Abstract
Background
We previously reported that acute functional tolerance (AFT) to the hypnotic effects of alcohol was significantly correlated to drinking in the dark (DID) in the LXS RI panel, but only in mice that had been pretreated with alcohol. Here we have conducted QTL mapping for AFT. DNA sequencing of the progenitor ILS and ISS strains and microarray analyses were also conducted to identify candidate genes and functional correlates.
Methods
LXS mice were given either saline or alcohol (5 g/kg) on day one and then tested for loss of righting reflex (LORR) AFT on day two. QTLs were mapped using standard procedures. Two microarray analyses from brain were conducted: 1) naïve LXS mice and 2) an alcohol treatment time course in the ILS and ISS. The full genomes of the ILS and ISS were sequenced to a depth of ~30×.
Results
A significant QTL for AFT in the alcohol pretreatment group mapped to distal chromosome 4; numerous suggestive QTLs were also mapped. Preference drinking and DID have previously been mapped to the chromosome 4 locus. The credible interval of the significant chromosome 4 QTL spanned 23 Mb and included 716 annotated genes of which 150 had at least one non-synonymous SNP or small indel that differed between the ILS and ISS; expression of 48 of the genes was cis-regulated. Enrichment analysis indicated broad functional categories underlying AFT including proteolysis, transcription regulation, chromatin modification, protein kinase activity, apoptosis, and others.
Conclusions
The chromosome 4 QTL is a key region containing possibly pleiotropic genes for AFT and drinking behavior. Given that the region contains many viable candidates and a large number of the genes in the interval fall into one or more of the enriched functional categories, we postulate that many genes of varying effect size contribute to the observed QTL effect.
Keywords: Ethanol, Genetics, Quantitative Trait Locus, Mouse, Microarray, Deep Sequencing, Alcoholism, AFT
Introduction
Twin studies and other areas of research have identified genetics as an important factor in alcohol use disorders (AUDs) accounting for as much as 50% or more of the phenotypic variance (Dick and Bierut, 2006). One well described genetic effect on AUDs is innate sensitivity to alcohol. Schuckit (1980) observed that individuals who were “family history positive” for AUDs compared to those who were “family history negative” were reliably less sensitive to an acute alcohol challenge, a phenomenon that Schuckit termed “low level of response” (LR) which has been shown to be a heritable trait (Heath et al., 1999). LR was found to be a reliable predictor of future drinking problems and it is therefore thought to be a valid and useful construct for investigating the genetics of AUD risk (Schuckit, 1994).
Newlin and Thomson (1990) argued that differences in LR were due, in part, to differences in the development of acute functional tolerance (AFT). The concept of AFT is first credited to Mellanby (1919) who observed that dogs that had been administered alcohol were more ataxic on the rising limb of alcohol distribution than they were at the same blood ethanol concentration (BEC) on the falling limb indicating the development of AFT. AFT has been shown to occur in humans as well as in rodents and also that it has an important genetic component (Cromer et al., 2010, Erwin and Deitrich, 1996, Hu et al., 2008, Radcliffe et al., 2006a, Radcliffe et al., 2013). Thus, differences in LR and its association to AUDs may be due in part to genetic differences in AFT.
The Inbred Long and Short Sleep mouse strains (ILS, ISS) were selectively bred for extreme differences in the duration of the loss of the righting reflex (“sleep time”; ST), an acute response to a hypnotic dose of alcohol (McClearn and Kakihana, 1981). AFT typically has not been considered in studies of ST because it is technically difficult to obtain an accurate estimate of BEC at the loss of function on the rising limb of alcohol distribution. To overcome this shortcoming, Ponomarev and Crabbe (2002) developed a variation of the standard test with which it is possible to measure AFT more accurately. Using this method, it was shown that a substantial portion of the enormous ILS/ISS difference in ST sensitivity was mediated by AFT (Radcliffe et al., 2006a).
The LXS recombinant inbred (RI) mouse strain panel was created from pairs of ILS/ISS-derived F2 offspring that were subsequently bred through brother-sister matings for more than 20 generations resulting in a panel of inbred strains, each of which contains a random assortment of alleles from the progenitors (Williams et al., 2004). RI panels are often referred to as “reference” populations with which phenotypic, genetic, and genomic results can be compared and co-analyzed in distinct cohorts that have been tested at different times and in different labs (Belknap, 1998). We have been investigating the genetics of AFT with the LXS using a procedure in which mice were administered a pretreatment of alcohol or saline 24 hours before being tested using the modified method of Ponomarev and Crabbe (2002). This procedure was originally employed to investigate rapid (one day) tolerance and we subsequently discovered that the pretreatment altered AFT in a genotype-dependent manner (Radcliffe et al., 2006a, ; for further discussion of the relationship between AFT and RT, see Radcliffe et al., 2013). Importantly, a significant genetic correlation was found between drinking in the dark (DID), a model of binge drinking, and AFT in the alcohol-pretreated mice, but not in the saline-pretreated group. The correlation was in the predicted direction and was consistent with human studies; i.e., higher drinking strains tended to have higher AFT (Radcliffe et al., 2013). This observation suggests that one’s initial AFT may be less important to risk for pathological drinking behavior than the way it changes in response to prior experience with alcohol.
