SUMMARY
The replisome is a large protein machine containing multiple enzymatic activities needed to complete DNA replication. In addition to helicase and polymerases needed for copying the DNA, the replisome also contains proteins like DNA methyltransferases, histone chaperone and chromatin modifying enzymes to couple DNA replication with chromatin deposition and establishment of the epigenetic code. In addition, since template DNA strands often contain DNA damage or other roadblocks to the replication machinery, replication stress response proteins associate with the replisome to stabilize, repair and restart stalled replication forks. Hundreds of proteins are needed to accomplish these tasks. Identifying these proteins, monitoring their post-translational modifications, and understanding how their activities are coordinated is essential to understand how the genome and epigenome are duplicated rapidly, completely, and accurately every cell division cycle. Here we describe an updated iPOND (isolation of proteins on nascent DNA) method to facilitate these analyses.
Keywords: DNA replication, chromatin, DNA repair, iPOND, EdU, click chemistry, replication stress, PCNA, DNA damage
1. INTRODUCTION
Studying proteins at active or damaged replication forks and monitoring the deposition of proteins on newly synthesized DNA requires a method of purifying these protein-DNA complexes. Chromatin fractionation and chromatin-immunoprecipitation (ChIP) provide two methods to accomplish this task. However, chromatin fractionation does not provide any spatial information about the location of where a protein is bound in the genome. ChIP provides spatial information but its utility in studying DNA replication or replication stress responses in mammalian cells is limited by difficulties in synchronizing replication in any specific genomic region. Super-resolution immunofluorescence imaging or analysis of protein-protein interactions using methods like proximity ligation assays can provide spatial information with respect to a known reference control. However, these methods require highly specific antibodies and are not compatible with unbiased approaches to identify new proteins or protein modifications.
We developed iPOND to overcome these experimental challenges, providing a method to examine protein recruitment and modification at replication forks as well as the processes of chromatin deposition and maturation (1, 2). iPOND is essentially a reverse chromatin immunoprecipitation in which newly replicated DNA is purified and its associated proteins analyzed by immunoblotting or mass spectrometry (3, 4).
iPOND relies on incorporation of a nucleoside analog 5-ethynyl-2′-deoxyuridine (EdU) into newly synthesized DNA. EdU is incorporated instead of thymidine and contains an alkyne functional group permitting a high-efficiency cycloaddition reaction to tether biotin to the newly synthesized DNA fragment (5). Biotin facilitates a single-step, streptavidin-based affinity purification of the DNA protein complexes, which can then be analyzed using standard DNA and protein detection methods.
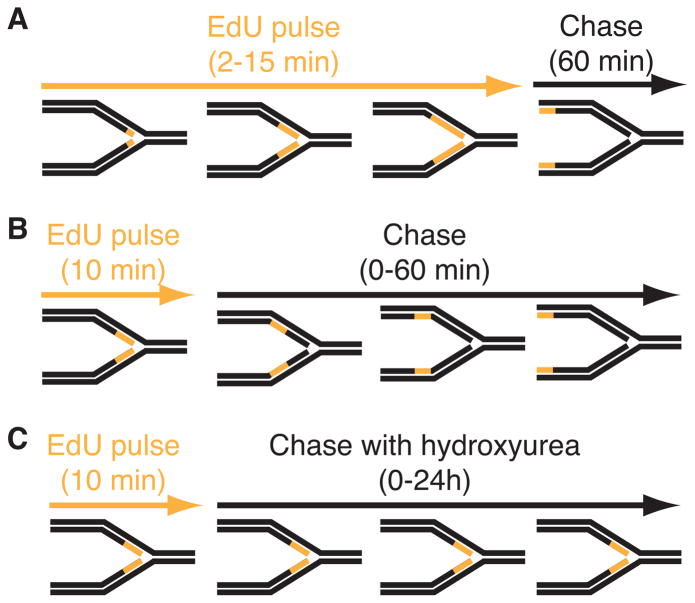
The procedure is most useful when cells are labeled with EdU for very short periods (2–15 minutes) so it is incorporated into only small DNA fragments immediately adjacent to elongating replication forks. Fixation of protein-DNA complexes using formaldehyde prior to performing the biotin-conjugation reaction and purification permits isolation of replisome components (Figure 1A). Combining the short EdU labeling period with increasing culture times in the absence of label (a chase period), is useful to study chromatin deposition and maturation as a function of time and distance from the elongating replication fork (Figure 1B). Finally, combining the EdU labeling with addition of drugs that damage DNA or otherwise stall replication forks such as hydroxyurea provides a method to study DNA repair and replication stress responses (Figure 1C).
