Abstract
Angiogenesis is the process of generating new blood vessels based on existing ones, which is involved in many diseases including cancers, cardiovascular diseases and diabetes mellitus. Recently, great efforts have been made to explore the mechanisms of angiogenesis in various diseases and many angiogenic factors have been discovered as therapeutic targets in anti- or pro-angiogenic drug development. However, the resulted information is sparsely distributed and no systematical summarization has been made. In order to integrate these related results and facilitate the researches for the community, we conducted manual text-mining from published literature and built a database named as PubAngioGen (http://www.megabionet.org/aspd/). Our online application displays a comprehensive network for exploring the connection between angiogenesis and diseases at multilevels including protein–protein interaction, drug-target, disease-gene and signaling pathways among various cells and animal models recorded through text-mining. To enlarge the scope of the PubAngioGen application, our database also links to other common resources including STRING, DrugBank and OMIM databases, which will facilitate understanding the underlying molecular mechanisms of angiogenesis and drug development in clinical therapy.
INTRODUCTION
Angiogenesis, the process of generating new blood vessels based on pre-existing ones (1), is a complex multistep dynamic process that comprises matured vessel changes, pericyte detachment, extracellular matrix (ECM) degradation and remodeling, proliferation, migration and assembly of endothelial cells (ECs) into tubule structures (2). The recruitment and differentiation of circulating endothelial progenitor cells (EPCs) also contributes to the vascularization process (3,4).
Angiogenesis itself is controlled by a fine balance between pro- and anti-angiogenic factors (5) involved in many signals such as vascular endothelial growth factor (VEGF), fibroblast growth factor (FGF), platelet-derived growth factor (PDGF) and endothelial nitric oxide synthase (eNOS) pathway (6). If the balance is tipped, it leads to some pathological situations, either in deficit conditions (e.g. cardiovascular diseases, diabetic tissue ischemia) or in excess ones (e.g. cancer growth and metastases, atherosclerotic plaque development and diabetic retinopathy). Therefore, angiogenesis plays a key role in main diseases, including cancers (7,8), cardiovascular diseases (9) and complications of diabetes mellitus (10).
The angiogenic switch is a critical progression point in a range of etiology and pathogenesis; therefore, it was assumed that angiogenesis inhibition or promotion of regulating pathway and special target might play a fundamental role in related diseases therapy. Since the first work of Folkman that highlighted the role of angiogenesis as a crucial determinant for tumor development and progression (11), the idea of anti-angiogenesis as a therapeutic strategy has been accounted for several decades. In fact, several anti-angiogenic therapies have recently been approved by Food and Drug Administration (FDA) for cancers, including the humanized antibody bevacizumab (Avastin), which targets VEGF-A, the tyrosine kinase inhibitor sorafenib (Nexavar), which targets Raf and VEGF and PDGF receptors, and the tyrosine kinase inhibitor sunitinib (Sutent), which targets VEGF and PDGF receptors (12,13). In addition, promoting blood vessel growth has been recognized as a potential therapeutic approach for the treatment of ischemic diseases. To date, therapeutic benefits have also been achieved with anti-angiogenic therapy in the treatment of life-threatening tumors.
Besides drugs such as small molecule inhibitors, researchers have also focused on gene therapy using pro-angiogenic factors and/or cell-based therapy using several types of cells, including bone marrow cells and EPCs, to achieve therapeutic angiogenesis. Therefore, the future of research and drug development involved in anti- or pro- angiogenesis depends on the comprehensive understanding of the mechanisms and relationships between objectives (e.g. compounds or proteins), cells, diseases as well as signaling pathways and targets. Fully integration of those published results will surely facilitate the researches of angiogenesis and diseases.
To this end, for the first time, we developed PubAngioGen, a knowledge-based database, to record those results related to angiogenesis. During data collection, we manually extracted those published relevant results at multilevels including molecular, cellular, animal model and disease. We then comprehensively summarized the relationships between angiogenesis and major diseases, which help users to obtain the latest information of angiogenesis involved in main diseases and signaling pathways. In addition, we also collected those potential drug targets and related compounds, which have been experimentally proven to be effective on angiogenesis. Moreover, those candidates of angiogenic factors that are under clinical trials and the therapeutic targets for anti-angiogenic or pro-angiogenic drug development also have been collected (Table 1).
