Abstract
NrichD (http://proline.biochem.iisc.ernet.in/NRICHD/) is a database of computationally designed protein-like sequences, augmented into natural sequence databases that can perform hops in protein sequence space to assist in the detection of remote relationships. Establishing protein relationships in the absence of structural evidence or natural ‘intermediately related sequences’ is a challenging task. Recently, we have demonstrated that the computational design of artificial intermediary sequences/linkers is an effective approach to fill naturally occurring voids in protein sequence space. Through a large-scale assessment we have demonstrated that such sequences can be plugged into commonly employed search databases to improve the performance of routinely used sequence search methods in detecting remote relationships. Since it is anticipated that such data sets will be employed to establish protein relationships, two databases that have already captured these relationships at the structural and functional domain level, namely, the SCOP database and the Pfam database, have been ‘enriched’ with these artificial intermediary sequences. NrichD database currently contains 3 611 010 artificial sequences that have been generated between 27 882 pairs of families from 374 SCOP folds. The data sets are freely available for download. Additional features include the design of artificial sequences between any two protein families of interest to the user.
INTRODUCTION
Efficiency of protein remote homology detection has been dependent on the availability of sequences which can convincingly connect distantly related proteins (1,2). With improvements in homology detection methods, natural linker sequences often serve as ‘tools’ that facilitate hops between proteins to connect them (3–6). Despite these advancements, identification of relationships, such as those between Lipocalins, Pleckstrin homology domains and Immunoglobins, solely from sequence information, remains a non-trivial and challenging task (2). It is now well accepted that the dispersion of sequences is not uniform in protein sequence space and such folds have a large number of sequences that cluster into pockets or subfamilies (7,8). Owing to such tight clusters and large sequence divergence between the pockets, commonly applied sequence search procedures fail in searches with, for instance, a novel member of such families (1,2,9,10).
In a recent publication, we have shown that the paucity of such sequences in many folds may be circumvented through the strategic computational design of artificial intermediary sequences capable of linking distantly related proteins in remote homology detection (11). The sequence design process described in this approach mainly attempts to populate sequence space in a meaningful manner and is purposefully biased to generate sequences that have residue preferences and structural features of the fold for which they are designed. This is possible because the design procedure, to begin with, represents members of protein families within a fold as multiple sequence alignments and multiple profiles and performs all-pairwise comparisons between the profiles. Where convincing alignments are achieved, sequence properties of both parents are captured in a hybrid matrix. A random number-based roulette-wheel selection procedure is then applied to guide sequence design for the protein families in consideration (12). When performed iteratively for all possible pairs within the fold, this gradually fills the observed sequence voids in the protein fold space. Several non-obvious relationships that become evident only after detailed structural and biochemical studies could be established using such computationally designed intermediate sequences (11).
While many computational methods have been devised in the past to explore the sequence space compatible with a fold, or examined such sequences for the purpose of improved stability or novel activities, our intention is solely to extend their applicability to improve homology detection (13–16). Although methods to design sequences with a similar motivation have been reported elsewhere (17–19), this is the first time that such intermediate sequences have been generated on a large scale, encompassing all known multi-membered folds. To this end, we now report the availability of annotated ready-to-use designed sequences generated for all known families in SCOP (20,21) through the publicly available NrichD database. Further, we enrich commonly employed SCOP and Pfam (22) data sets with designed sequences and provide various subsets of the data for easy use. An additional feature to facilitate users to design sequences between protein families or for specific families has been provided. This feature provides an access point to design sequences that specifically interest the user.
