Abstract
Long non-coding RNAs (lncRNAs) perform a diversity of functions in numerous important biological processes and are implicated in many human diseases. In this report we present lncRNAWiki (http://lncrna.big.ac.cn), a wiki-based platform that is open-content and publicly editable and aimed at community-based curation and collection of information on human lncRNAs. Current related databases are dependent primarily on curation by experts, making it laborious to annotate the exponentially accumulated information on lncRNAs, which inevitably requires collective efforts in community-based curation of lncRNAs. Unlike existing databases, lncRNAWiki features comprehensive integration of information on human lncRNAs obtained from multiple different resources and allows not only existing lncRNAs to be edited, updated and curated by different users but also the addition of newly identified lncRNAs by any user. It harnesses community collective knowledge in collecting, editing and annotating human lncRNAs and rewards community-curated efforts by providing explicit authorship based on quantified contributions. LncRNAWiki relies on the underling knowledge of scientific community for collective and collaborative curation of human lncRNAs and thus has the potential to serve as an up-to-date and comprehensive knowledgebase for human lncRNAs.
INTRODUCTION
In mammals, a small fraction of the genome (e.g. in human) is transcribed into messenger RNAs, whereas the most represents a transcribed ‘dark matter’ that does not encode for proteins. Among them, long non-coding RNAs (lncRNAs) are prevalently transcribed from mammalian genomes and are present in large amounts in mammalian cells (1–4). Evidence has accumulated that lncRNAs play significant roles in numerous fundamental biological processes such as transcription, translation, cell cycle, imprinting, splicing and protein localization (5–7) and are highly implicated in cancer progression (8–14) and development of many other human diseases such as mendelian disorders, cardiovascular diseases and neurological disorders (14–17).
Advances in studies of non-coding RNA and consequently the increasing number of lncRNAs identified have resulted in the development of several lncRNA-related databases. Among them, GENCODE, which aims at annotating all functional elements in the human genome (4), has made a comprehensive annotation of gene structure (gene loci, transcript loci, exon number and splicing boundary) of 23 898 human lncRNA transcripts (Version 19). NONCODE (18) collects 95 135 lncRNA transcripts in human obtained from published literatures and databases (Version 4.0). LNCipedia (19) contains a total of 32 181 human lncRNA transcripts and incorporates related statistics such as protein-coding potential, secondary structure information and microRNA binding sites (Version 2.1). lncRNAdb (20) focuses on collecting function annotations based on the published literatures and, to date, only about 200 lncRNAs have been included in lncRNAdb. Another database, Rfam, centers on non-coding RNA families and thus does not provide specialized information for an individual lncRNA (21). It can be seen that although these existing databases offer valuable information on different aspects and different level of coverage of the lncRNA universe, there is a lack of a dedicated database for human lncRNAs that provides lncRNA transcript details coupled with the capability to make data update and curation in a smooth and easy way. Specifically, existing databases are most dependent on expert curation and thus laborious to comprehensively update the fast growing number of newly discovered lncRNAs. Wikipedia (http://www.wikipedia.org), an online encyclopedia, is an extraordinarily successful example that relies on the community knowledge in information integration and allows people from all over the world to create/edit any content. Wikipedia features collaborative information integration, huge coverage, up-to-date content as well as low-maintenance cost. Many attempts have been made in application of wiki for biological data integration (22–25). For example, Rfam, dedicated to RNA families, has adopted the wiki technology for community curation. According to its name, LNCipedia looks like a wiki resource, but in fact it is not fully open to the scientific community for data provision/edit. Considering the exponentially accumulated volume of lncRNAs, it is desirable to exploit the knowledge of the broad scientific community for collaborative integration and curation of lncRNA information (26–30).
Here we present our developed lncRNAWiki (http://lncrna.big.ac.cn). This platform is wiki-based and open-content, publicly editable and aimed at community curation of human lncRNAs. Unlike existing relevant databases, lncRNAWiki features comprehensive integration of information on human lncRNAs, cataloging 105 255 non-redundant lncRNA transcripts obtained from multiple different resources. Moreover, it harnesses collective knowledge for collecting, editing and annotating information on human lncRNAs and rewards community-curated efforts by quantifying contributions of users and providing explicit authorship based on their quantified contributions, aiming to exploit the knowledge of broad scientific community in addressing collectively collaborative curation of human lncRNAs. Therefore, lncRNAWiki has the potential to serve as an up-to-date and comprehensive knowledgebase for human lncRNAs.