Table 1. Data statistics for PubAngioGen.
| Data field | Data source | Amount of data |
|---|---|---|
| Compounds/drugs | Text-mining, DrugBank | 6693 |
| Gene/protein targets | Text-mining | 963 |
| Signal pathways | Text-mining, CST pathways | 179 |
| Diseases | Text-mining, OMIM | 2364 |
MATERIALS AND METHODS
PubAngioGen provides information on three different areas related to angiogenesis, proteins, drugs, diseases and the links between them. The data integrated from related resources and manual text-mining from published articles, as well as the relation models were depicted in Figure 1.
Figure 1.
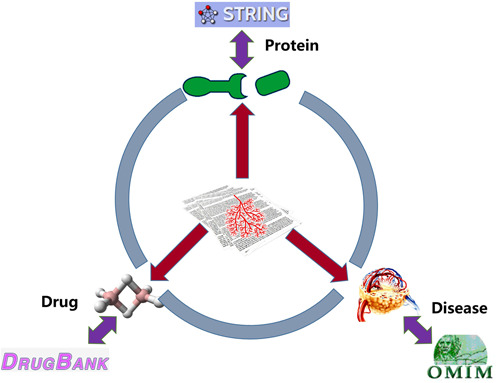
The data sources and relation models.
The core components were collected mainly through text-mining from published articles and protein–protein interaction was extended based on STRING database (14). Information for drugs and diseases was extended based on DrugBank (15,16) and Online Mendelian Inheritance in Man (OMIM) (17,18), respectively.
The main goal of our system is to build the connections between angiogenesis and diseases through disease, proteins as well as related drugs. Users can query our system easily according to their interests to obtain the information about angiogenesis, diseases, targets, drugs and the mechanisms of drug interactions.
First of all, through manual text-mining, we collected results published in the past decades related to angiogenesis at multilevels including molecular, cellular, animal model and disease. We extracted the information of signaling pathways involved in angiogenesis and summarized the relationships between angiogenesis and major diseases. The information on drugs that FDA approved to treat angiogenesis-related diseases or candidates that are in clinical trials was also extracted. All the information recorded was curated by the experts from National Consortium of GPCR of China and could be traced back to the original publications. Then, we used the well-defined data sources to extend the interaction between proteins or proteins and compounds.
DATABASE ACCESS AND NETWORK DISPLAY
Database query
Users can identify proteins, drugs or diseases when they would like to know their roles in angiogenesis at PubAngioGen through the quick search form (Figure 2A) or the query page (Figure 2B). PubAngioGen allows several types of query keywords consistent with other databases, including UniProtKB/Swiss-Prot ID (19), Entrez Gene name (20,21), Refseq provisional ID (NCBI) (22,23), symbols, disease name or drug name.
Figure 2.
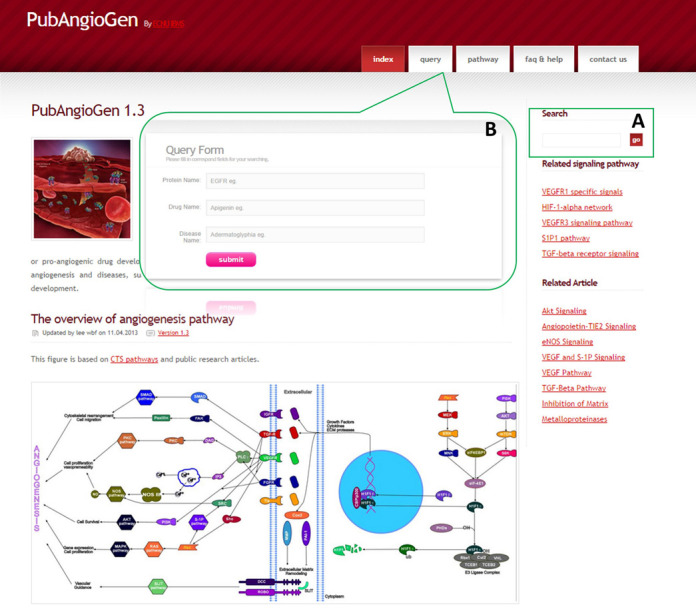
PubAngioGen homepage and query interface. (A) Quick search form on homepage. (B) Detail search form on query page.
