Abstract
The first ncRNA found was an alanine tRNA in baker's yeast, and the first detected microRNAs (miRNAs) promoted ncRNA research to a whole new level. Research on ncRNAs in animals has focused on the medical field, while in plant scientists are more concerned with improving agronomic traits. In 2010, we constructed a plant miRNA database named PMRD to meet the demand for miRNA research in plants. To provide a way to do fundamental research on plant ncRNAs and take full advantage of tremendous public resources, we designed an updated platform called plant ncRNA database (PNRD) based on its predecessor PMRD, which is accessible at http://structuralbiology.cau.edu.cn/PNRD. We collected a total of 25739 entries of 11 different types of ncRNAs from 150 plant species. Targets of miRNAs were extended to 178138 pairs in 46 species, while the number of miRNA expression profiles reached 35. Improvements in PNRD are not only the larger amounts of data, but also better service, such as a more user-friendly interface, more multifunctional and browsing options and more background data for users to download. We also integrated currently prevalent technologies and toolkits to strengthen the capability of the database and provide a one-stop service for scientific users.
INTRODUCTION
Transcripts that are not functional as templates for protein synthesis are called non-coding RNAs (ncRNAs). It has been more than half a century since the first ncRNA was found in baker's yeast and characterized as an alanine tRNA (1). Since then tens of thousands of regulatory ncRNAs of different shapes and sizes have been discovered. In 1993, a small ncRNA molecule named microRNA (miRNA) was first observed in Caenorhabditis elegans, and is known to be responsible for inhibiting translation of the lin-14 gene by bounding to its 3′-untranslated region (2). Based on this discovery, numerous complex mechanisms for translational control by small RNAs have been reported. For example, in medical research, some miRNAs act as tumor suppressors for cancer prevention and therapeutics while others are oncogenes that promote cancer development (3). The first miRNA in plants was discovered in 2002 (4), and these miRNAs have multiple functions, including not only regulation of mRNA expression or small interfering RNA (siRNA) biogenesis, but also involvement in various stress responses.
In addition to miRNA, efforts are underway to study other regulatory RNAs, such as ribosomal RNAs, transfer RNAs, small nucleolar RNAs and long ncRNAs (lncRNAs), which are transcripts longer than 200 bp (5) with non-protein coding potential. As research goes deeper, important roles of lncRNA in complex regulatory networks are constantly observed. Such classic lncRNA genes in humans as Xist (6,7), HOTAIR (8) and H19 (9) were discovered and their functions identified as associated with epigenetic regulation.
Further study of ncRNA function has shown some differences between plants and animals. For example, in miRNA biogenesis, plants differ from animals by using a different ribonuclease, as well as other aspects, such as genomic arrangement of miRNA genes, target recognition and contrasting modes of evolutionary emergence (10). For another example, the mechanism of interaction between lncRNA and siRNA in targeting DNA methylation differs between plants and mammals (11). The fact is that more plant-specific ncRNAs are found thanks to rapidly developed biological technologies, and more key regulatory networks ncRNAs involved are studied and applied to agricultural production. Zhang et al. constructed a plant miRNA database, PMRD, in 2010 to meet the demand of miRNA research in plants (12). Since then, some useful databases and tools focusing on plant miRNAs have been built: PmiRKB (13) is a plant miRNA knowledge base, which provides four major functional modules of miRNAs for two species (Arabidopsis and rice); SmiRFN (14) is a comprehensive web tool about soybean miRNA functional analysis; PMTED (15) is a plant miRNA target expression database which is designed to analyze expression profiles of miRNA targets using microarray data; and miRNEST (16) is a comprehensive database of animal, plant and virus miRNAs containing new miRNAs, targets, mirtrons and miRNA gene structures, which has been updated to version 2.0.
These databases are all limited to miRNAs and their functional analysis is restricted to only one or two aspects. As fundamental research on ncRNAs covers more than one aspect, while corresponding data are scattered everywhere, we decided to construct a platform to integrate this valuable information and provide a way to acquire novel knowledge from big data mining. What makes our platform special is the multiple resources used to annotate these ncRNAs, especially literature-based information that is rarely used and epigenetic information represented by ChIP-seq data. We also built a connection between miRNAs and other ncRNAs, such as lncRNAs to help illustrate complex biological phenomena.
PNRD is a comprehensive integrated web resource for ncRNA searching, browsing, predicting, visualizing and downloading, which can be accessed at http://structuralbiology.cau.edu.cn/PNRD. The improvements to the PNRD database include not only more data sources and a more user-friendly interface, but also the introduction of more useful toolkits to provide a one-stop service for researchers. With the help of PNRD, users can predict coding potential of sequences of interest using the CPC toolkit, utilize our improved miRNA prediction toolkit for new miRNA discovery or use any basic tools provided, such as keyword and profile searches. Two genome browsers are provided for users to explore ncRNA location along with coding genes and their relationship with epigenetic modification nearby. By integrating information from the literature, high-throughput sequencing data and useful methods and tools, we hope that PNRD will be a useful resource and integrated platform for plant ncRNA researchers.