Figure 1.
Diagram of three types of iPOND experiments. In these schematics, the black lines represent unlabeled DNA and orange lines represent DNA labeled with EdU. (A) Experiment to detect proteins that localize at elongating replication forks. Cells are incubated for increasing times in EdU prior to fixation. A single chase sample in which the EdU is removed for one hour prior to fixation is included as a control. True replication proteins will no longer be enriched in this chase control. (B) Experiment to monitor chromatin deposition and maturation. A single short EdU labeling period is followed by increasing chase times to monitor how the proteins associated with the EdU-labeled DNA fragment change as a function of time and distance from the fork. (C) Experiment to assess replication stress responses. Cells are labeled with EdU and then chased into media containing a replication stress-inducing drug like hydroxyurea. Variants of this procedure in which the replication stress agent is added prior to EdU or EdU remains in the growth media during the replication stress period are possible. All iPOND experiments should include an additional control sample that either lacks EdU labeling or in which the biotin-azide is omitted from the procedure (a “no-click” control).
2. MATERIALS
Culture mammalian cells in appropriate growth media and conditions. The protocol described was optimized for HEK293T cells grown in DMEM containing 7.5% fetal bovine serum. However other cell types including U2OS, hTERT-RPE, and mouse embryonic fibroblasts have been used successfully (6). All reagents are prepared and used at room temperature unless otherwise specified.
2.1. EdU cell labeling and fixation
EdU: 10 mM 5-ethynyl-2′-deoxyuridine (Life Technologies, Carlsbad, CA, USA) stock prepared in DMSO. Store at −20°C protected from light for up to 1 year.
Thymidine: 10 mM thymidine (Sigma, St. Louis, MO, USA) stock prepared in water and stored at −20°C.
Fixative and permeabilization buffer: Immediately prior to use, dilute 37% formaldehyde (Sigma, St. Louis, MO, USA) in phosphate buffered saline (PBS) solution (Thermo Fisher Scientific, Waltham, MA, USA) to a final concentration of 1%. Prepare 1.25 M glycine (Sigma, St. Louis, MO, USA) in water. Permeabilization buffer is prepared by diluting triton X-100 (Sigma, St. Louis, MO, USA) to 0.25% in PBS.
Cell wash solution: 0.5% (w/v) bovine serum albumin (Sigma, St. Louis, MO, USA) dissolved in PBS.
2.2 Click reaction Components
1 mM biotin-azide (Life Technologies, Carlsbad, CA, USA) stock is prepared in DMSO and stored at −20°C.
Prepare stock 100mM CuSO4 (Thermo Fisher Scientific, Waltham, MA, USA) in water and store at RT.
Freshly prepare 20mg/ml of (+) sodium L-ascorbate (Sigma, St. Louis, MO, USA) in water, limit exposure to air, and store on ice until needed.
2.3 Cell Lysis and Purification Components
-
1
Lysis buffer: 1% SDS (Sigma, St. Louis, MO, USA) in 50 mM Tris-HCl, pH 8.0
-
2
90 micron nylon mesh (Small Parts Inc., Logansport, IN, USA)
-
3
Streptavidin agarose (EMD Millipore, Darmstadt, Germany)
-
2
High salt wash: 1 M NaCl
2.4 Elution and Protein Detection Components
SDS sample buffer (2x): Mix 0.4 g SDS, 2 ml 100% Glycerol, 1.25 ml 1 M Tris pH 6.8, and 0.01g Bromophenol blue in 8 ml water. Prior to use, add 1M DTT to a final concentration of 200 mM.
