Abstract
The DNA sequence of the O-antigen gene clusters of Escherichia coli serogroups O62, O68, O131, O140, O142, and O163 was determined, and primers based on the wzx (O-antigen flippase) and/or wzy (O-antigen polymerase) genes within the O-antigen gene clusters were designed and used in PCR assays to identify each serogroup. Specificity was tested with E. coli reference strains, field isolates belonging to the target serogroups, and non-E. coli bacteria. The PCR assays were highly specific for the respective serogroups; however, the PCR assay targeting the O62 wzx gene reacted positively with strains belonging to E. coli O68, which was determined by serotyping. Analysis of the O-antigen gene cluster sequences of serogroups O62 and O68 reference strains showed that they were 94% identical at the nucleotide level, although O62 contained an insertion sequence (IS) element located between the rmlA and rmlC genes within the O-antigen gene cluster. A PCR assay targeting the rmlA and rmlC genes flanking the IS element was used to differentiate O62 and O68 serogroups. The PCR assays developed in this study can be used for the detection and identification of E. coli O62/O68, O131, O140, O142, and O163 strains isolated from different sources.
Keywords: PCR, Escherichia coli, serogroups, DNA sequence, O-antigen gene cluster, detection, identification
1. Introduction
In Escherichia coli and other Gram-negative bacteria, the major component of the outer membrane is lipopolysaccharide, which consists of three components: lipid A embedded in the membrane, an oligosaccharide core, and the lateral polysaccharide O-antigen. The O-antigen confers antigenic variability to the bacteria due to differences in the sugar components, the linkages, and the structure of the repeat O-units. Traditional serotyping of E. coli is based on agglutination reactions of the bacteria with antisera raised in rabbits immunized with different O-group reference strains. The test is performed in tubes, 96-well plates, or on slides [1,2]. The E. coli O-antigen is released by heating the bacteria for 2 h at 100 °C, and agglutination or clumping occurs when the O-antigen reacts with its specific antiserum. However, if the E. coli is capsulated or rough (does not carry O-antigen), agglutination does not occur. Furthermore, cross-reactions may occur with other E. coli O-groups, resulting in equivocal results, and serotyping is generally only performed in a few reference laboratories that have antisera against all of the E. coli O-groups. There are currently over 184 different E. coli O-groups classified as O1–O187 except for six (O31, O47, O67, O72, O94, O122) that have not been designated. The O-group defines the serogroup, and the combination of the O-antigen and the H-flagellar antigen defines the E. coli serotype.
Genes required for synthesis of the E. coli O-antigen are located on the chromosomal O-antigen gene cluster, which is located between a conserved 39-bp JUMPstart sequence (upstream) and downstream by the gnd gene that encodes for 6-phosphogluconate dehydrogenase [3,4]. Due to the differences in the composition of the O-antigens, the genes that encode for enzymes required for O-antigen synthesis vary among the different E. coli serogroups. Many E. coli O-antigen gene clusters have been sequenced, and the information has been deposited in GenBank. The sequence information can be used to identify unique regions that can be targeted, for example by PCR assays or other DNA-based methods, to identify the E. coli O-group. Furthermore, the sequence information can be used to study the evolution of E. coli O-antigens that can occur through gene deletion, acquisition, or inactivation [5].
Genes found in the O-antigen gene clusters that show genetic variability among the different serogroups include the wzx (O antigen flippase) and wzy (O antigen polymerase) genes, and PCR assays targeting these genes have been developed to identify different E. coli serogroups [4,6,7,8,9]. A DNA array approach was developed to identify E. coli O-groups using either representative oligonucleotides or PCR products to spot the array and labeled long PCR products for hybridization [10]. Lin et al. [11] performed PCR assays targeting the wzx and wzy genes of ten Shiga toxin-producing E. coli (STEC) serogroups, and then used the Luminex system to identify the ten serogroups through binding of the PCR products to fluorescent microspheres conjugated to specific DNA probes for each of the ten serogroups. Furthermore, multiplex assays can be designed to detect specific pathogenic E. coli serogroups targeting O-antigen gene cluster sequences and virulence genes [7,12]. Use of the Luminex system (Luminex, Austin, TX, USA) employing monoclonal antibodies coated to carboxylated magnetic microbeads to simultaneously detect Shiga toxin serogroup O157, as well as Shiga toxin 1 and Shiga toxin 2 has also been reported [13]. A review by DebRoy et al. [7] provides information on E. coli O-antigen gene clusters and methods used for O-group determination.
There are a number of E. coli pathotypes, consisting of various E. coli O-groups, that have been isolated from animals and that can cause illness in humans and animals. Enteropathogenic E. coli (EPEC) O142 has been isolated from infant stools, patients with diarrhea, and piglets [14,15,16]. Shiga toxin-producing E. coli (STEC) O62 was isolated from pork, and this serogroup has also been described as an enteroaggregative E. coli [17,18]. Verocytotoxin producing E. coli (VTEC, also known as STEC) O163 has been associated with cases of hemolytic uremic syndrome [19,20]. In addition, E. coli O163 was isolated from animals, including cows [21], lamb [22], goats, sheep [23], and pigs [24]. E. coli O131 was associated with pigs with post-weaning diarrhea in China [25], and E. coli O140 was associated with broiler chickens with dermatitis [26] and piglets with diarrhea [27].
Various molecular serotyping approaches could be used to identify E. coli O-groups, including the use of the Luminex® system, DNA microarrays, or the BioMarkTM real-time PCR array system (Fluidigm Corporation, South San Francisco, CA, USA), and others. Using some of these approaches, O-group determination could be coupled with simultaneous identification of virulence genes specific for certain E. coli pathotypes. However, to accomplish this, definitive determination of the O-antigen gene cluster sequences of all of the E. coli O-groups and of strains identified as untypeable by serotyping is needed. The objectives of this study were to determine the DNA sequence of the O-antigen gene clusters of E. coli serogroups O62, O68, O131, O140, O142, and O163, analyze the sequence data, and identify unique regions that are suitable targets for PCR assays to identify these serogroups. This work provides essential information for the application of molecular methods to differentiate E. coli serogroups, which is critically needed for accurate identification of E. coli and for epidemiological investigations of disease outbreaks.
2. Experimental Section
2.1. Bacterial Strains and Culture Conditions
E. coli O62 (F 10524-41, K-:H30), O68 (P 7d, K-:H4), O131 (S 239, K-:H26), O140 (CDC 149-51, K-:H43), O142 (C 771, H6) and O163 (SN3B/1, K-:H19) reference standard strains were obtained from the World Health Organization [1]. These strains were used for DNA sequencing of the O-antigen gene clusters. Bacterial strains used to validate the specificity of the PCR assays were from the culture collection of the E. coli Reference Center at the Pennsylvania State University. The following strains were included in the PCR assays: 148 field strains from E. coli serogroups O62, O68, O131, O140, O142, and O163 isolated from humans, animals, food, and water, and 174 E. coli standard reference strains belonging to serogroups O1-O187, but excluding O31, O47, O67, O72, O94, and O122, since these serogroups have not been designated [1]. In addition, 16 strains representative of other bacterial genera used to test the specificity of the PCR assays included Staphylococcus aureus ATCC13709, Staphylococcus aureus ATCC 29213, Klebsiella pneumoniae ATCC 27736, Serratia marcescens ATCC 13880, Shigella boydii ECRC 15.0055, Salmonella enterica sv. Typhi ECRC 15.0056, Enterobacter cloacae ECRC 15.0057, Salmonella enterica sv. Arizonae ECRC15.0058, Salmonella enterica sv. Choleraesuis ATCC 14028, Salmonella enterica sv. Choleraesuis ATCC 51741, Salmonella enterica sv. Anatum ATCC 9270, Citrobacter freundii ATCC 8090, Hafnia alvei ATCC 29926, Shigella flexneri ECRC 15.0059, Yersinia enterocolitica ECRC 15.0060, and Listeria innocua ATCC 51742. All of the bacteria were grown in Luria Bertani (LB) broth or on LB agar plates at 37 °C.
