Abstract
Gathering information about associations between methylated genes and diseases is important for diseases diagnosis and treatment decisions. Recent advancements in epigenetics research allow for large-scale discoveries of associations of genes methylated in diseases in different species. Searching manually for such information is not easy, as it is scattered across a large number of electronic publications and repositories. Therefore, we developed DDMGD database (http://www.cbrc.kaust.edu.sa/ddmgd/) to provide a comprehensive repository of information related to genes methylated in diseases that can be found through text mining. DDMGD's scope is not limited to a particular group of genes, diseases or species. Using the text mining system DEMGD we developed earlier and additional post-processing, we extracted associations of genes methylated in different diseases from PubMed Central articles and PubMed abstracts. The accuracy of extracted associations is 82% as estimated on 2500 hand-curated entries. DDMGD provides a user-friendly interface facilitating retrieval of these associations ranked according to confidence scores. Submission of new associations to DDMGD is provided. A comparison analysis of DDMGD with several other databases focused on genes methylated in diseases shows that DDMGD is comprehensive and includes most of the recent information on genes methylated in diseases.
INTRODUCTION
Epigenetics is the study of changes in genes expression and its relation to specific phenotypes using mechanisms that do not modify the underlying DNA sequence (1). DNA methylation is one of the common epigenetic modifications in eukaryotic organisms (2). In humans, DNA methylation has been found associated with many diseases, such as Beckwith–Wiedemann and Silver–Russell syndromes, type 2 diabetes, schizophrenia and autoimmune disease, as well as various cancers (3). In addition to humans, DNA methylation has been studied in other species such as in plants (4), drosophila (5) and mice (6) in association with diseases or other phenotypes.
Advancements in quantitative assessment of DNA methylation technologies facilitated large-scale studies of genes methylated in diseases (7). However, the results of such studies are scattered in a large number of publications and across several specialized databases, and thus it is difficult to search for it manually. Therefore, it is useful to develop efficient and accurate systems to facilitate extracting information on genes methylated in diseases in an automatic fashion. Recently, two methods were developed for automatic extraction of information on genes methylated in diseases from textual documents: one is part of MeInfoText 2.0 database (8) and the other one is DEMGD (9). MeInfoText 2.0 is used to extract such associations between methylated human genes and cancers from PubMed (http://www.ncbi.nlm.nih.gov/pubmed) abstracts using two maximum entropy models (8). DEMGD (Dragon Extractor of Methylated Genes and Diseases), which we developed earlier, extracts associations between methylated human genes and any disease from any free text submitted by users (9). DEMGD uses two machine learning models based on document-term matrix and position weight matrix characterization of text (9). A detailed description of machine learning models development can be found in (9), and the DEMGD system is available for online text mining at: www.cbrc.kaust.edu.sa/demgd/.
Some manually curated databases that focused on genes methylated in diseases are limited in scope, e.g. MethyCancer (10) relates only to cancers, MethylomeDB (11) contains genes methylated only in brain tissues, while some other databases contain information only for specific species, such as DiseaseMeth (12) with information only for humans. Moreover, some databases used limited data sources, such as PubMeth (13) that includes information extracted only from PubMed abstracts. The least restricted of these databases is MethDB (14,15) that aims to include manually curated information on genes methylated in any diseases and from any species. On the other hand, automated compilation of information allows for broader coverage of processed text and makes updating relevant information easier. So far, MeInfoText 2.0 has been the only available automatically compiled database, but it is restricted to methylated human genes in cancers and to information extracted from PubMed abstracts (8).
The goal of our study is to contribute a resource to support epigenetic research and biomedical community by providing a comprehensive repository for sharing text-mined information from both abstracts and full-text articles about methylation of genes in any disease and not restricted to specific species. We used the DEMGD text mining system (9) to extract the associations from PubMed Central full-text articles and PubMed abstracts. This text is processed and used to populate DDMGD database. This resulted in a comprehensive database of automatically compiled associations of genes methylated in diseases collected from many species. The integrated text mining system, the broad coverage of DDMGD, the high accuracy of extracted information and various implemented features in the friendly web-interface make this database useful and unique. DDMGD is free for academic and non-profit use and can be accessed at: www.cbrc.kaust.edu.sa/ddmgd/.