DATABASE CONTENT
Data collection, generation and annotation
For a practical and realistic extension of the utility of designed sequences and their amenability for search using any tool, combinations of some of the popularly used databases that have been integrated with the designed sequences have been made available through the NrichD database. These data sets are provided as a flat file and may be downloaded locally by the user, for use in stand-alone search procedures using any method, from the download link (http://proline.biochem.iisc.ernet.in/NRICHD/download). Typically, download of each of the data sets should take between 5–20 min at a download speed of 1.5 Mbps. However, if this is not desired, the user has the option to perform jackhammer searches online using the various data sets provided. These are as follows:
SCOP-DB: We collected 16 712 domain family sequences from 3901 SCOP families that were clustered at 95% sequence identity in the ASTRAL database (SCOP version 1.75) (23). Using these sequences from all SCOP classes A-G, PSI-BLAST searches were performed against the UniRef90 database [search criteria: E-value = 0.0001, H-value = 0.0001, iteration = 5 and 80% query coverage] (24). To ensure non-redundant homologs for each family, sequences were clustered at 90% sequence identity using CD-HIT. Homologs identified for each query sequence were suitably annotated with their SCOP definitions. All query sequences and their homologs were pooled together to create a SCOP-DB of 4 694 921 sequences (16 712 SCOP domain sequences + 4 678 209 sequence homologs from UniRef90 databases).
AS-DB: Artificial Sequence database—We grouped members of families into multiple alignments and profiles, all along guided by the SCOP codes. We then applied rigorous profile–profile alignments and generated sequences between as many family pairs within a fold as possible (see (11) for detailed methods). A total of 3 611 010 artificial protein-like intermediate sequences were designed between 27 882 pairs of families belonging to 374 SCOP domain folds. Each designed sequence was annotated with details of profile and fold of the parent family for which they were designed. This AS-DB (Artificial Sequence database) is also made publicly available.
SCOP-NrichD: (Natural sequences from SCOP database enriched with Designed sequences, in addition to natural linkers): The database of SCOP domain sequences integrated with natural and artificially designed sequences was generated with a total of 8 305 931 sequences in the database (4 694 921 sequences [SCOP-DB] and 3 611 010 designed sequences).
Pfam-DB: The Pfam database groups protein sequences into families based on similarities involving the functional domain (25). The grouping into protein families brings together diverse members that share similarities at this level and therefore association with any of the families is valuable to functional annotation of novel proteins. It must be noted that there are some families which may not be associated with any structural information. Note that 10 626 097 non-redundant protein sequences covering 14 831 Pfam families were retrieved from Pfam database (Pfam-A).
Pfam-NrichD: (Natural sequences from Pfam database enriched with Designed sequences). The designed sequences generated using SCOP fold definitions were plugged into the Pfam-DB to create this data set with a total of 14 237 107 sequences (10 626 097 sequences from Pfam augmented with 3 611 010 sequences).
RESULTS AND DISCUSSION
Extent of sequence space expansion: NrichD database statistics
Sequences were designed successfully for 374 of the 419 multi-membered SCOP folds belonging to SCOP class A–G. Sequences could not be designed for 45 folds due to the various eligibility criteria and stringent fidelity checks applied in the sequence design process (see (11) for more details). The percentage enrichment (ratio of the number of designed sequences to the natural members) in the four major classes ranges from 1.30 to 0.7. For some folds, such as the GINS helical bundle-like, restriction endonuclease-like, yeast killer toxins and others, the number of designed sequences is much larger than the number of naturally occurring homologs because we had employed a multiple PSSM-based approach to represent the families and a majority of the profile–profile alignments within these folds were satisfactory. For 56 folds, the number of designed sequences is nearly equal to the number of natural sequence members in those folds. For a majority of folds in the data set (120/374), the number of designed sequences is over half of the number of natural members in the folds. For 247 folds, fewer than half of the natural members could be generated due to poor alignments between the family members.
In general, it was observed that the number of designed sequences for each SCOP fold was directly proportional to the number of associated protein families within the fold. Thus, we measured the success rate of designed sequences based on how many possible pairs of families could, in theory, be aligned for the fold and how many of the folds among them had qualified sequences. Out of the total estimated possible 44 675 pairs of families, we could design sequences between 27 882 pairs of families. Details of success rates observed in 374 folds are shown in Figure 1(a). About 88% of the total number of folds, have a success rate of more than 50%, i.e. for these folds, at least half of the total theoretically possible pairs of families resulted in designed intermediate sequences.