Result page and network display
To virtually display the connection between proteins, drugs or diseases and angiogenesis, we developed network-display tools, which provide more detailed network information. Our network display includes protein–protein, compound–protein interaction and signaling pathways involved in angiogenesis, which help users to check the potential combinational effect. All returned pages inform the user of related protein annotations by texts, graphs and tables (Figure 3). For example, if the user is interested in the epidermal growth factor receptor (EGFR) protein, the ‘Related Information For EGFR’ shows a summary network of the compounds, peptides, proteins, complexes and diseases nodes with different colors that connected with EGFR (Figure 3A) and users can select a node to see its directed partners (Figure 3B). The first table (Figure 3C) lists the basic information of EGFR and the second table (Figure 3D) presents the detailed related information of EGFR such as interaction type, reaction type and so on. The underlined red parts are hyperlinks that include literature references from PubMed and experimental detection methods extracted from text-mining. Moreover, to explore the distinctive mechanisms of each angiogenesis-related gene, the gene expression levels in different statuses were recorded in our databases based on the experiments from Gene Expression Omnibus (GEO) (24); more experiments are under evaluation and the results will be processed and stored in PubAngioGen soon.
Figure 3.
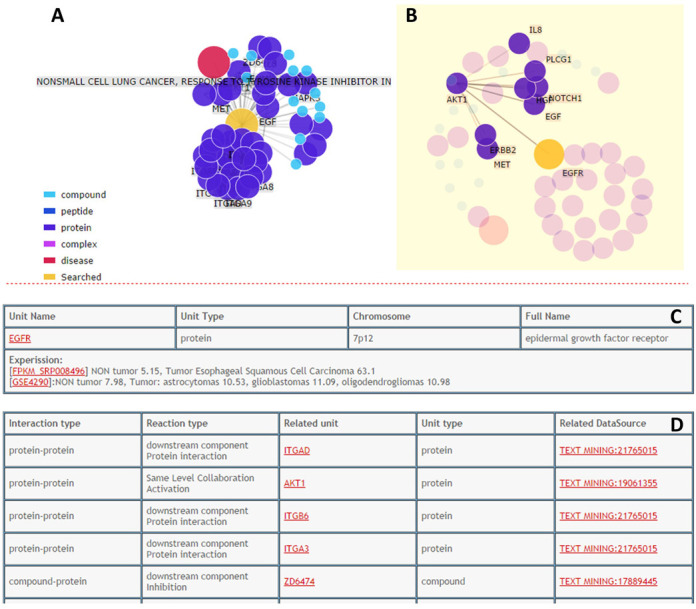
Query result for EGFR. (A) Network viewer to show the proteins, drugs and diseases related to EGFR and the interactions among all of them. (B) When the node in the network is selected, all the directed partners are highlighted to facilitate users to investigate the function of specific node; double click will return to the original network. (C) The annotation table for queried entry. (D) The interaction table for all related information.
Signaling pathway
During the process of angiogenesis-related diseases, the signaling pathway stimulation or repression acts in the different ways. To incorporate those diversities of signaling pathways, our database displays the latest singling pathways involved in angiogenesis from our collected data and other relevant databases (e.g. Cell Signaling Technology Pathway Database, http://www.cellsignal.com/). Users can submit those interested proteins, diseases or other components to view their roles in the process of diseases of angiogenesis. For example, when users enter the ‘pathway’ in home page of our website and click ‘EGFR-dependent Endothelin signaling events’, the returned page displays the network and components involved in this pathway.
DISCUSSION
In order to better elucidate the pathological angiogenesis in these diseases, such as cancers, cardiovascular diseases and diabetes mellitus, we extracted abundant angiogenesis-related results published in past decades and comprehensively summarized their relationships at multilevels including molecular, cellular, animal model and disease in our database. On one hand, we provided the latest information of angiogenesis involved in main diseases and signaling pathways like HIF-1 (symbol: SETD2) network, VEGFR1 (symbol: FLT1), VEGFR2 (symbol: KDR), VEGFR3 (symbol: FLT4) specific signals, transforming growth factor beta (TGFβ) (symbol: TGFB1) receptor signaling pathway and S1P1 (symbol: S1PR1) pathway (25–27). On the other hand, we provided the first integrated resources for exploring the potential drug targets, clinical compounds that are effective on angiogenesis as well as related research progresses for drug development. Therefore, PubAngioGen database would be a promising angiogenic online searcher to some extent.
