Abstract
PhosphoSitePlus® (PSP, http://www.phosphosite.org/), a knowledgebase dedicated to mammalian post-translational modifications (PTMs), contains over 330 000 non-redundant PTMs, including phospho, acetyl, ubiquityl and methyl groups. Over 95% of the sites are from mass spectrometry (MS) experiments. In order to improve data reliability, early MS data have been reanalyzed, applying a common standard of analysis across over 1 000 000 spectra. Site assignments with P > 0.05 were filtered out. Two new downloads are available from PSP. The ‘Regulatory sites’ dataset includes curated information about modification sites that regulate downstream cellular processes, molecular functions and protein-protein interactions. The ‘PTMVar’ dataset, an intersect of missense mutations and PTMs from PSP, identifies over 25 000 PTMVars (PTMs Impacted by Variants) that can rewire signaling pathways. The PTMVar data include missense mutations from UniPROTKB, TCGA and other sources that cause over 2000 diseases or syndromes (MIM) and polymorphisms, or are associated with hundreds of cancers. PTMVars include 18 548 phosphorlyation sites, 3412 ubiquitylation sites, 2316 acetylation sites, 685 methylation sites and 245 succinylation sites.
INTRODUCTION
Discovery mode tandem mass spectrometry (MS) has transformed the landscape of cell biology and cell signaling research over the past 10 years. PhosphoSite® began its life just as this new era was in its infancy. When launched in 2003, it encompassed 1200 modification sites on 500 human and mouse proteins. Now, 11 years later, PhosphoSitePlus® (PSP) contains one-third of a million non-redundant modification sites on over 20 000 protein groups, and behind the scenes it hosts 1.5 million peptides. The high-throughput MS2 (HTP) data in PSP now dwarfs the high-quality data aggregated by manual curation of published low-throughput (LTP) scientific literature: only 4.5% of the sites in PSP are derived from LTP studies. While the number of sites in PSP has expanded nearly 350-fold since 2003, it is not the site count that matters most—data quality and reliability are top priority. In this paper we will review changes made to PSP since 2012, including the measures taken to ensure that the information in PSP is as reliable as possible.
Since 2003, over 340 000 non-redundant MS2 PTM sites have been curated into PSP from over 8000 separate experiments performed at CST and from 202 publications. The algorithms used for site assignment, as well as the mass accuracy of the commonly used spectrometers, have significantly improved over the last 10 years. Much of the HTP data from 2003 to 2008 came from less sensitive instruments using an older generation of software. Beginning in 2011–12, much of the older phosphorylation data in PSP has been re-analyzed using contemporary algorithms including Ascore (1).
Since PSP relies increasingly on MS2 data, and 68% of all sites are associated with only 1 or 2 HTP records, it is important to understand what the limits of interpretation are for MS data. Specifically, how reliable are PTMs that have been reported only once or twice by HTP MS? This question will be addressed in the Results and Discussion section.
On another front, the extraordinary advances in genome sciences and sequencing are uncovering a prodigious number of genetic variants and disease mutations. One of the most exciting challenges in this domain of Big Data is at the intersection of genetic variation and PTMs. The observation that missense mutations can rewire signaling pathways (2,3,4,5) has provided the inspiration for the production and monthly updating of PTMVar, a dataset that maps missense mutations onto the PTM sequence space. PTMVar is available on the PSP Download page (www.phosphosite.org/staticDownloads.do). This data is expected to be of value to genetic researchers in understanding molecular mechanisms underlying frank disease mutations and the association between polymorphic variants and genetic risks, and to cell biologists investigating the mechanisms through which mutations can rewire signaling networks.
A third focus of this paper is an examination of the in vitro and in vivo kinase-substrate interactions (KSIs) curated into PSP, and their use in generating sequence logos derived from reactions between protein kinases and intact protein substrates. Kinase specificity profiles generated using short peptides (6,7), methods that provide excellent profiles of the intrinsic affinities of kinase substrate binding pockets for peptides, cannot capture any additional information that might be generated when two surfaces collide, especially when the acceptor site is on the surface of a globular domain. The number of high-quality mammalian KSIs reported in PSP is 12 180 in 2014; 84% of these include human kinases, and many of the KSIs have been corroborated using kinases or substrates from multiple species. There are adequate numbers of KSIs in PSP to generate up to 100 sequence preference profiles of kinases interacting with substrate proteins. The utility of this data in generating kinase substrate sequence logos can be seen in Figure 5.
Figure 5.
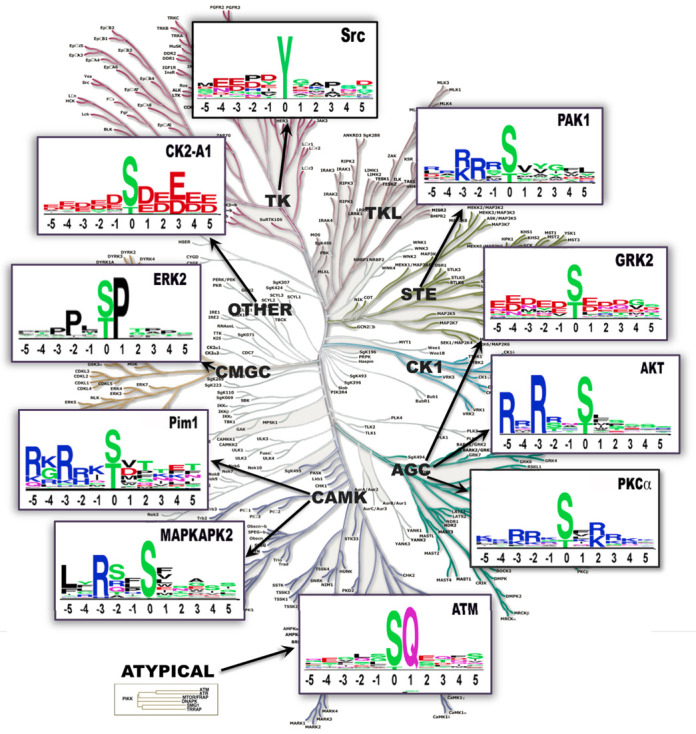
Sequence logos of 10 different kinases from various kinase groups. The number of substrate sequences used to generate each logo: SRC, 505; Akt, 200; GRK2, 58; ATM, 177; PKCα, 472; MAPKAPK2, 53; PIM1, 26; ERK2, 338; CK2-α1, 506 and PAK1, 63. Logos generated at www.phosphosite.org/siteSearchSelectAction.do using the Frequency Change logo graph method (31).