IMPLEMENTATION
LncRNAWiki is built based on MediaWiki version 1.19.1 (http://www.mediawiki.org), which is an open source wiki engine, MySQL version 5.1.58 (http://www.mysql.org) a popular and free relational database management system, and PHP version 5.2.17 (http://www.php.net), which is a scripting language. These were implemented on a Red Hat Enterprise Linux Server. In order to make lncRNAWiki more attractive for participants from the broader scientific community in tasks of collaborative curation of lncRNAs, we installed ‘AuthorReward’ (http://www.mediawiki.org/wiki/Extension:AuthorReward), an extension to MediaWiki that allows for obtaining customized functionalities (31). ‘AuthorReward’ quantifies participants’ contribution considering both edit quality and edit quantity, and provides explicit authorship based on these quantified contributions. It has been successfully demonstrated in RiceWiki (32), where it has attracted more than 800 participants in collaborative curation of ∼600 genes. All extensions and software implemented in lncRNAWiki are accessible at http://lncrna.big.ac.cn/index.php/Special:Version. We also integrated JBrowse (version 1.11.4) (33,34) into lncRNAWiki to facilitate visualization of the genomic context and transcript structure for each lncRNA.
We integrated lncRNA sequences and annotation information (e.g. genomic location, transcript structure) from three data sources: GENCODE (version 19; 23 898 human lncRNA transcripts), NONCODE (version 4.0; 95 135 human lncRNA transcripts) and LNCipedia (version 2.1; 32 181 human lncRNA transcripts). A process of error and redundancy elimination was performed on the integrated data set. First, we removed sequences containing ‘N’ in each data source, and as a result, a total of eight lncRNAs in LNCipedia were removed. Second, we excluded lncRNAs with ambiguous naming scheme; in each data source, two or more lncRNA transcripts having 100% sequence identity on the whole transcript length (based on blastn results) and occupying the same genomic location but having different IDs are considered as questionable lncRNAs. Consequently, 14, 20 and eight lncRNAs were removed from GENCODE, NONCODE and LNCipedia, respectively. Lastly, since different databases may have different naming schemes and a given lncRNA transcript may accordingly have different identifiers in different databases, we performed blastn across these three data sources. LncRNA transcripts having 100% sequence identity (based on blastn results) and occupying the same genomic location were regarded as the same lncRNA. Finally, we obtained a total of 105 255 non-redundant lncRNA transcripts (Figure 1). We also blasted these 105 255 lncRNAs against lncRNA sequences in lncRNAdb (223 lncRNAs in total as of July 21, 2014) and found only 103 lncRNAs have been functionally annotated (Supplementary Table S1), indicating that a large number of human lncRNAs are poorly annotated and need a platform for community annotation of lncRNAs.
Figure 1.
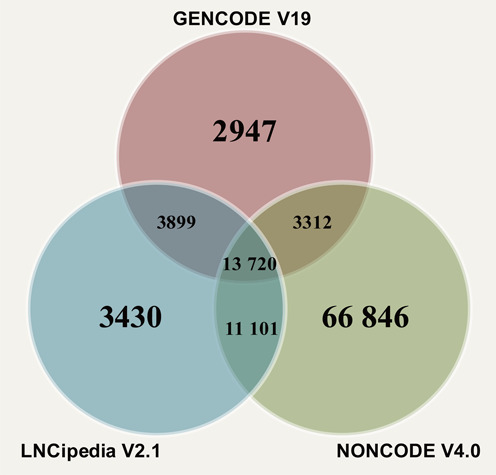
Data source of lncRNAWiki.
Based on our previous study (7) and categories of Derrien et al. (35), we classified the 105 255 non-redundant lncRNA transcripts into seven groups according to their genomic location in respect to protein-coding genes, viz., Intergenic, Intronic (S), Intronic (AS), Overlapping (S), Overlapping (AS), Sense and Antisense (Figure 2). The difference between our classification and Derrien's (35) is that we classified lncRNAs that intersect protein-coding genes into Sense or Antisense by considering the whole transcript sequence instead of exonic region only.
