Abstract
A triplex target DNA site (TTS), a stretch of DNA that is composed of polypurines, is able to form a triple-helix (triplex) structure with triplex-forming oligonucleotides (TFOs) and is able to influence the site-specific modulation of gene expression and/or the modification of genomic DNA. The co-localization of a genomic TTS with gene regulatory signals and functional genome structures suggests that TFOs could potentially be exploited in antigene strategies for the therapy of cancers and other genetic diseases. Here, we present the TTS Mapping and Integration (TTSMI; http://ttsmi.bii.a-star.edu.sg) database, which provides a catalog of unique TTS locations in the human genome and tools for analyzing the co-localization of TTSs with genomic regulatory sequences and signals that were identified using next-generation sequencing techniques and/or predicted by computational models. TTSMI was designed as a user-friendly tool that facilitates (i) fast searching/filtering of TTSs using several search terms and criteria associated with sequence stability and specificity, (ii) interactive filtering of TTSs that co-localize with gene regulatory signals and non-B DNA structures, (iii) exploration of dynamic combinations of the biological signals of specific TTSs and (iv) visualization of a TTS simultaneously with diverse annotation tracks via the UCSC genome browser.
INTRODUCTION
Triplex-forming oligonucleotides (TFOs) are short sequences of nucleic acids that can bind to the major groove of duplex nucleic acids and promote the formation of triple-helix (triplex) structures (1,2).
Recently, antigene technology (AT) that can control and/or modulate cellular functions at the genomic DNA level via triplex-based approaches (3) has attracted substantial interest due to the development of novel TFO strategies (4) and the increasing evidence that TFOs could be important in the regulation of gene expression and the modification of a specific gene region both in vitro and in vivo (5–9). It has been shown that TFOs (e.g. anti-IGF-I) can effectively suppress the development of tumors in animal models and in human malignancies (9,10). Moreover, an antisense anti-IGF-I TFO has been used in Phase I and II clinical trials to treat patients with glioblastoma and other types of cancers (9). These applications of AT should be considered promising models of the specific antigene therapy of cancers and other genetic disorders.
TFO-based treatment approaches have been developed to modulate gene expression, to initiate site-specific mutagenesis and to modify specific genomic DNA regions (4–5,11–13). Although the number of successful applications of AT in animal models has increased (8,13–15), the possible off-target binding sites in these cases is still relatively poorly understood. Off-target binding limits the specificity of therapeutic applications and is a great challenge for functional genomic studies. Indeed, with some probability, TFOs could bind at other genomic locations that have the same (or a similar) sequence structure as that of triplex target DNA sites (TTSs), resulting in both unexpected off-target effects of the TFO binding and the modification of DNA conformation. For example, Rogers et al. (15) showed that a TFO (PNA-Antp conjugate) could target almost 20 000 identical TTSs across the mouse genome (16); hence, the presence of triplex-forming structures at off-target genomic locations could result in unexpected downstream physical, chemical and biological effects.
Recently, Buske et al. (16) developed Triplex-Inspector, a program that takes a genomic locus of interest and searches for all putative triplex target sites via the Triplexator algorithm (17). Each of these putative targets is subsequently examined for its uniqueness by searching the genome for any locus with high-sequence similarity to that site. Triplex-Inspector could address the off-target issue by automatically determining theoretical possible off-target sites defined by Triplexator algorithm for each TTS candidate. By contrast, for other web-service tools, users must investigate potential off-target sites on a candidate-by-candidate basis (18,19).
However, only identifying theoretical possible off-target sites is not sufficient for finding suitable TTS candidates for AT applications. In the context of the modulation of gene expression via a TFO-based treatment approach, the genome cartography of a specific TTS should also be an important consideration when identifying relevant TTS candidates because a TTS co-localizes with relevant gene regions (such as the gene promoter) and their regulatory signals (also termed potential gene function-associated TTSs or pGFA-TTSs; e.g. transcription factor binding sites (TFBSs) and chromatin accessibility regions). Fortunately, the human genome has the most comprehensive lists of such functional regulatory signals or regions provided by the Encyclopedia of DNA Elements (ENCODE) Consortium (20,21). We propose that an integration of publicly available and well-organized data from the ENCODE project with TTS mapping could provide valuable information for the reliable selection of appropriate pGFA-TTSs for any given AT application.