DATA INTEGRATION
The PNRD database supports 11 kinds of known ncRNAs from multiple sources, including public data repositories, published references and in-house data. Among them, miRNA comprise 58% of the database while other ncRNAs (represented by tRNA and lncRNA) are the other 42%. PNRD integrates miRNA target information without being limited to the mRNA sequence and extends to other ncRNAs, such as endogenous target mimics (eTMs). Using text-mining, we collected papers describing ncRNA functions with experimental evidence and developed a mining pool for functional data mining. All collected data sets in PNRD are summarized in Supplementary Table S1.
Sequence information
So far, 25739 ncRNA entries are available supporting 150 plant species in PNRD (Table 1). PNRD integrates sequence information from multiple resources to expand the miRNA data set, mainly in the miRBase (release 21) (17) and PMRD databases (12). We used the union of sequences from the above two databases for miRNA updating. The number of miRNAs reached 15041 compared to 10898 in PMRD. Among these newly added miRNAs, 301 miRNAs of Setaria italica (foxtail millet) and 265 of Triticum aestivum L. (wheat) were collected from published literature (18,19).
Table 1. The number of different categories of ncRNAs collected from different sources for some widely studied plant species.
| Species | Number of ncRNAs | ||||
|---|---|---|---|---|---|
| miRNA | lncRNA | tasiRNA | Others | All | |
| Arabidopsis thaliana | 1581 | 2579 | 23 | 1432 | 5615 |
| Oryza sativa | 2819 | 752 | 9 | 1313 | 4893 |
| Populus trichocarpa | 2944 | 538 | N/A | 647 | 4129 |
| Zea mays | 325 | 1704 | 4 | N/A | 2033 |
| Glycine max | 664 | N/A | 6 | 860 | 1530 |
| Medicago truncatula | 810 | N/A | 3 | 684 | 1497 |
| Brachypodium distachyon | 537 | N/A | N/A | N/A | 537 |
| Triticum aestivum | 384 | N/A | 2 | N/A | 386 |
| Arabidopsis lyrata | 384 | N/A | N/A | N/A | 384 |
| Solanum tuberosum | 364 | N/A | N/A | N/A | 364 |
| Physcomitrella patens | 286 | N/A | 42 | N/A | 328 |
| Setaria italica | 301 | N/A | N/A | N/A | 301 |
| Gossypium raimondii | 296 | N/A | N/A | N/A | 296 |
| Malus domestica | 208 | N/A | 40 | N/A | 248 |
| Sorghum bicolor | 241 | N/A | N/A | N/A | 241 |
| Prunus persica | 224 | N/A | N/A | N/A | 224 |
| Vitis vinifera | 186 | N/A | 37 | N/A | 223 |
| Brassica rapa | 173 | N/A | N/A | N/A | 173 |
| Nicotiana tabacum | 164 | N/A | 5 | N/A | 169 |
| Manihot esculenta | 153 | N/A | N/A | N/A | 153 |
| Brassica napus | 137 | N/A | 2 | N/A | 139 |
| Linum usitatissimum | 124 | N/A | N/A | N/A | 124 |
| Cucumis melo | 120 | N/A | N/A | N/A | 120 |
| Solanum lycopersicum | 110 | N/A | 4 | N/A | 114 |
| Vigna unguiculata | 93 | N/A | N/A | N/A | 93 |
| Chlamydomonas reinhardtii | 87 | N/A | N/A | N/A | 87 |
| Theobroma cacao | 82 | N/A | N/A | N/A | 82 |
| Carica papaya | 81 | N/A | N/A | N/A | 81 |
| Gossypium hirsutum | 81 | N/A | N/A | N/A | 81 |
| Other species (121) | 1082 | N/A | 12 | N/A | 1094 |
| All species | 15041 | 5573 | 189 | 4936 | 25739 |
‘N/A’ means data are not available for now.
Other ncRNAs are mainly from a number of well-known databases: NONCODEv4 (20), Rfam (21), tasiRNAdb (22), GtRNAdb (23), TAIR (24) and RGAP (25). In addition to these public data, we obtained ncRNAs from published literature that had not been included in the PMRD database although some are reported as important factors in pivotal biological processes. When comparing data coverage of PNRD to other similar platforms, such as RNAcentral (26) and ncRNAdb (27), ours has significantly more ncRNAs categories (PNRD 11 categories and RNAcentral 6 categories), or more organisms (PNRD 150 organisms and ncRNAdb 99 organisms), which manifest the advantage of data size of our database.