CONTENT: GROWTH AND CHANGES SINCE 2012
Site statistics
The total number of modification sites in PSP has increased from 128 943 to 338 948 over the past 3 years (Table 1). These include 274 396 modification sites on 20 021 human proteins, 139 066 sites on 15 389 mouse proteins and 42 465 sites on 7204 rat proteins.
Table 1. Non-redundant numbers of proteins and post-translationally modified sites in PSP: 2011 vs. 2014.
| 2011 | 2014 | |
|---|---|---|
| Proteins | 14 256 | 20 368 |
| Mod type: | Sites | Sites |
| phospho-Ser | 65 511 | 144 899 |
| phospho-Thr | 19 609 | 61 654 |
| ubiquityl | 18 996 | 51 258 |
| phospho-Tyr | 15 053 | 41 273 |
| acetyl | 7869 | 27 660 |
| mono-methyl | 303 | 5000 |
| di-methyl | 378 | 2556 |
| galNAc | 0 | 2118 |
| glcNAc | 602 | 1393 |
| sumoyl | 622 | 816 |
| tri-methyl | 0 | 321 |
| Total Sites: | 128 943 | 338 948 |
User interfaces
The four main user interfaces (Homepage, Protein Page, Modification Site Page and Curated Information Page) have kept close to their original structure and function in order to provide a stable and recognizable milieu for user interactions. These were described in the 2012 article (8), are described in the online tutorial (http://www.phosphosite.org/staticTrainingTutorial.do) and will not be addressed here.
Protein group and site group—essential concepts
The concepts of Protein Group and Modification Site Group provide essential unique identifiers (UIDs) underlying the ontology and systematics of proteins and modification sites in PSP. A Protein Group includes all individual proteoforms and orthologs from other species. A Modification Site Group provides a critical classifier for the unambiguous identification of homologous and orthologous sites within proteoforms and between species. Other possible site-specific identifiers, including residue number and surrounding sequence for specific sites, are inadequate. Residue numbers cannot be used because they can vary between isoforms or orthologs. For example, in an alignment of all Tau protein group members curated into PSP (9 human isoforms of Tau plus mouse, rat and cow orthologs), many of the corresponding modification sites across all protein group members have different residue numbers, yet would be uniquely joined together under a single Site Group ID. Residue numbering can also vary over time in proteins from the same repository as sequences are refined or as terminal methionines come and go. Nor can the flanking sequences uniquely identify a site, as sequences surrounding a site can vary between proteoforms of the same individual (e.g. splice variants or somatic mutations), between individuals within a population (e.g. somatic or germline variants) and between orthologs. Additionally, sequences are often repeated throughout the same protein multiple times, rendering sequences alone unsuitable as modification site UIDs. For example, the tryptic peptide K.FS*MPGFKAEGPEVDVNLPK is found five times in human protein AHNAK (Table 2). The systematics underlying PSP require each repeat to have a different Site Group ID—within any specific sequence, a site group can only have a one-to-one correspondence. This example illustrates a conundrum faced while curating such peptides—how to assign it? Any single assignment has a four-fifths chance of being incorrect. Perhaps some sort of fuzzy logic could handle such situations. In any case, throughout this paper, as in our previous 2012 article (8), the terms ‘protein’ and ‘site’ shall apply to a Protein Group and a Site Group, respectively, unless otherwise stated. Note that the Site Group ID is now included with each site in downloads from PSP, enabling the user to identify, sort and consolidate site-specific information when desired.
Table 2. Five internal sequence duplications within the human protein AHNAK. Each site requires an unique Site Group ID.
| Residue | Tryptic peptide |
|---|---|
| S886 | K.Fs*MPGFKAEGPEVDVNLPK |
| S1721 | K.Fs*MPGFKAEGPEVDVNLPK |
| S2057 | K.Fs*MPGFKAEGPEVDVNLPK |
| S2519 | K.Fs*MPGFKAEGPEVDVNLPK |
| S2903 | K.Fs*MPGFKAEGPEVDVNLPK |
The evolution of sequence space
The processing of peptides from proteomic MS experiments has evolved over the years. We have always considered UniPROTKB SwissProt(9), followed by NCBI RefSeq NP (10), to be the most reliable protein sequence resource. Thus, we wanted to use them to define our sequence space, but we encountered many peptides reported in the literature associated with IPI, TrEMBL, RefSeq XP and Ensembl that mapped to neither. We wanted to represent what the authors reported so we imported protein sequences from these other resources when necessary. These additional sequences proved unstable (11), making the curation process time consuming and unsustainable, especially as we moved further into the Big Data era. We are now transitioning to a stable human and mouse sequence space that will be exclusively populated with UniPROT SwissProt sequences updated once per month. Over 99.9% of the human proteins in PSP are from UniPROTKB, 97.1% from SwissProt and 2.8% from TrEMBL; 94.7% of the mouse sequences are from UniPROT, 88.8% of which are SwissProt. Sequences and accession numbers are synchronized with UniPROTKB every month.
Molecular structures
Since 2011, the number of PDB files (protein structural files from Protein Data Bank, http://www.rcsb.org/) corresponding to proteins in PSP has increased from 22 958 (4500 proteins) to 33 204 (5989 proteins). Nearly 27% of the 22 241 proteins in PSP have associated PDB files corresponding to some segment of the protein's sequence. It is notable that 69 388 sites, 19.1% of all sites in PSP, are located within the structures included in PDBs. Thus, a significant fraction of post-translationally modified residues are amenable to some level of structural analysis, enabling, for example, research into the effect of phosphorylation on protein-protein interactions (12), or upon the binding of drug inhibitors to target proteins (13).
LTP data
The total number of curated papers that used LTP technology to characterize modification sites is nearly 17 000, an increase of 4000 over the past 3 years. The ordinal rank of the top four journals represented PSP is the same as reported in 2012 (J. Biol. Chem., Mol. Cell. Biol., Oncogene and Proc. Natl. Acad. Sci. U.S.A.) with the digital, open access journal PLoS now in fifth place. The use of I2E (14), a powerful natural language processing software application, to identify articles and highlight information for manual curation, has significantly increased the efficiency and throughput of our LTP curation efforts, and made recuration of selected information a realistic and less time-consuming option.