2.2. DNA Sequencing and Gene Annotation
Genomic DNA was isolated using the DNeasy Tissue Kit (Qiagen Inc., Valencia, CA, USA) according to the manufacturer’s instructions. Long PCR assays were performed to amplify the O-antigen gene clusters using the Expand Long Template PCR system (Roche Applied Science, Mannheim, Germany) and the JUMPSTART (named for Just Upstream of Many Polysaccharide-associated gene STARTs) and GND (6-phosphogluconate dehydrogenase gene) primer set targeting sequences that flank the E. coli O-antigen gene clusters as described previously [12]. However, some modifications were made to the JUMPSTART and GND primer sequences, and they are the following: JUMPSTART primer 5'-CATGGTAGCTGTAAAGCCAGGGGCGGTAGCGTG-3'; GND primer 5'-CATGCTGCCATACCGACGACGCCGATCTGTTGCTTKGACA-3' (Integrated DNA Technologies, Coralville, IA, USA). The long PCR conditions were as described previously [9]. The long PCR products were verified on 0.8% agarose gels and purified according to instructions in the QIAquick PCR Purification Kit (Qiagen Inc., Valencia, CA, USA). The long PCR products were sequenced by the methods described below.
DNA integrity was verified using a Bioanalyzer 2100 (Agilent, Palo Alto, CA, USA), and DNA concentration was quantified using a QuantiFluor fluorometer (Promega, Madison, WI, USA). For sequencing with the Roche/454 GS FLX instrument (Roche, 454 Life Sciences, Branford, CT, USA), the O-antigens were amplified from 40 ng of genomic DNA isolated as described above, with eight-bp sample-specific bar coded primers using 2.5 units of AccuPrime Taq DNA Polymerase High Fidelity (Invitrogen, Carlsbad, CA, USA) in a 50-μL reaction buffer containing 200 nM primers, 200 nM dNTP, 60 mM Tris-SO4, 18 mM (NH4)2SO4, 2.0 mM MgSO4, 1% glycerol, and 100 ng/µL bovine serum albumin (New England BioLabs, Ipswich, MA, USA). PCR was performed using the following cycling profile: initial denaturing at 95 °C for two min followed by 25 cycles of 95 °C 30 s, 50 °C 30 s, and 72 °C 120 s. Bar-coded amplicons were generated from each sample separately, purified using an Agencourt AMPure XP kit (Beckman Coulter Genomics, Danvers, MA, USA), and quantified using a QuantiFluor fluorometer. Bar-coded amplicons from individual samples were pooled in equal mass (molar) ratios. The purified bar-coded amplicon library was further verified and quantified using a BioAnalyzer 2100 (Agilent) and subjected to genome sequencing using the Roche/454 GS FLX. Illumina HiSeq 2000 (San Diego, CA, USA) sequencing was performed as described by Djikeng et al. [28] using long PCR products. The sequence reads generated from the Illumina, Roche/454 GS FLX, and the Sanger sequencing method using the 3730 DNA Analyzer (Applied Biosystems, Foster City, CA, USA) (see below) were each first assembled separately. The sequence data and the generated contigs were then combined and assembled into the final O-antigen clusters using CLC Genomics Workbench 4.6.1 (CLC bio, Aarhus, Denmark) and Sequencher version 5.1 software (Gene Codes Corporation, Ann Arbor, MI, USA). Some additional details on the sequencing strategy and contig assembly were as described by Djikeng et al. [28]. To confirm the sequences of each of the O-antigen gene clusters, the long PCR products were resequenced using a 3730 DNA Analyzer (Applied Biosystems, Foster City, CA, USA) using primers designed from different regions along the gene clusters, and gene annotation was performed as described previously [9]. The HMMTOP program [29] was used to identify potential transmembrane helices from the amino acid sequences.
2.3. PCR Specificity Testing
E. coli reference strains [1] and field strains belonging to serogroups O62, O68, O131, O140, O142 and O163, and non-E. coli bacteria were grown overnight on tryptic soy agar (TSA) plates at 37 °C. Single colonies were picked and resuspended in 100 µL of Tris-EDTA buffer (pH 8.0) and heated at 100 °C for 10 min. The suspension was centrifuged at 10,000× g, and the supernatant containing genomic DNA was used for the PCR reactions.
The PCR primers (Table 1) were designed from the wzx, wzy, and rmlA/rmlC region of the targeted O-serogroups. The PCR reaction mix (20 µL total volume) was comprised of template DNA (1 µL), 300 nM of each primer, and 10 µL of the Power SYBR® Green PCR master mix containing Taq Polymerase (Life Technologies, Carlsbad, CA, USA). RT-PCR reactions were conducted using an AB 7300 Real-Time PCR system (Applied Biosystems). The PCR cycling conditions consisted of an initial denaturation for 10 min at 95 °C followed by 40 cycles at 95 °C for 15 s and 60 °C for 1 min. Reaction mixtures without template DNA and without primers served as negative controls. Data were analyzed using 7300 system SDS software (Applied Biosystems, Foster City, CA, USA).
Table 1.
PCR primers targeting the wzx, wzy and rmlA/C genes of E. coli O62, O68, O131, O140, O142 and O163.
| Target Gene | Sequence | Amplicon Size (bp) |
|---|---|---|
| O62/O68 wzx | F: 5' ATGCTGCATTAGCGTTAGCA 3' | 288 |
| R: 5' CCTGTTGAATTGGCACGTAA 3' | ||
| O131 wzx | F: 5' TCGTGAGAAGGCTTTTTGGT 3' | 290 |
| R: 5' CCCTATCCAATGCGCTTAAA 3' | ||
| O140 wzx | F: 5' TTGGATAGCCGCGTTAATTC 3' | 294 |
| R: 5' GCCTGAGTTAGCGGATTGAG 3' | ||
| O142 wzx | F: 5' TCTCCATCCCCGTTTATTTG 3' | 285 |
| R: 5' CCCCAAACATTAGCATTCGT 3' | ||
| O163 wzy | F: 5' GCAATCTTGAAGCCAGAACC 3' | 262 |
| R: 5' GATAAACCCAGCCACCAAA 3' | ||
| O62/O68 rmlA/C | F: 5' CTACACTGATGTTAGCGGGTATT 3' | 1969 (for O62) |
| R: 5' CCGCTTCAAATTCAGGACAATAA 3' | 1172 (for O68) |
2.4. Nucleotide Sequence Accession Numbers
DNA sequences of the O-antigen gene clusters of E. coli O62, O68, O131, O140, O142 and O163 were deposited into GenBank with the following accession numbers: JX501334, KJ534585, JX501336, JX501338, JX501337, and JX501339, respectively.