Target information
To enrich the functional information concerning miRNAs, we collected target information from both the online web-server psRNATarget (26) and curated targets from published literature (27,28) which are called eTMs. Some studies found that miRNAs could also potentially target lncRNAs to participate in a regulatory network among ncRNA classes (29), so we took lncRNAs into consideration as putative targets of miRNA. We collected 178138 pairs of miRNA targets supporting 46 plant species, containing protein-coding genes, ncRNAs from literature and lncRNAs from NONCODE. PNRD also has an online cytoscape service to facilitate users to better understand miRNA-mediated gene regulatory networks.
Expression profile information
The expression profile data were collected in two ways, extracting from literature and in-house computation. The data are either next-generation sequencing data or microarray data. We collected a total of 35 profile projects from 11 different species, 18 from Arabidopsis and 8 from rice. PNRD collected 18 groups of small RNA sequencing data of six different species from a superseries (Accession number: GSE28755) in the Gene Expression Omnibus (GEO) database to extract new miRNA expression profiles using a customized miRanalyzer (30) toolkit. Figure 1 shows the expression profile of the MIR156 family in different tissues of five species. Previous research has shown that miR156 can control flowering time and root development in both Arabidopsis and rice (31). MIR156 family shows different expression patterns in plants, such as maize and tomato, with its expression significantly higher in leaves than in two other tissues (Figure 1).
Figure 1.
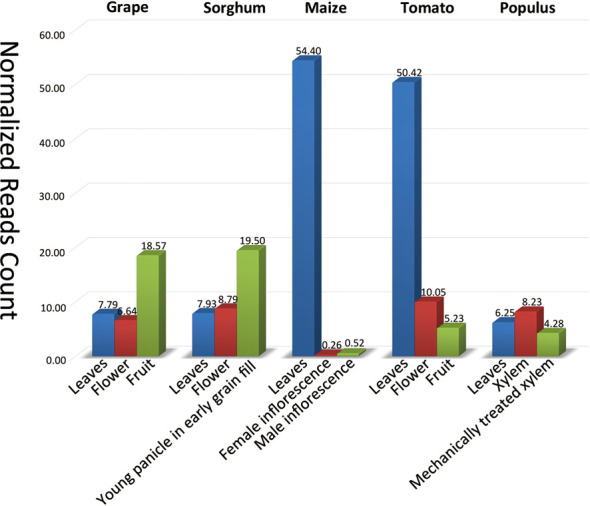
The expression profile of the MIR156 family in five species: grape, sorghum, maize, tomato and Populus, respectively. The x-axis represents different tissues in one species while the y-axis represents the normalized reads count of miR156 in corresponding tissue. The read counts were calculated by the localized miRanalyzer toolkit.
SERVICE IMPROVEMENT
The PNRD database was constructed on a standard LAMP (Linux+Apache+Mysql+PHP) system. Compared with the former PMRD database, PNRD has a more sophisticated interface, wider search range, more search options and more useful contents for users to download. From the website, users can obtain: (i) detailed information about every ncRNA using ID search or browse; (ii) miRNA target information using customized cytoscape; (iii) literature information using text-mining; (iv) sequence coding potential score using the CPC toolkit; (v) miRNA candidates using a customized miRanalyzer toolkit; (vi) miRNA expression profile by next-generation sequencing technology extracted from published literature and (vii) graphs displaying relationships between ncRNAs and histone modifications nearby. From the download page, users can get sequences of ncRNAs in FASTA format, their target information in tabular format and all literature used in the text-mining toolkit.
Browse and search
While keeping the two previous browse patterns, browse by species in taxonomic sorting and alphabetic sorting, PNRD included a new browse pattern ‘browse by category’ because there is more than one kind of ncRNA in the database. Considering that users may have a situation with more than one kind of ncRNA in a species in ‘browse by species’, users can choose the ncRNA category before they get the final results. In the ID search page, ‘batch search’ allows users to submit a list of unique IDs and the database returns the matching results. The ‘keyword search’ will find the literature with the best match depending on the typed-in ‘keywords’; while ‘exprofile search’ is added to help users find expression profile data of their miRNA of interest as quickly as they can by entering the miRNA name. We added another search engine into the search page called ‘miRNA-epigenomics search’ to help users search miRNA of interest and get their relationship with selected histone modification which have been include into localized University of California Santa Cruz (UCSC) Genome Browser. Putative relationship between miRNAs and nearby histone marks are extracted according to their genomic location.