HTP data: peptides and sites
Peptides and sites from 98 new proteomic MS publications and 6560 curation sets from Cell Signaling Technology (CST) have been curated since 2012. A majority of sites in PSP have only been observed using MS: of 344 413 sites in PSP, only 15 596 (4.5%) have been reported in the LTP literature. Of the MS-only sites, 55% have been reported only once, 25% between 2 and 5 times, 9% between 6 and 25 times, and 2.4% more than 25 times. MS peptide sequences that lie behind the assignments in PSP are now curated into PSP: nearly 1 700 000 redundant and 315 000 non-redundant peptides have been incorporated into PSP from CST curation sets and publications. This data will be made available in the coming months via download and in the longer term via an API.
This dominance of HTP data has made it critical to understand what the limitations of MS data are, to improve the quality of information already in PSP, and to ensure that data going into PSP is of the highest quality. Towards this end, we have reprocessed previously curated papers and CST curation sets to exclude lower quality site assignments.
Recuration of HTP data: applying a higher standard
A new threshold was implemented in 2012 for inclusion of new sites into PSP, either from publications or from CST experiments: sites had to have localization scores of P = /< 0.05 or Ascore > 13 to qualify for inclusion in PSP. There were, however, thousands of curation sets from CST and close to 120 proteomic MS papers already curated into PSP prior to establishing this threshold. Indeed, much of this data was curated into PSP prior to the existence of the Ascore algorithm (1) or the PTM localization probability score (15). Papers already curated into PSP that included site assignment scores were recurated, excluding sites below the thresholds. The site assignments from 4002 phosphorylation curation sets already in PSP were removed and reanalyzed on the new CST MS2 server. Note that 94 000 sites meeting the new standard were curated into PSP. Many thousands of sites previously in PSP did not meet the new standard, and similar numbers of new sites were introduced. A little over 4800 new curation sets, containing 139 600 filtered sites, have been added since January 2012.
As of October 2014, none of the 168 ubiquityl or 551 acetyl CST curation sets curated prior to January 2012 has been rescored. This process has, however, been started and should be completed within 6 months.
New links to useful resources
A number of new links that take the user to corresponding pages in other outstanding resources have been added to PSP Protein Pages. These include: the Human Protein Atlas (http://www.proteinatlas.org/) for protein expression levels and subcellular localization based on immunohistochemistry, and transcript expression levels for a large number of human tissues, cancers and cell lines (16); BioGPS (http://biogps.org/) for tissue-specific pattern of mRNA expression (17); Wikipedia pages include information about protein function, RNA expression, interactions, clinical significance, references and further reading (e.g. http://en.wikipedia.org/wiki/ANXA2); neXtProt (18), a human protein resource from Swiss Institute of Bioinformatics and GeneBio that integrates information from the UniProtKB/Swiss-Prot Knowledgebase, the HUPO Human Proteome Project (19) and other post-proteomic resources; and Reactome for the visualization, interpretation and analysis of pathway knowledge (20). Also, the chromosomal location of the gene encoding each human protein in PSP is now included on Protein Pages and in many downloads.
Downloads and their content
Two types of download are available through PSP: those for downloading results of user specified searches initiated interactively from search pages (Figure 1, top), and those available via the download page (Figure 1, bottom). Downloads from Site Search Result Pages include the results of user-defined searches for modification sites that fulfill one or more of 12 criteria including sites that are: responsive to specific treatments; observed within specific domains, and proteins of a specific type; within proteins associated with specific functions, processes, cellular components, or molecular weight ranges; observed within a specific sequence or motif; and observed in specific cell lines, cell types or tissues. Since 2012, Site Group IDs, LTP and HTP counts have been added to all downloads containing site-specific information. This is in addition to standard names, accession numbers, molecular weights, modified residues and their flanking sequences. The number of associated LTP and HTP records may be of particular use in evaluating the strength of evidence associated with the modification site (for exmple, see the section "Evidence for neutral mutation..." in the Results and Discussion section).
Figure 1.
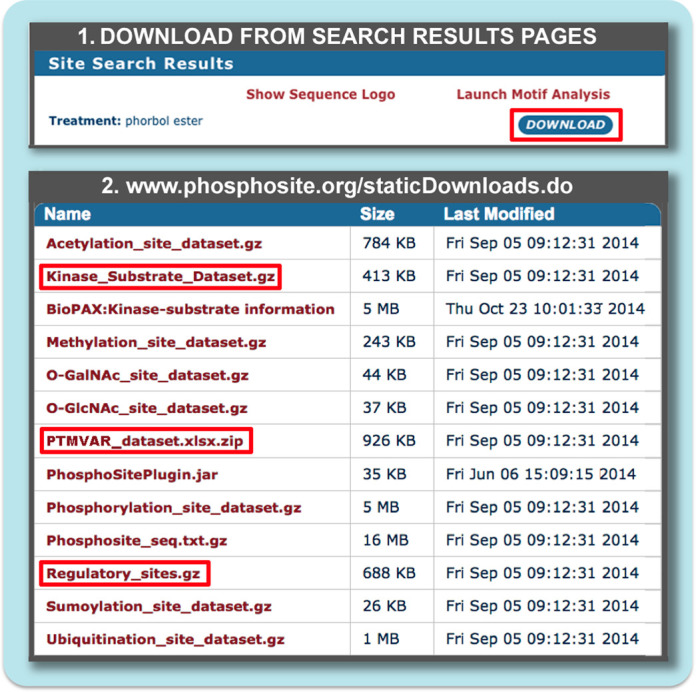
Downloads available from PSP. (1) The results pages of user-initiated searches for proteins and sites can be imported with the download button at the top of the page. (2) Static Downloads (www.phosphosite.org/staticDownloads.do) are updated monthly. The ‘Phosphosite_seq’ download contains all sequences of proteins currently in PSP (FASTA); All site datasets provide information including names and UIDs, modsite sequence, chromosomal location, Site Group ID, the number of HTP and LTP records, etc.; Disease-associated sites all have curated evidence linking them directly to a disease; BioPAX and Plugin provide pathway informatics extracted from PSP; ‘Kinase Substrate’, ‘PTM-VAR’ and ‘Regulatory sites’ all described within the text.
Static downloads described in this section are available via the Download Page (www.phosphosite.org/staticDownloads.do), and are complete, pre-compiled datasets of various categories of information contained in PSP (Figure 1, bottom). While the information in the database is curated and updated daily, the downloads are prepared only once a month. In contrast, queries through the UI interact in real time with the PSP DB. Three datasets containing functional metadata are described in more detail below. ‘PTMVar’ and ‘Regulatory Sites’ are new to this version of PSP. The Kinase Substrate Dataset is analyzed in a later section to evaluate its information content and utility.