3. Results and Discussion
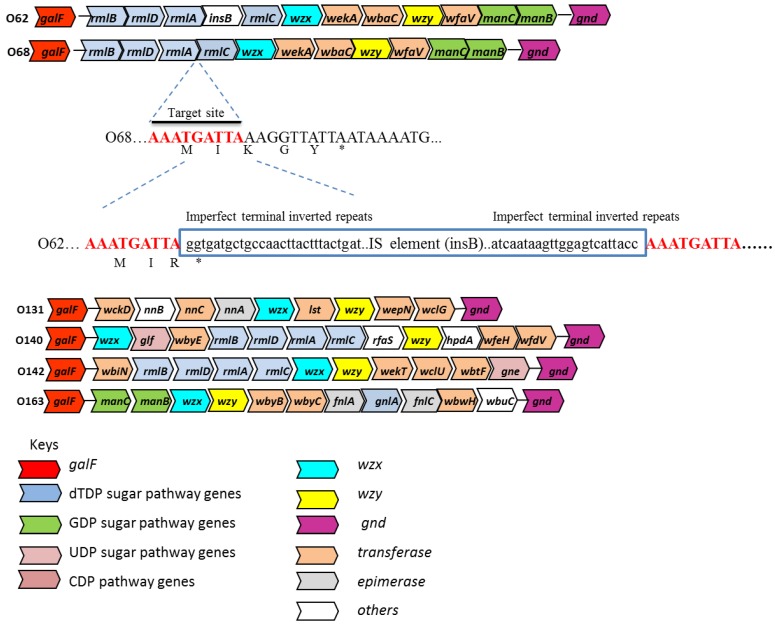
DNA sequences obtained from the E. coli O antigen gene clusters of serogroup O62, O68, O131, O140, O142 and O163 contained 9 to 12 ORFs (Figure 1 and Appendix Table A1, Tables A2, Tables A3, Tables A4, Tables A5 and Tables A6), all in the same transcriptional direction from galF to gnd. The deduced amino acid sequences from these ORFs were used to search the NCBI database for an indication of their possible functions. Gene names were assigned on the basis of the Bacterial Polysaccharide Gene Nomenclature system (http://sydney.edu.au/science/molecular_bioscience/BPGD/).
Figure 1.
Organization of the O-antigen gene clusters for E. coli O62 O68, O131, O140, O142 and O163. The insertion sequence (IS) insertion and its flanking sequences in E. coli O62 is shown relative to the O68 O-antigen gene cluster. The arrows represent the location and direction of translation for putative genes in the clusters. The genes are not drawn in scale. The 9-bp target duplications are shown in red. The 23-bp imperfect terminal repeats in the IS element (boxed) are shown in bold. Deduced amino acids sequences (shown in one-letter code) of the IS insertion site are also shown. galF is known to be upstream of the O-antigen gene clusters [7].
3.1. Sequence Analysis of the E. coli O-Antigen Gene Clusters of Serogroups O62, O68, O131, O140, O142 and O163
The genes coding for proteins within the E. coli O-antigen gene clusters primarily consist of three categories: nucleotide sugar biosynthesis, glycosyl transferase, and O-antigen processing. Nucleotide sugar biosynthesis genes encode for proteins that are involved in the synthesis of the nucleotide sugar precursors of the O-antigen, which occurs in the cytoplasm. Genes coding for glycosyl transferases are responsible for transferring the various precursor sugars to form an oligosaccharide on a carrier lipid, undecaprenyl phosphate (UndP), which is located on the inner membrane facing the cytoplasmic side. The O-antigen processing proteins include a flippase (Wzx) and the O-antigen polymerase (Wzy). These proteins are involved in translocation of the O-units across the membrane and in O-antigen polymerization, respectively. The O-antigen is synthesized by sequential transfer of sugars and other components to the first sugar, which is then translocated and flipped across the membrane by Wzx. They are further polymerized by Wzy. Both Wzx and Wzy are hydrophobic proteins with transmembrane helices that show high variation in sequence among different microorganisms [7].
3.1.1. Sugar Biosynthetic Pathway Genes
The four genes involved in the biosynthesis of dTDP-L-rhamnose [30] are clustered together in O68, O140 and O142 in the gene order of rmlBDAC (Appendix Tables A2, Tables A4, Tables A5). The rmlB (dTDP-glucose 4,6-dehydratase), rmlD (dTDP-4-dehydrorhamnose reductase), rmlA (glucose-1-phosphate thymidylyltransferase), rmlC (dTDP-4-dehydrorhamnose 3,5-epimerase) genes are also present in O62 with the same gene order except there is a transposase (insB) between the rmlA and rmlC genes (Table 2). The two genes (fnlA and fnlC) involved in the biosynthesis of UDP-L-FucNAc are present in the O-antigen gene cluster of E. coli O163 (Appendix Tables A6). The fnlA (UDP-glucose 4-epimerase) and fnlC (UDP-N-acetylglucosamine 2-epimerase) genes are present in several other reported gene clusters coding for UDP-L-FucNAc containing structures [30]. The manB and manC genes present in the O antigen gene clusters of O62, O68 (Appendix Tables A1 and Tables A2), and O163 (Appendix Tables A6) have been identified to be responsible for the biosynthesis of GDP-D-mannose [30]. manB and manC encode phosphomannomutase and mannose-1-phosphate guanyltransferase, respectively.
Table 2.
Specificity of the PCR assays for O groups tested.
| O Group Tested | Strains Tested | Specificity |
|---|---|---|
| O62/O68 (wzx) PCR | Reference strains (O1–O181) | All negative except O62 and O68 positive control strains |
| O62 field isolates (n = 2) | 2/2 positive (100%) a | |
| O68 field isolates (n = 6) | 6/6 positive (100%) a | |
| non-E. coli (n = 16) | 100% negative | |
| O131 (wzx) PCR | Reference strains (O1–O181) | All negative except O131 positive control strain |
| O131 field isolates (n = 15) | 15 positive (100%) | |
| non-E. coli (n = 16) | 100% negative | |
| O140 (wzx) PCR | Reference strains (O1–O181) | All negative except O140 positive control strain |
| O140 field isolates (n = 28) | 28 positive (100%) | |
| non-E. coli (n = 16) | 100% negative | |
| O142 (wzx) PCR | Reference strains (O1–O181) | All negative except O142 positive control strain |
| O142 field isolates (n = 50) | 50 positive (100%) | |
| Non-E. coli (n = 16) | 100% negative | |
| O163 (wzy) PCR | Reference strains (O1–O181) | All negative except O163 positive control strain |
| O163 field isolates (n = 47) | 47 positive (100%) | |
| non-E. coli (n = 16) | 100% negative |
a Although two strains were positive using both O62 and O68 antisera similar to the O62 reference strain, one strain did not show the presence of the insertion element found in the O62 reference strain by PCR, therefore, one strain could be either O62 or O68.
