Abstract
Many text-mining studies have focused on the issue of named entity recognition and normalization, especially in the field of biomedical natural language processing. However, entity recognition is a complicated and difficult task in biomedical text. One particular challenge is to identify and resolve composite named entities, where a single span refers to more than one concept(e.g., BRCA1/2). Most bioconcept recognition and normalization studies have either ignored this issue, used simple ad-hoc rules, or only handled coordination ellipsis, which is only one of the many types of composite mentions studied in this work. No systematic methods for simplifying composite mentions have been previously reported, making a robust approach greatly needed. To this end, we propose a hybrid approach by integrating a machine learning model with a pattern identification strategy to identify the antecedent and conjuncts regions of a concept mention, and then reassemble the composite mention using those identified regions. Our method, which we have named SimConcept, is the first method to systematically handle most types of composite mentions. Our method achieves high performance in identifying and resolving composite mentions for three fundamental biological entities: genes (89.29% in F-measure), diseases (85.52% in F-measure) and chemicals (84.04% in F-measure). Furthermore, our results show that, using our SimConcept method can subsequently help improve the performance of gene and disease concept recognition and normalization.
General Terms: Algorithms
Keywords: Mention simplification, conditional random field, natural language processing, name entity recognition, name entity normalization
1. Introduction
In biomedical text mining, many studies have focused on automatically extracting relevant information from published literature [1]. The relevant information is commonly focused on a specific topic, such as protein-protein interactions [2, 3]; protein transport and localization [4-6]; drug-disease associations [7-9], or gene function extraction [10, 11]. Most of the common retrieval methods apply natural language processing or machine learning to identify relations in text. One crucial step towards this goal is automatically recognizing bioconcept mentions (e.g., gene/protein) – the task of named entity recognition (NER) – and mapping the bioconcept to a specific database identifier (e.g., NCBI EntrezGene) – the task of normalization. Many international biomedical text mining competitions (e.g., BioCreative) have therefore focused on these tasks [12-16]. Genes, diseases and chemicals are particularly notable for not only being important concepts, but also being the most popular concepts in biomedical literature search [17]. Most normalization studies of different concepts face two challenges: term variation and ambiguity [18-22]. Many previous studies have defined individual strategies (e.g., machine learning, statistical inference and rule-based methods) to deal with these two issues. However, a particular type of error which has not been handled well is composite mentions, where a single span refers to more than one concept (e.g., “SMADs 1, 5, and 8”). We observe that in our datasets, approximately 10% of gene, disease and chemical mentions are composite mentions, hence it is important to handle them properly. This study presents a new method for bio-concept mention simplification in a systematical fashion.
Most previous related studies have focused on text (including document/paragraph [23-28] and sentence [29-35] levels) simplification. An early text simplification paper was introduced by Chandrasekar et al. [26, 27], which used a syntactic-parsing method (i.e., parse trees) to improve the performance of text simplification. In the past several years, most of the studies have focused on sentence simplification using three methodologies: lexical, syntactic and discourse simplification [29]. Leroy et al. [23] designed an algorithm that uses term familiarity to identify difficult text and select easier alternatives from lexical resources. Peng et al. [30] developed iSimp which is an alternative method by using shallow parsing to detect the various simplification structures in text in linear time. Most of the studies on text simplification have focused on documents or sentences. The few studies which have concentrated on mention simplification have only addressed coordination ellipsis. Buyko et al., [36] developed a CRF-based method with three states: conjunction, conjuncts, and ellipsis antecedent. For an example “human and mouse cells”, “human” and “mouse” are conjuncts, “and” is conjunction, and “cells” is ellipsis antecedent. In their evaluation on GENIA [37] corpus, they obtained 86% accuracy on elliptical entity expression evaluation. Due to the lower performance of this method on complex ellipsis (e.g., “recombinant human nm23- H1, -H2, mouse nm23-M1, and -M2”), Chae et al., [38] developed a pattern based method and used lexicons to identify the region of each component (i.e., conjunction, conjuncts, and ellipsis antecedent) for each mention. However, these previous studies have focused on only one type of composite mention: mentions with coordination ellipsis. Due to the demand for improving bioconcept identification and lack of related studies, we propose a hybrid method to handle most of the composite mention types.
This study proposes a CRF-based model and a pattern identification strategy to detect the antecedent, suffix/strain and conjunction from a composite mention, which refers to more than one concept. Instead of building heuristic rules to match all possible variants, we propose a machine learning approach to simplify the mention. For example, after identifying all concepts from “SMADs 1, 5, and 8”, three individual concepts “SMAD1”, “SMAD5” and “SMAD8” will be generated. Some mentions, such as abbreviation pairs (e.g., “Estrogen receptor (ER) alpha”), include more than one reference to the same concept. These therefore appear very similar to composite mentions, complicating the normalization of this type of mention to a specific database identifier. To make this model more robust and able to handle more types of term variation, this study considers abbreviation pairs as a type of composite mention. After simplifying the mentions, it is easier to map the resulting concept mentions to a database or controlled vocabulary identifier. In this study, composite mentions can be divided into 5 distinct types (including abbreviation pair) and a mixed type of mentions.
Mention with coordination ellipsis: the concepts in this type of mentions share part of the mention region, such as the token “SMAD” in the composite mention “SMADs 1, 5, and 8”, which is shared to “SMAD1”, “SMAD5”, “SMAD8”.
Range mention: Like mentions with coordination ellipsis, these mentions share part of the mention region, however this type represents a range of entities rather than a discrete set. For example, the range “SMAD 2 to 4” can be decomposed to “SMAD2”, “SMAD3”, “SMAD4”.
Individual mention: this is an independent composite mention. All concepts can be separated into non-overlapping spans (e.g., “BTK/ITK/TEC/TXK” can be decomposed to “BTK”, “ITK”, “TEC” and “TXK”).