Figure 2.
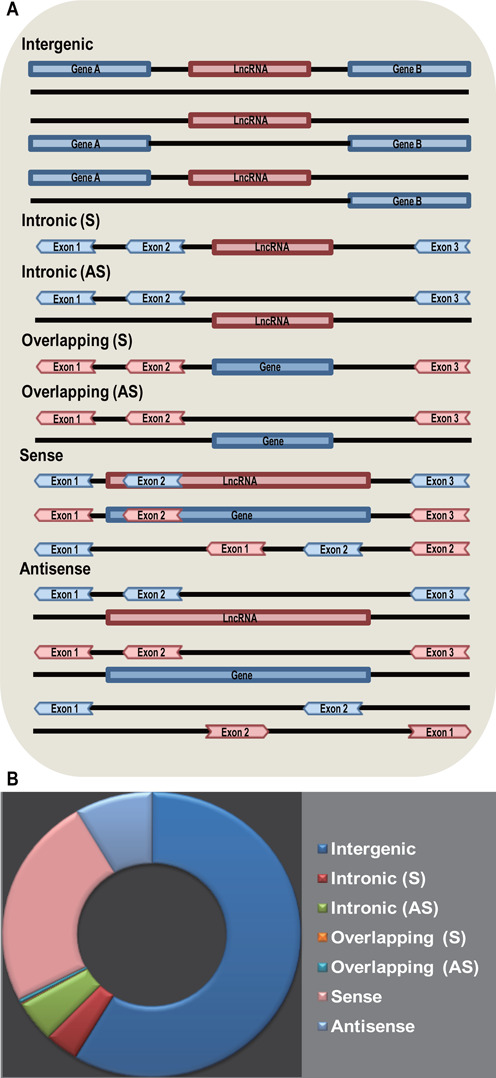
Classification of lncRNAs based on their genomic location in respect of protein-coding genes. (A) lncRNA categories. Intergenic: lncRNAs are transcribed from intergenic regions; Intronic (S): lncRNAs are transcribed entirely from introns of protein-coding genes; Intronic (AS): lncRNAs are transcribed from antisense strand of protein-coding genes and the entire sequences are covered by introns of protein-coding genes; Overlapping (S): lncRNAs that contain coding genes within an intron on the sense strand; Overlapping (AS): lncRNAs that contain coding genes within an intron on the antisense strand; Sense: lncRNAs are transcribed from the sense strand of protein-coding genes and the entire sequence of lncRNAs are covered by protein-coding genes (Intronic lncRNAs are not included), or the entire sequence of protein-coding genes are covered by lncRNAs (Overlapping lncRNAs are not included), or both lncRNAs and protein-coding genes intersect each other partially; Antisense: lncRNAs are transcribed from the antisense strand of protein-coding genes and the entire sequence of lncRNAs are covered by protein-coding genes (Intronic lncRNAs are not included), or the entire sequence of protein-coding genes are covered by lncRNAs (Overlapping lncRNAs are not included), or both lncRNAs and protein-coding genes intersect each other partially. (B) Proportions of human lncRNAs in different categories.
DATABASE CONTENT
The central entities of lncRNAWiki are human lncRNA transcripts. Thus, each transcript has a corresponding wiki page (Figure 3). Meanwhile, transcripts are grouped together by classification categories, as well as by genes. This information can be accessed at the homepage and at the bottom of the transcript page. Transcript-specific pages are generated based on transcript identifiers. The content of every transcript in lncRNAWiki is structured into two parts: ‘user-edit’ part and ‘Basic Information’ part. The ‘user-edit’ part allows users to add or delete annotations. On the contrary, the ‘Basic Information’ portion is organized as a table and will be regularly updated by the lncRNAWiki team based on the annotation information integrated from multiple different lncRNA-associated sources. The lncRNA information in lncRNAWiki was seeded from GENCODE, NONCODE and LNCipedia, yielding a comprehensive integration of 105 255 non-redundant lncRNA transcripts (Figure 1).
Figure 3.