Genome browser displaying
Two genome browsers are provided in PNRD. Gbrowse was kept for miRNA location checking and the corresponding information was updated. The other browser, UCSC Genome Browser, is a customized toolkit for further analysis based on miRNA location and histone mark location. We uploaded ChIP-seq data sets of three species (Arabidopsis, rice and maize), including in-house and public data, to our localized UCSC Genome Browser and displayed them on it. Using Arabidopsis as an example, we downloaded two groups of ChIP-seq data of four histone modifications: H3K4me1, H3K4me2 and H3K4me3 from GEO (GSE11657); and H3K4me3 and H3K27me3 from GEO (GSE50636). We then calculated epigenetic enriched peaks using our own analysis pipeline and displayed the results on the UCSC Genome Browser. For rice, we used our in-house experimental data of four published histone marks (H3K9ac, H3K27ac, H3K4me2 and H3K4me3) (32). For maize, we downloaded data of four histone marks (H3K9ac, H3K4me3, H3K27me3 and H3K36me3) in root and shoot tissues (GSE15286) and displayed them as for Arabidopsis. Users can search any ID of ncRNAs of interest identified by PNRD to look for the location of miRNA and try to determine any potential relationship between miRNA expression and histone modifications.
ncRNA FUNCTIONAL ANALYSIS
PNRD has several key characteristics to promote its capacity for functional search and analysis. Figure 2 shows the organizational structure of our database. Each ncRNA in PNRD has at most seven parts of annotated information: (i) basic information, (ii) sequence information, (iii) gene structure, (iv) function-related publication, (v) target information, (vi) RNA secondary structure and (vii) expression profile information. The literature-based search allows users to look up functional descriptions of related ncRNAs in the collected literature and three searching methods are provided to help users get what they need. Two prevalent tools focusing on ncRNA analysis were introduced into PNRD: miRanalyzer (30) and CPC (33) toolkits.
Figure 2.
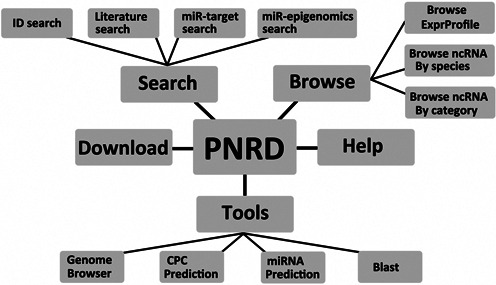
Structure of PNRD database, composed of five parts: Search section, Browse section, Tools section, Download page and Help page.
Function searching through text-mining
Published literature always includes much experimental data, and so is a valuable resource to help infer regulatory mechanisms and biological processes that ncRNA may participate in. According to a review, text-mining is being applied to identify molecular causes of diseases using information from literature (34), and so we decided to take full advantage of them for plant ncRNA research.
In PNRD, users can type in any phrase or sentence containing keyword(s) that they are interested in, choose the search manner they want and get literature with the most matched content. The three search manners are: ncRNA ID, in reference title and in reference abstract. Currently, PNRD has 148 references containing 36 miRNA families and 6 lncRNAs whose functions are mostly related to tissue development or response to stress (Table 2).
Table 2. Background literature used in text-mining technology.
| ncRNA family | Related function | No. supported references |
|---|---|---|
| miRNA | ||
| MIR156 | Flowering time control; phage changing modulation; later embryonic maturation and root development | 44 |
| MIR157 | Flowering time control; phage changing modulation; later embryonic maturation and root development | 7 |
| MIR159 | Flowering time control in short-day photoperiods; leaf development; seed size and shape determination | 27 |
| MIR160 | Root cap structure and lateral root development; seed development during embryogenesis; flower and leaf development | 9 |
| MIR162 | Regulation of somatic embryogenesis, environmental stress response, and fiber differentiation and development | 7 |
| MIR164 | Petal number control; development of leaf and shoot buds; lateral root development; normal embryonic development | 19 |
| MIR165 | Embryo development; leaf primordial, shoot apical meristem and floral stem cell development; root and nodule development | 5 |
| MIR166 | Embryo development; leaf primordial, shoot apical meristem and floral stem cell development; root and nodule development | 11 |
| MIR167 | Gynoecium and stamen maturation; seed development | 9 |
| MIR168 | Stress response and signal transduction in plant development | 6 |
| MIR169 | Nodule development control; leaf development | 5 |
| MIR170 | Leaf development | 2 |
| MIR171 | Negative regulation of shoot branching; root colonization regulation | 11 |
| MIR172 | Flowering time control; seed development; phase change in shoot | 23 |
| MIR319 | Leaf morphogenesis, complexity and senescence; flower development; male and female gametophyte development | 13 |
| MIR390 | Lateral root development; leaf development | 5 |
| MIR393 | Auxin-related leaf, root and shoot development; leaf development | 11 |
| MIR394 | Regulation of leaf curling-related morphology | 1 |
| MIR396 | Cell proliferation control during leaf and root development | 8 |