Overlap abbreviation pair mention: The long form and short form share some tokens, like “COUP (chicken ovalbumin upstream promoter) transcription factor” which can be decomposed to “COUP transcription factor” and “chicken ovalbumin upstream promoter transcription factor”. But the two concepts indicate the same database identifier.
Individual abbreviation pair mention: this is an independent composite mention. The same to overlap abbreviation pair mention, the two concepts indicate the same database identifier. For example, “ectodermal dysplasia (EDA)” can be decomposed to “ectodermal dysplasia” and “EDA”.
Mixed mention: It is a mixed mention of any two above types, like “high mobility group (HMG) protein 1 and 2” which can be decomposed to “high mobility group protein 1”, “HMG protein 1”, “high mobility group protein 2” and “HMG protein 2”.
The 3rd and 4th types of mentions do not overlap and therefore should be identified as separate by the mention recognition model. However, since these are easily confused with composite mentions, the mention recognition model may identify the wrong boundaries. It is therefore important for the mention simplification model to also identify these correctly and separate them into individual concepts. However, not all mentions which follow these patterns should be separated. For example, “ubiquitin-activating enzyme (E1)” and “metalloprotease/disintegrin/cysteine-rich protein 9” both represent single concepts and should not be separated. These cases might not be easy for pattern identification method. Therefore, a robust method should be able to deal with these exceptions.
In this study, we developed SimConcept, a simplification method for composite mentions, which aims to help concept normalization in the end. It detects various concepts of a composite mention. Here are the three main contributions: 1) SimConcept can handle six types of composite mentions, more than any other methods previously reported; 2) When applied to the three bio-concepts (i.e., gene, disease, and chemical), our method achieved state-of-the-art performance and 3) Based on our success on more than one entity type, our approach is shown to be robust and generalizable.
2. Methods
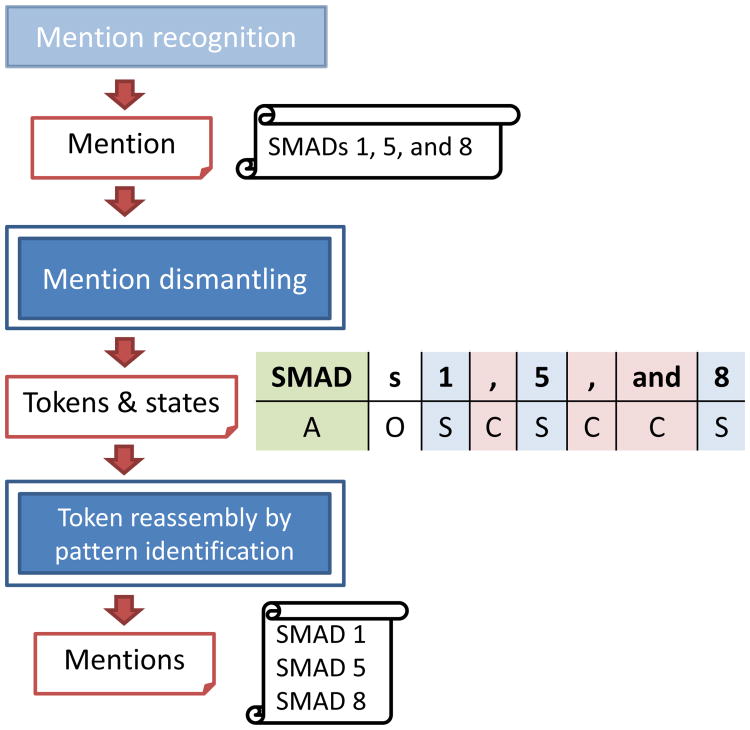
Our method consists of two modules as shown in Figure 1. The first module consists of a conditional random field model. In this module, the input mention is separated into tokens and each token assigned labels according to the most likely sequence of states through the model. The second module reassembles the tokens into individual mentions using a pattern identification method.
Figure 1. Architecture of the SimConcept mention simplification method.
2.1 Conditional random fields model
To recognize the composite mentions, we observed the composition of those mentions and defined nine states for building conditional random fields (CRF) model [39]: Antecedent (A); Strain/Suffix(S); Conjunction of mentions with coordination ellipsis (C); Conjunction of range mentions (CR); Left parentheses of abbreviation pair (L); Right parentheses of abbreviation (R); Right parentheses of abbreviation, but the abbreviation and long form cannot be separated (Ro); Conjunction of individual mentions (I); Redundant (O). The states “C”, “CR”, “L”, “R”, “Ro” (L and R/Ro occur in pairs), and “I” are conjunction states which can use to recognize the mention types. If one mention includes two or more conjunction states, this mention would be identified as a mixed mention.
As mentioned above, we regarded this mention simplification problem as a sequence labeling task, as illustrated in a simple example in Table 1. Our implementation uses a linear chain Conditional Random Fields (CRF) [39] provided by CRF++(http://crfpp.googlecode.com/svn/trunk/doc/index.html). The CRF model defines the conditional probability distribution P(Y|X) of label sequence Y given observation sequence X. The lengths of random variable sequences X(x1,…,xn) and Y(y1,…,yn) are the same.
Table 1. An example of label sequence Y and observation sequence X of gene mention “BRCA1/2”.The “BRCA” is antecedent (A). “1” and “2” are both suffixes (S). “/” is conjunction (C).
| X | BRCA | 1 | / | 2 |
| Y | A | S | C | S |
The CRF model on (X, Y) is specified by a vector F of global features and a corresponding weight vector λ(λ1,…,λl). is a global feature vector for label sequence Y and observation sequence X. In our research, the y1,…,yn indicate the label for the corresponding tokens. To identify the antecedent and conjuncts part of mention.
The weight λ presents the importance of the feature fi(yj, yj−1, X) and can be obtained from the training data. CRF++ applies L-BFGS [40] which is a quasi-newton algorithm for large scale numerical optimization problem.