Screenshots of the lncRNAWiki page for the lncRNA transcript MALAT-1, available at http://lncrna.big.ac.cn/index.php/ENST00000534336.1. This lncRNA was collaboratively curated by four researchers, yielding 125 versions as of October11, 2014. (A) Whole page, containing multiple different sections. (B) Brief authorship information in reward for community curation and a one-sentence summary for the description of this lncRNA. (C) Annotated information. It is organized as free text and links to several sub-sections. (D) Labs working on this lncRNA. It is a list of laboratories working on this lncRNA derived from references and provided by the community. (E) References, which are automatically generated and formatted with the help of the ‘Cite’ extension. (F) Basic Information. It is organized structurally in the form of a table, including transcript ID, source, transcript structure, genome context, sequence information, etc. (G) Authorship details generated by the ‘AuthorReward’ extension that quantifies researchers’ contributions and provides explicit authorship according to their quantitative contributions. The cutoff core for awarding authorship is configurable and set to 1 (by default) in lncRNAWiki.
‘Basic Information’ provides users with the basic details of lncRNA such as genomic location, transcript structure and sequence. There are 10 sub-sections in ‘Basic Information’, including ‘Transcript ID’, ‘Source’, ‘Same with’, ‘Classification’, ‘Length’, ‘Genomic location’, ‘Exon number’, ‘Exons’, ‘Genome context’ and ‘Sequence’. ‘Transcript ID’ refers to the lncRNA ID in the data source. ‘Source’ indicates the source database, as well as its version, from which this lncRNA is obtained. ‘Same with’ provides IDs of lncRNAs that are considered to be the same entry in other lncRNA sequence databases. Taking into account that the genomic context of lncRNAs may offer insights into their function, the classification of lncRNAs based on genomic location is of great biological significance in in-depth mining and analysis (7). We classified lncRNAs into seven categories considering their genomic location in respect of protein-coding genes, i.e. Sense, Antisense, Overlapping (S), Overlapping (AS), Intergenic, Intronic (S) and Intronic (AS) (Figure 2A). According to the present data set, it is shown that the majority of human lncRNAs belong to the categories of Intergenic (59.2%) and Sense (24.4%) (Figure 2B). The sub-section ‘Genome context’ facilitates visual inspection of the transcript structure and genomic context.
The ‘user-edit’ part includes three sections: ‘Annotated Information’, ‘Labs working on this lncRNA’ and ‘References’ (Figure 3). ‘Annotated Information’ is represented in a form of free text. It is helpful for users who do not have training in wiki techniques or curation to contribute edits and share knowledge, which simplifies editing significantly and lowers technological requirements for participation in curation of a wider community. ‘Annotated Information’ links to several sub-sections including ‘Function’, ‘Expression’, ‘Regulation’, ‘Diseases’ and ‘Evolution’, providing convenience of directing users to other sub-section(s) of interest. Considering the importance and the necessity of lncRNA nomenclature, we added the sub-section of ‘Transcriptomic Nomeclature’. As most of the lncRNAs have not been functionally studied, we named lncRNAs based on their biological features such as genomic location, alternative splicing and expression level by basically following the rules of HGNC (36) (Figure 3). Users can add new sub-sections if necessary, while those that are irrelevant can be deleted. This enables intuitive editing of the information through an ‘edit’ link available to each sub-section and, moreover, can be made by using application programming interface for automatic entry of information. ‘Labs working on this lncRNA’ contains a worldwide list of laboratories that work on this lncRNA, thus facilitating collaboration and interaction in curation of this lncRNA. ‘References’ provides publications related to the lncRNA and they are automatically formatted using the ‘Cite’ extension (http://www.mediawiki.org/wiki/Extension:Cite).
In lncRNAWiki, community-curated efforts are quantified and rewarded by explicit authorship, aiming to encourage more participants from the wider scientific community in collective and collaborative curation of lncRNAs. In any given lncRNA page, curation efforts for all participated contributors are quantified as contribution score which evaluates both quality and quantity of edits, and consequently, authorship is awarded to any contributor from the scientific community whose contribution score is greater than a cutoff score (by default, it is 1) (Figure 3). Each page at the top displays the brief authorship information, including contributor name(s), lncRNA ID, hyperlink to this lncRNA and last update time. The detailed authorship information is presented at the bottom of each page. This information includes a pie chart to depict contribution scores for all involved contributors, edit quality and quantity for each contributor are illustrated by a histogram, and contributor names are listed in a table with contribution score, edit count, edit quality, edit quantity, last edit time and edit details.