The polysaccharide structure of the E. coli O142 and O68 O-antigens has been determined [31,32,33]. The proposed function of the genes in the O-antigen gene clusters of E. coli O142 and O68 correlates well to the identified O142 and O68 polysaccharide structure [32,33].
3.1.2. Sugar Transferase Genes
Genes encoding for sugar transferases were identified based on their similarity to known sugar transferases. As shown in Figure 1 and Appendix Tables A1, Tables A2, Tables A3, Tables A4, Tables A5 and Tables A6, O62, O68, O140, and O163 each contained three sugar transferases, whereas O131 and O142 contained five and four sugar transferases, respectively. These ORFs have a high degree of sequence variation (30%–60% amino acid similarity), which is consistent with previous studies [30].
3.1.3. O Antigen Processing Genes
All of the six O-antigen gene clusters contained the wzx and wzy genes located in different regions within the gene clusters (Appendix Tables A1, Tables A2, Tables A3, Tables A4, Tables A5 and Tables A6). Analysis using the HMMTOP program [29] indicated that all six Wzx proteins contained 12 transmembrane helices, whereas the Wzy proteins contained 10 transmembrane helices, with the exception of the Wzy protein from O142 that contained 13 transmembrane helices.
3.2. Development of PCR Assays to Identify E. coli O62/O68, O131, O140, O142, and O163 Serogoups
Primers were designed targeting the wzx and/or wzy genes from the above E. coli serogroups (Table 1), and they were used in PCR assays to determine specificity for each serogroup against 174 E. coli standard strains, as well as field E. coli strains serotyped as O62, O68, O131, O140, O142 and O163 isolated from humans, animals, food, or water. Sixteen non-E. coli strains (see Experimental Section for the list of non-E. coli strains) were also included as negative controls for specificity testing. PCR assays targeting the wzx/wzy genes showed high specificity for each serogroup with no amplification of wzx/wzy genes from other E. coli serogroups and no amplification of DNA of other bacterial genera. All of the field isolates serogrouped as E. coli O131, O140, O142 and O163 were positive by PCR for the corresponding serogroup with 100% accuracy. However, the O62 wzx PCR assay also gave a positive result with the O68 reference strain (Table 2). This is not surprising, since our sequencing data alsodemonstrated that the wzx sequences of O62 were identical with those of O68 (Appendix Tables A1 and Tables A2). The field strains of E. coli O62 (n = 2) and O68 (n = 6) also exhibited positive PCR results with the wzx primers of O62.
3.3. Acquisition of the IS1 Element in E. coli O62 and Evolutionary Implications and Differentiation of Serogroups O62 and O68
Analysis of the O-antigen gene clusters of E. coli O62 and O68 showed that they are almost identical, except that E. coli O62 contained an IS element insertion (ORF4), 748 bp in size at the end of the rmlA gene. ORF4 (insB) encodes for a transposase that is identical to IS1 transposition proteins in Shigella flexneri 2b (Appendix Table A1). The IS1 element in E. coli O62 is inserted within the third codon from the end of the rmlA gene, resulting in a truncated protein ending with an R (arginine) in place of K (lysine), and in comparison with E. coli O68 the last two amino acids are missing (Figure 1). IS1 is a common mobile genetic element that usually generates a 8 to 9-bp target duplication upon integration [34]. In addition, the IS1 element contains 23-bp imperfect terminal repeats that is a characteristic of an IS element [35,36]. The IS1 element is widely distributed in prokaryotic genomes, is highly mobile, and can be a source of genome rearrangements [37,38]. The IS elements present in E. coli O24 seemed to play important roles for the assembly of the O24 O-antigen gene cluster by mediating lateral gene transfer and gene inactivation [5]. The high level of similarity between the O-antigen gene clusters of the E. coli O62 and O68 reference strains suggests that the O-antigen gene clusters are very closely related and may be derived from a common ancestor.
To differentiate E. coli O62 and O68, primers were designed targeting the rmlA and rmlC region flanking the IS element from O62 (Table 1). The predicted PCR products for O62 and O68 are 1969 bp and 1172 bp, respectively. These primers were used in PCR assays to differentiate two O62 and six O68 (determined by serotyping) field strains in our strain collection. Of the strains tested, six O68 strains were positive for O68 using the rmlA/C PCR (i.e., lacked the IS element), and they were positive only for O68 by serotyping (Table 3). One of the two strains that were originally serotyped as O62 strains was positive for O68 according to the PCR assay targeting rmlA/C (i.e., lacked the IS element) (Table 3); however, this strain was also positive for O68 by serotyping, similar to the pattern for O62 strains, which are positive by serotyping for both O62 and O68. Therefore, this strain should either be re-assigned as a variant of O68, or it is possible that it is actually O62, but does not carry the IS element. Because there are so few field strains belonging to serogroup O62 in a collection of approximately 70,000 strains at the E. coli Reference Center at the Pennsylvania State University, collected over the last fifty years, this suggests that this O-group is not commonly found in animals, humans, and the environment. Our data show that the O-antigen gene clusters of E. coli O62 and O68 are very similar. The high similarities between O62 and O68 likely result in antisera cross reaction, which is an important problem in traditional serotyping. It is puzzling, however, the antiserum prepared against O62 does not cross react with O68, whereas antiserum against O68 cross reacts with O62. To accurately serotype O62/68 strains, it is important to first perform serotyping followed by the PCR assay using the rmlA/C primers flanking the IS element for O62 positive (by serotyping) strains.
Table 3.
Serotyping results and PCR using rmlA/C primers for O62/O68 field strains.
| O62/68 Field Strain Designation (ECRC#) a | Serotyping Using O62 Antiserum | Serotyping Using O68 Antiserum | Serogroup by PCR Using rmlA/C Primers |
|---|---|---|---|
| 12.0591 | Positive | Positive | O62 (1969 bp) |
| 94.0296 | Positive | Positive | O68/O62 (1172 bp) b |
| 1.2557 | Negative | Positive | O68 |
| 3.1263 | Negative | Positive | O68 |
| 4.0175 | Negative | Positive | O68 |
| 4.2378 | Negative | Positive | O68 |
| 5.1791 | Negative | Positive | O68 |
| 6.2334 | Negative | Positive | O68 |
a ECRC#—E. coli Reference Center strain designation; b Although this strain was positive using both O62 and O68 antisera similar to O62 strain 12.0591 and the O62 reference strain, it did not show the presence of the insertion element by PCR.
4. Conclusions
The O-antigen gene cluster sequences for six E. coli serogroups have been determined, and thus PCR primers can be designed for unique regions within the gene cluster sequences to develop genetic-based methods for serotyping, which are more specific than traditional serotyping. The PCR assays designed in the current study could potentially be used for rapid diagnostic screening for the E. coli serogroups. Since serotyping results are often ambiguous and sometimes may not be able to distinguish the serogroups, PCR assays in conjunction with serotyping may be able to circumvent these problems and distinguish the serogroups more accurately.
Acknowledgments
We thank Amy Ream for assistance in sequencing and gene annotation, and we are grateful to Robert Tebbs for assistance with sequencing and data analysis of O62/O68. We are also grateful to James Smith for critical reading of the manuscript.
