Abstract
We present MethHC (http://MethHC.mbc.nctu.edu.tw), a database comprising a systematic integration of a large collection of DNA methylation data and mRNA/microRNA expression profiles in human cancer. DNA methylation is an important epigenetic regulator of gene transcription, and genes with high levels of DNA methylation in their promoter regions are transcriptionally silent. Increasing numbers of DNA methylation and mRNA/microRNA expression profiles are being published in different public repositories. These data can help researchers to identify epigenetic patterns that are important for carcinogenesis. MethHC integrates data such as DNA methylation, mRNA expression, DNA methylation of microRNA gene and microRNA expression to identify correlations between DNA methylation and mRNA/microRNA expression from TCGA (The Cancer Genome Atlas), which includes 18 human cancers in more than 6000 samples, 6548 microarrays and 12 567 RNA sequencing data.
INTRODUCTION
DNA methylation is one of the epigenetic mechanisms that can control gene expression without incurring any change to genomic sequence during development and cell proliferation (1). Many studies have shown that DNA methylation plays an important role in cancer biology and provides promising biomarkers for cancer diagnosis and prognosis evaluation (2,3). Recent studies have shown that epigenetics plays an important role in cancer biology, viral infections, somatic gene therapy, transgenic technologies, developmental abnormalities and genomic imprinting (4,5). In recent decades, attention has focused on DNA methylation in cancer. For example, TP53 (6) and RB1 (7) are hypermethylated in glioma, WNT5A is hypomethylated in prostate cancer (8), SMN2 is hypermethylated in spinal muscular atrophy and HSPB1 methylation is a biomarker in prostate cancer as well as in human breast cancer, colorectal cancer and malignant melanoma (9). Furthermore, DNA methylation may serve as a tumor marker or diagnosis and therapy index in colorectal cancer (10,11) and ovarian cancer (12).
A wide range of techniques is used to detect DNA methylation, including mass spectrometry, methylation specific PCR, CHIP-on-chip assays, methylated DNA immunoprecipitation, microarray assay, pyrosequencing of bisulfite treated DNA, whole genome bisulfite sequencing, etc. These methods can be divided into two categories, gene-specific approaches and genome-wide approaches. Studies in recent years have produced copious amounts of DNA methylation data, especially epigenome-wide array data and next-generation sequencing (NGS) data. Collection of DNA methylation and mRNA/microRNA expression data related to diseases produced by high-throughput approaches based on array and NGS will facilitate the discovery of potential methylation events in human diseases (13). Hence, several DNA methylation databases had been constructed. MethDB is a database that stores manually curated experimental data on DNA methylation and environmental epigenetic effects (14). PubMeth is an annotated and reviewed database of methylation in cancer based on text mining of the published literature (15). MethyCancer (16) contains DNA methylation, cancer-related gene and cancer information from public data sources and large-scale experimental data sets produced from the Cancer Epigenome Project in China. NGSMethDB (17) provides DNA methylation data for humans, chimpanzees, mice and Arabidopsis thaliana from publicly available data sets using the NGS bisulfite sequencing technique. DiseaseMeth (18) provides both locus-specific and high-throughput data sets for many human diseases. MENT (19) is the first database to provide both DNA methylation and gene expression information for diverse tumor tissues.
MicroRNA, a small non-coding RNA, functions in RNA silencing and post-transcriptional regulation of gene expression, and is frequently associated with cancers and may be an important causal factor. Aberrant DNA methylation has been found to silence microRNA genes in breast cancer, such as mir-31, mir-130a, mir-155, mir-137 and mir-34b/mir-34c genes (20), indicating the important role of DNA methylation in microRNA deregulation in cancer. Recently, various high-throughput approaches based on array and NGS have been developed and applied in combination with bisulfite conversion of the DNA for genome-wide DNA methylation analysis in mammals and plants (21), and a large number of DNA methylation data has been rapidly accumulated. However, there are fewer databases that provide both DNA methylation and gene expression information. Thus, there is a great need for an integrated database that provides both DNA methylation and mRNA/microRNA expression information in normal and tumor tissues. This paper describes a DNA methylation and gene expression database in human cancer, MethHC (database of DNA methylation and gene expression in human cancer). MethHC is a web-based resource focused on the aberrant methylomes of human diseases. MethHC integrates data including DNA methylation, gene expression, microRNA methylation, microRNA expression and the correlation between DNA methylation and gene expression from TCGA (The Cancer Genome Atlas).