| MIR397 | Seed development | 4 |
| MIR399 | Regulation of cellular response to local phosphate increase during arbuscular mycorrhizal symbiosis | 3 |
| MIR402 | Accelerate seed germination and seedling growth under stress conditions | 1 |
| MIR408 | Root development under many stress conditions | 7 |
| MIR444 | Floral patterning and development control | 3 |
| MIR482 | Nodule numbers | 2 |
| MIR528 | Root development under many stress conditions; seed development | 2 |
| MIR538 | Moss development | 2 |
| MIR824 | Stomatal development | 2 |
| MIR828 | Anthocyanin biosynthesis, trichome initiation and root hair patterning | 2 |
| MIR858 | Anthocyanin biosynthesis | 3 |
| MIR902 | Rhizoid development in moss | 2 |
| MIR1218 | Organ separation | 1 |
| MIR1219 | Moss development | 1 |
| MIR1223 | Organ separation | 1 |
| MIR1446 | Formation of specialized woody tissue in trees; cambium differentiation in the stem; DNA damage repair | 1 |
| MIR1512 | Nodule numbers | 1 |
| lncRNA | ||
| AtR8 | Hypoxic stress | 1 |
| cis-NATPHO1;2 | Phosphate homeostasis and plant fitness | 1 |
| IPS1 | Phosphate starvation | 1 |
| TARs | Responsive to biotic stress | 1 |
| asHSFB2a | Heat-inducible | 1 |
| TER2 | Telomere repeat synthesis | 1 |
The three columns indicate the ncRNA family, related function and the corresponding numbers of references. TARs include TAR-191, TAR-197 and TAR-224.
New miRNA prediction
Nowadays, online web-servers for next-generation sequencing data analysis are convenient and efficient. To provide a better service for miRNA analysis, we decided to incorporate the well-known miRNA prediction toolkit miRanalyzer (30) in the PNRD platform. We made some improvements to extend its practicability. First, the supported species were increased to eight: Arabidopsis thaliana, Chlamydomonas reinhardtii, Oryza sativa, Populus trichocarpa, Sorghum bicolor, Solanum lycopersicum, Vitis vinifera and Zea mays. Second, we included alternative alignment libraries for known miRNA detection: the official library and our home-made library containing more newly discovered miRNAs. Third, we added an email notification mode to ensure that users actually get the results. Some of the results are displayed on the website, while all results can be downloaded for further analysis.
Example of using PNRD
Here, we show a specific example demonstrating how to use the PNRD database for some useful scientific exploration. Initially, users may not know which ncRNA is important for their research, but do know which biological process they are interested in. The keyword used here is ‘flowering time control’, while the species is ‘Arabidopsis thaliana’, and this produces four families: MIR156, MIR157, MIR159 and MIR172 (Figure 3A). To determine more about the relationship between miRNAs and flowering-related biological processes, users can either click reference ID in the ‘supportive reference’ column, or use ‘literature related search’ to find more detailed references. When looking at the literature concerning the miR172 family, one study presents that miR172 serves as a translational repressor of APETALA2, a floral homeotic gene, to regulate its expression in Arabidopsis flower development (35).
Figure 3.

Analysis pipeline and corresponding results of ath-miR172 using PNRD database. We use a pipeline to demonstrate how to do research with PNRD using different functional tools. (A) Results of keyword search on search page using ‘flowering time control’ as input. PNRD gives users the most matched ncRNAs with supportive literature containing their input. (B) The alignment results of ath-miR172c and its targets using psRNAtarget software and text-mining, which includes not only coding mRNAs, but also target mimics. (C) Information on literature from the text-mining pool with an intimate association with ath-miR172c and some important relevant knowledge. (D) Expression profiles of ath-miR172c. The left frame is the profile projects containing ath-miR172c using ‘search in exprofile’ section of the Search page. The other two histograms summarize results of the expression profile as searched above. The upper histogram reflects the MIR172 family expression profile derived from a project named ‘MicroRNA activity in the Arabidopsis male germline’. The x-axis represents five members of the MIR172 family while the y-axis is normalized read counts expressed in three different tissues, identified in three different colors. The lower histogram was derived from six different projects. The x-axis represents different experimental conditions and the y-axis represents normalized read counts. Different colors are used to identify different projects. (E) Basic information about ath-miR172c, such as the genomic location, sequence and secondary structure. (F) Customized UCSC Genome Browser screenshot of ath-miR172c. Five histone marks from two data sets of GEO (GSE50636: H3K27me3, H3K4me3; and GSE657: H3K4me1, H3K4me2 and H3K4me3) are near ath-miR172c. Data were downloaded from GEO database and calculated in-house to display in UCSC.