Figure 1.
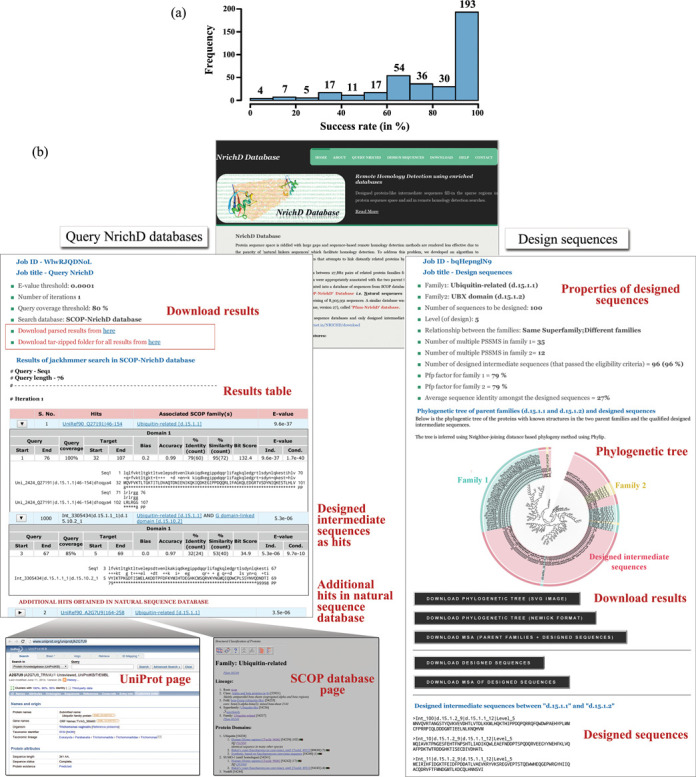
(a). Histogram representation of success rate (in percentage) in designing sequences for 374 folds. (b) Screenshots of Results pages of the database query and protein design interface.
Data access
Database search interface
The user may search the SCOP-NrichD and Pfam-NrichD database by providing an input sequence in FASTA format or by uploading the sequence file (Figure 1b). It must be noted that although these two databases have been made available for searching purpose, any submission always entails that the searches are also performed, in parallel, in the related data sets of SCOP-DB/Pfam-DB in the background. This way, it is ensured that hits from both searches are pooled together and homologs identified in both the searches are reported. Filters for evaluating the hits, such as E-value cut-off, number of jackhmmer (26) iterations and extent of query coverage to be imposed in the searches, etc., may be defined by the user. When results are presented to the user, the results from both searches are consolidated and checked for redundancy with results from searches in the SCOP-DB or Pfam-DB listed at the end. Designed sequences are annotated with the term ‘Int’ and qualified further with the multiple PSSM from which they were derived as also the SCOP codes of the parent families between which they were designed. These details serve as important cues in tracking the direction of searches and to determine the natural and designed intermediates that mediated the hops. They are especially useful to establish roundabout relationships in which they mediate hops between protein families that were not involved in their design (for details see results in (11)). Searches typically take between 5 and 30 min. A link is provided to view the results which are stored online for 14 days.
The interface also provides the results in tabular format for quick and easy view. Each row corresponds to a hit that contains clickable information with links to Uniref90 and the associated SCOP fold. Each hit can be expanded by clicking the radio links which lead to a view of the alignment and result statistic for each hit. The hits are hyperlinked to their respective entries in both the UniProt and SCOP databases. Two options to download the results are provided. Either in the form of parsed results, which contains hits which pass the E-value and query coverage cut-offs in searches in the NrichD database and additional hits (if any) that were observed in searches in the control databases (such as SCOP-DB/ Pfam-DB) or as tar-zipped folders of hits in jackhmmer searches in the control database and NrichD database separately.