In addition, when a newly identified human lncRNA is reported, any user can create a new page to add specific information for this lncRNA, enabling lncRNAWiki to become an up-to-date and comprehensive knowledgebase for human lncRNAs. For example, we added to lncRNAWiki (http://lncrna.big.ac.cn/index.php/LUNAR1) LUNAR1, a recently discovered human lncRNA, immediately after the relevant paper was published online. LUNAR1 is Notch-regulated and it enhances mRNA expression of IGF1R (insulin-like growth factor receptor 1) aiming at maintenance of human T-cell acute lymphoblastic leukemia (T-ALL) (37).
DISCUSSION
LncRNAWiki is a wiki-based database dedicated for human lncRNAs, comprehensively integrating information on human lncRNAs from multiple different resources and exploiting the wide scientific community to collect, edit and annotate human lncRNAs. It allows not only existing lncRNAs to be edited, updated and curated by different users but also newly identified lncRNAs to be added by any user. As the number of lncRNAs grows fast and is contrasted by the small number of expert curators focused on lncRNA, lncRNAWiki has the potential to serve as an up-to-date and comprehensive knowledgebase for human lncRNAs. It should be noted, however, that lncRNAWiki does not aim to replace traditionally expert-curated databases, but represents their important complement. LncRNAWiki relies on community intelligence to curate a wide range of lncRNA-related topics, which thus can significantly reduce efforts and time of expert curators. With explicit authorship as a reward for community curation, lncRNAWiki bears the promise to attract more people (especially field experts) to share their expertise and to provide edits on lncRNAs of their interest. In addition, it is of great significance for authors of recent publications to curate their newly reported lncRNAs and to submit their functional descriptions to lncRNAWiki, facilitating information dissemination and maximizing the scope of knowledge sharing. Together, based on community curation, lncRNAWiki has a potential to grow into an lncRNA encyclopedia by the community, of the community and for the community. Future directions for lncRNAWiki include integrating more types of data in the section of ‘Basic information’ (e.g. expression level, tissue-specific expression, orthologs) from different sources and improving links to existing relevant databases, such as the lncRNA-associated disease database LncRNADisease (38) and lncRNA expression database NRED (39). To facilitate the detection and annotation of lncRNAs, we will also integrate analysis tools into lncRNAWiki for lncRNA detection and classification and employ automatic text mining in aid of lncRNA-related literature curation.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
We thank anonymous reviewers for their valuable comments and members of the Zhang Lab for reporting bugs and sending comments.
Footnotes
The authors wish it to be known that, in their opinion, the first two authors should be regarded as Joint First Authors.
FUNDING
Strategic Priority Research Program of the Chinese Academy of Sciences [XDB13040500 to Z.Z.]; National Natural Science Foundation of China [31200978 to L.M.]; Base Research Fund of King Abdullah University of Science and Technology [to V.B.B.]; ‘100-Talent Program’ of Chinese Academy of Sciences [Y1SLXb1365 to Z.Z.]. Funding for open access charge: Strategic Priority Research Program of the Chinese Academy of Sciences [XDB13040000].
Conflict of interest statement. None declared.