The download ‘PhosphoSitePlugin’ (Figure 2) is a Cytoscape plugin (21) that contains site-specific information abstracted from PSP. Initially, only KSIs were included in the plugin; subsequently, treatments that regulate the modification status of specific residues have been added to the graph. More than 1390 treatments and 29 700 downstream targets that they regulate have been curated into PSP and are available for incorporation into the Cytoscape plugin. Note, however, that the site-centric information in PSP is represented only at the protein level in Cytoscape, reducing graph complexity but also losing valuable information.
Figure 2.
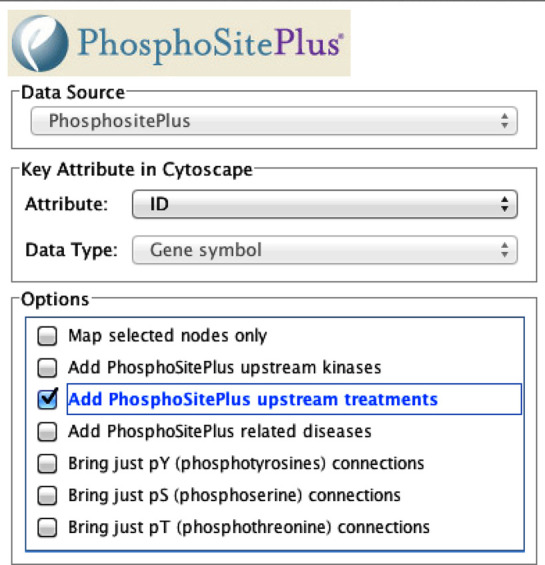
The interactive popup page of the Cytoscape Plugin for importing data from PSP into Cytoscape.
The ‘Regulatory Sites’ dataset provides curated information mainly from LTP papers about 7211 sites on 2374 proteins that regulate downstream cellular processes, molecular functions and protein-protein interactions: 3630 sites regulate 47 downstream cellular processes including 1743 that regulate transcription, 778 that influence cytoskeletal organization, 781 that regulate apoptosis, 252 that regulate cell adhesion and 539 that regulate cell motility; 6368 sites control 8 different molecular functions including 1788 that regulate intracellular localization, 1178 that induce enzymatic activity, 1045 that regulate protein stabilization or degradation and 399 that inhibit enzymatic activity; and 2845 sites on 1238 proteins that regulate protein-protein interactions with 821 binding partners. The top five binding partners are Src, Grb2, SHP-2, Shc1 and 14–3–3 β.
The ‘PTMVar’ dataset identifies modification sites that are either at or within five residues of a site that has been mutated, either in germline or somatically. PTMVar currently contains variant data from monthly downloads of ‘humsavar.txt’ from UniProtKB (http://www.uniprot.org/docs/humsavar). Cancer somatic mutations, identified in reference 4, are from TCGA (http://cancergenome.nih.gov/), cBIO (22), COSMIC (http://cancer.sanger.ac.uk/cancergenome/projects/cosmic/) and other sources (23). The current PTMVar dataset includes 17 551 modification sites associated with 24 592 mutations, of which 39% are classified as Disease mutations (including 1250 discreet diseases and genetic disorders from Online Mendelian Inheritance in Man (http://omim.org/)), 44% as polymorphisms, and 17% as unclassified. Our goal is to include somatic mutations from cancer genomes only when observed in three to five independent cancer patient samples.
The current Kinase Substrate’ dataset contains 12 180 high quality manually curated KSIs, 10 194 of which have been demonstrated using human kinases. It is notable that 9% of the kinases and 8% of the substrate proteins have been corroborated using kinases and/or substrates from multiple species. While in vivo and in vitro results are curated into PSP, in vitro results outscore in vivo by 11 396 to 6231. Note that 368 mammalian kinases have at least one experimentally reported substrate in PSP. A total of 232 have 5 or more non-redundant substrates, 170 have 10 or more and 100 have 20 or more and are listed in Table 3.
Table 3. The numbers of non-redundant substrate sites (N) for 100 protein kinases curated into PSP, September 2014.
| Kinase | N | Kinase | N | Kinase | N | Kinase | N | Kinase | N | Kinase | N | Kinase | N |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PKACα | 921 | PKCβ | 162 | Lyn | 86 | InsR | 59 | CDK7 | 39 | FAK | 27 | ZAP70 | 23 |
| PKCα | 667 | Fyn | 149 | PKCζ | 82 | LRRK2 | 58 | CK1ε | 37 | FGFR1 | 27 | Met | 22 |
| CK2α1 | 601 | JNK1 | 147 | ATR | 80 | GSK3α | 57 | LKB1 | 36 | MSK1 | 27 | PKCτ | 22 |
| CDK2 | 550 | Abl | 146 | PAK1 | 75 | ChaK1 | 55 | IKKα | 34 | PAK2 | 27 | PKG2 | 22 |
| ERK2 | 526 | PLK1 | 143 | p90RSK | 71 | IKKβ | 54 | CAMK1α | 33 | TBK1 | 27 | PKR | 22 |
| Src | 444 | PKCδ | 140 | GRK2 | 69 | CDK4 | 50 | CDK6 | 33 | AMPKα2 | 26 | Btk | 21 |
| CDK1 | 438 | mTOR | 137 | EGFR | 67 | PKCγ | 49 | IRAK4 | 33 | HIPK2 | 26 | DAPK3 | 21 |
| ERK1 | 348 | AurB | 133 | JNK2 | 65 | RSK2 | 49 | Akt2 | 30 | JNK3 | 26 | IRAK1 | 21 |
| CAMK2α | 259 | CK1α | 133 | ROCK1 | 65 | SGK1 | 48 | CAMK2β | 30 | Arg | 24 | MST2 | 21 |
| GSK3β | 243 | CK1δ | 111 | MK2 | 64 | p70S6K | 47 | ERK5 | 30 | MST1 | 24 | P38β | 21 |
| Akt1 | 230 | PKG1 | 105 | Syk | 64 | DYRK1α | 44 | MELK | 30 | PKN1 | 24 | ||
| ATM | 188 | DNAPK | 100 | AurA | 63 | PDK1 | 43 | PLK3 | 30 | ROCK2 | 24 | ||
| P38α | 184 | PKCε | 96 | Chk2 | 63 | TTK | 42 | PDGFRβ | 29 | DYRK2 | 23 | ||
| Chk1 | 170 | Lck | 94 | JAK2 | 62 | CDK9 | 40 | Pim1 | 29 | Hck | 23 | ||
| CDK5 | 167 | AMPKα1 | 93 | PKD1 | 60 | PKCτ | 40 | Ret | 29 | IKKε | 23 |
RESULTS AND DISCUSSION
Evidence for neutral mutation and the non-functional nature of rarely phosphorylated sites
Only 4.5% of the sites reported in PSP are associated with LTP literature records. All others are based on MS data alone. Of these, 55% have been reported in only one record, 13% in two, and 6% in three. Apparently none of these MS-only sites have yet been corroborated in publications using LTP methodologies. Are these 'low hitters' real sites or artifact? Is there any evidence that they represent functional sites? This first question can be gauged by evaluating the phosphosites in PSP, many of which have been reanalyzed, keeping only site assignments with p < 0.05 or Ascore =/> 13. Although many of the 'one hitters' were removed during this process, a similar number were added back, keeping their overall numbers relatively constant, indicating that as many as 90-95% of the rescored 'one-hit' phosphosites from CST may be real, i.e. are not spurious artifacts of MS. Furthermore, the numbers associated with these ‘one hitters’ do not increase systematically over time. In contrast, the number of hits for sites known to be bona fide cell signaling nodes tend to increase as the number of experiments increases. This evidence suggests that these 'low hit' sites may represent random non-functional phosphorylation sites that are rarely phosphorylated, play no role in cell signaling (24), and might be considered a sort of biological noise that occurs sporadically as kinases come into contact with non-functional, possibly low-affinity, sites on proteins scattered throughout the cell.