MATERIALS AND METHODS
System structure
Figure 1 shows the structure of the system that comprises four main components: Data Sources, DEMGD text mining system, DDMGD database and DDMGD graphical user interface (GUI). Here we provide a brief description of the system. More details are available in the Supplementary material.
Figure 1.
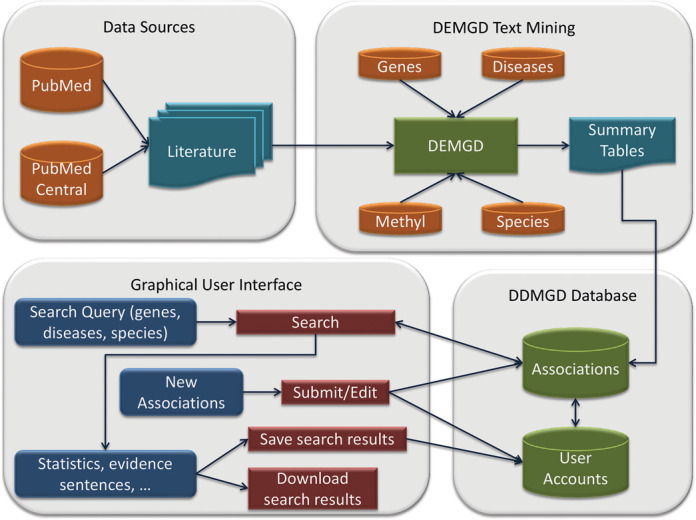
DDMGD system structure overview. The system consists of four main components: (i) free text sources, (ii) DEMGD text mining system, (iii) DDMGD database (iv) DDMGD graphical user interface.
Data sources
We used scientific literature (PubMed and Open Access Subset (OAS) of PubMed Central (http://www.ncbi.nlm.nih.gov/pmc/)) as the main sources of information about genes methylated in diseases in any species. We searched OAS and found 23 572 full-text articles related to genes methylated in diseases. In addition to these, we found 27 395 abstracts from PubMed, which are related to genes methylated in diseases but were different from the abstracts of the full-text articles in the analyzed OAS.
DEMGD text mining system
DEMGD is a text mining system that can extract in its original version (9) associations between methylated human genes and diseases from any free text without restriction to specific diseases. For the purpose of this study, we extended DEMGD to extract associations between genes methylated in diseases from various species as follows:
The gene dictionary from (9) is extended using NCBI Gene Database (16) to include genes from any species.
The disease dictionary from (9) is extended using various resources (see the Supplementary material) to include diseases from any species.
We included a species dictionary from NCBI Taxonomy Database (17) in addition to common names and synonyms of the species used in MethDB.
- We implemented three post-processing steps performed after an association between a methylated gene and a disease is extracted from a sentence. These steps are:
- Extraction of species names.
- Extract gene expression and disease progression information using pattern matching rules.
- Filter out false positive associations (entries that are wrongly predicted to be associations) using filtering rules.
After the associations were extracted, we manually curated and removed ambiguous extracted genes, diseases and species from the database. Additionally, we manually assessed information from 1000 entries (500 predicted to be associations and 500 predicted to be non-associations) relative to the sentences from which these entries were extracted. These 1000 entries we name SET1 (SET1 is available at: www.cbrc.kaust.edu.sa/ddmgd/download.php). We used SET1 to identify wrongly extracted genes, diseases and species and they were removed from their corresponding dictionaries. We also identified genes, diseases and species that were mentioned in sentences, but were not extracted by the text mining system, and we added them to their corresponding dictionaries. Then, we re-processed the full-text articles and the abstracts using the modified dictionaries and post-processing filtering rules.