2.2 CRF Features
We adapted tmVar [41], our previous study on mutation recognition, to this task. We used tmVar's tokenization and part of its features in SimConcept development. Like tmVar, our tokenization separates uppercase characters, lowercase characters and digits. For example, “SMADs 2 to 4” is separated to “SMAD”, “s”, “2”, “to” and “4”. We adapted tmVar's features to reflect the difference in input between tmVar (i.e., documents) and SimConcept (i.e., individual mentions). After reviewing the evidence for different token types of a mention, we defined several suffixes, prefixes and some semantic types for identifying bioconcepts (i.e., gene, disease, and chemical) mention characteristics. In particular, most mention suffixes for disease and chemical mentions are not digits, for example “breast and ovarian cancer” (disease) and “b-sitosteryl and stigmasteryl linoleates” (chemical), which might be difficult to recognize without any semantic evidence. Therefore, we collected the semantic features used in some previous studies [41-43] and grouped the suffixes/prefixes we defined into semantic feature types such as those shown below.
Chemical Suffix: yl, ylidyne, oyl, sulfony, one, ol, carboxylic, amide, ate, acid, ium, ylium and etc.
Chemical Alkane Stem: meth, eth, prop, tetracos
Chemical Trivial Ring: benzene, pyridine, toluen
Chemical Simple Multiplier: di, tri, tetra and etc.
Chemical elements: hydrogen, helium, lithium, beryllium, boron, carbon and etc.
Disease Suffix: cancer, disease, symptom and etc.
Gene/Protein Suffix: gene, protein, receptor, factor, element, unit and etc.
Family, Complex : family, subfamily, superfamily, complex
We also continue to use three of tmVar's features types (i.e., Character features, Case pattern features and Contextual features). Character features inlcude number of digits, number of uppercase and lowercase letters, number of all characters, and specific characters (; , . -> + _ / ?). Case pattern features are created by replacing uppercase alphabetic character to “A”and any lower case to ‘a’. Likewise any number (0-9) is replaced by ‘0’. Moreover, we also merged consecutive letters and numbers to generate additional features, such as “AAA” to “A”.In order to take advantage of contextual information, for a given token we included the token and semantic features of 3 neighboring tokens from each side.
2.3 Token reassembly through pattern identification
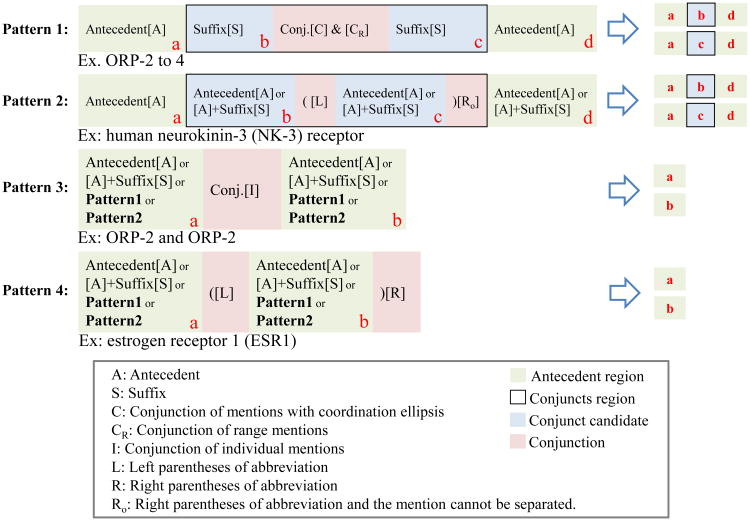
By observing the characteristics of composite mentions in our training data, we manually defined four patterns to model the six types of composite bioconcept mentions, as shown in Figure 2. To simplify mentions, we distinguish between the antecedent region (green), conjuncts region (frame), conjunct candidate (blue) and conjunctions (red). The tokens in antecedent region should be present in all possible mentions. The tokens in conjuncts region should be replaced by all possible conjunct candidates in this region. Every conjuncts region consists of at least one conjunction. Conjunctions are used to separate individual conjunct candidates.
Figure 2. Patterns for formulating bioconcept mentions. Note that in Patterns 1 & 2, it is common to have Antecedent appearing either at the beginning or the end but not both locations (i.e. component a and d may not appear together in one mention).
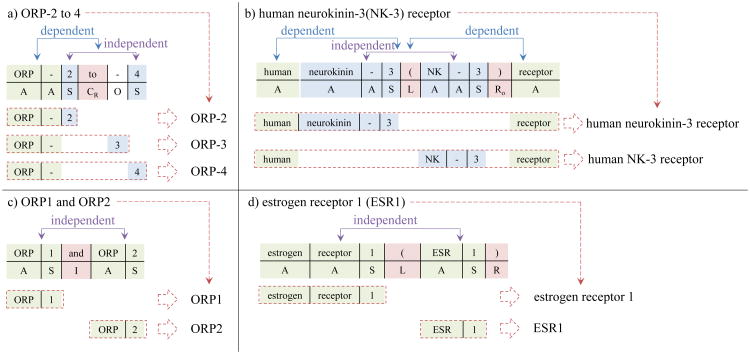
In our definition, every mention can map to one of the patterns. Range mentions and mentions with coordination ellipsis map to Pattern 1. As shown in Figure 3a, the “ORP-2 to -4” is a range mention which can be separated to “ORP-” (antecedent region), and “2 to -4” (conjuncts region). In conjuncts region, all possible candidates (i.e., 2, 3 and 4 in “2 to -4”) belong to one of the possible mentions. Therefore, “ORP-2 to -4” is reassembled to “ORP-2”, “ORP-3” and “ORP-4”. In another similar case, the “ORP-1 and -2” is similar to “ORP-2 to -4”. The major difference is the conjunction (i.e., “and”). In this case, “-1” and “-2” in conjuncts region are independent. Therefore “ORP-1 and -2” becomes “ORP-1” and “ORP-2”.