DATABASE CONTENT
Figure 1 shows the data generation flow of the MethHC database, which comprises two parts: (i) integration of experimental data source and (ii) integration of annotated databases. A detailed description is provided below.
Figure 1.
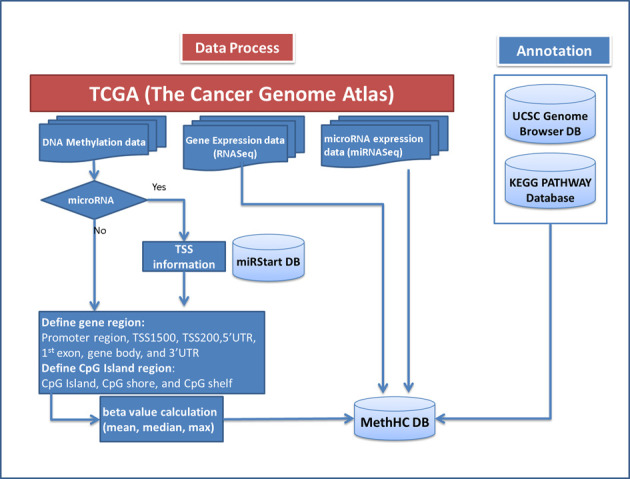
Data generation flow of MethHC.
The DNA methylation data sets used in MethHC are mainly taken from TCGA which was launched in 2006 with an investment of US$50 million from the National Cancer Institute and the National Human Genome Research Institute. The goal of TCGA is to improve the ability to diagnose, treat and prevent cancer. Therefore, TCGA collects and analyzes high-quality tumor samples such as clinical information about participants, metadata about the samples, histopathology slide images of the sample and molecular information derived from the samples (e.g. copy number variation, DNA methylation, single-nucleotide polymorphism, protein expression, DNA sequencing and mRNA/microRNA expression etc.). A pilot project developed and tested the research framework to systematically explore the entire spectrum of genomic changes involved in more than 20 types of human cancer. The resulting data are freely available through the TCGA Data Portal and the Cancer Genomics Hub (CGHub). TCGA collects three types of array-based DNA methylation platform data: Illumina DNA Methylation Cancer Panel I, Illumina HumanMethylation27 BeadChip and Illumina HumanMethylation450K BeadChip. To obtain human DNA methylation and gene expressions, we integrated the heterogeneous data from TCGA. First, DNA methylation data are specific to the Illumina HumanMethylation450K BeadChip platform, because of the more probes in HumanMethylation450K BeadChip. Second, Illumina HiSeq 2000 RNA Sequencing platform data are for gene expression. Third, microRNA expression profiles were done using Illumina HiSeq 2000 miRNA Sequencing platform.
MethHC provides not only DNA methylation and gene expression data for protein-coding genes, but also miRNAs. The information for gene transcription start sites (TSS) and microRNA TSS was taken from the UCSC Genome Browser (22) and the miRStart database (23). miRStart integrates data from many TSS-relevant experiments, offering reliable human microRNA TSSs for the further decipherment of microRNA transcription regulation.
To study DNA methylation patterns, different regions and CpG islands are considered. Hypermethylation in the promoter region would induce the down-regulation of gene expression. However, hypermethylation in the gene body may not block and might even stimulate transcription elongation and exciting new evidence suggests that gene body methylation may have an impact on splicing. CpG island (CGI) hypermethylation of the TSS is associated with long-term silencing (24). The differential methylation level at the CpG island shore is tissue-specific (25). Most DNA methylation-cancer studies have focused on promoters, CpG islands and sometimes a bit further out (e.g. CpG shores), but mostly overlap with putative enhancer regions, and they are enriched in consensus binding sequences for important renal transcription factors (26). Therefore, we provide several different gene regions (promoter, TSS, untranslated region, exon, gene body and enhancer region) and CGI regions (CpG Island, shore and shelf).
Finally, to complete the gene information, we also integrate the UCSC Genome Browser, miRStart and KEGG PATHWAY databases. The KEGG (Kyoto Encyclopedia of Genes and Genomes) project was initiated in 1995 (27) and the KEGG PATHWAY database is a collection of manually drawn KEGG pathway maps representing experimental knowledge for metabolism, genetics information processing, cellular processes, organism systems and human diseases. MethHC allows users to select a particular KEGG pathway and identify differentially methylated genes in a set of tumors.