Members of the MIR172 family can be examined more closely, for example, using the selected member ‘ath-miR172c’. From the detail page, basic information about ath-miR172c is displayed, such as the sequence, genomic location and secondary structure of its precursor (Figure 3E). The ‘function-related publication’ presents all literature that is highly associated with the MIR172 family in the text-mining literature pool, which will help in understanding the biological function of MIR172 (Figure 3C). There are two transcripts of the AP2 gene in the target information region: AT4G36920.1 and AT4G36920.2 (Figure 3B). Studies show that miR172 regulates the initiation of flowering when ambient environment changes (36) and it has high sequence complementarities with APETALA2 (AP2) (35). Another two targets of ath-miR172c, SMZ (locus ID: AT2G39250) and SNZ (locus ID: AT3G54990) are paralogs. SMZ is short for SCHLAFMUTZE and is a potent repressor of flowering (37), while SNZ is short for SCHNARCHZAPFEN and is a paralog of SMZ and has been shown to repress flowering when expressed at high levels (38). Their functions of encoding an AP2 domain transcription factor that can repress flowering suggested that they may participate in a miR172-mediated network and indicated that the results are biologically meaningful. Other members of the MIR172 family (e.g. ath-miR172a, ath-miR172b-3p, ath-miR172d, ath-miR172e and ath-miR172m) share almost the same targets as ath-miR172c, with one exception being ath-miR172b-5p due to a different sequence.
The section ‘search in exprofile’ in the ID search page helps users to find more about the dynamic expression level of miRNAs of interest. Typing ‘miR172c’ in the query box and choosing the species ‘Arabidopsis thaliana’ gives users 11 different profile projects concerning ath-miR172c (Figure 3D). Clicking on the ‘detail’ link of the project named ‘MicroRNA activity in the Arabidopsis male germline’ helps users to find that ath-miR172c is significantly more expressed in pollen compared to the other two tissues, with the same pattern as for other three MIR172 members (ath-miR172a, ath-miR172b and ath-miR172d; Figure 3D), meaning that this miRNA may have tissue specificity at transcript level. Another histogram (Figure 3D) represents an overview of the expression profile of ath-miR172c under the different conditions of six projects, which are all derived from the above 11 projects. The ‘search in exprofile’ gives users a better and quicker understanding of the characteristics of ath-miR172c expression.
The UCSC Genome Browser can help users to further understand epigenetic effects on miRNA expression. There are five tracks on the UCSC Genome Browser representing five histone modifications from two different GEO datasets (Figure 3F), which is also an example displayed after using our ‘miRNA-epigenomics search’ tool to search its putative relationship with histone modification. It is interesting that only one histone mark, H3K27me3, had a significant peak near ath-miR172c and this histone mark were reported to be related with transcript repression (39), which suggests a putative transcriptional regulation mechanism of miRNA expression through changing epigenetic modification.
CONCLUSION AND OUTREACH
Compared with PMRD, PNRD has greatly progressed in many aspects, especially in functional analysis and service improvement. PNRD inherited the virtue of PMRD in supporting a large number of plant species with detailed annotation information from multiple sources. PNRD also integrates in-house experimental data to provide greater support for results. To meet demands of more users, we greatly enlarged the background data sets to include as many kinds of ncRNA as possible and enhanced the search range to not only ncRNA IDs, but also published literature, ncRNA targets and miRNA expression profiles. In addition, we introduced some innovation to the new platform, such as the application of text-mining technology to look at confirmed biological functions of ncRNAs, integrating currently prevalent analysis toolkits to help with next-generation sequencing data and the use of localized UCSC Genome Browser to study relationships between miRNA expression and epigenetic modification. In summary, PNRD is a comprehensive online platform for plant ncRNA analysis, providing a one-stop service for researchers.
In terms of ncRNA categories, only Arabidopsis and rice have multiple types of ncRNAs, while others have at most two or three types, so continuous updating to enlarge ncRNA data is required. Immediately following the updating, work will focus on tRNA, snRNA and snoRNA because of their widely available data resources. The amount of literature in the text-mining pool is also limiting for large-scale data mining, so the text-mining service will be actively updated. A high-throughput technology called parallel analysis of RNA ends, also referred to as the degradome sequencing method, has been widely applied to identify small RNA cleavage sites and reveal new miRNA targets. Those data are of great value but have not been widely applied yet, especially in plants. One of our likely future tasks is to make full use of these Degradome-Seq data to help study miRNA-mediated gene regulation. We hope that PNRD will become a useful resource and multifunctional platform to help scientists in plant fundamental research field with research on ncRNA characteristics and that numerous researchers will benefit from it in the future.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
We thank Dr. Zhou Du for comments during manuscript preparation.
FUNDING
Funding for open access charge: Ministry of Science and Technology of China [31371291 and 31171276].
Conflict of interest statement. None declared.