To test the possibility that 'low-hitters' are non-functional vis-a-vis critical signaling networks, we measured the amount of evolutionary divergence of phosphorylation sites (Ser, Thr and Tyr) between human and mouse, and between mouse and rat, reasoning that sites that play no functional role within cell signaling networks will diverge more quickly than sites that are functional nodes within cellular communication systems. Phosphosites were sorted into 5 bins according to the number of ‘hits’ (i.e., the number of records in which they were reported) from 1 to >100 (Figure 3). A total of 203 161 human-mouse and 148 441 mouse-rat pairs were tested. In both sets of pairs, the divergence is highest for sites with only 1 hit (20.6% and 8.8% for the human/mouse and the mouse/rat pairs, respectively), and lowest for the sites with >100 hits (5.4% and 3.0% for the human/mouse and the mouse/rat pairs, respectively). These results demonstrate that the more frequently sites are observed, the more likely they are to be conserved.
Figure 3.
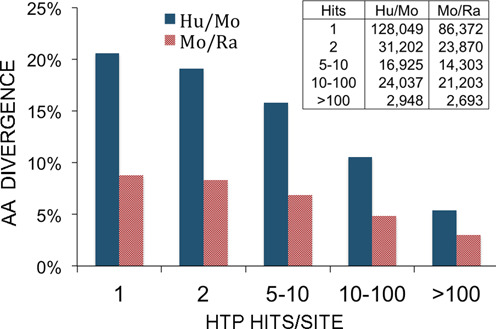
Comparison of the % of amino acid divergence (Y axis) of phosphorylation sites (pSer/pThr/pTyr) with varying numbers of associated hits (X axis). Ser, Thr or Tyr variants were excluded from counting. A total of 203,161 human/mouse and 148,441 mouse/rat phosphorylation sites were compared. The inset indicates the number of phosphosites included in each comparison. Blue, fraction of amino acid substitutions between human phosphosites and orthologous mouse residues; red, fraction of amino acid substitutions between mouse phosphosites and orthologous rat residues.
It is notable that the divergence is on average 2.3-fold higher for the human-mouse pairs than for the mouse-rat pairings, numbers that are consistent with estimated divergence times between humans and mice and between mice and rats of about 96 and 33 million years ago, respectively (25). These results indicate that these low-scoring sites, while most are almost certainly “good” assignments, should not be taken as strong evidence of any biological role for the site without additional evidence for functionality.
PTMVar: mutations that can rewire signaling circuits
Non-synonymous mutations are missense mutations that change one amino acid for another in a protein's coding region. If the mutation is at or near the site of a PTM that participates in cell signaling, it can significantly impact signaling networks in which the affected site participates. Class I PTMVars are those in which a PTM site is lost by altering the relevant amino acid (Figure 4). In the Class I example in Figure 4, Ser-16 of phenylalanine hydroxylase (PAH) is changed to Pro. PAH, requiring phosphorylation of Ser-16 for activation, is defective with the S16P mutation, leading to the buildup of phenylalanine and subsequently to phenylketonuria (2). Class Ia PTMVars are those in which the variant AA can still be enzymatically modified with the same side group as the wt. The variant in this case, TIE2 Y897S (VAR_008716), is associated with venous malformations (VMCM; OMIM 600195).
Figure 4.
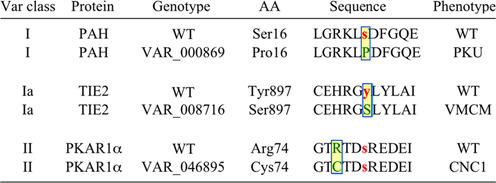
Classification of modification sites in the PTMVar dataset. Class I PTMVars are those in which a site is lost by an amino acid substitution of the modified residue. The variant in this case, PAH S16P (VAR_000869), causes phenylketonuria (2). Class Ia PTMVars are those in which the variant AA can still be enzymatically modified with the same side group as the wt substrate. The variant in this case, TIE2 Y897S (VAR_008716), is associated with venous malformations (VMCM; OMIM 600195) (29). Class II PTMVars are those in which the mutation occurs on a flanking residues ±5 amino acids from the modification site. The variant in this case, PKAR1α R74C (VAR_046895), is associated with Carney complex (CNC), a familial multiple neoplasia syndrome (30).
Class II PTMVars are those in which a mutation occurs on a flanking residue ±5 amino acids from the modification site. The variant in this example, PKAR1α R74C (VAR_046895), is associated with Carney complex (CNC), a familial multiple neoplasia syndrome. Many Class II PTMVars can dramatically alter the properties of substrates, changing or ablating their capacity to be phosphorylated by their wt upstream kinases. There are a handful of flanking residues that are critical for catalysis and are of special interest, for example R at −3, P at +1 and Q at +1. The PTMVar dataset includes a column indicating the relative position of the variant relative to the site of the PTM, enabling users to rapidly identify changes at specific positions in flanking sequences. For example, Arg at the −3 position, critical for phosphorylation by Akt and related kinases, is mutated in 439 substrate sequences; Pro at +1, required for most CMGC kinases, is lost in 266 substrate sequences; and Gln at +1, essential for catalysis by the DNA damage kinases ATM, ATR and DNAPK, is lost in 66 sequences in the current release.