Here, we developed the TTS Mapping and Integration (TTSMI) database, which provides an integrated and comprehensive catalog of TTSs and pGFA-TTSs that are unique to the human genome. TTSMI was designed as a user-friendly bioinformatics tool that facilitates the following: (i) fast searching and selecting of TTSs using several search terms (e.g. genomic location, gene ID, NCBI RefSeq ID, TTS ID, gene symbol and gene description keywords) and criteria associated with sequence stability (e.g. percent guanine content and dinucleotide content) and specificity (e.g. number of potential off-target sites); (ii) interactive filtering of appropriate TTSs that co-localize with specific gene regions, genome regulatory signals and even predicted non-B DNA structures; (iii) exploration of the dynamic combinations of structural and functional annotations of specific TTSs and (iv) visualization of the TTS with diverse and biologically relevant annotation tracks via the UCSC genome browser. TTSMI also incorporates experimental data from the ENCODE project, which includes computationally integrated ChIP-seq data (chromatin accessibility, TFBS and chromatin state) from several dozen cell lines. The consolidation of all this information in an integrated database can increase the feasibility of identifying pGFA-TTSs that could be appropriate candidates as therapeutic targets for antigene therapy.
MATERIALS AND METHODS
New TTS mapping algorithm
To identify genomic TTSs with the potential to function as a therapeutic target, the TTS mapping algorithm (18) was re-designed to search for all possible polypurine sequences subject to the following criteria: (i) length ranging from 15 to 30 nucleotides, (ii) guanine content ranging from 20% to 90% and (iii) number of pyrimidine interruptions no greater than 1.
Each TTS was tagged by a unique ID. Each TTS ID fully describes the location of the TTS in the genome. For example, the information fields in the TTS ID ‘TTS.17.25.48279135’ are separated by the dot character. This particular TTS is located on chromosome 17, its length is 25 nucleotides and the start position of the TTS is at coordinate 48279135.
Data sources
TTSs were identified from the TTS mapping algorithm by searching against the human reference genome sequence hg19 (NCBI build GRCh37). The short sequence alignment program Bowtie (22) was used to assess the uniqueness of each TTS in the human genome.
Several genome annotation tracks from the UCSC (http://genome.ucsc.edu) genome browser (20) were chosen and retrieved for integration with the TTS information. These tracks were retrieved from several annotation groups: (i) gene annotation (refGene table); (ii) gene regulation tracks containing CpG Islands (cpgIslandExt table) and ENCODE Integrated ChIP-seq gene regulation (wgEncodeRegDnaseClusteredV2, wgEncodeRegTfbsClusteredV3, wgEncodeBroadHmm tables), as well as TFBSs conserved in the human/mouse/rat alignment (tfbsConsSites table); (iii) evolutionary conservation across 100 vertebrate species (this track is collected in the phastConsElements100way table and refers to the predictions of conserved elements produced by the phastCons program based on multiple alignments of 100 vertebrate species) and (iv) variation and repeats containing SNPs, i.e. single-nucleotide polymorphisms (snp138, snp138Common, snp138CodingDbSnp and snp138Flagged tables) and DNA repeat elements (rmsk table).
Furthermore, we also integrated information about putative non-B DNA structures (23), including A-phased repeats, G-quadruplex motifs, Z-DNA motifs, direct repeats, inverted repeats, mirror repeats and short tandem repeats, because these structures could potentially compete with triplex formation and/or modify triplex stabilization.
Construction and implementation of the TTSMI database
The detailed construction of the TTSMI database is shown in Supplementary Figure S1. The dynamic web interface of TTSMI was developed using the PHP server-side scripting language and the JavaScript language along with jQuery, jQuery Slider (http://egorkhmelev.github.io/jslider) and DataTables (https://datatables.net). TTSMI data are stored in an optimized MySQL relational database, which run on a Linux-Apache web server.