DATABASE STATISTICS
MethHC is a web-based resource focused on the aberrant methylomes of human diseases. MethHC integrates DNA methylation data, gene expression data and microRNA expression data from TCGA. MethHC currently comprises 6548 DNA methylation data generated by Illumina HumanMethylation450K BeadChip and 12 567 mRNA/microRNA expression data generated by RNA-seq/ miRNA-seq in 18 human cancers. Table 1 provides a detailed list of the cancer and sample numbers, and the data include a greater number of samples in the breast invasive carcinoma cohort, thyroid carcinoma and head and neck squamous cell carcinoma. MethHC contains 20 500 genes and 1040 miRNAs, and each gene includes eight gene regions and five CGI regions.
Table 1. MethHC statistics for samples in each cancer.
| Cancer | DNA methylation | microRNA expression | Gene expression |
|---|---|---|---|
| Bladder urothelial carcinoma | 276 | 260 | 260 |
| Breast invasive carcinoma | 839 | 798 | 1159 |
| Cervical squamous cell carcinoma and endocervical adenocarcinoma | 217 | 204 | 188 |
| Colon adenocarcinoma | 315 | 246 | 305 |
| Head and neck squamous cell carcinoma | 569 | 520 | 542 |
| Kidney renal clear cell carcinoma | 458 | 314 | 593 |
| Kidney renal papillary cell carcinoma | 229 | 230 | 203 |
| Liver hepatocellular carcinoma | 256 | 250 | 244 |
| Lung adenocarcinoma | 471 | 476 | 540 |
| Lung squamous cell carcinoma | 406 | 377 | 451 |
| Pancreatic adenocarcinoma | 102 | 88 | 76 |
| Prostate adenocarcinoma | 387 | 376 | 342 |
| Rectum adenocarcinoma | 105 | 94 | 106 |
| Sarcoma | 178 | 138 | 109 |
| Skin cutaneous melanoma | 380 | 356 | 353 |
| Stomach adenocarcinoma | 330 | 316 | 318 |
| Thyroid carcinoma | 565 | 566 | 541 |
| Uterine corpus endometrial carcinoma | 465 | 419 | 209 |
| Total | 6548 | 6,028 | 6539 |
COMPARISON
Table 2 compares MethHC with other methylation databases including MethDB, PubMeth, MethyCancer, NGSMethDB, DiseaseMeth and MENT. MethDB only stores DNA methylation data and makes these data readily available to the public. PubMeth is based on text mining of Medline/PubMed abstracts and combined with manual reading and annotation of preselected abstracts. MethyCancer provides large-scale methylation data, cancer-related genes, mutation, cancer information and methylation status in CpG islands and promoter regions from multiple public sources. NGSMethDB provides sequence-based methylation data of six species samples in CpG islands and promoter regions obtained by NGS. DiseaseMeth is a human methylation database that provides both locus-specific and high-throughput data sets in 72 human diseases. MENT was developed by collecting over 6000 samples from Gene Expression Omnibus (GEO) and TCGA, and provides regression and correlation analysis. MethHC collects DNA methylation and mRNA/microRNA expression profiles in 18 human normal and tumor tissues. Illumina HumanMethylation450K BeadChip data includes more than 480 000 CpG sites. RNA-Seq and small RNA-Seq are applied to calculate the abundance of mRNA/microRNA expressions. To help investigators reveal new potential epigenetic biomarkers for cancer, MethHC provides additional functions and multiplicity information, including several different gene regions (promoter, TSS, untranslated region, exon, gene body and enhancer region), CGI regions (CpG island, shore and shelf) and estimate of the DNA methylation level methods (average, maximum and median).
Table 2. Comparing MethHC with other resources.