Crosslink reversal solution: Mix 0.5 M EDTA, 1 M Tris-HCl, pH 6.7, Proteinase K (500units/mL) (Sigma, St. Louis, MO, USA) in a 2:4:1 volume ratio immediately prior to use.
3. METHODS
3.1. Labeling of Cells
Expand HEK293T cells such that there are 4–6 × 107 cells per 150mm dish. Three dishes are needed per sample. This protocol is written to assume three samples (see Note 1). Also prepare one extra dish of cells so 10 dishes are needed in total.
Harvest the extra dish of cells and count viable cells. This cell number is used to calculate the volume of reagents to use in subsequent steps.
Remove sample dishes from incubator and add EdU to the culture media to a final concentration of 10 μM for all three samples (see Note 2). Replace into incubator rapidly and incubate for 10 minutes.
For samples #1 and #2 remove the cells from the incubator and fix on the dish by adding 10 mL of 1% formaldehyde solution. Incubate 20 minutes at room temperature.
For sample #3 remove cells from incubator after 10 minutes of labeling, decant growth media, carefully wash cells with 5 mL of growth media containing 10 μM thymidine, decant, and replace with 20 mL of growth media containing 10 μM thymidine before replacing cells into incubator for a further 60 minutes prior to fixing as in step 4 (see Note 3).
Quench crosslinking reaction by adding 1 mL of 1.25M glycine
Collect cells by scraping with cell lifter and transfer into 50 ml conical tube. Place on ice. Combine the three plates per sample into a single tube. Record the volume.
Centrifuge 5 mins at 900g, 4°C.
Decant supernatant.
Wash cell pellets with PBS and centrifuge 5 mins at 900g, 4°C. PBS wash volume is same as fixation volume noted in step 7. Vortex to resuspend pellets in PBS.
Decant PBS wash and repeat step 10 two additional times.
After final wash, decant PBS (see Note 4).
Resuspend cells in permeabilization buffer at a concentration of 1 × 107 cells/mL for 30 minutes at room temperature.
Centrifuge cells 5 mins at 900g, 4°C. Decant supernatant
Wash cells once with 0.5% BSA in PBS at 4°C.
Centrifuge cells 5 mins at 900g, 4°C. Decant supernatant
Wash cells once with PBS at 4°C.
Centrifuge cells 5 mins at 900g, 4°C. Decant supernatant. Place on ice.
3.2 Click chemistry reaction to conjugate biotin to EdU
Prepare click reaction cocktail to contain a final concentration of 10 mM sodium ascorbate, 2 mM CuSO4, 10 μM biotin-azide, in PBS. Prepare sufficient cocktail for 5 mL per 108 cells (see Note 5). Omit the biotin-azide from cocktail prepared for sample #1. This is the “no click” negative control. (see Note 6).
Resuspend cell pellets in click reaction cocktails at a final concentration of 2 × 107 cells/mL.
Rotate reactions at room temperature for 1–2h
Centrifuge cells 5 mins at 900g, 4°C. Decant supernatant.
Wash cells once with 0.5% BSA in PBS at 4°C with a PBS volume equal to the click reaction cocktail volume used in step 2.
Centrifuge cells 5 mins at 900g, 4°C. Decant supernatant.
Wash cells once with PBS at 4°C.
Centrifuge cells 5 mins at 900g, 4°C. Decant supernatant. (see Note 7).
3.3 Cell Lysis and Purification
-
1
Resuspend cells in lysis buffer containing aprotinin and leupeptin (1 μg/mL) at a concentration of 1.5 × 107 cells per 100 μL. Transfer to 1.5 mL microfuge tubes. Place on ice.
-
2
Sonicate using a microtip sonicator at a power of 13–16 Watts, 20 second pulse.
-
3
Pause 40 seconds with samples on ice to prevent over-heating.
-
4
Repeat sonication steps 2 and 3 four times (see Note 8).