DDMGD database
Once the associations are extracted, DEMGD organizes the associations in summary tables that include names/symbols of gene, gene ID, disease, disease ID, methylation words, species, species ID, gene expression, disease progression, evidence sentences that include the association, PubMed Central ID of the article where the sentence is mentioned, PubMed ID of the abstract where the association is mentioned and a confidence score generated by the DEMGD system. The confidence score, which ranges from 0 to 1, is given by the DEMGD classification model, and it indicates the likelihood that the gene found in the sentence is methylated in the disease found in the same sentence. A confidence score of ‘1’ suggests the highest confidence that an association exists between the methylated gene and the disease, while the smaller the confidence score, the less likely it is that the association exists. Moreover, the database stores users’ accounts if users want to have an account (more details are available in the next section).
We used Gene IDs, disease IDs and species IDs to allow normalizing search queries that involve genes, diseases and species, respectively. For example, if the user restricts search to a specific disease (e.g. breast cancer), the user will get results for all diseases in our database that have the same disease ID as the selected disease (e.g. breast tumor, breast carcinoma and breast tumors). We used NCBI Taxonomy Database, NCBI Gene Database and Comparative Toxicogenomics Database (18) for species IDs, genes IDs and diseases IDs, respectively.
DDMGD GUI
We provided a user-friendly web interface that facilitates searching, exploring information and updating the database. The instructions page (http://www.cbrc.kaust.edu.sa/ddmgd/instructions.php) is included for users who want to know more about the system usage and interpretation of search results. The web interface can be accessed from any web browser and provides the following features.
Search
Users can search using individual names/symbols of genes, diseases and/or species, in addition to unlimited batch searching using several genes, diseases and/or species at the same time. Users can either enter gene names in the text box or select from the list of genes. In addition, diseases are organized in a hierarchy to allow users to narrow down the list of diseases using disease category, disease type or disease sub-type. The diseases hierarchy was developed using the Comparative Toxicogenomics Database. If users want to retrieve associations for all genes, diseases and species contained in our database, users can click the submit button directly without selecting any gene, disease or species.
Statistics, graphs and additional information
DDMGD provides basic statistics about genes, diseases, species and associations. Figure 2 shows partial output of query ‘RASSF1A, lung cancer, human’. For example, for each gene, DDMGD lists the number of diseases and species with which the gene is associated, as well as the number of sentences, PubMed abstracts and PubMed Central articles that include the gene. DDMGD provides bar graphs that show the frequency of the associations found and downloadable connectivity graphs. The table at the bottom of the page allows the user to select associations of their interest to get more information such as color-highlighted evidence sentences and confidence scores (Figure 3). If a sentence contains associations between several genes, diseases and/or species, each association is stored as a separate record as shown in Figure 3.
Figure 2.
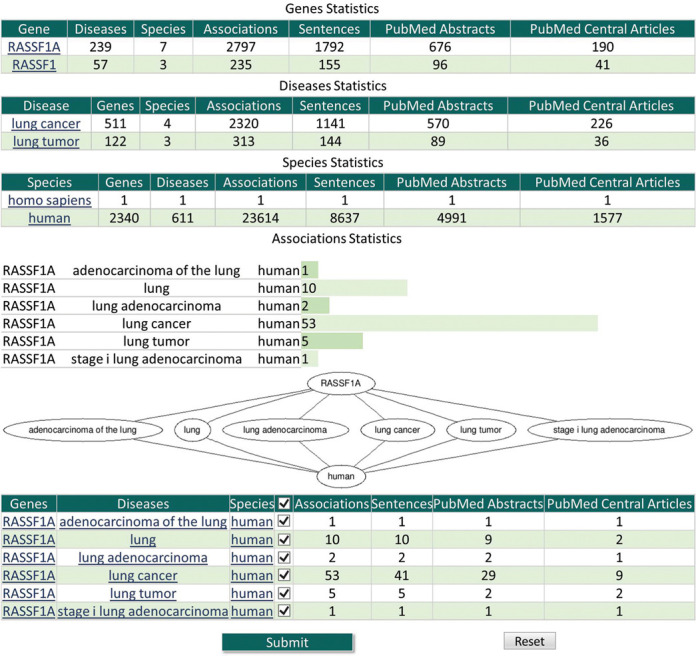
Example of search results. The system shows statistics about genes, diseases and species that were selected in users’ queries and the associations, links to other databases and graphs.