The current download includes 1622 Class I, 181 Class Ia and 23 178 Class II PTMVars. All modification types are affected by variants and include 18 548 phosphorlyation sites, 3412 ubiquitylation sites, 2316 acetylation sites, 463 mono-methylation sites and 245 succinylation sites (Table 4).
Table 4. The numbers of PTMVars for each of 18 modification types in the PTMVar download, September 2014.
| Mod type | AA | N | Mod type | AA | N | Mod type | AA | N |
|---|---|---|---|---|---|---|---|---|
| Phospho | Ser | 9871 | Mono-methyl | Arg | 382 | Sumoyl | Lys | 77 |
| Phospho | Thr | 4754 | Mono-methyl | Lys | 76 | O-GlcNAcetyl | Ser | 49 |
| Phospho | Tyr | 3923 | Di-methyl | Arg | 174 | O-GalNAcetyl | Ser | 48 |
| Ubiquityl | Lys | 3412 | Di-methyl | Lys | 24 | O-GlcNAcetyl | Thr | 36 |
| Acetyl | Lys | 2316 | Tri-methyl | Lys | 22 | Neddyl | Lys | 35 |
| Succinyl | Lys | 244 | O-GalNAc | Thr | 84 | Caspase cleavage | Asp | 26 |
Searching the PTMVar dataset can be further enhanced by cross-dataset joins made by keying on Site Group IDs. This enables users to aggregate information about specific modsites from multiple datasets. For example, a join of the disease mutations from the PTMVar with and the Regulatory Site datasets identifies 272 PTMVars at sites known to regulate downstream cellular processes, molecular functions or protein-protein interactions. Disruptions of the phosphorylation of these ‘regulatory’ sites may well be expected to rewire the circuits in which they participate (Supplemental Table 1).
Generating sequence logos and PSSMs from whole protein KSIs
PSP contains enough high-quality, expert edited and curated, KSIs to generate up to 100 substrate preference profiles based on intact protein-protein interactions alone. This stands in contrast to resources that generate KSIs based on in vitro reactions using short peptides only (6,7) and modified variously with predictions or adjustments of one sort or another (26). Figure 5 shows 10 sequence logos of kinases from different major groups built with data from PSP with tools available at www.phosphosite.org/sequenceLogoAction.do. Included are (i) three kinases with exceptionally strong and nearly absolute substrate preferences: ERK2, with a strong preference for P at +1, and secondary preference for P at −2; ATM, with a preference for Q at +1, and no observable secondary preference; and the basophilic kinase AKT with a strong preference for R at −3 and a secondary preference for R at −5; (ii) kinases with moderate or slight preferences: PKCα: [RK] at +2, −3 and −2; Src: E at −4 and −3, [DP] at −2, D at −1 and P at +3; and (iii) preferences for a general quality but no outstanding single residue: GRK2, acidic residues throughout the kinase binding region.
How many sequences does it take to make a logo that reflects specificity? That depends on the topography and strength of the interactions between the residues surrounding the phosphate-accepting group on the substrate and the corresponding contact residues on the kinase. This is illustrated in Figure 6, where logos are built for Akt and PKCα from graded numbers of substrate sequences. In dozens of repetitions of randomly selected sequences, the preference of Akt for R at −3 and −5 showed up strongly for all 5-mers examined. In contrast, the preference of PKCα for [RK] at +2, −3 and −2 did not show up consistently until the input sequences numbered somewhere between 10 and 50. The conclusion drawn from this example is that as few as five input sequences can reveal strong preferences, but kinases with less selective requirements will require 20 or more sequences to reveal their substrate predilictions; visualizing fine specificities would require significantly more than these numbers. A review of 100 kinase logos generated with data from PSP suggests that most kinases exhibit only moderate substrate sequence specificities (data not shown).
Figure 6.
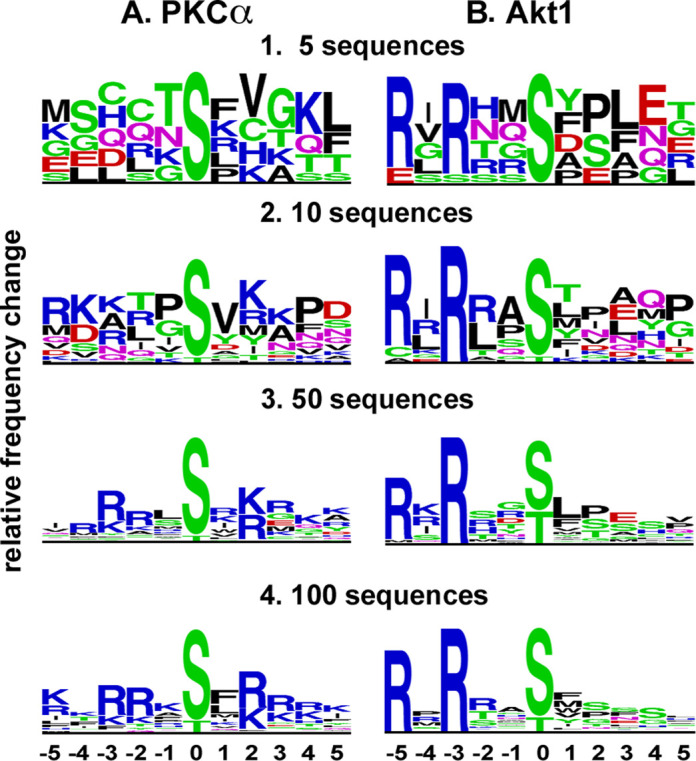
Sequence logos generated using increasing numbers of input sequences. At 5 input Akt sequences, the dominance of R at −3 and −5 was already evident. The preference of PKCα for R or K at +2 was evident using 10 input sequences, but the preference for R at −2 and −3 required between 10 and 50 input sequences. Logos were generated using the Frequency Change algorithm at http://www.phosphosite.org/sequenceLogoAction.do.