Meanwhile, the PubAngioGen is supported by National Consortium of GPCR of China; many experts in the field are involved in curating angiogenesis-related articles. While the amount of data entries is increasing daily, many exploring experiments are performed in our collaborators’ labs including high-throughput experiments like RNA-Seq (28) and LC MS/MS (29) to validate or extend our findings in PubAngioGen. Along with the accumulation of data, in the future, we will further extend and redefine angiogenesis-related pathways to help other researchers to better understand the mechanism of it.
In conclusion, PubAngioGen database systemically proposes the relationships between angiogenesis and diseases. Our data retrieve and display system makes researchers access the latest information efficiently. All of these will promote PubAngioGen to be referred by modern medicine researchers for novel discovery in clinical medicine and research work in the near future.
Acknowledgments
We are also grateful for the permission from ‘Cell Signaling Technology’ for the related pathway information and the help from MegaSci Inc. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Footnotes
The authors wish it to be known that, in their opinion, the first four authors should be regarded as Joint First Authors.
FUNDING
Funding for open access charge: Major State Basic Research Development Program of China [2015CB910400]; National Natural Science Foundation of China [81272463, 81472788, 81330049, 31401133]; Science and Technology Commission of Shanghai Municipality [14YF1404400].
Conflict of interest statement. None declared.
REFERENCES
- 1.Song Y., Dai F., Zhai D., Dong Y., Zhang J., Lu B., Luo J., Liu M., Yi Z. Usnic acid inhibits breast tumor angiogenesis and growth by suppressing VEGFR2-mediated AKT and ERK1/2 signaling pathways. Angiogenesis. 2012;15:421–432. doi: 10.1007/s10456-012-9270-4. [DOI] [PubMed] [Google Scholar]
- 2.Rundhaug J.E. Matrix metalloproteinases and angiogenesis. J. Cell. Mol. Med. 2005;9:267–285. doi: 10.1111/j.1582-4934.2005.tb00355.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fam N.P., Verma S., Kutryk M., Stewart D.J. Clinician guide to angiogenesis. Circulation. 2003;108:2613–2618. doi: 10.1161/01.CIR.0000102939.04279.75. [DOI] [PubMed] [Google Scholar]
- 4.Hristov M., Erl W., Weber P.C. Endothelial progenitor cells: mobilization, differentiation, and homing. Arterioscler. Thromb. Vasc. Biol. 2003;23:1185–1189. doi: 10.1161/01.ATV.0000073832.49290.B5. [DOI] [PubMed] [Google Scholar]
- 5.Anand S. A brief primer on microRNAs and their roles in angiogenesis. Vasc. Cell. 2013;5:2. doi: 10.1186/2045-824X-5-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dong Y., Lu B., Zhang X., Zhang J., Lai L., Li D., Wu Y., Song Y., Luo J., Pang X., et al. Cucurbitacin E, a tetracyclic triterpenes compound from Chinese medicine, inhibits tumor angiogenesis through VEGFR2-mediated Jak2-STAT3 signaling pathway. Carcinogenesis. 2010;31:2097–2104. doi: 10.1093/carcin/bgq167. [DOI] [PubMed] [Google Scholar]
- 7.Zhang X., Song Y., Wu Y., Dong Y., Lai L., Zhang J., Lu B., Dai F., He L., Liu M., et al. Indirubin inhibits tumor growth by antitumor angiogenesis via blocking VEGFR2-mediated JAK/STAT3 signaling in endothelial cell. Int. J. Cancer. 2011;129:2502–2511. doi: 10.1002/ijc.25909. [DOI] [PubMed] [Google Scholar]
- 8.Li J., Xu Y., Jiao H., Wang W., Mei Z., Chen G. Sumoylation of hypoxia inducible factor-1alpha and its significance in cancer. Sci. China Life Sci. 2014;57:657–664. doi: 10.1007/s11427-014-4685-3. [DOI] [PubMed] [Google Scholar]
- 9.Deveza L., Choi J., Yang F. Therapeutic angiogenesis for treating cardiovascular diseases. Theranostics. 2012;2:801–814. doi: 10.7150/thno.4419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tahergorabi Z., Khazaei M. Imbalance of angiogenesis in diabetic complications: the mechanisms. Int. J. Prev. Med. 2012;3:827–838. doi: 10.4103/2008-7802.104853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Folkman J. Tumor angiogenesis: therapeutic implications. N Engl. J. Med. 1971;285:1182–1186. doi: 10.1056/NEJM197111182852108. [DOI] [PubMed] [Google Scholar]