Figure 3. The strategy of reassembly for mention with coordination ellipsis, range mention and abbreviation.
Observing these two cases, it becomes clear that the difference between range mentions and mentions with coordination ellipsis is the conjunctions. In case the conjunction is recognized as a conjunction of range mentions (CR state), all values in the range of these two suffixes should be considered as conjunct candidates. Otherwise, once the conjunction is recognized as a conjunction of mentions with coordination ellipsis (C state), the candidates in the conjuncts region are independent to each other, and dependent to the antecedent region. Therefore, the reassembly mentions are the combinations of antecedent region and each conjunct candidate.
As shown in Figure 3b, individual and overlap abbreviation pair mention belong to Pattern 2. The pair (long form “neurokinin-3” and abbreviation “NK-3”) of abbreviation mentions is in the conjuncts region. Therefore, the two candidates, long form and abbreviation, are reassembled with antecedent region individually. We detected the long form region by applying the Ab3P abbreviation identification tool [44]. Thus, we are able to identify the conjuncts region in these mentions. Patterns 3 and 4 in Figure 2 are relatively easier than Pattern 1 and 2. Since the patterns do not contain a conjuncts region, assembling the individual mentions only requires splitting conjunctions and parentheses. As shown in Figure 3c/d, the mentions can be separated individually.
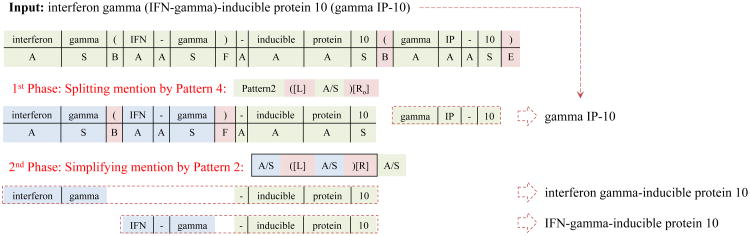
In addition to the above types, mixed mention is more complicated. We defined a two phase strategy to divide concepts. In the 1st phase we split the mention using Patterns 3 or 4, which do not contain any conjuncts region. In this phase, all conjunctions of range (CR), mention with coordination ellipsis (C) and abbreviation parentheses (L/Ro) are considered as part of antecedent region. In the 2nd phase, the mention is decomposed by Pattern 1 or 2. In this phase we start to face the conjuncts region. As shown in Figure 4, “interferon gamma (IFN-gamma)-inducible protein 10 (gamma IP-10)” is split to “interferon gamma (IFN-gamma)-inducible protein 10” and “gamma IP-10” by cutting in 1st phase. According the states L/Ro which have been identified in “interferon gamma (IFN-gamma)- inducible protein 10”, the 2th phase should choose Patten 2 for simplifying. We therefore obtained “interferon gamma-inducible protein 10” and “IFN-gamma-inducible protein 10” from the 2th phase simplification.
Figure 4. The strategy of reassembly for mixed mention.
In other words, the main idea of this two phase strategy is to retain all sub-mentions with a conjuncts region in the second phase. Since the sub-mentions which map to Patterns 1 & 2 are more complicated and cannot be separated individually, those sub-mentions will be processed in the second step.
2.4 The SimConcept Corpus
The SimConcept corpus was compiled using 5 datasets: three for genes, one for diseases and one for chemicals. For genes, we integrated the BioCreative II gene normalization task training (281 abstracts) and test (262 abstracts) corpora and the 151 GIA test collection (http://ii.nlm.nih.gov/DataSets/index.shtml#GIA). In addition, we also collected disease mention corpus from NCBI Disease corpus [18, 45] with 793 abstracts, and sampled Chemical mention corpus from BioCreative IV CHEMDNER task [46] training dataset for 937 abstracts. As shown in Table 2, we collected 2,424 abstracts in total. For each article, in addition to the annotations of all described bioconcept mentions, we appended following annotations: 1) the decomposed mentions, such as “BRCA1” and “BRCA2” of “BRCA1/2”; 2) the five types of composite mentions (e.g., “mention with coordination ellipsis”); 3) the states of tokens (“ ”). We used PubTator [47-49], a web-based annotation tool to annotate the corpus. The distributions of the five composite mention types (CR: Range mention, C: mention of coordination ellipsis, I: individual mentions, IA: individual abbreviation, and OA: overlap abbreviation.) in Table 2 are different between the three sets. Chemicals contain significantly more range mentions than either disease or genes, and diseases contain more individual abbreviations than chemicals or genes. The distribution for genes is more even across all types than either diseases or chemicals.
Table 2. Descriptive statistics for the SimConcept corpus. The numbers of composite mentions (of different types) are first listed followed by the numbers of individual mentions after decomposition in parentheses.
| Concept | # of abstracts | Five types of composite mentions | |||||
|---|---|---|---|---|---|---|---|
| All | CR | C | I | IA | OA | ||
| Gene | 694 | 810 (1895) | 14 (60) | 101 (246) | 442 (1089) | 253 (534) | 41 (107) |
| Disease | 793 | 1012 (2293) | 2 (18) | 245 (583) | 303 (809) | 486 (1045) | 52 (123) |
| Chemical | 937 | 1012 (2944) | 99 (505) | 201 (771) | 496 (1389) | 302 (716) | 0 (0) |
3. Experimental Results and Discussion
To evaluate our method, we used leave-one-out cross validation on the three sets (i.e., gene, disease and chemical). Table 3 shows the results of our evaluation, where we see that the overall performance is high for all three entity types.