DATABASE DESCRIPTION
To systematically study the structural and functional features of TTSs, computational genomics and integrative bioinformatics approaches could be helpful. In silico-defined TTSs that co-localize with genes and their regulatory elements will be termed potential gene function-associated TTSs (pGFA-TTSs). Here, we developed a tool called the TTSMI database, which helps users to search for both TTSs and pGFA-TTSs in the human genome. Unlike the TTS mapping performed in 2009 (18), the TTS mapping algorithm described in the present study was re-designed and implemented in the Python language to identify potential therapeutic targets (see in Materials and Methods). Using the new algorithm, we predicted more than 170 million TTSs in the human genome. Only 36 million (of 170 million) TTSs are uniquely located in the human genome, and they are distributed across 23 665 RefSeq genes (94% of RefSeq genes) and/or their putative promoters (2 kb upstream of transcription start sites) and downstream regions (2 kb downstream of transcription end sites). These unique TTSs were integrated with the genomic locations of genes, gene regulatory elements and non-B DNA structure elements (genome annotation tracks).
TTSs were separated into two groups according to percentage of guanine content including TTSs with ≥40% G (GA TFOs form more stable triplex where higher %G) and TTSs with <40% G (TC TFOs form more stable triplex where higher %A) (24). In each group, TTSs were further grouped according to number of pyrimidine interruption where 0 determines no pyrimidine base interruption and 1 determine 1 pyrimidine base interruption. Moreover, the number of GpGpG is another factor that could discriminate TTSs into two sub-groups including ≥4 GpGpG and <4 GpGpG. The TTSs containing ≥4 GpGpG tend to form alternative structures (e.g. G-quadruplex). Table 1 shows the distribution of the number of TTSs for a particular criterion and the number of pGFA-TTSs for several genome annotation tracks. In particular, this table shows that according to the conditions (%G <40%, no pyrimidine interruptions, #GpGpGG<4) the 2,349 × 103 TTSs could be found in the human genome and the 236 × 103 of these TTSs are pGFA-TFBSs co-localized with TFBS defined by Chip-seq. Interestingly, the number TTSs in the case of high G rich TTS subset (%G>=40%, no pyrimidine interruptions, #GpGpG<4) as well as the corresponding number of pGTA-TTSs is more enriched in comparison to previous case. All TTSs are stored in the MySQL relational database on our server and can be searched and filtered using the dynamic web interface of the TTSMI database.
Table 1. Statistics for the TTSMI database.
| TTS search criteria | Number of TTSs (thousands) | Number of pGFA-TTSs (thousands) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| %G | pyr | #GpGpG | with chromatin accessibility | with chromatin state | with TFBS (ChIP-seq) | with CpG Island | with G4 | with non-B DNA | with TFBS (Motif) | with evolutionarily conserved regions | with repeat elements | with SNPs (138) | |
| <40 | 0 | <4 | 2349 | 119 | 506 | 236 | 2 | 57 | 2003 | 13 | 66 | 2117 | 1331 |
| <40 | 1 | <4 | 9259 | 820 | 2043 | 1134 | 17 | 89 | 5164 | 138 | 503 | 6313 | 4166 |
| ≥40 | 0 | <4 | 6095 | 798 | 1661 | 1042 | 28 | 1178 | 5072 | 48 | 192 | 5483 | 2993 |
| ≥40 | 1 | <4 | 17047 | 3579 | 5238 | 3417 | 195 | 2088 | 9136 | 337 | 971 | 11 287 | 7086 |
| ≥40 | 0 | ≥4 | 634 | 137 | 243 | 103 | 6 | 588 | 628 | 6 | 23 | 617 | 357 |
| ≥40 | 1 | ≥4 | 930 | 270 | 350 | 198 | 26 | 838 | 894 | 25 | 62 | 809 | 506 |
%G, guanine content; pyr, #pyrimidine interruptions; #GpGpG, number of three consecutive guanines; TTSs, triplex target DNA sites; pGFA-TTSs, potential gene function-associated TTSs; G4, G-quadruplex.
To use our TTSMI database
To use the TTSMI database, users must know the gene or chromosome location that is a candidate for finding TFO targets or TTSs. The TTS search options can be used to identify a list of TTSs for the gene or chromosome location; then, TTS filtering options can be used to narrow the list of TTSs according to AT applications, e.g. for applications that modulate gene expression, the list can be limited to TTSs that overlap with the gene regulatory signals. Detailed information about the individual TTSs can be viewed via either the TTS information web page or the chromosome view.