Figure 3.
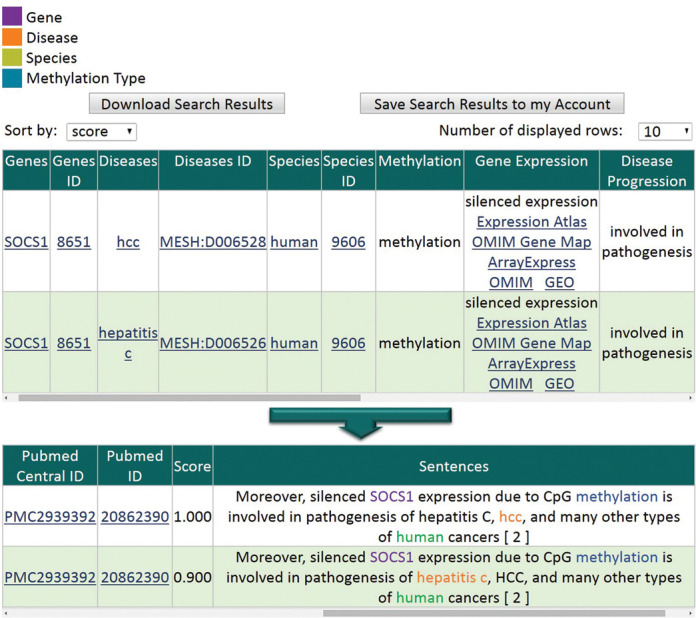
Details about associations. The system shows color-highlighted evidence sentences, gene expression, disease progression, confidence scores and links to other databases.
Gene expression and disease progression
Gene expression column provides associations between gene methylation and gene expression. For example, Figure 3 shows that gene methylation silences gene expression. However, disease progression column provides associations between gene methylation/expression and disease progression, pathogenesis and/or prognosis. Figure 3 shows that gene methylation/expression is involved in disease pathogenesis.
Links to other databases
Figures 2 and 3 show how DDMGD is linked to many databases to facilitate access to additional information. For each association, in addition to links to PubMed abstracts and PubMed Central articles from which the information is compiled, DDMGD provides links to NCBI Gene, Comparative Toxicogenomics Database and NCBI Taxonomy databases to provide more information about genes, diseases and species, respectively. In addition, we provide links to Expression Atlas (19), OMIM Gene Map (20), ArrayExpress (21), OMIM (20) and GEO (22) for additional gene expression information.
Sorting
Users can sort the search results using the gene, gene ID, disease, disease ID, methylation type, species, species ID, sentence, PubMed ID, PubMed Central ID or confidence score (Figure 3).
Download and save search results
Users can download the search results as a comma separated values (CSV) file. Alternatively, users can save the search results and the search queries in their accounts (Figure 3). Additionally, users can download the whole database from http://www.cbrc.kaust.edu.sa/ddmgd/download.php.
Submit new information
The scientific community can contribute to DDMGD, keep DDMGD up-to-date and expand it by submitting new information about genes methylated in diseases through the web interface. We provide a checking mechanism for the submitted data (see www.cbrc.kaust.edu.sa/ddmgd/instructions.php#Q4). Although a fully automated system and minimal human intervention is desirable, we believe that performing a periodic manual verification of the submitted data will ensure that the quality of the database's content remains consistent over time.