Sequence design interface
A feature that might interest a user who is interested in working with a limited set of designed sequences is the generation of protein-like sequences for a protein family or alternatively between any two families specified by the user. Here, a choice of either designing sequences for known SCOP domain family/families (such as a.1.1.1) or a multiple sequence alignment of protein family/families (in ClustalW/Stockholm format) is expected as input. A maximum of 500 sequences may be designed with an additional handle on the level of design. This feature controls the extent of sequence dispersion of the designed sequences from the parent family (for details, refer Methods in Reference (11)). This typically takes 2–5 min.
The Results page lists the number of multiple PSSMs in the parent families specified by the user and the percentage of designed sequences that passed various eligibility criteria, such as the ability to detect parent family profiles for the associated fold at E-value better than 0.0001 and at least 80% query coverage and not detect hits from any other fold (Figure 1b). These metrics become relevant when the user wishes to evaluate the quality of the designed sequences. Higher values indicate proximity to the parent family. Ideally, sequence dispersion in the pool of designed sequences should be uniform so as to facilitate its function as an intermediary. Indicators such as average sequence identity and ease factor (number of designed sequences that qualify all imposed criteria) are useful if the user would like to increase the dispersion of the designed sequences by modifying the ‘level’ of sequence design. The purpose of the designed sequences is to restore continuity in protein sequence space by filling the voids. A visual representation of their spread with respect to the parent family or families for which they are designed is captured through a phylogenetic tree derived using neighbor-joining in Phylip suite of programs (27). The tree may be downloaded either as an image or in Newick format for easy representation in other tools. Multiple sequence alignments of the designed sequences, either alone or in combination with the parent family sequences for which they were designed, are also provided. Here again, the designed intermediate sequences are appropriately annotated with details pertaining to parent families and level of design.
CONCLUSIONS
The goal of NrichD database is to relate proteins that have diverged extensively in sequence space and whose relationship might become evident through the availability of structure. For this purpose, it relies on artificially/computationally generated intermediate sequences that have been derived, in part, from structure. But its use should be seen, in our opinion, as a step in connecting distantly related proteins through methods that are not limited by the lack of structural information. Therefore, the foldability/stability of such intermediary proteins although interesting is immaterial for the question on hand. Further, protein sequence sampling in evolutionary time scales does not preclude the exploration of residues that might ultimately be selected out since they did not fold suitably. However, to ensure that theoretical estimations of its ability to fold appropriately are accounted for, in future versions of the method, we plan to incorporate and implement many filters to evaluate these aspects.
The success rate of searches in well established databases is reliant on the nature of the query (7,28). Therefore, if outliers of the family are employed as queries then ‘bona fide relatives’ may remain undetected even in simple searches. For example, searches initiated with a query (d1poca_: a.133.1.1) that belonged to the Insect phospholipase A2 fold, always picked up hits from the parent family and additionally Vertebrate phospholipase A2 family (a.133.1.2) as hits using HHPred (29). However, hits from Prokaryotic phospholipase A2 family (a.133.1.3) could not be detected. On the other hand, searches in the SCOP-NrichD data set could find all these relationships through the designed sequences. (11). Clearly, each method has its own strengths in detecting homologs and an integrated assessment involving multiple tools is recommended to resolve the veracity of hits obtained in such searches.
The AS-DB and the enriched data sets provided here can be easily downloaded and plugged into other data sets and queried through any sequence search method. Further, updates of the database will be made in line with the updates of the underlying SCOP and PFAM databases. Intermediary sequences that are generated between different families of a fold are annotated with family and fold details of the parent fold. Such detailed annotation is very useful to track the searches and determines the proximity of the intermediately related sequences, which act as ‘connectors’ between protein families, to their parent PSSMs and families. Therefore, a handle on the route/direction of search that mediated the connection is provided to the user.