TTS search options
The TTSMI database provides several search options for TTS searches (Figure 1A). The user can search TTSs by TTS ID, Entrez gene ID, NCBI reference sequence (refSeq) ID, chromosome location, gene symbol or gene description keywords. For example, Figure 1A illustrates a TTS search for Gene ID 4609, which represents the c-Myc gene. In addition, to increase the speed of the search, TTSs were categorized into groups based on basic search criteria for TTSs, including the guanine content (%G), number of pyrimidine interruptions and number of three consecutive guanines (GpGpG) (see Supplementary Figure S2 and Table 1).
Figure 1.
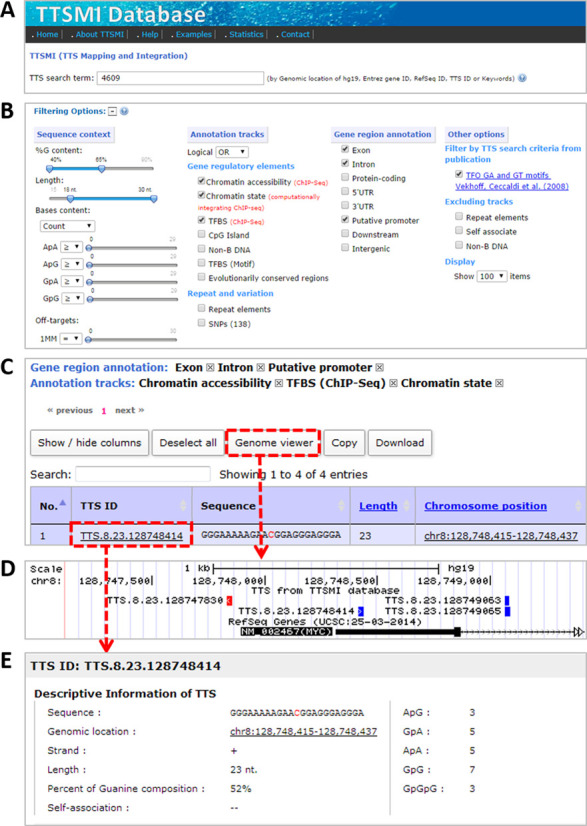
A screenshot of the TTSMI database and the search results. (A) TTS search terms and basic search criteria (e.g. Gene ID 4609). (B) TTS filtering options, which are categorized into four groups: ‘Sequence context’ (sequence content of the TTS), ‘Gene region annotation’ (the TTS is located in a specific gene annotation regions), ‘Annotation tracks’ (the TTS shares a common nucleotide with the annotation tracks) and ‘Other options’ (other helpful options). (C) A close-up screenshot of TTS search results showing the current active filtering options, including gene regions and annotation tracks, as well as the paging, genome visualization and downloading buttons at the bottom. (D) Chromosome view of TTS locations (Watson strand in blue and Crick strand in red) in the human genome via the UCSC genome browser. (E) TTS information page of TTS.8.23.128748414 (the detailed information is available in Supplementary Figure S3).
TTS filtering options
In addition to the TTS search options, one of the main features of the TTSMI database is the ability to filter TTSs and/or pGFA-TTSs using TTS filtering options (Figure 1B). The TTS filtering options were designed to use a combination of filters to narrow the search results and find a biologically relevant TTS. The TTS filtering options are categorized into four groups: ‘Sequence context’, ‘Annotation tracks’, ‘Gene region annotation’ and ‘Other options’.
The first option, ‘Sequence context’, allows users to filter TTS results by specifying a range of nucleotide properties, such as%G content, length, number or frequency of dinucleotides (ApA, GpA, ApG and GpG), and number of potential off-target TTSs with one base mismatch.
The second option, ‘Annotation tracks’, allows users to filter the pGFA-TTS results by selecting TTSs that are integrated with several annotation tracks (an annotation track is the list of genomic locations for a given type of known biological information), i.e. a TTS that has at least one nucleotide that overlaps with the annotation track. In total, the TTSMI database has information about 10 annotation tracks, which include gene regulatory elements, repeat elements and variations.
The third option, ‘Gene region annotation’, allows users to filter the TTS and/or pGFA-TTS results by whether the TTSs are located in specific gene annotation regions, including exon, intron, protein-coding, 5′UTR, 3′UTR, putative promoter, downstream and intergenic regions.