Appendix
Table A1.
Open reading frames (ORFs) in the O-antigen gene cluster of E. coli serogroup O62.
| ORF | Proposed Gene Name | Location | No. of Amino Acids | Putative Function | Most Significant Similarity (Accession No.) | % Amino Acid Identity/% Similarity |
|---|---|---|---|---|---|---|
| 1 | rmlB | 106:1191 | 361 | dTDP-glucose 4,6-dehydratase | dTDP-glucose 4,6-dehydratase [Escherichia coli] Sequence ID: ref|WP_001723646.1| | 99/100 |
| 2 | rmlD | 1191:2090 | 299 | glucose-1-phosphate thymidylyltransferase | dTDP-4-dehydrorhamnose reductase [Escherichia coli] Sequence ID: ref|WP_001723645.1| | 100/100 |
| 3 | rmlA | 2148:3020 | 290 | glucose-1-phosphate thymidylyltransferase | glucose-1-phosphate thymidylyltransferase [Escherichia coli] Sequence ID: ref|WP_021516244.1| | 99/100 |
| 4 | insB | 3265:3768 | 167 | Transposases IS1 | IS1 transposition protein [Shigella flexneri 2b] Sequence ID: ref|NP_052905.1| | 100/100 |
| 5 | rmlC | 3808:4350 | 180 | dTDP-4-dehydrorhamnose 3,5-epimerase | dTDP-4-dehydrorhamnose 3,5-epimerase [Escherichia coli] Sequence ID: ref|WP_001723643.1| | 100/100 |
| 6 | wzx | 4365:5576 | 403 | O antigen flippase | polysaccharide biosynthesis family protein [Escherichia coli] Sequence ID: ref|WP_001723642.1| | 100/100 |
| 7 | wekA | 5584:6534 | 316 | glycosyl transferase | hypothetical protein [Escherichia coli] Sequence ID: ref|WP_001607665.1| | 100/100 |
| 8 | wbcC | 6515:7603 | 362 | glycosyl transferase | hypothetical protein [Escherichia coli] Sequence ID: ref|WP_001607663.1| | 99/100 |
| 9 | wzy | 7593:8711 | 372 | O antigen polymerase | putative membrane protein [Escherichia coli] Sequence ID: ref|WP_001723640.1| | 99/100 |
| 10 | wfaV | 8713:9894 | 393 | glycosyl transferases group 1 family | glycosyl transferases group 1 family protein [Escherichia coli] Sequence ID: ref|WP_001723639.1| | 99/99 |
| 11 | manC | 9891:11315 | 474 | mannose-1-phosphate guanyltransferase | mannose-1-phosphate guanylyltransferase/mannose-6-phosphate isomerase [Escherichia coli] Sequence ID: ref|WP_001723638.1| | 99/99 |
| 12 | manB | 11405:12775 | 456 | phosphomannomutase | phosphoglucomutase/phosphomannomutase, C-terminal domain protein [Escherichia coli] Sequence ID: ref|WP_001723637.1| | 99/100 |
Table A2.
Open reading frames (ORFs) in the O-antigen gene cluster of E. coli serogroup O68.
| ORF | Proposed Gene Name | Location | No. of Amino Acids | Putative Function | Most Significant Similarity (Accession No.) | % Amino Acid Identity/% Similarity |
|---|---|---|---|---|---|---|
| 1 | rmlB | 106:1191 | 361 | dTDP-glucose 4,6-dehydratase | dTDP-glucose 4,6-dehydratase [Escherichia coli] Sequence ID: ref|WP_001723646.1| | 99/100 |
| 2 | rmlD | 1191:2090 | 299 | glucose-1-phosphate thymidylyltransferase | dTDP-4-dehydrorhamnose reductase [Escherichia coli] Sequence ID: ref|WP_001723645.1| | 100/100 |
| 3 | rmlA | 2148:3026 | 292 | glucose-1-phosphate thymidylyltransferase | glucose-1-phosphate thymidylyltransferase [Escherichia coli] Sequence ID: ref|WP_021516244.1| | 99/100 |
| 4 | rmlC | 3031:3573 | 180 | dTDP-4-dehydrorhamnose 3,5-epimerase | dTDP-4-dehydrorhamnose 3,5-epimerase [Escherichia coli] Sequence ID: ref|WP_001723643.1| | 100/100 |
| 5 | wzx | 3588:4799 | 403 | O antigen flippase | polysaccharide biosynthesis family protein [Escherichia coli] Sequence ID: ref|WP_001723642.1| | 100/100 |
| 6 | wekA | 4807:5757 | 316 | glycosyl transferase | hypothetical protein [Escherichia coli] Sequence ID: ref|WP_001607665.1| | 100/100 |
| 7 | wbcC | 5738:6826 | 362 | glycosyl transferase | hypothetical protein [Escherichia coli] ref|WP_001607663.1| | 99/100 |
| 8 | wzy | 6816:7934 | 372 | O antigen polymerase | putative membrane protein [Escherichia coli] Sequence ID: ref|WP_001723640.1| | 99/100 |
| 9 | wfaV | 7936:9117 | 393 | glycosyl transferases group 1 family | glycosyl transferases group 1 family protein [Escherichia coli] Sequence ID: ref|WP_001723639.1| | 100/100 |
| 10 | manC | 9114:10538 | 474 | mannose-1-phosphate guanyltransferase | mannose-1-phosphate guanylyltransferase/mannose-6-phosphate isomerase [Escherichia coli] Sequence ID: ref|WP_001723638.1| | 100/100 |
| 11 | manB | 10628:11998 | 456 | phosphomannomutase | phosphoglucomutase/phosphomannomutase, C-terminal domain protein [Escherichia coli] Sequence ID: ref|WP_001723637.1| | 99/100 |
Table A3.
Open reading frames (ORFs) in the O-antigen gene cluster of E. coli serogroup O131.
| ORF | Proposed Gene Name | Location | No. of Amino Acids | Putative Function | Most Significant Similarity (Accession No.) | % Amino Acid Identity/% Similarity |
|---|---|---|---|---|---|---|
| 1 | wckD | 589–1209 | 206 | sialic acid O-acetyltransferase NeuD family sugar O-acyltransferase | ref|YP_002403330.1| WckD [Escherichia coli 55989] | 91/98 |
| 2 | nnaB | 1211–2251 | 346 | N-acetylneuraminic acid synthetase | ref|ZP_02904231.1| NnaB [Escherichia albertii TW07627] | 94/97 |
| 3 | nnaC | 2254–3516 | 420 | N-acylneuraminate cytidylyltransferase | ref|ZP_02904216.1| N-acylneuraminate cytidylyltransferase [Escherichia albertii TW07627] | 90/96 |
| 4 | nnaA | 3513–4694 | 393 | UDP-N-acetylglucosamine 2-epimerase | ref|ZP_02904222.1| UDP-N-acetylglucosamine 2-epimerase [Escherichia albertii TW07627] | 91/95 |
| 5 | wzx | 4691–5959 | 422 | O antigen flippase | ref|ZP_02904256.1| Lsg [Escherichia albertii TW07627] | 87/93 |
| 6 | lst | 5966–6940 | 324 | UDP-glucose:glucosyl LPS a1,2-glucosyltransferase | ref|ZP_02904182.1| putative Lst [Escherichia albertii TW07627] | 79/88 |
| 7 | wzy | 7024–8265 | 413 | O antigen polymerase | ref|YP_002310938.1| unnamed protein product [Shewanella piezotolerans WP3] | 31/52 |
| 8 | wepN | 8262–9122 | 286 | glycotransferase | ref|ZP_03611741.1| hypothetical protein AM202_0156 [Actinobacillus minor 202] | 29/52 |
| 9 | wclG | 9587–10057 | 156 | glycosyl transferase | ref|ZP_07136721.1| glycosyltransferase, group 2 family protein [Escherichia coli MS 115-1] | 64/80 |
Table A4.