User accounts
Creating user accounts is optional. DDMGD allows users to manage, view and edit information they submitted, by creating private accounts. Also, users can save search queries and search results using their accounts. Searching the database and downloading the search results do not require accounts.
RESULTS AND DISCUSSION
In this section, we evaluate and compare the capabilities of DDMGD with existing databases. Definitions of evaluation metrics are available in the Supplementary material.
Performance of association extraction and named entity recognition
It is shown in (9) that the original version of DEMGD achieved accuracy and F-score of 83.5 and 84.7%, respectively, as assessed based on 10-fold cross-validation on a manually curated data set that includes ∼3200 associations. Additionally, we obtained the accuracy and F-score of 87.5 and 88.7%, respectively, on an independent testing set that contains 200 associations (9). However, it is reported in (8) that the MeInfoText 2.0 text mining system achieved Precision-Recall for two machine learning models of 94.7–90.1% and 91.8–90%, respectively. However, we discussed in (9) that such results are incomparable to DEMGD for the following reasons.
Differences in the scopes of the text mining systems: DEMGD extracts associations from all diseases, but MeInfoText is limited to cancer.
Differences in the test sets: The test set that the authors of MeInfoText 2.0 used to evaluate their text mining system was not available during the time of our study. So we could not compare their text mining system with DEMGD using the same test sets.
The text mining system used to compile MeInfoText 2.0 was not available during our study, so we could not compare it with DEMGD using our test sets.
In this study, we evaluated the accuracy of named entity recognition (NER) and extracted associations as used for population of DDMGD. This is done because the dictionaries are changed, and we also implemented additional post-processing. To evaluate this, we used another set, SET2, made up of randomly selected 2500 entries from DDMGD that include 1250 entries predicted as associations and 1250 entries predicted as non-associations. These 2500 entries were different from entries that made SET1 used in the DEMGD Text Mining System section (SET 2 test data are available at: www.cbrc.kaust.edu.sa/ddmgd/download.php). Note that we did not make any change to the DEMGD models from (9).
With the new dictionaries, DEMGD achieved Precision-Recall-F-score of 94.06–98.55–96.25%, 97.59–97.93–97.76%, 100.00–100.00–100.00% and 100.00–100.00–100.00% for NER of gene, disease, species and methylation words, respectively. The high performance of NER demonstrates the good quality of the dictionaries we used. Additionally, the high performance achieved using the species and methylation words dictionaries can be explained by minimal ambiguities occurring in these terms as compared to the genes and diseases names. For the extracted associations DEMDG with the post-processing achieved Precision, Recall, Specificity, F-score and Accuracy of 81.85, 81.92, 82.43, 81.89 and 82.18%, respectively. Although the accuracy of extracted associations obtained using a manually curated large data set of 2500 entries is high, it only serves as an assessment of the quality of information in our database.
Evaluation of database's content
As benchmark lists, we used three published lists about DNA methylation. Each benchmark list has different characteristics, which allows for evaluating DDMGD from different perspectives. The first benchmark list includes a set of 58 genes that are methylated in colorectal cancer (the genes are mentioned in a review article (Table 1 from (23))). First, we looked at how many genes were found in the database. Out of the 58 genes, DDMGD found 56. Then we looked for the genes in the benchmark list found to be methylated in colorectal cancer. DDMGD contains information on 56 out of 58 methylated genes in colorectal cancer.
The second benchmark list is a recent survey about genes methylated in various autoimmune diseases (Table 1 from (24)). The survey was published in 2013 and lists 14 genes that are associated with seven autoimmune diseases. There are 20 associations between the methylated genes and these diseases (24). DDMGD contains all 14 genes, all seven diseases and 15 out of the 20 associations. Finally, the third benchmark list is a recent list that was published in 2010, which includes genes methylated in various cancers in mice (Table 1 from (25)). The list contains 16 associations that include 12 genes, which are found to be associated with 10 types of cancers in mice. DDMGD contains nine genes, nine types of cancers and six associations in mice.