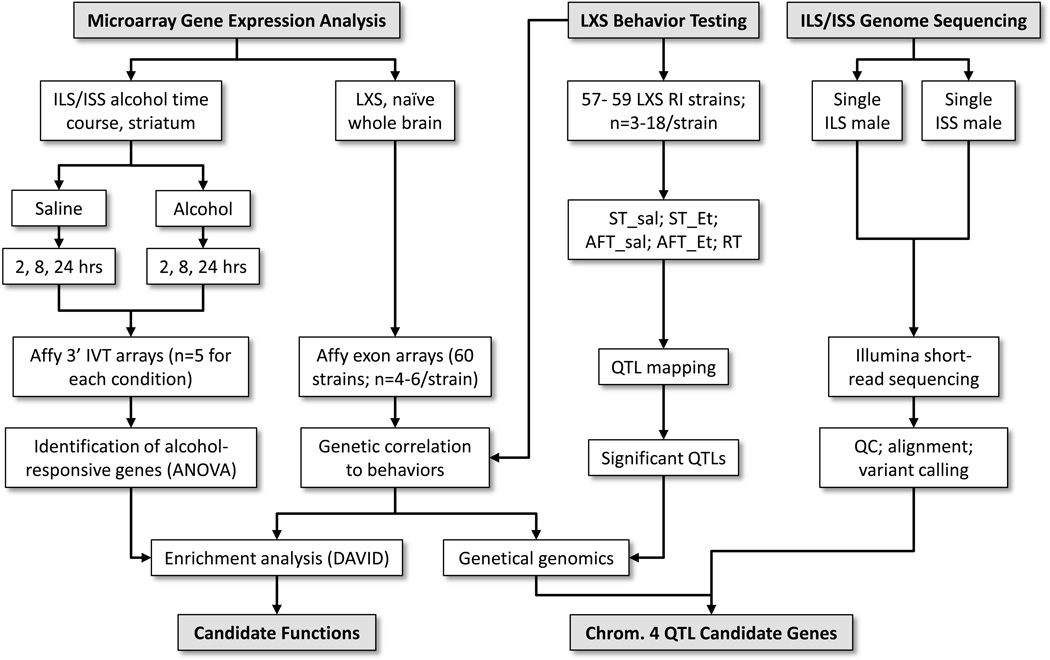
In a continuing effort to understand the genetic factors that contribute to variation in AFT, we report here quantitative trait locus (QTL) mapping of AFT and ST in the LXS panel using the same pretreatment procedure as described above. Multiple suggestive QTLs and a single significant chromosome 4 QTL for AFT were mapped. This was followed by two microarray gene expression experiments: one on whole brain of the LXS RIs and the second in striatum from alcohol-treated ILS and ISS. These experiments were conducted to identify candidate functions that might provide insight into the mechanistic underpinnings of AFT and for the application of a “genetical genomics” approach to identify cis-regulated candidate genes for the chromosome 4 AFT QTL. Finally, the genomes of the ILS and ISS were sequenced to examine DNA variants in the chromosome 4 QTL. The overall goal of these experiments was to identify candidate genes in the chromosome 4 QTL as well as to gain insight into the molecular basis of the behavioral results. Figure 1 provides an overview of the design and integration of the various experiments.
Figure 1.
Overview of the experimental design. The LXS RI strains were tested for LORR-related behaviors followed by QTL mapping (center). Two microarray experiments were conducted: expression profiling of a time course in the striatum of ILS and ISS that had been administered saline or alcohol and expression profiling of whole brain from 60 of the LXS RI strains (left). Alcohol-responsive genes were identified by ANOVA in the ILS/ISS time course experiment and the LXS profiles were used to identify genes that were genetically correlated to the behaviors. The gene lists derived from those two analyses were independently subjected to functional enrichment analysis. Genes that correlated to behavior were additionally tested for cis-regulation within the significant AFT_Et QTL; these genes were considered to be candidates. Finally, DNA sequencing of the ILS and ISS provided coding region candidates for the AFT_Et QTL (right). Strains in common among the LXS RI experiments can be found in the Venn diagram shown in supplemental file S1. See text for additional details.