An important point to note is that we recommend that searches in the NrichD database are always performed in addition to searches in natural database. The main driver for this being that while searches in natural databases are able to cover a large number of ‘obvious’ relationships, such as finding relatives within the same family as the query, etc., the strength of searches in databases such as NrichD is the number of ‘non-obvious’ connections. This refers to its ability to detect relationships between families from diverse superfamilies within the fold. We have therefore chosen to present all relationships, those within the NrichD database as also relationships within natural databases hosted on our server, with the expectation that the user will be able to recognize obvious false positives using detailed annotation of the designed sequences.
Acknowledgments
R.M. is grateful to the IISc. Mathematics Initiative (IMI) for PhD research fellowship.
FUNDING
Mathematical Biology program of Department of Science and Technology as well as by the Department of Biotechnology [BT/01/COE/09/01], Government of India. Financial support through the Women Scientist scheme-A, Department of Science and Technology, Government of India [to S.S.]. Funding for open access charge: Department of Biotechnology, Government of India.
Conflict of interest statement. None declared.
REFERENCES
- 1.Park J., Teichmann S.A., Hubbard T., Chothia C. Intermediate sequences increase the detection of homology between sequences. J. Mol. Biol. 1997;273:349–354. doi: 10.1006/jmbi.1997.1288. [DOI] [PubMed] [Google Scholar]
- 2.Salamov A.A., Suwa M., Orengo C.A., Swindells M.B. Combining sensitive database searches with multiple intermediates to detect distant homologues. Protein Eng. 1999;12:95–100. doi: 10.1093/protein/12.2.95. [DOI] [PubMed] [Google Scholar]
- 3.Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aravind L., Koonin E.V. Gleaning non-trivial structural, functional and evolutionary information about proteins by iterative database searches. J. Mol. Biol. 1999;287:1023–1040. doi: 10.1006/jmbi.1999.2653. [DOI] [PubMed] [Google Scholar]
- 5.Bateman A., Finn R.D. SCOOP: a simple method for identification of novel protein superfamily relationships. Bioinformatics. 2007;23:809–814. doi: 10.1093/bioinformatics/btm034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jung I., Kim D. SIMPRO: simple protein homology detection method by using indirect signals. Bioinformatics. 2009;25:729–735. doi: 10.1093/bioinformatics/btp048. [DOI] [PubMed] [Google Scholar]
- 7.Bhadra R., Sandhya S., Abhinandan K.R., Chakrabarti S., Sowdhamini R., Srinivasan N. Cascade PSI-BLAST web server: a remote homology search tool for relating protein domains. Nucleic Acids Res. 2006;34:W143–W146. doi: 10.1093/nar/gkl157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sandhya S., Kishore S., Sowdhamini R., Srinivasan N. Effective detection of remote homologues by searching in sequence dataset of a protein domain fold. FEBS Lett. 2003;552:225–230. doi: 10.1016/s0014-5793(03)00929-3. [DOI] [PubMed] [Google Scholar]
- 9.Margelevicius M., Venclovas C. PSI-BLAST-ISS: an intermediate sequence search tool for estimation of the position-specific alignment reliability. BMC Bioinformatics. 2005;6:185–194. doi: 10.1186/1471-2105-6-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holm L. Unification of protein families. Curr. Opin. Struct. Biol. 1998;8:372–379. doi: 10.1016/s0959-440x(98)80072-9. [DOI] [PubMed] [Google Scholar]
- 11.Mudgal R., Sowdhamini R., Chandra N., Srinivasan N., Sandhya S. Filling-in void and sparse regions in protein sequence space by protein-like artificial sequences enables remarkable enhancement in remote homology detection capability. J. Mol. Biol. 2014;426:962–979. doi: 10.1016/j.jmb.2013.11.026. [DOI] [PubMed] [Google Scholar]