REFERENCES
- 1.Okazaki Y., Furuno M., Kasukawa T., Adachi J., Bono H., Kondo S., Nikaido I., Osato N., Saito R., Suzuki H., et al. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature. 2002;420:563–573. doi: 10.1038/nature01266. [DOI] [PubMed] [Google Scholar]
- 2.Carninci P., Kasukawa T., Katayama S., Gough J., Frith M.C., Maeda N., Oyama R., Ravasi T., Lenhard B., Wells C., et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 3.Johnson J.M., Edwards S., Shoemaker D., Schadt E.E. Dark matter in the genome: evidence of widespread transcription detected by microarray tiling experiments. Trends Genet. 2005;21:93–102. doi: 10.1016/j.tig.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 4.Harrow J., Frankish A., Gonzalez J.M., Tapanari E., Diekhans M., Kokocinski F., Aken B.L., Barrell D., Zadissa A., Searle S., et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clark M.B., Mattick J.S. Long noncoding RNAs in cell biology. Semin. Cell Dev. Biol. 2011;22:366–376. doi: 10.1016/j.semcdb.2011.01.001. [DOI] [PubMed] [Google Scholar]
- 6.Ponting C.P., Oliver P.L., Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 7.Ma L., Bajic V.B., Zhang Z. On the classification of long non-coding RNAs. RNA Biol. 2013;10:925–933. doi: 10.4161/rna.24604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Perez D.S., Hoage T.R., Pritchett J.R., Ducharme-Smith A.L., Halling M.L., Ganapathiraju S.C., Streng P.S., Smith D.I. Long, abundantly expressed non-coding transcripts are altered in cancer. Hum. Mol. Genet. 2008;17:642–655. doi: 10.1093/hmg/ddm336. [DOI] [PubMed] [Google Scholar]
- 9.Huarte M., Rinn J.L. Large non-coding RNAs: missing links in cancer. Hum. Mol. Genet. 2010;19:R152–R161. doi: 10.1093/hmg/ddq353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spizzo R., Almeida M.I., Colombatti A., Calin G.A. Long non-coding RNAs and cancer: a new frontier of translational research. Oncogene. 2012;31:4577–4587. doi: 10.1038/onc.2011.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tano K., Akimitsu N. Long non-coding RNAs in cancer progression. Front. Genet. 2012;3:219. doi: 10.3389/fgene.2012.00219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gibb E.A., Brown C.J., Lam W.L. The functional role of long non-coding RNA in human carcinomas. Mol. Cancer. 2011;10:38. doi: 10.1186/1476-4598-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsai M.C., Spitale R.C., Chang H.Y. Long intergenic noncoding RNAs: new links in cancer progression. Cancer Res. 2011;71:3–7. doi: 10.1158/0008-5472.CAN-10-2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maass P.G., Luft F.C., Bahring S. Long non-coding RNA in health and disease. J. Mol. Med. 2014;92:337–346. doi: 10.1007/s00109-014-1131-8. [DOI] [PubMed] [Google Scholar]
- 15.Qureshi I.A., Mattick J.S., Mehler M.F. Long non-coding RNAs in nervous system function and disease. Brain Res. 2010;1338:20–35. doi: 10.1016/j.brainres.2010.03.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wapinski O., Chang H.Y. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21:354–361. doi: 10.1016/j.tcb.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 17.Johnson R. Long non-coding RNAs in Huntington's disease neurodegeneration. Neurobiol. Dis. 2011;46:245–254. doi: 10.1016/j.nbd.2011.12.006. [DOI] [PubMed] [Google Scholar]
- 18.Xie C., Yuan J., Li H., Li M., Zhao G., Bu D., Zhu W., Wu W., Chen R., Zhao Y. NONCODEv4: exploring the world of long non-coding RNA genes. Nucleic Acids Res. 2014;42:D98–D103. doi: 10.1093/nar/gkt1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Volders P.J., Helsens K., Wang X., Menten B., Martens L., Gevaert K., Vandesompele J., Mestdagh P. LNCipedia: a database for annotated human lncRNA transcript sequences and structures. Nucleic Acids Res. 2012;41:D246–D251. doi: 10.1093/nar/gks915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Amaral P.P., Clark M.B., Gascoigne D.K., Dinger M.E., Mattick J.S. lncRNAdb: a reference database for long noncoding RNAs. Nucleic Acids Res. 2010;39:D146–D151. doi: 10.1093/nar/gkq1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burge S.W., Daub J., Eberhardt R., Tate J., Barquist L., Nawrocki E.P., Eddy S.R., Gardner P.P., Bateman A. Rfam 11.0: 10 years of RNA families. Nucleic Acids Res. 2013;41:D226–D232. doi: 