Materials and Methods
Animals
LXS RI breeders were obtained from The Jackson Laboratory (Bar Harbor, ME) and bred in-house in the UCAMC vivarium, a pathogen-free facility. Offspring were weaned and sex-separated at 21 days of age. All experiments were conducted with males that were group-housed in standard housing containing from 2 to 5 mice per cage. Only male mice were used for reasons of economy and to avoid variance associated with the estrus cycle. To our knowledge, female ILS, ISS, and LXS have never been tested for DID or ST AFT. Sex differences have been noted for ST in the LXS lines (Bennett and Carosone-Link, 2006) and DID (Rhodes et al., 2005), but not for ST AFT among 21 inbred mouse strains (Ponomarev and Crabbe, 2004). The mice were between 65 and 89 days of age at the time of their use. They were maintained in a constant temperature (22–23°C), humidity (20–24%), and light (14L/10D) environment. The procedures described in this report have been established to ensure the absolute highest level of humane care and use of the animals, and have been reviewed and approved by the UCAMC IACUC.
Loss of Righting Reflex
Mice were administered a single intraperitoneal (i.p.) injection of vehicle (saline) or alcohol (ethanol; 5 g/kg, 20% v/v in saline) on day one and then placed back into their home cage; this is referred to as the pretreatment dose. The 5 g/kg dose was selected because it was found to be not overtly toxic, and it elicited a robust rapid tolerance (see below) compared with other doses (Radcliffe et al., 2005). The loss of righting reflex (LORR) was assessed in both groups using the Ponomarev and Crabbe (2002) method 24 hours following the pretreatment. The LORR test dose was 4.1 g/kg which had been found to elicit a desirable range of sleep times among the LXS; i.e., approximately 30 to 180 minutes (Bennett et al., 2006); the range of sleep times was similar in the current study (see supplemental file S1). Immediately after alcohol injection (ip), the mouse was placed in a closed Plexiglas cylinder that was rotated 90° every 2–3 s. Loss of righting was defined as the time at which the mouse remained supine for at least 5 seconds; at this time, the mouse was removed from the tube and a retro-orbital blood sample was drawn for determination of BEC at the loss of righting reflex (BEC1). The mouse was then tested for recovery of LORR every 3 to 6 minutes thereafter. When the animal was able to right itself within a 5 second period, it was removed from the tube and a second blood sample was drawn (BEC2). Duration of LORR (“sleep time”; ST) was defined as the elapsed time between collection of BEC1 and BEC2. An increase in BEC2 from BEC1 was interpreted as development of AFT and was quantified as the difference between the two. Mice that had not recovered by 180 minutes were given a ST score of 180 and a blood sample for determination of BEC2 was taken at that time. Approximately 3% of the mice reached this criterion, but based on a detailed analysis (not shown), it was determined that none of the results were substantially affected by inclusion of these mice (see supplemental file S1 for a table of means). BEC values were determined using an enzymatic assay with spectrophotometry (Lundquist, 1959). Rapid tolerance (RT) for ST was a within-strain measure calculated as the difference between mean ST in the alcohol pretreatment group and mean ST in the saline pretreatment group. In the remainder of the text, the pretreatment group is designated with either “_sal” (saline pretreatment) or “_Et” (alcohol pretreatment) following the variable name. An average of 12 mice per strain and pretreatment condition (range: 3 to 18) was obtained resulting in 57 strains for ST_sal, 59 for ST_Et, 57 for AFT_sal, 59 for AFT_Et, and 57 for RT. Many of the mice (63%) were used in our previous study (Radcliffe et al., 2013). We have since tested an additional 535 mice which resulted in an increased n for 37 of the 44 original strains and in the addition of 15 new strains. The complete set of behavioral data can be found in supplemental file S1.
QTL Mapping
QTL mapping was conducted at the Phenogen site (http://phenogen.ucdenver.edu/Phenogen) (Bennett et al., 2011) using a weighted marker regression on strain means (Carlborg et al., 2005). Genotypes were collected by Dr. Gary Churchill and colleagues at The Jackson Laboratory using the Affymetrix Mouse Diversity Genotyping Array (http://cgd.jax.org/mda/v1). A total of 43,870 informative SNPs were used for QTL mapping (see Saba et al., 2011). Significance levels were empirically determined using the method of Churchill and Doerge (1994) (10,000 permutations) and 90% Bayesian credible intervals were determined using methods implemented in R/qtl (Broman et al., 2003). The proportion of genetic variance contributed by a QTL (VG) was calculated following Belknap (1998).