Table 3. The Statistic of SimConcept corpus.
| Precision | Recall | F-measure | |
|---|---|---|---|
| Gene | 88.41% | 90.19% | 89.29% |
| Disease | 87.03% | 84.07% | 85.52% |
| Chemical | 85.36% | 82.78% | 84.04% |
The chemical corpus includes two types of mentions which are not addressed by our patterns. The first exception is a joint mention which the second mention uses coreference to indicate the previous mention, such as “3-0-propargylated betulinic acid and its 1,2,3-triazoles”. The pronoun “its” represents the previous mention “3-0-propargylated betulinic acid”. Therefore, this composite mention contains two individual mentions “3-0-propargylated betulinic acid” and “1,2,3-triazoles of 3-0-propargylated betulinic acid”. The other exception mention is a variant of the continuous type. For example, “tenofovir mono- or diphosphate”, “phosphate” is the postcedent region. Even though our applied tokenization splits on special characters, digits, lowercase and uppercase letters, which is more specific than general tokenization, it is still unable to split the conjuncts region and postcedent region. We have ignored these two types of mentions since they are rare.
To assess the performance on each composite mention type, we computed results shown in Table 4. There are only two range mentions in the disease set, and we therefore ignored these. There are also no overlap mentions in the chemical set. Since two exception mentions belong to continuous mention type in chemical corpus, the performance of continuous mention becomes lower.
Table 4. The evaluation of individual mention types. Scores are f-measures.
| Gene | Disease | Chemical | |
|---|---|---|---|
| Individual abbreviation | 91.11% | 84.21% | 84.15% |
| Overlap abbreviation | 80.92% | 91.53% | N/A |
| Mention with coordination ellipsis | 75.88% | 77.46% | 61.96% |
| Range mention | 91.34% | N/A | 94.14% |
| Individual mention | 90.24% | 87.13% | 87.34% |
| Mixed mention | 81.64% | 81.25% | 83.76% |
| All composite mentions | 89.29% | 85.88% | 84.04% |
As mentioned in introduction, this study is aimed for helping bioconcept normalization. We therefore applied SimConcept in GenNorm [21] and DNorm [18], and evaluated on the test sets of BioCreative II gene normalization task [12] and NCBI disease corpus [50], respectively (no normalized chemical corpus is available). To avoid training on the test set, the training set for SimConcept excluded the test corpora for GenNorm and DNorm. As shown in Table 5 and Table 6, using SimConcept can further improve the state-of-the-art performance for 1.17% in F-measure (P-value=0.02) for gene normalization and 1.34% in F-measure (P-value=0.03) for disease normalization. We also applied the heuristic rules used in previous gene normalization studies [51, 52] and showed the result in second row of Table 5. Our set of heuristics includes 9 rules. Those rules are defined by regular expressions to recognize the conjuncts at the end of the mention (e.g., detecting “1” and “2” in “BRCA1/2”) and handle some mentions containing coordination ellipsis and ranges. However, the composite mentions which are not considered in the refinement of heuristic rules cannot be recognized. This comparison shows that using heuristic rules is not as robust as SimConcept. As also shown in Table 5, using heuristic rules raises performance about half as much as SimConcept.
Table 5. The SimConcept contribution on gene normalization performance.
| Method | Precision | Recall | F-measure |
|---|---|---|---|
| GenNorm + SimConcept | 87.01% | 86.13% | 86.57% |
| GenNorm + heuristic rules | 86.78% | 85.23% | 86.00% |
| GenNorm | 86.72% | 84.09% | 85.38% |
Table 6. The SimConcept contribution on disease normalization performance.
| Method | Precision | Recall | F-measure |
|---|---|---|---|
| DNorm + SimConcept | 80.91% | 79.23% | 80.06% |
| DNorm | 80.69% | 76.85% | 78.72% |
In order to examine the contribution of individual feature types, we performed a feature ablation study where different feature types were removed from the entire set of features one at a time. As shown in Table 7, the largest drop in performance was due to the removal of token features, followed by semantic and character features. The removal of case pattern or contextual features had little effect on final performance. In addition to removing features, we also changed the order of CRF model from order 2 to order 1. The result shows order 2 performs better than order 1.
Table 7. Performance decrease when removing features. (Evaluated on gene corpus).
| Precision | Recall | F-measure | |
|---|---|---|---|
| SimConcept | 88.41% | 90.19% | 89.29% |
| - Token features | 86.94% (-1.47%) | 88.50% (-1.69%) | 87.71% (-1.58%) |
| - Semantic features | 87.90% (-0.51%) | 89.29% (-0.90%) | 88.58% (-0.71%) |
| - Character features | 87.78% (-0.63%) | 89.87% (-0.32%) | 88.81% (-0.48%) |
| - Pattern features | 88.30% (-0.11%) | 90.10% (-0.09%) | 89.19% (-0.10%) |
| Order 2 → Order 1 | 86.52% (-1.89%) | 88.39% (-1.80%) | 87.44% (-1.85%) |
4. Conclusion
In this study, we present SimConcept – a method to handle the task of composite named entity simplification. We integrated a CRF-based method with a pattern identification strategy to systematically decompose the six types of composite mentions. To handle the three most fundamental bioconcepts, we re-annotated the composite mentions in five existing corpora for gene (BioCreative 2 GN task train/test corpus and NLM GIA corpus), disease (NCBI disease corpus) and chemicals (BioCreative IV ChemDNER task corpus), and used these to evaluate SimConcept. The results show that SimConcept handles composite mention simplification issue effectively.
We further used SimConcept to assist the bioconcept normalization task. The result suggests that SimConcept is helpful for improving normalization performance. Our approach should generalize to other entity types in addition to the three concepts that were the focus of this study: genes, diseases and chemicals.
Acknowledgments
This research was supported by the NIH Intramural Research Program, National Library of Medicine.
Footnotes
Categories and Subject Descriptors: I.2.7 [ARTIFICIAL INTELLIGENCE]: Natural Language Processing – Language parsing and understanding, Speech recognition and synthesis, Text analysis.
Contributor Information
Chih-Hsuan Wei, Email: Chih-Hsuan.Wei@nih.gov, 8600 Rockville Pike, National Center for Biotechnology Information (NCBI), Bethesda, Maryland, USA, 20894.