In summary, DDMGD was able to extract 94.04, 94.44 and 81.91% of genes, diseases and associations, respectively, from all the three benchmark lists combined. Although the genes, diseases and species dictionaries in DEMGD include all the genes, diseases and species in the benchmark lists, as much as we were able to ascertain, the genes or diseases that DDMGD could not recover were not mentioned together with the methylation words in the same sentences and this seems to be the reason why our system was not able to extract them. More details about the missing genes, diseases/cancer and associations from the three benchmark lists are in the Supplementary material.
Comparison of DDMGD with other databases
We did thorough comparison of the features of DDMGD and other databases that include information about genes methylated in diseases to highlight the current properties of DDMGD with respect to these databases. We compared nine databases with DDMGD using 23 features. Some of the features are general so that they can be applicable to all the databases. We used only features that can be objectively assessed for all databases. Details of the comparison are available in the Supplementary material. The comparison results are split into two tables. Supplementary Table S1(A) shows the comparison between DDMGD and DiseaseMeth, MeInfoText 2.0, MethDB and MethyCancer databases, while Supplementary Table S1(B) shows the comparison of DDMGD with MethylomeDB, NGSmethDB (26), PubMeth, MENT (27) and CMS (28) databases.
Additionally, we compared the content of DDMGD with the content of other relevant databases to demonstrate that DDMGD provides additional and more comprehensive information not provided by other resources. This also represents one way of evaluating the utility and quality of DDMGD. First, we compared DDMGD with another automatically compiled database, MeInfoText 2.0, using the first benchmark list because this is the same list that was used to evaluate MeInfoText 2.0 in (8). MeInfoText 2.0 includes all the 58 genes, but it contains only 42 out of the 58 associations between the 58 genes and colorectal cancer in the first benchmark list, while DDMGD contains 56 of these associations.
Second, considering that MeInfoText 2.0 is restricted to DNA methylation information specifically in cancers, we also compared DDMGD with another database, DiseaseMeth, that includes information about other diseases. DiseaseMeth is the most comprehensive manually curated database for DNA methylation information in human diseases and we used the second benchmark list to make this comparison. DiseaseMeth did not contain any of the 20 associations, although it contains two and four of the genes and diseases, respectively. Contrary to this, DDMGD contains all the genes and diseases and 15 out of 20 associations.
Finally, we compared DDMGD with MethDB, because MethDB has the most similar scope to DDMGD to include DNA methylation information in different species. We used the third benchmark list that includes DNA methylation information in mice for the comparison. MethDB contains two of the 10 cancers in mice, however, none of the genes or associations. On the other hand, DDMGD contains nine of the 12 genes, nine of 10 cancers and six of 16 associations. More detailed analysis for these three databases is available in the Supplementary material.
MeInfoText 2.0 and DiseaseMeth were published in 2011 and 2012, respectively. MethDB was first published in 2001, and the last version of the database was published in 2006. We accessed them in August 2014 for comparison. The high coverage of DDMGD can be due to using a large set of abstracts and full-text articles that includes the recent literature, comprehensive dictionaries to extract genes, diseases and species and accurate machine learning models to extract the associations. However, we highlight that the purpose of this comparison is to compare the current status and capabilities of different databases related to associations between methylated genes and diseases.
CONCLUSION AND FUTURE WORK
The analysis in the previous sections demonstrates that DDMGD is, in general, more comprehensive than the existing manually curated or automatically compiled databases and thus can serve as a useful complement to these resources, significantly expanding the coverage that other databases have, while at the same time preserve high level of accuracy of the extracted information. Although DDMGD included most of the information in the benchmark lists, the remaining information from these lists was not captured by DDMGD mainly due to copyright restrictions and scarcity of full-text articles in a proper searchable format (e.g. XML). However, this can be overcome by using more full-text articles besides the PubMed Central OAS for future updates of data in DDMGD.