The last option, ‘Other options’, allows users to filter the TTS search results by some useful criteria; for example, the filter can be pre-set according to criteria similar to those in a previous publication (24). Furthermore, users can filter TTSs by their potential to fold into one of several distinct classes of non-B DNA structures (e.g. cruciform (type of inverted repeat); G-quadruplexes, triplexes or H-DNA (type of mirror repeat); hairpins (type of direct repeat) (23)) because the formation of these non-B DNA structures could modulate or obstruct the formation of a TFO and its TTS. For instance, in certain G-rich genome regions, G-triplexes might be intermediates that become stabilized in G-quadruplexes that compete with triplex formation (25). Each individual non-B structure class has unique sequence requirements and physical properties, and in TTSMI, the TTSs associated with these structures are categorized separately, providing specific structure-function information that might be used to find the most relevant TTS for AT applications and research aims.
TTS search results
The result report provided by the TTSMI database presents the descriptive information, gene region annotation and annotation tracks of the TTSs (Supplementary Figure S2). The TTS descriptive information shows basic information about a particular TTS, including the TTS ID, sequence content, length, chromosome location, strand, %G content, number or frequency of dinucleotides (ApA, GpA, ApG and GpG), number of GpGpG occurrences and number of off-target TTSs with one mismatch (Supplementary Figure S2A). The gene region annotation fields (Supplementary Figure S2B) shows genes and specific gene regions that overlap with the TTS. The complete set of TTSs with a particular search/filter criterion in a particular gene is provided via the gene name with an underlined hyperlink. The annotation track fields (Supplementary Figure S2C) show the number of overlaps between the TTS and each selected annotation track in columns. Detailed information about the overlap between the TTS and a given annotation track is provided via the number with an underlined hyperlink.
Furthermore, the TTSMI database provides additional useful features, such as paging, sorting, column reordering and searching, and data selection, downloading and visualization (Figure 1C) that allow users to interact with the search results. One of the highlighted features is the ‘Genome viewer’, where all TTSs or selected TTSs in the table can be displayed in a chromosome view via the UCSC genome browser (Figure 1D).
In addition, detailed information about a particular TTS is provided via the TTS ID with an underlined hyperlink, which links to the TTS information web page (Figure 1E and Supplementary Figure S3). The TTS information web page can be separated into four parts: (i) descriptive information about the TTS, (ii) statistics of the possible TTS off-target sites, (iii) alternative TTSs in the same locus and (iv) information about the genes and annotation tracks that overlap with the TTS. The first part (Supplementary Figure S3A) shows the descriptive information about the TTS that is shown in the result report, except for the ‘self-association’, i.e. the potential for the formation of an intra-molecular triplex structure in which more than 50% of the content is a simple repeat (e.g. GA, AG, GAA, AAG, GGA and AGG). The second part (Supplementary Figure S3B) shows TTS off-target sites in the human genome. Unlike the off-target sites defined by Buske et al. (16), here, the number of TTS off-target sites was defined by counting the number of aligned results from the alignment of the TTS against the human genome, allowing for one or two mismatches. These TTS off-target sites could potentially form a triplex structure with the TFO that has much less triplex stability than that for the TTS without mismatches (26,27). However, as the number of TTS off-target sites in the genome decreases, the TFO will bind to its target more specifically. In Supplementary Figure S3B, TTS off-target sites are shown in a bar graph, where the x-axis shows the sequence of the TTS from the 5′- to 3′-end, and the y-axis is the number of mismatched base(s) of the TTS off-target site at a given base position (see Supplementary Figure S4). In addition, if the number of TTS off-target sites is less than or equal to 30, a donut graph will be shown. This graph represents the frequency of the TTS off-target sites located in genic or chromatin accessibility regions, both genic and chromatin accessibility regions or in an intergenic region. Therefore, the TTS off-target sites report is useful for exploring the potential effect of off-target TTSs in the genome; thus, this report facilitates the development and optimization of a TFO design. The third part (Supplementary Figure S3C) represents a list of alternative TTSs, which the user might also consider as additional candidates in the same genomic locus. The last part (Supplementary Figure S3D) provides detailed information about the gene regions and annotation tracks that overlap with a given TTS.
BIOLOGICAL INTERPRETATION
To illustrate the performance of the TTSMI database, we employed the TTSMI database to analyze applications of triplex technology, e.g. the modulation of the gene expression of IGF1 (28) and TNF-α (7) (see also the ‘Examples’ page for the TTSMI database).