Robert Leaman, Email: Robert.Leaman@nih.gov, 8600 Rockville Pike, National Center for Biotechnology Information (NCBI), Bethesda, Maryland, USA, 20894.
Zhiyong Lu, Email: Zhiyong.Lu@nih.gov, 8600 Rockville Pike, National Center for Biotechnology Information (NCBI), Bethesda, Maryland, USA, 20894.
References
- 1.Zweigenbaum P, Demner-Fushman D, Yu H, Cohen KB. Frontiers of biomedical text mining: current progress. Briefings in Bioinformatics. 2007;8(5):358. doi: 10.1093/bib/bbm045. 2007. DOI= http://dx.doi.org/10.1093/bib/bbm045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baumgartner WA, Jr, Lu Z, Johnson HL, Caporaso JG, Paquette J, Lindemann A, White EK, Medvedeva O, Cohen KB, Hunter L. Concept recognition for extracting protein interaction relations from biomedical text. Genome biology. 2008;9(Suppl 2):S9. doi: 10.1186/gb-2008-9-s2-s9. 2008. DOI= http://dx.doi.org/0.1186/gb-2008-9-s2-s9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Krallinger M, Vazquez M, Leitner F, Salgado D, Chatr-aryamontri A, Winter A, Perfetto L, Briganti L, Licata L, Iannuccelli M. The Protein-Protein Interaction tasks of BioCreative III: classification/ranking of articles and linking bio-ontology concepts to full text. BMC bioinformatics. 2011;12(Suppl 8):S3. doi: 10.1186/1471-2105-12-S8-S3. 2011. DOI= http://dx.doi.org/10.1186/1471-2105-12-S8-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Poon H, Vanderwende L. Joint inference for knowledge extraction from biomedical literature; Proceedings of the Human Language Technologies: The 2010 Annual Conference of the North American Chapter of the Association for Computational Linguistics; Los Angeles, CA, USA. June 1-6, 2010; 2010. NAACL HLT 2010. [Google Scholar]
- 5.Hunter L, Lu Z, Firby J, Baumgartner WA, Johnson HL, Ogren PV, Cohen KB. OpenDMAP: an open source, ontology-driven concept analysis engine, with applications to capturing knowledge regarding protein transport, protein interactions and cell-type-specific gene expression. BMC bioinformatics. 2008;9(1):78. doi: 10.1186/1471-2105-9-78. 2008. DOI= http://dx.doi.org/10.1186/1471-2105-9-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bethard S, Lu Z, Martin JH, Hunter L. Semantic role labeling for protein transport predicates. BMC bioinformatics. 2008;9(1):277. doi: 10.1186/1471-2105-9-277. 2008. DOI= http://dx.doi.org/10.1186/1471-2105-9-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang CC, Yang H, Jiang L. Postmarketing Drug Safety Surveillance Using Publicly Available Health-Consumer-Contributed Content in Social Media. ACM Transactions on Management Information Systems (TMIS) 2014;5(1):2. 2014. DOI= http://dx.doi.org/10.1145/2576233. [Google Scholar]
- 8.Doğan RI, Névéol A, Lu Z. A context-blocks model for identifying clinical relationships in patient records. BMC bioinformatics. 2011;12(Suppl 3):S3. doi: 10.1186/1471-2105-12-S3-S3. 2011. DOI= http://dx.doi.org/10.1186/1471-2105-12-S3-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li J, Lu Z. Systematic identification of pharmacogenomics information from clinical trials. Journal of Biomedical Informatics. 2012;45(5):870–878. doi: 10.1016/j.jbi.2012.04.005. 2012. DOI= http://dx.doi.org/10.1016/j.jbi.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mao Y, Van Auken K, Li D, Arighi CN, Lu Z. The gene ontology task at biocreative IV; Proceedings of the Proceedings of the fourth BioCreative challenge evaluation workshop; Bethesda, Maryland, USA. October 7-9, 2013; 2013. BioCreative IV. [Google Scholar]
- 11.Blaschke C, Leon EA, Krallinger M, Valencia A. Evaluation of BioCreAtIvE assessment of task 2. BMC bioinformatics. 2005;6(Suppl 1):S16. doi: 10.1186/1471-2105-6-S1-S16. 2005. DOI= http://dx.doi.org/10.1186/1471-2105-6-S1-S16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morgan AA, Lu Z, Wang X, Cohen AM, Fluck J, Ruch P, Divoli A, Fundel K, Leaman R, Hakenberg J. Overview of BioCreative II gene normalization. Genome biology. 2008;9(Suppl 2):S3. doi: 10.1186/gb-2008-9-s2-s3. 2008. DOI= http://dx.doi.org/10.1186/gb-2008-9-s2-s3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arighi CN, Wu CH, Cohen KB, Hirschman L, Krallinger M, Valencia A, Lu Z, Wilbur JW, Wiegers TC. BioCreative-IV virtual issue. Database 2014. 2014 doi: 10.1093/database/bau039. 2014, bau039.DOI= http://dx.doi.org/10.1093/database/bau039. [DOI] [PMC free article] [PubMed]