Additionally, our system requires gene, disease and methylation words to be in the same sentence to extract the associations. Although this condition seems to limit the number of associations that can be extracted, this condition is necessary in order to preserve the accuracy of the system. However, cross-sentence association extraction is an active area of research, and we plan to extend our system to handle associations that exist in multiple sentences. Moreover, in order to extract species information more comprehensively for associations that do not have the species mentioned in the same sentence, we plan to develop a system that will try to infer species information from the full text or abstract. We plan to constantly improve DDMGD by expanding its content using the improved text mining system.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
King Abdullah University of Science and Technology. Funding for open access charge: King Abdullah University of Science and Technology.
Conflict of interest statement. None declared.
REFERENCES
- 1.Capell B.C., Berger S.L. Genome-Wide Epigenetics. J. Invest. Dermatol. 2013;133:e9. doi: 10.1038/jid.2013.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li P., Demirci F., Mahalingam G., Demirci C., Nakano M., Meyers B.C. An integrated workflow for DNA methylation analysis. J. Genet. Genomics. 2013;40:249–260. doi: 10.1016/j.jgg.2013.03.010. [DOI] [PubMed] [Google Scholar]
- 3.Reynolds R.M., Jacobsen G.H., Drake A.J. What is the evidence in humans that DNA methylation changes link events in utero and later life disease? Clin. Endocrinol. 2013;78:814–822. doi: 10.1111/cen.12164. [DOI] [PubMed] [Google Scholar]
- 4.Vanyushin B.F., Ashapkin V.V. DNA methylation in higher plants: past, present and future. Biochim. Biophys. Acta. 2011;1809:360–368. doi: 10.1016/j.bbagrm.2011.04.006. [DOI] [PubMed] [Google Scholar]
- 5.Mandrioli M., Borsatti F. DNA methylation of fly genes and transposons. Cell Mol. Life Sci. 2006;63:1933–1936. doi: 10.1007/s00018-006-6039-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meehan R.R. DNA methylation in animal development. Semin. Cell Dev. Biol. 2003;14:53–65. doi: 10.1016/s1084-9521(02)00137-4. [DOI] [PubMed] [Google Scholar]
- 7.Wilhelm-Benartzi C.S., Koestler D.C., Karagas M.R., Flanagan J.M., Christensen B.C., Kelsey K.T., Marsit C.J., Houseman E.A., Brown R. Review of processing and analysis methods for DNA methylation array data. Br. J. Cancer. 2013;109:1394–1402. doi: 10.1038/bjc.2013.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fang Y.C., Lai P.T., Dai H.J., Hsu W.L. MeInfoText 2.0: gene methylation and cancer relation extraction from biomedical literature. BMC Bioinformatics. 2011;21:471–478. doi: 10.1186/1471-2105-12-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Raies A.B., Monsour H., Incitti R., Bajic V.B. Combining position weight matrices and document-term matrix for efficient extraction of associations of methylated genes and diseases from free text. PLoS One. 2013;8:e77848. doi: 10.1371/journal.pone.0077848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He X., Chang S., Zhang J., Zhao Q., Xiang H., Kusonmano K., Yang L., Sun Z.S., Yang H., Wang J. MethyCancer: the database of human DNA methylation and cancer. Nucleic Acids Res. 2008;36:D836–D841. doi: 10.1093/nar/gkm730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xin Y., Chanrion B., O'Donnell A.H., Milekic M., Costa R., Ge Y., Haghighi F.G. MethylomeDB: a database of DNA methylation profiles of the brain. Nucleic Acids Res. 2012;40:D1245–D1249. doi: 10.1093/nar/gkr1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lv J., Liu H., Su J., Wu X., Liu H., Li B., Xiao X., Wang F., Wu Q., Zhang Y. DiseaseMeth: a human disease methylation database. Nucleic Acids Res. 2012;40:D1030–D1035. doi: 