Here, we provide an example of the gene regulation via AT using TFO recognition of the TTS located within the promoter region of the gene. Gene regulation via TFO recognition has largely been used to modulate gene expression. In studies of in vitro and in vivo systems, TFOs have been shown to modulate gene expression during the transcription process by interfering with the binding of transcription factors or the transcription initiation complex to the target DNA (5,11–12). One of the studies using this approach examined the tumor necrosis factor-α (TNF-α) gene (7). An anti-TNF-α TFO was used to target the TTS located within the promoter region of the TNF-α gene (rn4, chr20:3660925–3660945, –54 bases from the TSS, 5′-TCGAAAAGGGGTGGGAGAAGG-3′) and modulate its gene expression, resulting in the reduction of disease development in both acute and chronic rat arthritis models. Using the TTSMI database to search for TTSs in the human TNF gene promoter with defined parameter settings (G content higher than 40%, 1 pyrimidine interruption, fewer than 4 GpGpGs, TTS off-target site of 1 mismatch equal to zero and overlap between the TTS and ‘Putative promoter’, ‘Chromatin accessibility (ChIP-seq)’ and ‘TFBS (ChIP-seq)’), we identified two TTSs that passed the filtering criteria: TTS.6.21.31543279 (hg19, chr6:31543280–31543300, -43 bases from the TSS, 5′-GAGAGGAGGGCGGGGAAAGAA-3′) and TTS.6.20.31543279 (hg19, chr6:31543280–31543299, -44 bases from the TSS, 5′- AGAGGAGGGCGGGGAAAGAA-3′). Interestingly, these TTSs were found to be conserved between the human and rat genomes (Figure 2). In addition, these TTSs co-localize with chromatin accessibility regions defined by ChIP-seq in 56 cell types, 35 TFBSs defined by ChIP-seq and 2 TFBS motifs in MAZ- and EBF1-binding regions. This finding suggests that both TTSs (TTS.6.21.31543279 and TTS.6.20.31543279) could be good candidates for further investigation as part of a therapeutic tool for human TNF-dependent inflammatory disorders.
Figure 2.
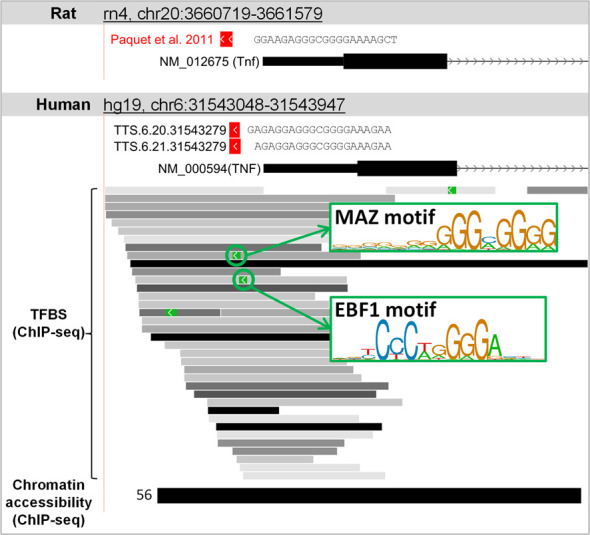
The TTS locations of anti-TNF-α targets in the mouse chromosome and two TTS candidates in the human chromosome.
CONCLUSIONS AND FUTURE WORK
With the development of the TTSMI database for the human genome, we provide the scientific and industrial communities with a new resource to identify the locations of unique TTSs; these TTSs can be integrated with genomic non-B DNA structures, NGS-detected hot spots and functionally annotated regulatory signals. The integration of a TTS and this relevant biological knowledge is key to finding appropriate therapeutic targets for developing antigene therapy via a triplex-based approach. The user can quickly search for and filter TTS and pGFA-TTS candidates using our large collection of unique TTS locations. Moreover, the specificity of a TTS is one of the most important factors for controlling off-target effects. Therefore, we provide not only TTSs that are located solely in the specified genome but simultaneously provide an off-target report to ensure that users will choose to target a TTS that minimizes off-target effects. We believe that the TTSMI database will help non-bioinformaticians to easily find suitable TTS candidates for antigene therapy development via triplex technology. Future work entails the extension of the database to the mouse and rat genomes, as well as providing a list of the evolutionary conserved locations for the human TTSs in the mouse and rat genomes. This extension will help researchers find appropriate animal model TTSs that could be translated into human antigene therapy.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
Thanks to Mr. Ow Ghim Siong and Mr. Arsen Batagov for reading the manuscript and their very useful comments and suggestions, and Bioinformatics Institute for providing the web server.