10.1093/nar/gks1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McIntosh B.K., Renfro D.P., Knapp G.S., Lairikyengbam C.R., Liles N.M., Niu L., Supak A.M., Venkatraman A., Zweifel A.E., Siegele D.A., et al. EcoliWiki: a wiki-based community resource for Escherichia coli. Nucleic Acids Res. 2012;40:D1270–D1277. doi: 10.1093/nar/gkr880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Good B.M., Clarke E.L., de Alfaro L., Su A.I. The Gene Wiki in 2011: community intelligence applied to human gene annotation. Nucleic Acids Res. 2012;40:D1255–D1261. doi: 10.1093/nar/gkr925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stehr H., Duarte J.M., Lappe M., Bhak J., Bolser D.M. PDBWiki: added value through community annotation of the Protein Data Bank. Database. 2010;2010:baq009. doi: 10.1093/database/baq009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Z., Sang J., Ma L., Wu G., Wu H., Huang D., Zou D., Liu S., Li A., Hao L., et al. RiceWiki: a wiki-based database for community curation of rice genes. Nucleic Acids Res. 2014;42:D1222–D1228. doi: 10.1093/nar/gkt926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Salzberg S.L. Genome re-annotation: a wiki solution. Genome Biol. 2007;8:102. doi: 10.1186/gb-2007-8-1-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hu J.C., Aramayo R., Bolser D., Conway T., Elsik C.G., Gribskov M., Kelder T., Kihara D., Knight T.F., Jr., Pico A.R., et al. The emerging world of wikis. Science. 2008;320:1289–1290. doi: 10.1126/science.320.5881.1289b. [DOI] [PubMed] [Google Scholar]
- 28.Howe D., Costanzo M., Fey P., Gojobori T., Hannick L., Hide W., Hill D.P., Kania R., Schaeffer M., St Pierre S., et al. Big data: the future of biocuration. Nature. 2008;455:47–50. doi: 10.1038/455047a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Finn R.D., Gardner P.P., Bateman A. Making your database available through Wikipedia: the pros and cons. Nucleic Acids Res. 2011;40:D9–D12. doi: 10.1093/nar/gkr1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Z., Cheung K.H., Townsend J.P. Bringing Web 2.0 to bioinformatics. Brief. Bioinform. 2009;10:1–10. doi: 10.1093/bib/bbn041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dai L., Tian M., Wu J., Xiao J., Wang X., Townsend J.P., Zhang Z. AuthorReward: increasing community curation in biological knowledge wikis through automated authorship quantification. Bioinformatics. 2013;29:1837–1839. doi: 10.1093/bioinformatics/btt284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Z., Sang J., Ma L., Wu G., Wu H., Huang D., Zou D., Liu S., Li A., Hao L., et al. RiceWiki: a wiki-based database for community curation of rice genes. Nucleic Acids Res. 2013;42:D1222–D1228. doi: 10.1093/nar/gkt926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Skinner M.E., Uzilov A.V., Stein L.D., Mungall C.J., Holmes I.H. JBrowse: a next-generation genome browser. Genome Res. 2009;19:1630–1638. doi: 10.1101/gr.094607.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Skinner M.E., Holmes I.H. Setting up the JBrowse genome browser. Curr. Protoc. Bioinform. 2010 doi: 10.1002/0471250953.bi0913s32. doi:10.1002/0471250953.bi0913s32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Derrien T., Johnson R., Bussotti G., Tanzer A., Djebali S., Tilgner H., Guernec G., Martin D., Merkel A., Knowles D.G., et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wright M.W. A short guide to long non-coding RNA gene nomenclature. Hum. Genomics. 2014;8:7. doi: 10.1186/1479-7364-8-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trimarchi T., Bilal E., Ntziachristos P., Fabbri G., Dalla-Favera R., Tsirigos A., Aifantis I. Genome-wide mapping and characterization of Notch-regulated long noncoding RNAs in acute leukemia. Cell. 2014;158:593–606. doi: 10.1016/j.cell.2014.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen G., Wang Z., Wang D., Qiu C., Liu M., Chen X., Zhang Q., Yan G., Cui Q. LncRNADisease: a database for long-non-coding RNA-associated diseases. Nucleic Acids Res. 2013;41:D983–D986. doi: 10.1093/nar/gks1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dinger M.E., Pang K.C., Mercer T.R., Crowe M.L., Grimmond S.M., Mattick J.S. NRED: a database of long noncoding RNA expression. Nucleic Acids Res. 2009;37:D122–D126. doi: 10.1093/nar/gkn617. [DOI] [PMC free article] [PubMed] [Google Scholar]