REFERENCES
- 1.Holley R.W., Apgar J., Everett G.A., Madison J.T., Marquisee M., Merrill S.H., Penswick J.R., Zamir A. Structure of a ribonucleic acid. Science. 1965;147:1462–1465. doi: 10.1126/science.147.3664.1462. [DOI] [PubMed] [Google Scholar]
- 2.Lee R.C., Feinbaum R.L., Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 3.Zhang B., Pan X., Cobb G.P., Anderson T.A. microRNAs as oncogenes and tumor suppressors. Dev. Biol. 2007;302:1–12. doi: 10.1016/j.ydbio.2006.08.028. [DOI] [PubMed] [Google Scholar]
- 4.Reinhart B.J., Weinstein E.G., Rhoades M.W., Bartel B., Bartel D.P. MicroRNAs in plants. Genes Dev. 2002;16:1616–1626. doi: 10.1101/gad.1004402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perkel J.M. Visiting ‘noncodarnia’. BioTechniques. 2013;54:303–304. doi: 10.2144/000114037. [DOI] [PubMed] [Google Scholar]
- 6.Brockdorff N., Ashworth A., Kay G.F., McCabe V.M., Norris D.P., Cooper P.J., Swift S., Rastan S. The product of the mouse Xist gene is a 15 kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell. 1992;71:515–526. doi: 10.1016/0092-8674(92)90519-i. [DOI] [PubMed] [Google Scholar]
- 7.Brown C.J., Hendrich B.D., Rupert J.L., Lafreniere R.G., Xing Y., Lawrence J., Willard H.F. The human XIST gene: analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell. 1992;71:527–542. doi: 10.1016/0092-8674(92)90520-m. [DOI] [PubMed] [Google Scholar]
- 8.Rinn J.L., Kertesz M., Wang J.K., Squazzo S.L., Xu X., Brugmann S.A., Goodnough L.H., Helms J.A., Farnham P.J., Segal E., et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brannan C.I., Dees E.C., Ingram R.S., Tilghman S.M. The product of the H19 gene may function as an RNA. Mol. Cell. Biol. 1990;10:28–36. doi: 10.1128/mcb.10.1.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Axtell M.J., Westholm J.O., Lai E.C. Vive la difference: biogenesis and evolution of microRNAs in plants and animals. Genome Biol. 2011;12:221. doi: 10.1186/gb-2011-12-4-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rinn J.L., Chang H.Y. Genome regulation by long noncoding RNAs. Ann. Rev. Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Z., Yu J., Li D., Liu F., Zhou X., Wang T., Ling Y., Su Z. PMRD: plant microRNA database. Nucleic Acids Res. 2010;38:D806–D813. doi: 10.1093/nar/gkp818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meng Y., Gou L., Chen D., Mao C., Jin Y., Wu P., Chen M. PmiRKB: a plant microRNA knowledge base. Nucleic Acids Res. 2011;39:D181–D187. doi: 10.1093/nar/gkq721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu Y., Guo M., Liu X., Wang C., Liu Y. Inferring the soybean (Glycine max) microRNA functional network based on target gene network. Bioinformatics. 2014;30:94–103. doi: 10.1093/bioinformatics/btt605. [DOI] [PubMed] [Google Scholar]
- 15.Sun X., Dong B., Yin L., Zhang R., Du W., Liu D., Shi N., Li A., Liang Y., Mao L. PMTED: a plant microRNA target expression database. BMC Bioinformat. 2013;14:174. doi: 10.1186/1471-2105-14-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Szczesniak M.W., Deorowicz S., Gapski J., Kaczynski L., Makalowska I. miRNEST database: an integrative approach in microRNA search and annotation. Nucleic Acids Res. 2012;40:D198–D204. doi: 10.1093/nar/gkr1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kozomara A., Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42:D68–D73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yi F., Xie S., Liu Y., Qi X., Yu J. Genome-wide characterization of microRNA in foxtail millet (Setaria italica) BMC Plant Biol. 2013;13:212. doi: 10.1186/1471-2229-13-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun F., Guo G., Du J., Guo W., Peng H., Ni Z., Sun Q., Yao Y. Whole-genome discovery of miRNAs and their targets in wheat (Triticum aestivum L.) BMC Plant Biol. 2014;14:142. doi: 10.1186/1471-2229-14-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xie C., Yuan J., Li H., Li M., Zhao G., Bu D., Zhu W., Wu W., Chen R., Zhao Y. NONCODEv4: exploring the world of long non-coding RNA genes. Nucleic Acids Res. 2014;42:D98–D103. doi: 10.1093/nar/gkt1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Griffiths-Jones S., Bateman A., Marshall M., Khanna A., Eddy S.R. Rfam: an RNA family database. Nucleic Acids Res. 2003;31:439–441. doi: 10.1093/nar/gkg006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang C., Li G., Zhu S., Zhang S., Fang J. tasiRNAdb: a database of ta-siRNA regulatory pathways. Bioinformatics. 2014;30:1045–1046. doi: 10.1093/bioinformatics/btt746. [DOI] [PubMed] [Google Scholar]