Open reading frames (ORFs) in the O-antigen gene cluster of E. coli serogroup O140.
| ORF | Proposed Gene Name | Location | No. of Amino Acids | Putative Function | Most Significant Similarity (Accession No.) | % Amino Acid Identity/% Similarity |
|---|---|---|---|---|---|---|
| 1 | wzx | 26–1273 | 415 | O antigen flippase | gb|ACA24898.1| Wzx [Escherichia coli] | 62/82 |
| 2 | glf | 1270–2379 | 369 | UDP-galactopyranose mutase | gb|EFZ69214.1| UDP-galactopyranose mutase [Escherichia coli OK1357] | 73/88 |
| 3 | wbyE | 2383–3375 | 330 | group 1 glycosyl transferase | ref|YP_002987178.1| hypothetical protein Dd703_1557 [Dickeya dadantii Ech703] | 45/63 |
| 4 | rmlB | 3420–4505 | 361 | dTDP-glucose 4,6-dehydratase | gb|AAZ85703.1| dTDP-glucose 4,6-dehydratase [Escherichia coli] | 98/96 |
| 5 | rmlD | 4505–5404 | 299 | dTDP-4-dehydrorhamnose reductase | gb|EGB44384.1| RmlD substrate binding domain-containing protein [Escherichia coli H120] | 99/98 |
| 6 | rmlA | 5462–6337 | 291 | glucose-1-phosphate thymidylyltransferase | gb|ABE98410.1| glucose-1-phosphate thymidylyltransferase [Escherichia coli] | 99/99 |
| 7 | rmlC | 6345–6878 | 177 | dTDP-4-dehydrorhamnose 3,5-epimerase | ref|YP_853147.1| rmlC gene product [Escherichia coli APEC O1] | 85/90 |
| 8 | rfaS | 6945–7913 | 322 | lipopolysaccharide core biosynthesis protein | gb|EHN67535.1| lipopolysaccharide core biosynthesis protein [Comamonas testosteroni ATCC 11996] | 35/57 |
| 9 | wzy | 7936–9126 | 396 | O antigen polymerase | gb|ACH97152.1| Wzy [Escherichia coli] | 26/49 |
| 10 | hpdA | 9137–9913 | 258 | unknown | ref|YP_001534197.1| hypothetical protein Dshi_2863 [Dinoroseobacter shibae DFL 12] | 40/56 |
| 11 | wfeH | 10117–10761 | 214 | glycosyl transferase | ref|ZP_06693546.1| predicted protein [Acinetobacter sp. SH024] | 39/56 |
| 12 | wfdV | 10758–11516 | 252 | glycosyl transferase | gb|AEH27518.1| WehL [Cronobacter muytjensii] | 63/77 |
Table A5.
Open reading frames (ORFs) in the O-antigen gene cluster of E. coli serogroup O142.
| ORF | Proposed Gene Name | Location | No. of Amino Acids | Putative Function | Most Significant Similarity (Accession No.) | % Amino Acid Identity/% Similarity |
|---|---|---|---|---|---|---|
| 1 | wbiN | 78–1094 | 338 | glycosyl transferase | ref|YP_002329693.1| wbiN gene product [Escherichia coli O127:H6 str. E2348/69] | 58/71 |
| ref|ZP_07222371.1| glycosyltransferase, group 1 family [Escherichia coli MS 78-1] | ||||||
| ref|ZP_10058779.1| hypothetical protein ESBG_00585 [Escherichia sp. 4_1_40B] | ||||||
| 2 | rmlB | 1114–2199 | 361 | dTDP-glucose-4,6-dehydratase | ref|YP_002391833.1| dTDP-glucose-4,6-dehydratase [Escherichia coli S88] | 85/92 |
| 3 | rmlD | 2109–3098 | 329 | dTDP-6-deoxy-L-mannose-dehydrogenase | gb|AAZ85704.1| dTDP-6-deoxy-L-mannose-dehydrogenase [Escherichia coli] | 82/91 |
| 4 | rmlA | 3129–4028 | 299 | glucose-1-phosphate thymidylyltransferase | gb|ABE98410.1| glucose-1-phosphate thymidylyltransferase [Escherichia coli] | 93/96 |
| 5 | rmlC | 4018–4590 | 190 | dTDP-4-dehydrorhamnose 3,5-epimerase | gb|ACA24817.1| RmlC [Escherichia coli] | 75/85 |
| 6 | wzx | 4587–5831 | 414 | O antigen flippase | ref|YP_541307.1| O-antigen transporter [Escherichia coli UTI89] | 58/79 |
| 7 | wzy | 5883–7061 | 392 | O antigen polymerase | gb|ADC54950.1| Wzy [Escherichia coli] | 49/68 |
| 8 | wekT | 7010–7978 | 322 | rhamnosyltransferase | ref|YP_541305.1| rhamnosyltransferase [Escherichia coli UTI89] | 59/74 |
| 9 | wclU | 7975–8763 | 262 | glycosyltransferase | ref|YP_541304.1| glycosyltransferase [Escherichia coli UTI89] | 56/70 |
| 10 | wbtF | 8760–9854 | 364 | glycosyltransferase | gb|AFI60269.1| WepF [Cronobacter sakazakii] | 53/73 |
| 11 | gne | 10098–11117 | 339 | UDP-glucose 4-epimerase | ref|ZP_07142276.1| UDP-glucose 4-epimerase [Escherichia coli MS 182–1] | 74/86 |
Table A6.