- 14.Lu Z, Kao HY, Wei CH, Huang M, Liu J, Kuo CJ, Hsu CN, Tsai RT, Dai HJ, Okazaki N. The gene normalization task in BioCreative III. BMC bioinformatics. 2011;12(Suppl 8):S2. doi: 10.1186/1471-2105-12-S8-S2. 2011. DOI= http://dx.doi.org/10.1186/1471-2105-12-S8-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arighi CN, Lu Z, Krallinger M, Cohen KB, Wilbur WJ, Valencia A, Hirschman L, Wu CH. Overview of the BioCreative III workshop. BMC bioinformatics. 2011;12(Suppl 8):S1. doi: 10.1186/1471-2105-12-S8-S1. 2011. DOI= http://dx.doi.org/10.1186/1471-2105-12-S8-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rebholz-Schuhmann D, Yepes AJ, Li C, Kafkas S, Lewin I, Kang N, Corbett P, Milward D, Buyko E, Beisswanger E. Assessment of NER solutions against the first and second CALBC Silver Standard Corpus. Journal of biomedical semantics. 2011;2(5):1–12. doi: 10.1186/2041-1480-2-S5-S11. 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Névéol A, Doğan RI, Lu Z. Semi-automatic semantic annotation of PubMed queries: A study on quality, efficiency, satisfaction. Journal of Biomedical Informatics. 2011;44(2):310–318. doi: 10.1016/j.jbi.2010.11.001. 2011. DOI= http://dx.doi.org/10.1016/j.jbi.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leaman R, Doğan RI, Lu Z. DNorm: disease name normalization with pairwise learning to rank. Bioinformatics. 2013;29(22):2909–2917. doi: 10.1093/bioinformatics/btt474. 2013. DOI= http://dx.doi.org/10.1093/bioinformatics/btt474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wei CH, Kao HY, Lu Z. SR4GN: a species recognition software tool for gene normalization. Plos one. 2012;7(6):e38460. doi: 10.1371/journal.pone.0038460. 2012. DOI= http://dx.doi.org/10.1371/journal.pone.0038460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rocktäschel T, Weidlich M, Leser U. ChemSpot: a hybrid system for chemical named entity recognition. Bioinformatics. 2012;28(12):1633–1640. doi: 10.1093/bioinformatics/bts183. 2012. DOI= http://dx.doi.org/10.1093/bioinformatics/bts183. [DOI] [PubMed] [Google Scholar]
- 21.Wei CH, Kao HY. Cross-species gene normalization by species inference. BMC bioinformatics. 2011;12(Suppl 8):S5. doi: 10.1186/1471-2105-12-S8-S5. 2011. DOI= http://dx.doi.org/10.1186/1471-2105-12-S8-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Torii M, Wagholikar K, Liu H. Detecting concept mentions in biomedical text using hidden Markov model: multiple concept types at once or one at a time? Journal of biomedical semantics. 2014;5:3. doi: 10.1186/2041-1480-5-3. 2014. DOI= http://dx.doi.org/10.1186/2041-1480-5-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leroy G, Endicott JE, Mouradi O, Kauchak a, Just Melissa L. Improving Perceived and Actual Text Difficulty for Health Information Consumers using Semi-Automated Methods; Proceedings of the American Medical Infomatics Association (AMIA) Annual Symposium; Chicago, USA. November 3-7, 2012; 2012. AMIA 2012. [PMC free article] [PubMed] [Google Scholar]
- 24.Ong E, Damay J, Lojico G, Lu K, Tarantan D. Simplifying text in medical literature. Journal of Research in Science, Computing and Engineering. 2007;4(1):37–47. 2007. DOI= http://dx.doi.org/10.3860/jrsce.v4i1.441. [Google Scholar]
- 25.Siddharthan A. Syntactic simplification and text cohesion. Research on Language and Computation. 2006;4(1):77–109. 2006. DOI= http://dx.doi.org/10.1007/s11168-006-9011-1. [Google Scholar]
- 26.Chandrasekar R, Srinivas B. Automatic induction of rules for text simplification. Knowledge-Based Systems. 1997;10(3):183–190. 1997. [Google Scholar]
- 27.Chandrasekar R, Doran C, Srinivas B. Motivations and methods for text simplification; Proceedings of the Proceedings of the 16th conference on Computational linguistics - Volume 2; Copenhagen, Denmark. Augest 5-9, 1996; 1996. COLING '. [Google Scholar]
- 28.Kauchak D. Improving Text Simplification Language Modeling Using Unsimplified Text Data; Proceedings of the Proceedings of the 51st Annual Meeting of the Association for Computational Linguistics; Sofia, Bulgaria. August 4-9, 2013; 2013. ACL 2013. [Google Scholar]
- 29.Silveira SB, Branco A. Enhancing multi-document summaries with sentence simplification; Proceedings of the ICAI 2012: International Conference on Artificial Intelligence; Las Vegas, Nevada, USA. July 19, 2012; 2012. ICAI 2012. [Google Scholar]
- 30.Peng Y, Tudor CO, Torii M, Wu CH, Vijay-Shanker K. iSimp: A Sentence Simplification System for Biomedical Text; Proceedings of the The 2012 IEEE International Conference on Bioinformatics and Biomedicine; Philadelphia, PA, USA. October 4-7, 2012; 2012. BIBM 2012. [Google Scholar]
- 31.Zhu Z, Bernhard D, Gurevych I. A monolingual tree-based translation model for sentence simplification; Proceedings of the Proceedings of the 23rd international conference on computational linguistics; Beijing, China. August 23–27, 2010; 2010. Association for Computational Linguistics. [Google Scholar]
- 32.Miwa M, Saetre R, Miyao Y, Tsujii Ji. Entity-focused sentence simplification for relation extraction; Proceedings of the Proceedings of the 23rd international conference on computational linguistics; Beijing, China. August 23–27, 2010; 2010. ACL 2010. [Google Scholar]
- 33.Jonnalagadda S, Gonzalez G. BioSimplify: an open source sentence simplification engine to improve recall in automatic biomedical information extraction; Proceedings of the AMIA Annual Symposium Proceedings; Washington, DC, USA. November 11-17, 2010; 2010. American Medical Informatics Association. [PMC free article] [PubMed] [Google Scholar]
- 34.Vickrey D, Koller D. Sentence Simplification for Semantic Role Labeling; Proceedings of the 22nd International Conference on. Computational Linguistics; Stroudsburg, PA, USA. August 23, 2008; 2008. Association for Computational Linguistics. [Google Scholar]