10.1093/nar/gkr1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ongenaert M., Neste L.V., Meyer T.D., Menschaert G., Bekaert S., Criekinge W.V. PubMeth: a cancer methylation database combining text-mining and expert annotation. Nucleic Acids Res. 2008;36:D842–D846. doi: 10.1093/nar/gkm788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Amoreira C.l., Hindermann W., Grunau C. An improved version of the DNA methylation database (MethDB) Nucleic Acids Res. 2003;31:75–77. doi: 10.1093/nar/gkg093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Negre V., Grunau C. The MethDB DAS server: adding an epigenetic information layer to the human genome. Epigenetics. 2006;1:101–105. doi: 10.4161/epi.1.2.2765. [DOI] [PubMed] [Google Scholar]
- 16.NCBI Resource Coordinators. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2014;42:D7–D17. doi: 10.1093/nar/gkt1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Federhen S. The NCBI Taxonomy database. Nucleic Acids Res. 2012;40:D136–D143. doi: 10.1093/nar/gkr1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davis A.P., Murphy C.G., Johnson R., Lay J.M., Lennon-Hopkins K., Saraceni-Richards C., Sciaky D., King B.L., Rosenstein M.C., Wiegers T.C., et al. The Comparative Toxicogenomics Database: update 2013. Nucleic Acids Res. 2013;41:D1104–D1114. doi: 10.1093/nar/gks994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Expression Atlas update–a database of gene and transcript expression from microarray- and sequencing-based functional genomics experiments. Nucleic Acids Res. 2014;42:D926–D932. doi: 10.1093/nar/gkt1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Amberger J., Bocchini C.A., Scott A.F., Hamosh A. McKusick's Online Mendelian Inheritance in Man (OMIM) Nucleic Acids Res. 2009;37:D793–D796. doi: 10.1093/nar/gkn665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rustici G., Kolesnikov N., Brandizi M., Burdett T., Dylag M., Emam I., Farne A., Hastings E., Ison J., Keays M. ArrayExpress update—trends in database growth and links to data analysis tools. Nucleic Acids Res. 2013;41:D987–D990. doi: 10.1093/nar/gks1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barrett T., Wilhite S.E., Ledoux P., Evangelista C., Kim I.F., Tomashevsky M., Marshall K.A., Phillippy K.H., Sherman P.M., Holko M., et al. NCBI GEO: archive for functional genomics data sets—update. Nucleic Acids Res. 2013;41:D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim M.S., Lee J., Sidransky D. DNA methylation markers in colorectal cancer. Cancer Metastasis Rev. 2010;29:181–206. doi: 10.1007/s10555-010-9207-6. [DOI] [PubMed] [Google Scholar]
- 24.Lu Q. The critical importance of epigenetics in autoimmunity. J. Autoimmun. 2013;41:1–5. doi: 10.1016/j.jaut.2013.01.010. [DOI] [PubMed] [Google Scholar]
- 25.Conerly M., Grady W.M. Insights into the role of DNA methylation in disease through the use of mouse models. Dis. Models Mech. 2010;3:290–297. doi: 10.1242/dmm.004812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Geisen S., Barturen G., Alganza A.n.M., Hackenberg M., Oliver J.L. NGSmethDB: an updated genome resource for high quality, single-cytosine resolution methylomes. Nucleic Acids Res. 2014;42:D53–D59. doi: 10.1093/nar/gkt1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baek S.J., Yang S., Kang T.W., Park S.M., Kim S.Y. MENT: methylation and expression database of normal and tumor tissues. Gene. 2013;518:194–200. doi: 10.1016/j.gene.2012.11.032. [DOI] [PubMed] [Google Scholar]
- 28.Gu F., Doderer M.S., Huang Y.W., Roa J.C., Goodfellow P.J., Kizer E.L., Huang T.H.M., Chen Y. CMS: a web-based system for visualization and analysis of genome-wide methylation data of human cancers. PLoS One. 2013;8:e60980. doi: 10.1371/journal.pone.0060980. [DOI] [PMC free article] [PubMed] [Google Scholar]