| MethDB | PubMeth | MethyCancer | NGSMethDB | DiseaseMeth | MENT | MethHC | |
|---|---|---|---|---|---|---|---|
| Publication | NAR. 2001 | NAR Database Issue (2008) | NAR Database Issue (2008) | NAR Database Issue (2010) | NAR Database Issue (2012) | GENE (2013) | |
| Last update | 2009 | – | 2008 | 2014 | – | – | 2014 |
| Support species | Four species | Homo sapiens | Homo sapiens | Six species | Homo sapiens 72 diseases | Homo sapiens 30 tissues | Homo sapiens 18 cancers |
| Number of samples | 20 236 fragments 6312 profiles | – | 11 561 (genes) | Human: 19 Monkey: 6 Tomato: 8 Mouse: 45 Arabidopsis: 27 | 175 data sets (3610 microarray data) | Methylation: 9243 microarray data Gene expression: 34 000 microarray data | Methylation: 6548 microarray data, Gene expression: 12 567 RNA sequencing data |
| Number of methylation sites | – | – | 10 486 | – | – | 27 578 | 482 481 |
| Number of gene | – | – | 11 561 | – | – | 14 476 | 20 500 genes |
| 1040 microRNAs | |||||||
| Data sources | Experiments | Medline/ PubMed | BIG, MethDB, HEP | GEO | GEO | GEO, TCGA | TCGA |
| Correlation analysis | No | No | Yes | Yes | Yes | Yes | Yes |
| microRNA expression | No | No | No | No | No | No | Yes |
| Gene region | No | No | CpG island, promoter region | CpG Promoter region | Promoter region (−1.5K to 0.5K) | Probe level only | Eight gene regions* five CpG island regions** |
| Other characteristic | Experimental data only | Automated text mining | Repeat sequence Search | CpG, CHG | Offline analysis, CpG | Differential methylation, Correlation analysis, Sample subtype | MicroRNA expression, Differential methylation, Correlation analysis |
INTERFACE
To facilitate access to data and further analyses for the identification of differentially methylated genes, MethHC provides a variety of interfaces and graphical visualizations. Users can utilize top 250 analysis to identify the 250 hypermethylated and hypomethylated genes, yielding genes which are respectively hypermethylated and hypomethylated in tumors. Figure 2A provides a detailed view of the interface. Hierarchical clustering is probably the most widely used clustering method for the detection of co-regulated methylation genes and the identification of cancer subtypes. In the hierarchical clustering graph, genes and samples with similar DNA methylation patterns are grouped together (Figure 2B). In addition, MethHC can submit a group of interested genes/miRNAs or the name of a KEGG pathway to identify differentially methylated genes in a set of tumors. A detailed view of each gene is demonstrated in the correlation of DNA methylation and gene expression. GSTM2 and PENK are hypermethylated in prostate cancer (3). Aberrant DNA methylation of mir-31, mir-130a, let-7a-3/let-7b and mir-155 gene promoters in breast cancer is linked to silencing of miRNA expression (20). Mir-9 family and mir-34 family were also reported to be silenced by DNA methylation in various cancer (28). We use PENK as a case study. Our system shows the differential methylation status of each transcript in selected tumors by boxplot (Figure 2C). The asterisk means significant differential methylation in selected tumors. The correlation between methylation levels and expression status was shown by scatter plot (Figure 2D).
Figure 2.
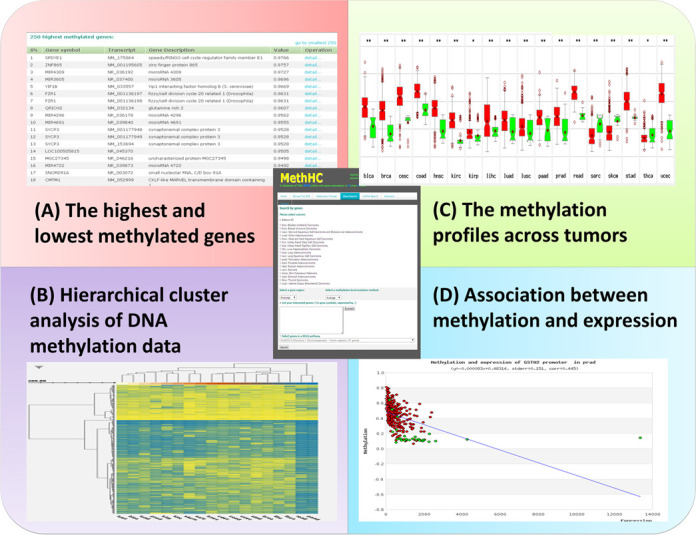
Illustrative screenshot of the MethHC interface.
CONCLUSIONS AND PERSPECTIVES
MethHC is an integrated and useful database comprising DNA methylation and mRNA/microRNA expression profiles in 18 human normal and tumor tissues. We use various textual and graphical interfaces to demonstrate the methylation patterns in normal and tumor tissues and to analyze the correlation between methylation and expression. Genome-wide analysis reveals differentially methylated genes and gene clusters. MethHC also provides more multiplicity information, such as several different gene regions, CGI regions and estimate of the DNA methylation level methods, to facilitate further investigation of genomic methylation status.
To help investigators reveal new potential epigenetic biomarkers for cancer diagnosis and prognosis, prospective works of the proposed database are listed as follows: (i) to support more data resources from the GEO and the International Cancer Genome Consortium, (ii) to integrate clinical–pathological indicators from TCGA and (iii) to collect bisulfate sequencing data from recent TCGA releases.