- 35.Vanderwende L, Suzuki H, Brockett C, Nenkova A. Beyond SumBasic: Task-focused summarization with sentence simplification and lexical expansion. Information Processing & Management. 2007;43(6):1606–1618. doi: 10.1016/j.ipm.2007.01.023. 2007. DOI= http://dx.doi.org/10.1016/j.ipm.2007.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Buyko E, Tomanek K, Hahn U. Resolution of coordination ellipses in biological named entities using conditional random fields; Proceedings of the 10th Conference of the Pacific Association for Computational Linguistics; Melbourne, Australia. September 19–21, 2007; 2007. Pacling. [Google Scholar]
- 37.Kim JD, Ohta T, Tateisi Y, Tsujii Ji. GENIA corpus—a semantically annotated corpus for bio-textmining. Bioinformatics. 2003;19(suppl 1):i180–i182. doi: 10.1093/bioinformatics/btg1023. 2003. DOI= http://dx.doi.org/10.1093/bioinformatics/btg1023. [DOI] [PubMed] [Google Scholar]
- 38.Chae J, Jung Y, Lee T, Jung S, Huh C, Kim G, Kim H, Oh H. Identifying non-elliptical entity mentions in a coordinated NP with ellipses. Journal of Biomedical Informatics. 2013;47:139–152. doi: 10.1016/j.jbi.2013.10.002. 2013. DOI= http://dx.doi.org/0.1016/j.jbi.2013.10.002. [DOI] [PubMed] [Google Scholar]
- 39.Lafferty J, McCallum A, Pereira F. Conditional Random Fields: Probabilistic Models for Segmenting and Labeling Sequence Data; Proceedings of the Proceedings of the International Conference on Machine Learning (ICML 01); Williamstown, MA, USA. June 28-July 1, 2001.2001. [Google Scholar]
- 40.Liu DC, Nocedal J. On the limited memory BFGS method for large scale optimization. Mathematical Programming B. 1989;45(3):503–528. 1989. [Google Scholar]
- 41.Wei CH, Harris BR, Kao HY, Lu Z. tmVar: A text mining approach for extracting sequence variants in biomedical literature. Bioinformatics. 2013;29(11):1433–1439. doi: 10.1093/bioinformatics/btt156. 2013. DOI= http://dx.doi.org/10.1093/bioinformatics/btt156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lowe DM, Corbett PT, Murray-Rust P, Glen RC. Chemical name to structure: OPSIN, an open source solution. Journal of chemical information and modeling. 2011;51(3):739–753. doi: 10.1021/ci100384d. 2011. DOI= http://dx.doi.org/10.1021/ci100384d. [DOI] [PubMed] [Google Scholar]
- 43.Tanabe L, Wilbur WJ. Tagging gene and protein names in biomedical text. Bioinformatics. 2002;18(8):1124–1132. doi: 10.1093/bioinformatics/18.8.1124. 2002. DOI= http://dx.doi.org/10.1093/bioinformatics/18.8.1124. [DOI] [PubMed] [Google Scholar]
- 44.Sohn S, Comeau DC, Kim W, Wilbur WJ. Abbreviation definition identification based on automatic precision estimates. BMC bioinformatics. 2008;9(1):402. doi: 10.1186/1471-2105-9-402. 2008. DOI= http://dx.doi.org/10.1186/1471-2105-9-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Doğan RI, Lu Z. An improved corpus of disease mentions in PubMed citations; Proceedings of the Proceedings of the 2012 Workshop on Biomedical Natural Language Processing; Montreal, Canada. June 8, 2012; 2012. Association for Computational Linguistics. [Google Scholar]
- 46.Krallinger M, Leitner F, Rabal O, Vazquez M, Oyarzabal J, Valencia A. Overview of the chemical compound and drug name recognition (CHEMDNER) task; Proceedings of the Proceedings of the fourth BioCreative challenge evaluation workshop; Bethesda, Maryland, USA. October 7-9, 2013; 2013. BioCreative IV. [Google Scholar]
- 47.Wei CH, Kao HY, Lu Z. PubTator: a Web-based text mining tool for assisting Biocuration. Nucleic acids research. 2013;41(Web Server Issue):W518–W522. doi: 10.1093/nar/gkt441. 2013. DOI= http://dx.doi.org/10.1093/nar/gkt44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wei CH, Kao HY, Lu Z. PubTator: A PubMed-like interactive curation system for document triage and literature curation; Proceedings of the proceedings of BioCreative; Washington DC, USA. April 5, 2012; 2012. BioCreative 2012 workshop. [Google Scholar]
- 49.Wei CH, Harris BR, Li D, Berardini TZ, Huala E, Kao HY, Lu Z. Accelerating literature curation with text-mining tools: a case study of using PubTator to curate genes in PubMed abstracts. Database: The Journal of Biological Databases & Curation. 2012;2012 doi: 10.1093/database/bas041. (2012) bas041.DOI= http://dx.doi.org/10.1093/database/bas041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Doğan RI, Leaman R, Lu Z. NCBI disease corpus: A resource for disease name recognition and concept normalization. Journal of Biomedical Informatics. 2014;47:1–10. doi: 10.1016/j.jbi.2013.12.006. 2014. DOI= http://dx.doi.org/10.1016/j.jbi.2013.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wermter J, Tomanek K, Hahn U. High-performance gene name normalization with GeNo. Bioinformatics. 2009;25(6):815–821. doi: 10.1093/bioinformatics/btp071. 2009. DOI= http://dx.doi.org/10.1093/bioinformatics/btp071. [DOI] [PubMed] [Google Scholar]
- 52.Hakenberg J, Plake C, Leaman R, Schroeder M, Gonzalez G. Inter-species normalization of gene mentions with GNAT. Bioinformatics. 2008;24(16):i126–i132. doi: 10.1093/bioinformatics/btn299. 2008. DOI= http://dx.doi.org/10.1093/bioinformatics/btn299. [DOI] [PubMed] [Google Scholar]