Domain location of various classes of PTMs
Serine and threonine phosphorylation sites modulate the structure and function of short linear motifs, regions that play important roles in cellular communication, and that are generally located between structured domains (27). We previously examined the location of various classes of PTMs relative to PFAM A domains and reported enrichment of pSer and pThr between domains (8). We repeated this analysis with substantially more data, and, in addition, analyzed the locations of arginine methylation sites (Figure 7). We confirmed the observations that 70–75% of all pSer/Thr sites occur in regions between structured domains, that tyrosine sites are more evenly distributed between structured and unstructured regions, and that acetylated lysine sites are enriched within domains. We observed significantly more ubiquitylation within domains, perhaps marking damaged domains bound for proteasomal destruction. Interestingly, nearly 78% of methylated arginine sites are located outside of domains, perhaps suggesting a role for methylation in the regulation of short linear domains or other unstructured regions.
Figure 7.
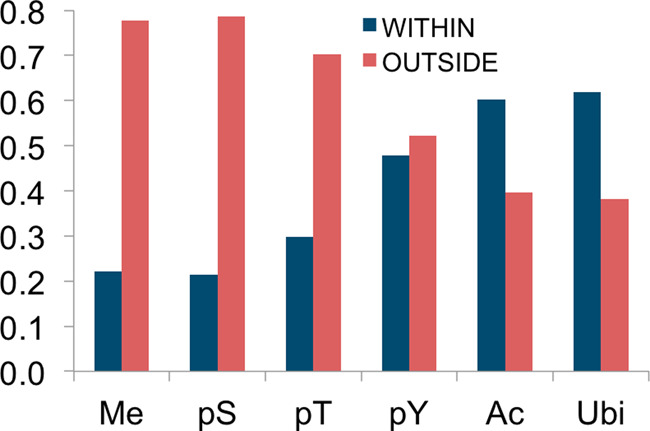
The locations of six types of PTM relative to Pfam-A domains. The modification types and number of modsites included in each group are: Me: 1- and 2-methyl-Arg, 3189;pS: phospho-Ser, 96570;pT: phospho-Thr, 40358;pY: phospho-Tyr, 31948;Ac: acetyl-Lys, 7403;Ubi: ubiquityl-Lys,17397.
CONCLUDING REMARKS
The rescoring of peptide and site assignments across archived CST experiments performed over the past nine years has increased the coherence and reliability of 85% of the phoshorylation sites from CST. The ongoing reanalysis of archived acetylation, methylation and ubiquitylation data in the coming months will similarly improve its coherence and accuracy. Users should not be alarmed when sites with 1 or a few hits disappear; they probably don't meet our new standards, and are being replaced with more accurate data. They should also be aware that the MS data generated at CST and stored in PSP can be periodically reanalyzed using more advanced algorithms as they are developed, a capability shared by the PTM resource PHOSIDA (28) but few others.
The demonstration that pSer, pThr and pTyr sites with very low numbers of MS hits have undergone relatively rapid evolutionary divergence, and that amino acids frequently observed to be phosphorylated have diverged more slowly, indicates that evolutionary pressure conserves sites with lots of phosphorylation activity, but exerts little pressure against neutral mutations at sites with only one or a few phosphorylation hits. These results imply two things. First, the low divergence observed between orthologous sites with high phosphorylation activity suggests that these sites are being actively conserved, perhaps because they play positive roles in cellular life. Second, that the apparent lack of selective pressure on sites with only one or a few MS hits suggests that their phosphorylation is of little consequence to the organism, and that neutral mutation is relatively unconstrained at these sites. This in turn should suggest to the experimental biologist that many of these 'low-hitter' sites are the result of stochastic processes and, in the absence of additional compelling evidence, may be risky targets for further research.
Lastly, PTMVar provides a proteome-wide snapshot of how thousands of disease-causing missense mutations can interact with the 300 000+ unique PTMs contained within PSP. There are almost certainly clues within this dataset that can provide actionable insights into the interplay between disease mutations and cell signaling mechanisms.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
With gratitude to Michael Comb for supporting this work since its inception.
FUNDING
National Institutes of Health [R43GM065768, R44AA014848m, R44CA126080 and U54-HL127624-01]. Funding for open access charge: Cell Signaling Technology.
Conflict of interest statement. None declared.
REFERENCES
- 1.Beausoleil S.A., Villen J., Gerber S.A., Rush J., Gygi S.P. A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat. Biotechnol. 2006;24:1285–1292. doi: 10.1038/nbt1240. [DOI] [PubMed] [Google Scholar]
- 2.Miranda F.F., Teigen K., Thorolfsson M., Svebak R.M., Knappskog P.M., Flatmark T., Martinez A. Phosphorylation and mutations of Ser(16) in human phenylalanine hydroxylase. Kinetic and structural effects. J. Biol. Chem. 2002;277:40937–40943. doi: 10.1074/jbc.M112197200. [DOI] [PubMed] [Google Scholar]
- 3.Gelmann E.P., Steadman D.J., Ma J., Ahronovitz N., Voeller H.J., Swope S., Abbaszadegan M., Brown K.M., Strand K., Hayes R.B., et al. Occurrence of NKX3.1 C154T polymorphism in men with and without prostate cancer and studies of its effect on protein function. Cancer Res. 2002;62:2654–2659. [PubMed] [Google Scholar]
- 4.Reimand J., Bader G.D. Systematic analysis of somatic mutations in phosphorylation signaling predicts novel cancer drivers. Mol. Syst. Biol. 2013;9:637. doi: 10.1038/msb.2012.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ryu G.-M., Song P., Kim K.-W., Oh K.-S., Park K.-J., Kim J.H. Genome-wide analysis to predict protein sequence variations that change phosphorylation sites or their corresponding kinases. Nucleic Acids Res. 2009;37:1297–1307. doi: 10.1093/nar/gkn1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Obenauer J.C., Cantley L.C., Yaffe M.B. Scansite 2.0: proteome-wide prediction of cell signaling interactions using short sequence motifs. Nucleic Acids Res. 2003;31:3635–3641. doi: 10.1093/nar/gkg584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Turk B.E., Hutti J.E., Cantley L.C. Determining protein kinase substrate specificity by parallel solution-phase assay of large numbers of peptide substrates. Nat. Protoc. 2006;1:375–379. doi: 10.1038/nprot.2006.57. [DOI] [PubMed] [Google Scholar]