AVAILABILITY
The MethHC database will be continuously maintained and updated. The database is now publicly accessible at http://MethHC.mbc.nctu.edu.tw/.
Acknowledgments
The authors would like to thank the Ministry of Science and Technology of the Republic of China for financially supporting this research. We also thank the UST-UCSD International Center of Excellence in Advanced Bio-engineering sponsored by the Taiwan Ministry of Science and Technology I-RiCE Program, Veterans General Hospitals and University System of Taiwan (VGHUST) Joint Research Program, and MOE ATU. We would like to thank The Cancer Genome Atlas for use of data.
FUNDING
Ministry of Science and Technology of the Republic of China [NSC 101–2311-B-009–005-MY3 and NSC 102–2627-B-009–001]. UST-UCSD International Center of Excellence in Advanced Bio-engineering sponsored by the Taiwan Ministry of Science and Technology I-RiCE Program [NSC 102–2911-I-009–101]; Veterans General Hospitals and University System of Taiwan (VGHUST) Joint Research Program [VGHUST103-G5–1–2]. MOE ATU. Funding for open access charge: Ministry of Science and Technology of the Republic of China, Taiwan [NSC101-2311-B-009-005-MY3].
Conflict of interest statement. None declared.
REFERENCES
- 1.Zerbini L.F., Libermann T.A. GADD45 deregulation in cancer: frequently methylated tumor suppressors and potential therapeutic targets. Clin. Cancer Res. 2005;11:6409–6413. doi: 10.1158/1078-0432.CCR-05-1475. [DOI] [PubMed] [Google Scholar]
- 2.Das P.M., Singal R. DNA methylation and cancer. J. Clin. Oncol. 2004;22:4632–4642. doi: 10.1200/JCO.2004.07.151. [DOI] [PubMed] [Google Scholar]
- 3.Ashour N., Angulo J.C., Andres G., Alelu R., Gonzalez-Corpas A., Toledo M.V., Rodriguez-Barbero J.M., Lopez J.I., Sanchez-Chapado M., Ropero S. A DNA hypermethylation profile reveals new potential biomarkers for prostate cancer diagnosis and prognosis. Prostate. 2014;74:1171–1182. doi: 10.1002/pros.22833. [DOI] [PubMed] [Google Scholar]
- 4.Laird P.W. The power and the promise of DNA methylation markers. Nat. Rev. Cancer. 2003;3:253–266. doi: 10.1038/nrc1045. [DOI] [PubMed] [Google Scholar]
- 5.Ballestar E., Esteller M. The impact of chromatin in human cancer: linking DNA methylation to gene silencing. Carcinogenesis. 2002;23:1103–1109. doi: 10.1093/carcin/23.7.1103. [DOI] [PubMed] [Google Scholar]
- 6.Amatya V.J., Naumann U., Weller M., Ohgaki H. TP53 promoter methylation in human gliomas. Acta Neuropathol. 2005;110:178–184. doi: 10.1007/s00401-005-1041-5. [DOI] [PubMed] [Google Scholar]
- 7.Nakamura M., Yonekawa Y., Kleihues P., Ohgaki H. Promoter hypermethylation of the RB1 gene in glioblastomas. Lab. Invest. 2001;81:77–82. doi: 10.1038/labinvest.3780213. [DOI] [PubMed] [Google Scholar]
- 8.Wang Q., Williamson M., Bott S., Brookman-Amissah N., Freeman A., Nariculam J., Hubank M.J., Ahmed A., Masters J.R. Hypomethylation of WNT5A, CRIP1 and S100P in prostate cancer. Oncogene. 2007;26:6560–6565. doi: 10.1038/sj.onc.1210472. [DOI] [PubMed] [Google Scholar]
- 9.Vasiljevic N., Ahmad A.S., Beesley C., Thorat M.A., Fisher G., Berney D.M., Moller H., Yu Y., Lu Y.J., Cuzick J., et al. Association between DNA methylation of HSPB1 and death in low Gleason score prostate cancer. Prostate Cancer Prostatic Dis. 2013;16:35–40. doi: 10.1038/pcan.2012.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim M.S., Lee J., Sidransky D. DNA methylation markers in colorectal cancer. Cancer Metastasis Rev. 2010;29:181–206. doi: 10.1007/s10555-010-9207-6. [DOI] [PubMed] [Google Scholar]
- 11.Carmona F.J., Azuara D., Berenguer-Llergo A., Fernandez A.F., Biondo S., de Oca J., Rodriguez-Moranta F., Salazar R., Villanueva A., Fraga M.F., et al. DNA methylation biomarkers for noninvasive diagnosis of colorectal cancer. Cancer Prev. Res. 2013;6:656–665. doi: 10.1158/1940-6207.CAPR-12-0501. [DOI] [PubMed] [Google Scholar]
