bennasser Y., Myers T.G., Jiang G., Benkirane M., Jeang K.T. Changes in microRNA expression profiles in HIV-1-transfected human cells. Retrovirology. 2005;2:81. doi: 10.1186/1742-4690-2-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Houzet L., Yeung M.L., de Lame V., Desai D., Smith S.M., Jeang K.T. MicroRNA profile changes in human immunodeficiency virus type 1 (HIV-1) seropositive individuals. Retrovirology. 2008;5:118. doi: 10.1186/1742-4690-5-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang Z., Zhu Q., Luo K., Zhou Q. The 7SK small nuclear RNA inhibits the CDK9/cyclin T1 kinase to control transcription. Nature. 2001;414:317–322. doi: 10.1038/35104575. [DOI] [PubMed] [Google Scholar]