Open reading frames (ORFs) in the O-antigen gene cluster of E. coli serogroup O163.
| ORF | Proposed Gene Name | Location | No. of Amino Acids | Putative Function | Most Significant Similarity (Accession No.) | % Amino Acid Identity/% similarity |
|---|---|---|---|---|---|---|
| 1 | manC | 379:1818 | 479 | mannose-1-phosphate guanylyltransferase manC | mannose-1-phosphate guanylyltransferase manC [Enterobacterc loacae] ref|WP_023300545.1| | 82/89 |
| 2 | manB | 1925:3325 | 466 | Phosphomanno-mutase | phosphomannomutase [Enterobacter cloacae subsp. cloacae ENHKU01] ref|YP_006579401.1| | 90/96 |
| 3 | wzx | 3318:4571 | 417 | O-antigen repeat unit transporter | hypothetical protein SARI_00795 [Salmonella enterica subsp. Arizonaeserovar 62:z4,z23:- str. RSK2980] Sequence ID: ref|YP_001569857.1| | 49/55 |
| 4 | wzy | 4568:5803 | 411 | O antigen polymerase | O-unit polymerase [Salmonella enterica subsp. Arizonae] Sequence ID: gb|ACJ26814.1| | 57/66 |
| 5 | wbyB | 5815:6828 | 337 | glycosyl transferase, group 1 | glycosyl transferase, group 1 [Rhodopirellula baltica] Sequence ID: ref|WP_007337184.1| | 42/44 |
| 6 | wbyC | 6848:7951 | 367 | glycosyl transferase, group 1 | hypothetical protein [Bacillus cereus] Sequence ID: ref|WP_000651756.1| | 40/46 |
| 7 | fnlA | 8076:9167 | 363 | UDP-glucose 4-epimerase | hypothetical protein SARI_00798 [Salmonella enterica subsp. Arizonae serovar 62:z4,z23:- str. RSK2980] Sequence ID: ref|YP_001569860.1| | 87/91 |
| 8 | qnlA | 9133:10035 | 300 | dTDP-4-dehydrorhamnose reductase | hypothetical protein SARI_00799 [Salmonella enterica subsp. Arizonae serovar 62:z4,z23:- str. RSK2980] Sequence ID: ref|YP_001569861.1| | 67/74 |
| 9 | fnlC | 10007:11164 | 385 | UDP-N-acetylglucosamine 2-epimerase | UDP-N-acetylglucosamine 2-epimerase [Salmonella enterica subsp. Arizonae] Sequence ID: gb|ACJ26818.1| | 68/84 |
| 10 | wbwH | 11149:12357 | 402 | glycosyl transferase | hypothetical protein SARI_00801 [Salmonella enterica subsp. Arizonae serovar 62:z4,z23:- str. RSK2980] Sequence ID: ref|YP_001569863.1| | 67/81 |
| 11 | wbuC | 12380:12877 | 165 | unknown | conserved LPS biosynthetic protein [Salmonella enterica subsp. Arizonae] Sequence ID: gb|ACJ26820.1| | 53/75 |
Author Contributions
Yanhong Liu designed the experiment and drafted the manuscript, extracted genomic DNA and performed long PCR experiments. David S. Needleman did PCR sequencing. PCR specificity testing was performed by Chitrita DebRoy and Narasimha Hegde. All of the authors contributed to DNA whole genome sequencing.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Ørskov I., Ørskov F., Jann B., Jann K. Serology, chemistry, and genetics of O and K antigens of Escherichia coli. Bacteriol. Rev. 1977;41:667–710. doi: 10.1128/br.41.3.667-710.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ørskov F., Ørskov I. Serotyping of Escherichia coli. Methods Microbiol. 1984;14:43–112. [Google Scholar]
- 3.Hobbs M., Reeves P.R. The JUMPstart sequence: A 39 bp element common to several polysaccharide gene clusters. Mol. Microbiol. 1994;12:855–856. doi: 10.1111/j.1365-2958.1994.tb01071.x. [DOI] [PubMed] [Google Scholar]
- 4.Wang L., Reeves P.R. Organization of Escherichia coli O157 O-antigen gene cluster and identification of its specific genes. Infect. Immun. 1998;66:3545–3551. doi: 10.1128/iai.66.8.3545-3551.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheng J., Wang Q., Wang W., Wang Y., Wang L., Feng L. Characterization of E. coli O24 and O56 O antigen gene clusters reveals a complex evolutionary history of the O24 gene cluster. Curr. Microbiol. 2006;53:470–476. doi: 10.1007/s00284-006-0032-7. [DOI] [PubMed] [Google Scholar]
- 6.Beutin L., Tao J., Feng L., Krause G., Zimmermann S., Gleier K., Xia Q., Wang L. Sequence analysis of the Escherichia coli O15 antigen gene cluster and development of a PCR assay for rapid detection of intestinal and extraintestinal pathogenic E. coli O15 strains. J. Clin. Microbiol. 2005;43:703–710. doi: 10.1128/JCM.43.2.703-710.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DebRoy C., Roberts E., Fratamico P.M. Detection of O antigens in Escherichia coli. Anim. Health Res. Rev. 2011;12:169–185. doi: 10.1017/S1466252311000193. [DOI] [PubMed] [Google Scholar]
- 8.DebRoy C., Roberts E., Valadez A.M., Dudley E.G., Cutter C.N. Detection of Shiga toxin producing Escherichia coli O26, O45, O103, O111, O113, O121, O145, and O157 serogroups by multiplex PCR of the wzx gene of the O-antigen gene cluster. Foodborne Pathog. Dis. 2011;8:651–652. doi: 10.1089/fpd.2010.0769. [DOI] [PubMed] [Google Scholar]
- 9.Fratamico P.M., Briggs C.E., Needle D., Chen C.Y., DebRoy C. Sequence of the Escherichia coli O121 O-antigen gene cluster and detection of enterohemorrhagic E. coli O121 by PCR amplification of the wzx and wzy genes. J. Clin. Microbiol. 2003;41:3379–3383. doi: 10.1128/JCM.41.7.3379-3383.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu Y., Fratamico P. Escherichia coli O antigen typing using DNA microarrays. Mol. Cell. Probes. 2006;20:239–244. doi: 10.1016/j.mcp.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 11.Lin A., Nguyen L., Lee T., Clotilde L.M., Kase J.A., Son I., Carter J.M., Lauzon C.R. Rapid O serogroup identification of the ten most clinically relevant STECs by Luminex microbead-based suspension array. J. Microbiol. Methods. 2011;87:105–110. doi: 10.1016/j.mimet.2011.07.019. [DOI] [PubMed] [Google Scholar]
- 12.Fratamico P.M., DebRoy C., Liu Y.H. The DNA sequence of the Escherichia coli O22 O-antigen gene cluster and detection of pathogenic strains belonging to E. coli serogroups O22 and O91 by multiplex PCR assays targeting virulence genes and genes in the respective O-antigen gene clusters. Food Anal. Methods. 2009;2:169–179. doi: 10.1007/s12161-008-9046-z. [DOI] [Google Scholar]
- 13.Clotilde L.M., Bernar Clay I.V., Hartman G.L., Lau D.K., Carter J.M. Microbead-based immunoassay for simultaneous detection of Shiga toxins and isolation of Escherichia coli O157 in foods. J. Food Prot. 2011;74:373–379. doi: 10.4315/0362-028X.JFP-10-344. [DOI] [PubMed] [Google Scholar]
- 14.Bockemühl J., Fricke G., Seeliger H.P. [Enteritis due to Escherichia coli O142 K86 H34 in a ward of premature infants. With a discussion on the problem of pathogenicity of “enteropathogenic serogroups of E. coli” (authors transl)] Zentralbl. Bakteriol. Orig. A. 1979;243:197–206. [PubMed] [Google Scholar]