- 12.Barton C.A., Hacker N.F., Clark S.J., O'Brien P.M. DNA methylation changes in ovarian cancer: implications for early diagnosis, prognosis and treatment. Gynecol. Oncol. 2008;109:129–139. doi: 10.1016/j.ygyno.2007.12.017. [DOI] [PubMed] [Google Scholar]
- 13.Feinberg A.P. Genome-scale approaches to the epigenetics of common human disease. Virchows Arch. 2010;456:13–21. doi: 10.1007/s00428-009-0847-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grunau C., Renault E., Rosenthal A., Roizes G. MethDB—a public database for DNA methylation data. Nucleic Acids Res. 2001;29:270–274. doi: 10.1093/nar/29.1.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ongenaert M., Van Neste L., De Meyer T., Menschaert G., Bekaert S., Van Criekinge W. PubMeth: a cancer methylation database combining text-mining and expert annotation. Nucleic Acids Res. 2008;36:D842–D846. doi: 10.1093/nar/gkm788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He X., Chang S., Zhang J., Zhao Q., Xiang H., Kusonmano K., Yang L., Sun Z.S., Yang H., Wang J. MethyCancer: the database of human DNA methylation and cancer. Nucleic Acids Res. 2008;36:D836–D841. doi: 10.1093/nar/gkm730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hackenberg M., Barturen G., Oliver J.L. NGSmethDB: a database for next-generation sequencing single-cytosine-resolution DNA methylation data. Nucleic Acids Res. 2011;39:D75–D79. doi: 10.1093/nar/gkq942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lv J., Liu H., Su J., Wu X., Liu H., Li B., Xiao X., Wang F., Wu Q., Zhang Y. DiseaseMeth: a human disease methylation database. Nucleic Acids Res. 2012;40:D1030–D1035. doi: 10.1093/nar/gkr1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baek S.J., Yang S., Kang T.W., Park S.M., Kim Y.S., Kim S.Y. MENT: methylation and expression database of normal and tumor tissues. Gene. 2013;518:194–200. doi: 10.1016/j.gene.2012.11.032. [DOI] [PubMed] [Google Scholar]
- 20.Vrba L., Munoz-Rodriguez J.L., Stampfer M.R., Futscher B.W. miRNA gene promoters are frequent targets of aberrant DNA methylation in human breast cancer. PLoS One. 2013;8:e54398. doi: 10.1371/journal.pone.0054398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laird P.W. Principles and challenges of genomewide DNA methylation analysis. Nat. Rev. Genet. 2010;11:191–203. doi: 10.1038/nrg2732. [DOI] [PubMed] [Google Scholar]
- 22.Karolchik D., Barber G.P., Casper J., Clawson H., Cline M.S., Diekhans M., Dreszer T.R., Fujita P.A., Guruvadoo L., Haeussler M., et al. The UCSC Genome Browser database: 2014 update. Nucleic Acids Res. 2014;42:D764–D770. doi: 10.1093/nar/gkt1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chien C.H., Sun Y.M., Chang W.C., Chiang-Hsieh P.Y., Lee T.Y., Tsai W.C., Horng J.T., Tsou A.P., Huang H.D. Identifying transcriptional start sites of human microRNAs based on high-throughput sequencing data. Nucleic Acids Res. 2011;39:9345–9356. doi: 10.1093/nar/gkr604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jones P.A. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 25.Irizarry R.A., Ladd-Acosta C., Wen B., Wu Z., Montano C., Onyango P., Cui H., Gabo K., Rongione M., Webster M., et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat. Genet. 2009;41:178–186. doi: 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ko Y.A., Mohtat D., Suzuki M., Park A.S., Izquierdo M.C., Han S.Y., Kang H.M., Si H., Hostetter T., Pullman J.M., et al. Cytosine methylation changes in enhancer regions of core pro-fibrotic genes characterize kidney fibrosis development. Genome Biol. 2013;14:R108. doi: 10.1186/gb-2013-14-10-r108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kanehisa M., Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suzuki H., Maruyama R., Yamamoto E., Kai M. DNA methylation and microRNA dysregulation in cancer. Mol. Oncol. 2012;6:567–578. doi: 10.1016/j.molonc.2012.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]