- 12.Carmeliet P., Jain R.K. Molecular mechanisms and clinical applications of angiogenesis. Nature. 2011;473:298–307. doi: 10.1038/nature10144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jain R.K. Normalizing tumor microenvironment to treat cancer: bench to bedside to biomarkers. J. Clin. Oncol. 2013;31:2205–2218. doi: 10.1200/JCO.2012.46.3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Franceschini A., Szklarczyk D., Frankild S., Kuhn M., Simonovic M., Roth A., Lin J., Minguez P., Bork P., von Mering C., et al. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41:D808–D815. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wishart D.S., Knox C., Guo A.C., Cheng D., Shrivastava S., Tzur D., Gautam B., Hassanali M. DrugBank: a knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res. 2008;36:D901–D906. doi: 10.1093/nar/gkm958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Law V., Knox C., Djoumbou Y., Jewison T., Guo A.C., Liu Y., Maciejewski A., Arndt D., Wilson M., Neveu V., et al. DrugBank 4.0: shedding new light on drug metabolism. Nucleic Acids Res. 2014;42:D1091–D1097. doi: 10.1093/nar/gkt1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hamosh A., Scott A.F., Amberger J., Bocchini C., Valle D., McKusick V.A. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 2002;30:52–55. doi: 10.1093/nar/30.1.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Amberger J., Bocchini C., Hamosh A. A new face and new challenges for Online Mendelian Inheritance in Man (OMIM(R)) Hum. Mutat. 2011;32:564–567. doi: 10.1002/humu.21466. [DOI] [PubMed] [Google Scholar]
- 19.UniProt C. Activities at the Universal Protein Resource (UniProt) Nucleic Acids Res. 2014;42:D191–D198. doi: 10.1093/nar/gkt1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maglott D., Ostell J., Pruitt K.D., Tatusova T. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 2011;39:D52–D57. doi: 10.1093/nar/gkq1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maglott D., Ostell J., Pruitt K.D., Tatusova T. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 2005;33:D54–D58. doi: 10.1093/nar/gki031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pruitt K.D., Tatusova T., Maglott D.R. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–D65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pruitt K.D., Brown G.R., Hiatt S.M., Thibaud-Nissen F., Astashyn A., Ermolaeva O., Farrell C.M., Hart J., Landrum M.J., McGarvey K.M., et al. RefSeq: an update on mammalian reference sequences. Nucleic Acids Res. 2014;42:D756–D763. doi: 10.1093/nar/gkt1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barrett T., Wilhite S.E., Ledoux P., Evangelista C., Kim I.F., Tomashevsky M., Marshall K.A., Phillippy K.H., Sherman P.M., Holko M., et al. NCBI GEO: archive for functional genomics data sets—update. Nucleic Acids Res. 2013;41:D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Veikkola T., Karkkainen M., Claesson-Welsh L., Alitalo K. Regulation of angiogenesis via vascular endothelial growth factor receptors. Cancer Res. 2000;60:203–212. [PubMed] [Google Scholar]
- 26.Maceyka M., Payne S.G., Milstien S., Spiegel S. Sphingosine kinase, sphingosine-1-phosphate, and apoptosis. Biochim. Biophys. Acta. 2002;1585:193–201. doi: 10.1016/s1388-1981(02)00341-4. [DOI] [PubMed] [Google Scholar]
- 27.Li J.M., Shen X., Hu P.P., Wang X.F. Transforming growth factor beta stimulates the human immunodeficiency virus 1 enhancer and requires NF-kappaB activity. Mol. Cell. Biol. 1998;18:110–121. doi: 10.1128/mcb.18.1.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen G., Shi T. Next-generation sequencing technologies for personalized medicine: promising but challenging. Sci. China Life Sci. 2013;56:101–103. doi: 10.1007/s11427-013-4436-x. [DOI] [PubMed] [Google Scholar]
- 29.Liu Y., Yin H., Chen K. Platelet proteomics and its advanced application for research of blood stasis syndrome and activated blood circulation herbs of Chinese medicine. Sci. China Life Sci. 2013;56:1000–1006. doi: 10.1007/s11427-013-4551-8. [DOI] [PubMed] [Google Scholar]