- 8.Hornbeck P.V., Kornhauser J.M., Tkachev S., Zhang B., Skrzypek E., Murray B., Latham V., Sullivan M. PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res. 2012;40:D261–D270. doi: 10.1093/nar/gkr1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu C.H., Apweiler R., Bairoch A., Natale D.A., Barker W.C., Boeckmann B., Ferro S., Gasteiger E., Huang H., Lopez R., et al. The Universal Protein Resource (UniProt): an expanding universe of protein information. Nucleic Acids Res. 2006;34:D187–D191. doi: 10.1093/nar/gkj161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pruitt K.D., Brown G.R., Hiatt S.M., Thibaud-Nissen F., Astashyn A., Ermolaeva O., Farrell C.M., Hart J., Landrum M.J., McGarvey K.M., et al. RefSeq: an update on mammalian reference sequences. Nucleic Acids Res. 2014;42:D756–D763. doi: 10.1093/nar/gkt1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Griss J., Martin M., O'Donovan C., Apweiler R., Hermjakob H., Vizcaino J.A. Consequences of the discontinuation of the International Protein Index (IPI) database and its substitution by the UniProtKB ‘complete proteome’ sets. Proteomics. 2011;11:4434–4438. doi: 10.1002/pmic.201100363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nishi H., Hashimoto K., Panchenko A.R. Phosphorylation in protein-protein binding: effect on stability and function. Structure. 2011;19:1807–1815. doi: 10.1016/j.str.2011.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith K.P., Gifford K.M., Waitzman J.S., Rice S.E. Survey of phosphorylation near drug binding sites in the Protein Data Bank (PDB) and their effects. Proteins. 2014 doi: 10.1002/prot.24605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bandy J., Milward D., McQuay S. Mining protein-protein interactions from published literature using Linguamatics I2E. Methods Mol. Biol. 2009;563:3–13. doi: 10.1007/978-1-60761-175-2_1. [DOI] [PubMed] [Google Scholar]
- 15.Olsen J.V., Blagoev B., Gnad F., Macek B., Kumar C., Mortensen P., Mann M. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 16.Uhlen M., Oksvold P., Fagerberg L., Lundberg E., Jonasson K., Forsberg M., Zwahlen M., Kampf C., Wester K., Hober S., et al. Towards a knowledge-based Human Protein Atlas. Nat. Biotechnol. 2010;28:1248–1250. doi: 10.1038/nbt1210-1248. [DOI] [PubMed] [Google Scholar]
- 17.Wu C., Macleod I., Su A.I. BioGPS and MyGene.info: organizing online, gene-centric information. Nucleic Acids Res. 2013;41:D561–D565. doi: 10.1093/nar/gks1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gaudet P., Argoud-Puy G., Cusin I., Duek P., Evalet O., Gateau A., Gleizes A., Pereira M., Zahn-Zabal M., Zwahlen C., et al. neXtProt: organizing protein knowledge in the context of human proteome projects. J. Proteome Res. 2013;12:293–298. doi: 10.1021/pr300830v. [DOI] [PubMed] [Google Scholar]
- 19.Legrain P., Aebersold R., Archakov A., Bairoch A., Bala K., Beretta L., Bergeron J., Borchers C.H., Corthals G.L., Costello C.E., et al. The human proteome project: current state and future direction. Mol. Cell Proteomics. 2011;10:M111.009993. doi: 10.1074/mcp.M111.009993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Croft D., Mundo A.F., Haw R., Milacic M., Weiser J., Wu G., Caudy M., Garapati P., Gillespie M., Kamdar M.R., et al. The Reactome pathway knowledgebase. Nucleic Acids Res. 2014;42:D472–D477. doi: 10.1093/nar/gkt1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saito R., Smoot M.E., Ono K., Ruscheinski J., Wang P.-L., Lotia S., Pico A.R., Bader G.D., Ideker T. A travel guide to Cytoscape plugins. Nature methods. 2012;9:1069–1076. doi: 10.1038/nmeth.2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cerami E., Gao J., Dogrusoz U., Gross B.E., Sumer S.O., Aksoy B.A., Jacobsen A., Byrne C.J., Heuer M.L., Larsson E., et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kan Z., Jaiswal B.S., Stinson J., Janakiraman V., Bhatt D., Stern H.M., Yue P., Haverty P.M., Bourgon R., Zheng J., et al. Diverse somatic mutation patterns and pathway alterations in human cancers. Nature. 2010;466:869–873. doi: 10.1038/nature09208. [DOI] [PubMed] [Google Scholar]
- 24.Lienhard G.E. Non-functional phosphorylations? Trends Biochem. Sci. 2008;33:351–352. doi: 10.1016/j.tibs.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 25.Nei M., Glazko G.V. The Wilhelmine E. Key 2001 Invitational Lecture. Estimation of divergence times for a few mammalian and several primate species. J. Heredity. 2002;93:157–164. doi: 10.1093/jhered/93.3.157. [DOI] [PubMed] [Google Scholar]
- 26.Miller M.L., Jensen L.J., Diella F., Jørgensen C., Tinti M., Li L., Hsiung M., Parker S.A., Bordeaux J., Sicheritz-Ponten T., et al. Linear motif atlas for phosphorylation-dependent signaling. Sci. signal. 2008;1:ra2. doi: 10.1126/scisignal.1159433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iakoucheva L.M., Radivojac P., Brown C.J., O'Connor T.R., Sikes J.G., Obradovic Z., Dunker A.K. The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res. 2004;32:1037–1049. doi: 10.1093/nar/gkh253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gnad F., Gunawardena J., Mann M. PHOSIDA 2011: the posttranslationalmodification database. Nucleic Acids Res. 2011;39:D253–D560. doi: 10.1093/nar/gkq1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Calvert J.T., Riney T.J., Kontos C.D., Cha E.H., Prieto V.G., Shea C.R., Berg J.N., Nevin N.C., Simpson S.A., Pasyk K.A., et al. Allelic and locusheterogeneity in inherited venous malformations. Hum. mol. genets. 1999;8:1279–1289. doi: 10.1093/hmg/8.7.1279. [DOI] [PubMed] [Google Scholar]
- 30.Veugelers M., Wilkes D., Burton K., McDermott D.A., Song Y., Goldstein M.M., Perle K.L., Vaughan C.J., O'Hagan A., Bennett K.R. Comparative PRKAR1Agenotype-phenotype analyses in humans with Carney complex and prkar1ahaploinsufficient mice. Proc. Natl Acad. Sci. U.S.A. 2004;101:14222–14227. doi: 10.1073/pnas.0405535101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vacic V., Iakoucheva L.M., Radivojac P. Two Sample Logo: a graphicalrepresentation of the differences between two sets of sequence alignments. Bioinformatics. 2006;22:1536–1537. doi: 10.1093/bioinformatics/btl151. [DOI] [PubMed] [Google Scholar]