- 15.Galane P.M., Le Roux M. Molecular epidemiology of Escherichia coli isolated from young South African children with diarrhoeal diseases. J. Health Popul. Nutr. 2001;19:31–38. [PubMed] [Google Scholar]
- 16.Garabal J.I., González E.A., Vázquez F., Blanco J., Blanco M., Blanco J.E. Serogroups of Escherichia coli isolated from piglets in Spain. Vet. Microbiol. 1996;48:113–123. doi: 10.1016/0378-1135(95)00150-6. [DOI] [PubMed] [Google Scholar]
- 17.Li M.C., Wang F., Li F. Identification and molecular characterization of antimicrobial-resistant Shiga toxin-producing Escherichia coli isolated from retail meat products. Foodborne Pathog. Dis. 2011;8:489–493. doi: 10.1089/fpd.2010.0688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith H.R., Cheasty T., Rowe B. Enteroaggregative Escherichia coli and outbreaks of gastroenteritis in UK. Lancet. 1997;350:814–815. doi: 10.1016/S0140-6736(05)62611-6. [DOI] [PubMed] [Google Scholar]
- 19.Chart H., Smith H.R., Scotland S.M., Rowe B., Milford D.V., Taylor C.M. Serological identification of Escherichia coli O157:H7 infection in haemolytic uraemic syndrome. Lancet. 1991;337:138–140. doi: 10.1016/0140-6736(91)90801-U. [DOI] [PubMed] [Google Scholar]
- 20.Scotland S M., Rowe B., Smith H.R., Willshaw G.A., Gross R.J. Vero cytotoxin-producing strains of Escherichia coli from children with haemolytic uraemic syndrome and their detection by specific DNA probes. J. Med. Microbiol. 1988;25:237–243. doi: 10.1099/00222615-25-4-237. [DOI] [PubMed] [Google Scholar]
- 21.Kobayashi H., Kanazaki M., Ogawa T., Iyoda S., Hara-Kudo Y. Changing prevalence of O-serogroups and antimicrobial susceptibility among STEC strains isolated from healthy dairy cows over a decade in Japan between 1998 and 2007. J. Vet. Med. Sci. 2009;71:363–366. doi: 10.1292/jvms.71.363. [DOI] [PubMed] [Google Scholar]
- 22.Kalchayanand N., Arthur T.M., Bosilevac J.M., Brichta-Harhay D.M., Guerini M.N., Shackelford S.D., Wheeler T.L., Koohmaraie M. Microbiological characterization of lamb carcasses at commercial processing plants in the United States. J. Food Prot. 2007;70:1811–1819. doi: 10.4315/0362-028x-70.8.1811. [DOI] [PubMed] [Google Scholar]
- 23.Cid D., Ruiz-Santa-Quiteria J.A., Marín I., Sanz R., Orden J.A., Amils R., de la Fuente R. Association between intimin (eae) and EspB gene subtypes in attaching and effacing Escherichia coli strains isolated from diarrhoeic lambs and goat kids. Microbiology. 2001;147:2341–2353. doi: 10.1099/00221287-147-8-2341. [DOI] [PubMed] [Google Scholar]
- 24.Vu-Khac H., Holoda E., Pilipcinec E., Blanco M., Blanco J.E., Dahbi G., Mora A., López C., González E.A., Blanco J. Serotypes, virulence genes, intimin types and PFGE profiles of Escherichia coli isolated from piglets with diarrhoea in Slovakia. Vet. J. 2007;174:176–187. doi: 10.1016/j.tvjl.2006.05.019. [DOI] [PubMed] [Google Scholar]
- 25.Chen X., Gao S., Jiao X., Liu X.F. Prevalence of serogroups and virulence factors of Escherichia coli strains isolated from pigs with postweaning diarrhoea in eastern China. Vet. Microbiol. 2004;103:13–20. doi: 10.1016/j.vetmic.2004.06.014. [DOI] [PubMed] [Google Scholar]
- 26.Glünder G. Dermatitis in broilers caused by Escherichia coli: Isolation of Escherichia coli from field cases, reproduction of the disease with Escherichia coli O78:K80 and conclusions under consideration of predisposing factors. Zentralbl. Veterinarmed. B. 1990;37:383–391. doi: 10.1111/j.1439-0450.1990.tb01073.x. [DOI] [PubMed] [Google Scholar]
- 27.Smyth C.J., Olsson E., Moncalvo C., Söderlind O., Orskov F., Orskov I. K99 antigen-positive enterotoxigenic Escherichia coli from piglets with diarrhea in Sweden. J. Clin. Microbiol. 1981;13:252–257. doi: 10.1128/jcm.13.2.252-257.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Djikeng A., Halpin R., Kuzmickas R., Depasse J., Feldblyum J., Sengamalay N., Afonso C., Zhang X., Anderson N.G., Ghedin E., et al. Viral genome sequencing by random priming methods. BMC Genomics. 2008 doi: 10.1186/1471-2164-9-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tusnády G.E., Simon I. The HMMTOP transmembrane topology prediction server. Bioinformatics. 2001;17:849–850. doi: 10.1093/bioinformatics/17.9.849. [DOI] [PubMed] [Google Scholar]
- 30.Samuel G., Reeves P. Biosynthesis of O-antigens: Genes and pathways involved in nucleotide sugar precursor synthesis and O-antigen assembly. Carbohydr. Res. 2003;338:2503–2519. doi: 10.1016/j.carres.2003.07.009. [DOI] [PubMed] [Google Scholar]
- 31.Datta A.K., Basu S. Chemical characterization of the lipopolysaccharides from enteropathogenic Escherichia coli O142 and O158. Indian J. Biochem. Biophys. 1999;36:55–58. [PubMed] [Google Scholar]
- 32.Landersjö C., Weintraub A., Widmalm G. Structural analysis of the O-antigenic polysaccharide from the enteropathogenic Escherichia coli O142. Eur. J. Biochem. 1997;244:449–453. doi: 10.1111/j.1432-1033.1997.t01-1-00449.x. [DOI] [PubMed] [Google Scholar]
- 33.Jiang L., Perepelov A.V., Filatov A.V., Liu B., Shashkov A.S., Senchenkova S.N., Wang L., Knirel Y.A. Structure and gene cluster of the O-antigen of Escherichia coli O68. Carbohydr. Res. 2014;397:27–30. doi: 10.1016/j.carres.2014.07.022. [DOI] [PubMed] [Google Scholar]
- 34.Iida S., Hiestand-Nauer R., Arber W. Transposable element IS1 intrinsically generated target duplications of variable length. Proc. Natl. Acad. Sci. USA. 1985;82:839–843. doi: 10.1073/pnas.82.3.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grindley N.D. ISI insertion generates duplication of a nine base pair sequence at its target target site. Cell. 1978;13:419–426. doi: 10.1016/0092-8674(78)90316-1. [DOI] [PubMed] [Google Scholar]
- 36.Mahillon J., Chandler M. Insertion sequences. Microbiol. Mol. Biol. Rev. 1998;62:725–774. doi: 10.1128/mmbr.62.3.725-774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Siguier P., Gourbeyre E., Chandler M. Bacterial insertion sequences: Their genomic impact and diversity. FEMS Microbiol. Rev. 2014;38:865–891. doi: 10.1111/1574-6976.12067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Touchon M., Rocha E.P. Causes of insertion sequence abundance in prokaryotic genomes. Mol. Biol. Evol. 2007;24:969–981. doi: 10.1093/molbev/msm014. [DOI] [PubMed] [Google Scholar]