- 12.Sandhya S., Mudgal R., Jayadev C., Abhinandan K.R., Sowdhamini R., Srinivasan N. Cascaded walks in protein sequence space: use of artificial sequences in remote homology detection between natural proteins. Mol. BioSyst. 2012;8:2076–2084. doi: 10.1039/c2mb25113b. [DOI] [PubMed] [Google Scholar]
- 13.Koehl P., Levitt M. De novo protein design. I. In search of stability and specificity. J. Mol. Biol. 1999;293:1161–1181. doi: 10.1006/jmbi.1999.3211. [DOI] [PubMed] [Google Scholar]
- 14.Dahiyat B.I., Mayo S.L. De novo protein design: fully automated sequence selection. Science. 1997;278:82–87. doi: 10.1126/science.278.5335.82. [DOI] [PubMed] [Google Scholar]
- 15.Dahiyat B.I., Sarisky C.A., Mayo S.L. De novo protein design: towards fully automated sequence selection. J. Mol. Biol. 1997;273:789–796. doi: 10.1006/jmbi.1997.1341. [DOI] [PubMed] [Google Scholar]
- 16.Socolich M., Lockless S.W., Russ W.P., Lee H., Gardner K.H., Ranganathan R. Evolutionary information for specifying a protein fold. Nature. 2005;437:512–518. doi: 10.1038/nature03991. [DOI] [PubMed] [Google Scholar]
- 17.Kumar A., Cowen L. Augmented training of hidden Markov models to recognize remote homologs via simulated evolution. Bioinformatics. 2009;25:1602–1608. doi: 10.1093/bioinformatics/btp265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cai W., Pei J., Grishin N.V. Reconstruction of ancestral protein sequences and its applications. BMC Evol. Biol. 2004;4:33–55. doi: 10.1186/1471-2148-4-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pei J., Dokholyan N.V., Shakhnovich E.I., Grishin N.V. Using protein design for homology detection and active site searches. Proc. Natl. Acad. Sci. U.S.A. 2003;100:11361–11366. doi: 10.1073/pnas.2034878100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murzin A.G., Brenner S.E., Hubbard T., Chothia C. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- 21.Andreeva A., Howorth D., Chandonia J.M., Brenner S.E., Hubbard T.J., Chothia C., Murzin A.G. Data growth and its impact on the SCOP database: new developments. Nucleic Acids Res. 2008;36:D419–D425. doi: 10.1093/nar/gkm993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Finn R.D., Bateman A., Clements J., Coggill P., Eberhardt R.Y., Eddy S.R., Heger A., Hetherington K., Holm L., Mistry J., et al. Pfam: the protein families database. Nucleic Acids Res. 2014;42:D222–D230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chandonia J.M., Hon G., Walker N.S., Lo Conte L., Koehl P., Levitt M., Brenner S.E. The ASTRAL Compendium in 2004. Nucleic Acids Res. 2004;32:D189–D192. doi: 10.1093/nar/gkh034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suzek B.E., Huang H., McGarvey P., Mazumder R., Wu C.H. UniRef: comprehensive and non-redundant UniProt reference clusters. Bioinformatics. 2007;23:1282–1288. doi: 10.1093/bioinformatics/btm098. [DOI] [PubMed] [Google Scholar]
- 25.Punta M., Coggill P.C., Eberhardt R.Y., Mistry J., Tate J., Boursnell C., Pang N., Forslund K., Ceric G., Clements J., et al. The Pfam protein families database. Nucleic Acids Res. 2012;40:D290–D301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eddy S.R. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- 27.Retief J.D. Phylogenetic analysis using PHYLIP. Methods Mol. Biol. 2000;132:243–258. doi: 10.1385/1-59259-192-2:243. [DOI] [PubMed] [Google Scholar]
- 28.Kaushik S., Mutt E., Chellappan A., Sankaran S., Srinivasan N., Sowdhamini R. Improved detection of remote homologues using cascade PSI-BLAST: influence of neighbouring protein families on sequence coverage. PloS ONE. 2013;8:e56449. doi: 10.1371/journal.pone.0056449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Soding J., Biegert A., Lupas A.N. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:W244–W248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]