- 23.Chan P.P., Lowe T.M. GtRNAdb: a database of transfer RNA genes detected in genomic sequence. Nucleic Acids Res. 2009;37:D93–D97. doi: 10.1093/nar/gkn787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lamesch P., Berardini T.Z., Li D., Swarbreck D., Wilks C., Sasidharan R., Muller R., Dreher K., Alexander D.L., Garcia-Hernandez M., et al. The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res. 2012;40:D1202–D1210. doi: 10.1093/nar/gkr1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kawahara Y., de la Bastide M., Hamilton J.P., Kanamori H., McCombie W.R., Ouyang S., Schwartz D.C., Tanaka T., Wu J., Zhou S., et al. Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice. 2013;6:4. doi: 10.1186/1939-8433-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dai X., Zhao P.X. psRNATarget: a plant small RNA target analysis server. Nucleic Acids Res. 2011;39:W155–W159. doi: 10.1093/nar/gkr319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu H.J., Wang Z.M., Wang M., Wang X.J. Widespread long noncoding RNAs as endogenous target mimics for microRNAs in plants. Plant Physiol. 2013;161:1875–1884. doi: 10.1104/pp.113.215962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shuai P., Liang D., Tang S., Zhang Z., Ye C.Y., Su Y., Xia X., Yin W. Genome-wide identification and functional prediction of novel and drought-responsive lincRNAs in Populus trichocarpa. J. Exp. Botany. 2014;65:4975–4983. doi: 10.1093/jxb/eru256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jalali S., Bhartiya D., Lalwani M.K., Sivasubbu S., Scaria V. Systematic transcriptome wide analysis of lncRNA-miRNA interactions. PLoS One. 2013;8:e53823. doi: 10.1371/journal.pone.0053823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hackenberg M., Sturm M., Langenberger D., Falcon-Perez J.M., Aransay A.M. miRanalyzer: a microRNA detection and analysis tool for next-generation sequencing experiments. Nucleic Acids Res. 2009;37:W68–W76. doi: 10.1093/nar/gkp347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bai Y., Dai X., Harrison A.P., Chen M. RNA regulatory networks in animals and plants: a long noncoding RNA perspective. Brief. Funct. Genom. 2014 doi: 10.1093/bfgp/elu017. doi: 10.1093/bfgp/elu017. [DOI] [PubMed] [Google Scholar]
- 32.Du Z., Li H., Wei Q., Zhao X., Wang C., Zhu Q., Yi X., Xu W., Liu X.S., Jin W., et al. Genome-wide analysis of histone modifications: H3K4me2, H3K4me3, H3K9ac, and H3K27ac in Oryza sativa L. japonica. Mol. Plant. 2013;6:1463–1472. doi: 10.1093/mp/sst018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kong L., Zhang Y., Ye Z.Q., Liu X.Q., Zhao S.Q., Wei L., Gao G. CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 2007;35:W345–W349. doi: 10.1093/nar/gkm391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rebholz-Schuhmann D., Oellrich A., Hoehndorf R. Text-mining solutions for biomedical research: enabling integrative biology. Nat. Rev. Genet. 2012;13:829–839. doi: 10.1038/nrg3337. [DOI] [PubMed] [Google Scholar]
- 35.Chen X. A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science. 2004;303:2022–2025. doi: 10.1126/science.1088060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jin D., Wang Y., Zhao Y., Chen M. MicroRNAs and their cross-talks in plant development. J. Genet. Genom. 2013;40:161–170. doi: 10.1016/j.jgg.2013.02.003. [DOI] [PubMed] [Google Scholar]
- 37.Mathieu J., Yant L.J., Murdter F., Kuttner F., Schmid M. Repression of flowering by the miR172 target SMZ. PLoS Biol. 2009;7:e1000148. doi: 10.1371/journal.pbio.1000148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schmid M., Uhlenhaut N.H., Godard F., Demar M., Bressan R., Weigel D., Lohmann J.U. Dissection of floral induction pathways using global expression analysis. Development. 2003;130:6001–6012. doi: 10.1242/dev.00842. [DOI] [PubMed] [Google Scholar]
- 39.Zhang X., Clarenz O., Cokus S., Bernatavichute Y.V., Pellegrini M., Goodrich J., Jacobsen S.E. Whole-genome analysis of histone H3 lysine 27 trimethylation in Arabidopsis. PLoS Biol. 2007;5:e129. doi: 10.1371/journal.pbio.0050129. [DOI] [PMC free article] [PubMed] [Google Scholar]