FUNDING
Biomedical Research Council of Agency for Science, Technology and Research (A*STAR), Singapore.
Conflict of interest statement. None declared.
REFERENCES
- 1.Doan T. Le., Perrouault L., Praseuth D., Habhoub N., Decout J.L., Thuong N.T., Lhomme J., Hélène C. Sequence-specific recognition, photocrosslinking and cleavage of the DNA double helix by an oligo-[alpha]-thymidylate covalently linked to an azidoproflavine derivative. Nucleic Acids Res. 1987;15:7749–7760. doi: 10.1093/nar/15.19.7749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mose H.E., Dervan P.B. Sequence-specific cleavage of double helical DNA by triple helix formation. Science. 1987;238:645–650. doi: 10.1126/science.3118463. [DOI] [PubMed] [Google Scholar]
- 3.Helene C. The anti-gene strategy: control of gene expression by triplex-forming-oligonucleotides. Anti-Cancer Drug Des. 1991;6:569–584. [PubMed] [Google Scholar]
- 4.Mukherjee A., Vasquez K.M. Triplex technology in studies of DNA damage, DNA repair, and mutagenesis. Biochimie. 2011;93:1197–1208. doi: 10.1016/j.biochi.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Duca M., Vekhoff P., Oussedik K., Halby L., Arimondo P.B. The triple helix: 50 years later, the outcome. Nucleic Acids Res. 2008;36:5123–5138. doi: 10.1093/nar/gkn493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rogers F.A., Hu R.H., Milstone L.M. Local delivery of gene-modifying triplex-forming molecules to the epidermis. J. Invest. Dermatol. 2013;133:685–691. doi: 10.1038/jid.2012.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paquet J., Henrionnet C., Pinzano A., Vincourt J.B., Gillet P., Netter P., Chary-Valckenaere I., Loeuille D., Pourel J., Grossin L. Alternative for anti-TNF antibodies for arthritis treatment. Mol. Ther. 2011;19:1887–1895. doi: 10.1038/mt.2011.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McNeer N.A., Chin J.Y., Schleifman E.B., Fields R.J., Glazer P.M., Saltzman W.M. Nanoparticles deliver triplex-forming PNAs for site-specific genomic recombination in CD34+ human hematopoietic progenitors. Mol. Ther. 2011;19:172–180. doi: 10.1038/mt.2010.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Trojan J., Pan Y.X., Wei M.X., Ly A., Shevelev A., Bierwagen M., Ardourel M.Y., Trojan L.A., Alvarez A., Andres C., et al. Methodology for Anti-Gene Anti-IGF-I Therapy of Malignant Tumours. Chemother. Res. Pract. 2012;2012:721873. doi: 10.1155/2012/721873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Trojan J., Ly A., Wei M.X., Bierwagen M., Kopinski P., Pan Y., Ardourel M.Y., Dufour T., Shevelev A., Trojan L.A., et al. Antisense anti IGF-I cellular therapy of malignant tumours: immune response in cancer patients. Biomed. Pharmacother. 2010;64:576–578. doi: 10.1016/j.biopha.2010.01.019. [DOI] [PubMed] [Google Scholar]
- 11.Jain A., Magistri M., Napoli S., Carbone G.M., Catapano C.V. Mechanisms of triplex DNA-mediated inhibition of transcription initiation in cells. Biochimie. 2010;92:317–320. doi: 10.1016/j.biochi.2009.12.012. [DOI] [PubMed] [Google Scholar]
- 12.Cooney M., Czernuszewicz G., Postel E.H., Flint S.J., Hogan M.E. Site-specific oligonucleotide binding represses transcription of the human c-myc gene in vitro. Science. 1988;241:456–459. doi: 10.1126/science.3293213. [DOI] [PubMed] [Google Scholar]
- 13.Vasquez K.M., Narayanan L., Glazer P.M. Specific mutations induced by triplex-forming oligonucleotides in mice. Science. 2000;290:530–533. doi: 10.1126/science.290.5491.530. [DOI] [PubMed] [Google Scholar]