-
5
Centrifuge samples for 10 mins at 16,100g, RT.
-
6
Filter supernatant through a 90 micron nylon mesh into new tube. Place on ice. (see Note 9).
-
9
Dilute lysate 1:1 (v/v) using PBS containing 1 μg/mL of aprotinin and leupeptin at 4°C.
-
10
Save 15 μL of lysate as an “input” sample.
-
11
Add 50 μL of packed streptavidin-agarose beads pre-washed with PBS (see Note 10).
-
12
Incubate 1–16h in cold room while rotating. (see Note 11)
-
13
Centrifuge the streptavidin-agarose beads containing the captured DNA-protein complexes for 3 minutes at 1,800g, RT.
-
14
Decant supernatant.
-
15
Wash beads with 1 mL of cold lysis buffer. Rotate for 5 minutes.
-
16
Centrifuge for 1min at 1,800g. Decant supernatant.
-
17
Wash with 1 mL of 1M NaCl. Rotate for 5 minutes.
-
18
Centrifuge for 1min at 1,800g. Decant supernatant.
-
19
Repeat washes with lysis buffer in steps 15 and 16 two additional times.
-
20
Centrifuge for 1min at 1,800g. Decant supernatant.
3.4 Elution and detection of proteins bound to captured DNA
Add SDS sample buffer (1:1 v/v of packed beads).
Incubate 25 minutes at 95°C. (see Note 12).
Centrifuge for 1min at 1,800g, RT.
Purified protein samples and input samples are ready for separation by standard SDS-PAGE. (see Note 13).
Transfer proteins to nitrocellulose and detect by standard immunoblotting procedures (see Notes 14 and 15).
As an alternative to immunoblotting, proteins can be detected and analyzed by mass spectrometry (see Note 16).
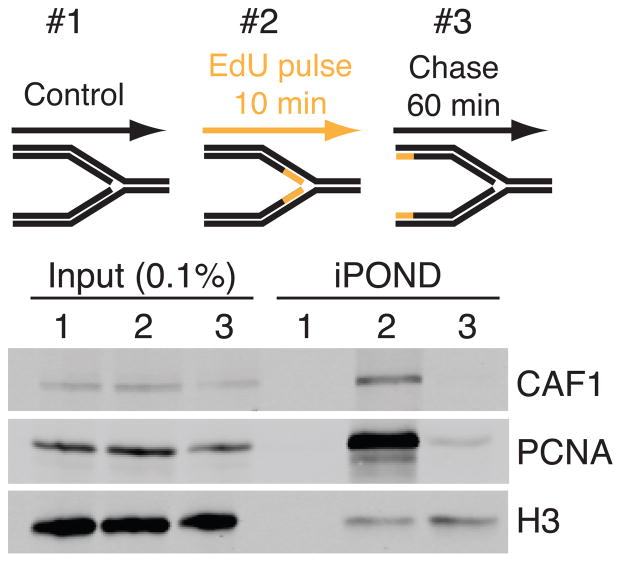
Figure 2.
An example of the results expected from a simple three-sample iPOND experiment. No-click control, 10 min EdU label, and 10 min EdU followed by one hour chase samples were prepared and immunoblotted for CAF1, PCNA and histone H3. Purified samples are compared to the inputs.
Acknowledgments
This work was supported by NCI grant R01CA136933 to D.C. We thank Bianca Sirbu and Frank Couch who contributed to the initial invention of iPOND.
Footnotes
A typical experiment to examine a protein at an elongating replication fork will require three samples (9 dishes). Sample #1 is used as a negative control for the purification. Sample #2 purifies proteins at or immediately adjacent to the replication fork. Sample #3 purifies general chromatin binding proteins. This protocol is written for an experiment with these three samples.
Stagger samples to ensure sufficient time for processing each sample throughout the experiment. EdU labeling times can vary and be as short as 2 minutes. Typically, we find that a 10 minute labeling time is a useful starting point to purify replication proteins.