- 14.Schleifman E.B., McNeer N.A., Jackson A., Yamtich J., Brehm M.A., Shultz L.D., Greiner D.L., Kumar P., Saltzman W.M., Glazer P.M. Site-specific genome editing in PBMCs with PLGA nanoparticle-delivered PNAs confers HIV-1 resistance in humanized mice. Mol. Ther. Nucleic Acids. 2013;2:e135. doi: 10.1038/mtna.2013.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rogers F.A., Lin S.S., Hegan D.C., Krause D.S., Glazer P.M. Targeted gene modification of hematopoietic progenitor cells in mice following systemic administration of a PNA-peptide conjugate. Mol. Ther. 2012;20:109–118. doi: 10.1038/mt.2011.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Buske F.A., Bauer D.C., Mattick J.S., Bailey T.L. Triplex-Inspector: an analysis tool for triplex-mediated targeting of genomic loci. Bioinformatics. 2013;29:1895–1897. doi: 10.1093/bioinformatics/btt315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Buske F.A., Bauer D.C., Mattick J.S., Bailey T.L. Triplexator: detecting nucleic acid triple helices in genomic and transcriptomic data. Genome Res. 2012;22:1372–1381. doi: 10.1101/gr.130237.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jenjaroenpun P., Kuznetsov V.A. TTS mapping: integrative WEB tool for analysis of triplex formation target DNA sequences, G-quadruplets and non-protein coding regulatory DNA elements in the human genome. BMC Genom. 2009;10(Suppl. 3):S9. doi: 10.1186/1471-2164-10-S3-S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gaddis S.S., Wu Q., Thames H.D., DiGiovanni J., Walborg E.F., MacLeod M.C., Vasquez K.M. A web-based search engine for triplex-forming oligonucleotide target sequences. Oligonucleotides. 2006;16:196–201. doi: 10.1089/oli.2006.16.196. [DOI] [PubMed] [Google Scholar]
- 20.Karolchik D., Barber G.P., Casper J., Clawson H., Cline M.S., Diekhans M., Dreszer T.R., Fujita P.A., Guruvadoo L., Haeussler M., et al. The UCSC Genome Browser database: 2014 update. Nucleic Acids Res. 2014;42:D764–D770. doi: 10.1093/nar/gkt1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Consortium E.P., Bernstein B.E., Birney E., Dunham I., Green E.D., Gunter C., Snyder M. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cer R.Z., Donohue D.E., Mudunuri U.S., Temiz N.A., Loss M.A., Starner N.J., Halusa G.N., Volfovsky N., Yi M., Luke B.T., et al. Non-B DB v2.0: a database of predicted non-B DNA-forming motifs and its associated tools. Nucleic Acids Res. 2013;41:D94–D100. doi: 10.1093/nar/gks955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vekhoff P., Ceccaldi A., Polverari D., Pylouster J., Pisano C., Arimondo P.B. Triplex formation on DNA targets: how to choose the oligonucleotide. Biochemistry. 2008;47:12277–12289. doi: 10.1021/bi801087g. [DOI] [PubMed] [Google Scholar]
- 25.Rogers F.A., Lloyd A. J., Tiwari M.K. Improved bioactivity of G-rich triplex-forming oligonucleotides containing modified guanine bases. Artificial DNA, PNA & XNA. 2014;5 doi: 10.4161/adna.27792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mergny J.L., Sun J.S., Rougee M., Montenay-Garestier T., Barcelo F., Chomilier J., Helene C. Sequence specificity in triple-helix formation: experimental and theoretical studies of the effect of mismatches on triplex stability. Biochemistry. 1991;30:9791–9798. doi: 10.1021/bi00104a031. [DOI] [PubMed] [Google Scholar]
- 27.Roberts R.W., Crothers D.M. Specificity and stringency in DNA triplex formation. Proc. Natl. Acad. Sci. U.S.A. 1991;88:9397–9401. doi: 10.1073/pnas.88.21.9397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trojan L.A., Kopinski P., Wei M.X., Ly A., Glogowska A., Czarny J., Shevelev A., Przewlocki R., Henin D., Trojan J. IGF-I: from diagnostic to triple-helix gene therapy of solid tumors. Acta Biochim. Pol. 2002;49:979–990. [PubMed] [Google Scholar]