Pre-equilibrate the growth media to 37°C and the proper CO2 content. Complete the wash step as rapidly as possible to minimize the length of time the cells are not in the incubator. The thymidine in this step is optional but may decrease the chance of residual EdU being incorporated into the DNA during the hour “chase” incubation.
We find it useful to flash freeze cell pellets and store at −80°C when multiple samples are prepared over an extended staggered time.
A typical sample of three 150mm dishes of HEK293T cells will have 1.5×108 cells and will need 7.5 ml of click reaction cocktail. In our growth conditions approximately 50% of cells in culture are actively synthesizing DNA. We recommend increasing cell numbers for cell types that have fewer cells in S-phase at the time of the experiment.
The “no click” negative control can also be replaced with a sample in which no EdU is used to label the cells.
Samples can be stored at −80°C after this step.
Lysate should clarify some during sonication. Cloudiness is an indicator of insufficient sonication or incorrect cell/lysis volume ratio. Larger volumes will require additional sonication steps.
Lysate should be clear after centrifugation and filtering. If not, repeat centrifugation to remove any insoluble material.
Streptavidin-bound paramagnetic beads can be substituted. We have found that purification with magnetic beads using standard magnetic bead separation procedures can yield less non-specific protein purification in some cases. Bead volumes may need to be adjusted based on the biotin binding capacity for each bead manufacturer.
We have found that incubation for shorter times (1 hour) typically yields good results. Extended incubations may increase sensitivity.
This step reverses the protein-DNA crosslinks and solubilizes the denatured proteins. The “input” samples saved in step 10, (Section 3.3) should also be mixed with equal volume of SDS sample buffer and heated for 25 minutes.
We usually use a 15% gel to detect PCNA and histone proteins as controls. The purified sample can be split onto 2 or 3 gels to detect these positive controls easily by standard immunoblotting methods. Load all the sample onto a single gel for detecting proteins found more rarely at replication forks. We recommend loading the equivalent of 0.1% input (v/v) to compare to the purified samples.
It may be difficult to fully reverse the crosslinks especially for large proteins. Thus, you should expect that some proteins will be detected at both their typical position and as higher molecular weight aggregates on SDS-PAGE gels.
See figure 2 for an example of the expected result from this simple, three sample experiment.
We typically will use a “short stack” method prior to mass spectrometry. In this procedure, proteins are electrophoresed for a short time into the top 0.5 cm of the polyacrylamide gel. This gel piece is excised prior to standard in-gel trypsinization and mass spectrometry methods.
References
- 1.Sirbu BM, Couch FB, Cortez D. Monitoring the spatiotemporal dynamics of proteins at replication forks and in assembled chromatin using isolation of proteins on nascent DNA. Nature protocols. 2012;7:594–605. doi: 10.1038/nprot.2012.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sirbu BM, Couch FB, Feigerle JT, et al. Analysis of protein dynamics at active, stalled, and collapsed replication forks. Genes Dev. 2011;25:1320–1327. doi: 10.1101/gad.2053211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lopez-Contreras AJ, Ruppen I, Nieto-Soler M, et al. A proteomic characterization of factors enriched at nascent DNA molecules. Cell Rep. 2013;3:1105–1116. doi: 10.1016/j.celrep.2013.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sirbu BM, McDonald WH, Dungrawala H, et al. Identification Of Proteins At Active, Stalled, And Collapsed Replication Forks Using Isolation Of Proteins On Nascent DNA (iPOND) Coupled With Mass Spectrometry. J Biol Chem. 2013 doi: 10.1074/jbc.M113.511337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Salic A, Mitchison TJ. A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proc Natl Acad Sci U S A. 2008;105:2415–2420. doi: 10.1073/pnas.0712168105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nagarajan P, Ge Z, Sirbu B, et al. Histone acetyl transferase 1 is essential for mammalian development, genome stability, and the processing of newly synthesized histones H3 and H4. PLoS Genet. 2013;9:e1003518. doi: 10.1371/journal.pgen.1003518. [DOI] [PMC free article] [PubMed] [Google Scholar]