Microarray Analysis
Two Affymetrix microarray datasets were generated as shown in Figure 1. The first was from naïve whole brain from 60 of the LXS using the GeneChip Mouse Exon 1.0 ST Array which was originally generated as a community resource (data are available at http://phenogen.ucdenver.edu/PhenoGen/). The second was an alcohol treatment time course from the striatum of the ILS and ISS using the GeneChip Mouse Genome 430 2.0 Array to examine the effects of alcohol on gene expression that might relate to previously observed ILS/ISS differences in DID (Saba et al., 2011).
LXS Whole Brain Gene Expression Analysis
For the LXS Exon Array dataset, total RNA was isolated from the whole brain homogenate of naive male mice using the QIAGEN RNeasy Midi kit followed by the RNeasy Mini kit. RNA preparation and array hybridization to the GeneChip Mouse Exon 1.0 ST Array was performed according to standard Affymetrix protocols. Sixty of the LXS strains were assayed with four to six mice per strain (see supplemental file S1). Brain samples from C57BL/6J mice were included in every batch (35 individual arrays), and DBA/2J mice were included in some of the batches (9 arrays) to control for batch effects.
A total of 341 individual arrays passed quality control standards (Affymetrix Power Tools). A probe mask was constructed to ensure probe specificity and to avoid SNP hybridization artifacts (Walter et al., 2007). Individual probes were eliminated if any of the following conditions were true: their sequence did not map to any part of the NCBI m38 build of the mouse genome; their sequence mapped to multiple locations in the mouse genome; or the probe contained one or more ILS/ISS SNPs. Probe sets also were eliminated if less than three of the original probes remained after masking. Masking removed 341,674 (27%) of the 1,256,831 original probe sets, leaving 915,157 informative sets.
Data from individual probes were normalized and summarized into transcript clusters using RMA (Irizarry et al., 2003). Presence or absence of probe sets was determined using the detection above background (DABG) method (Affymetrix, 2005). The data set was adjusted for batch effects using the empirical Bayes method (Johnson et al., 2007). Affymetrix housekeeping genes, probe sets detected in less than 50% of the samples, and probe sets with heritability less than 33% were removed prior to higher level analyses (Bennett et al., 2011, Saba et al., 2011). The final data set consisted of 8112 annotated genes. These genes were examined for genetic correlations between mean expression and mean behavioral score for the five behavioral phenotypes (p<0.05, uncorrected). The gene lists were used in a functional enrichment analysis using the DAVID bioinformatics resource (Huang et al., 2009).
ILS/ISS Striatum Alcohol Time Course Gene Expression Analysis
Male ILS and ISS mice were treated with either saline or alcohol (5 g/kg, 16% w/v in saline; i.p.) and sacrificed at 2, 8, or 24 hours. Total RNA was isolated from striatal homogenate using the QIAGEN RNeasy Mini Kit. RNA preparation and array hybridization to the GeneChip Mouse Genome 430 2.0 Array was performed according to standard Affymetrix protocols as previously described (Radcliffe et al., 2006b). There were 5 mice for each condition. Masks were applied as described above which eliminated 3571 out of 45,037 probe sets leaving a total of 41,466 informative probe sets. Probe sets that were not reliably expressed were eliminated based on their Affymetrix Microarray Suite 5.0 Absence/Presence call (MAS5; Hubbell et al., 2002). This eliminated another 17,204 probe sets for a final number of 24,262 probe sets. Data were analyzed independently within each time group by two-way ANOVA (strain-by-treatment). Genes that showed significant main effects of treatment (FDR ≤ 0.01) or interaction (FDR ≤ 0.05) were combined into a single list which was used in a functional enrichment analysis.
Whole Genome Sequencing
High molecular weight DNA was isolated from the livers of a single male ILS and ISS using standard phenol/chloroform extraction procedures. The Illumina TruSEQ DNA Library Sample Preparation kit was used as per the manufacturer’s instructions to prepare 2×100 sequencing libraries (200 bp expected insert size). Sequencing was performed on an Illumina HiSeq 2000 Sequencing System with 4 flow-cell lanes per strain. A total of 427×106 and 440×106 read pairs were generated for the ILS and ISS, respectively. Raw reads were assessed for quality using the FastX Toolkit (http://hannonlab.cshl.edu/fastx_toolkit/) and mapped to the m38 build using BWA (v. 0.5.9) (Li and Durbin, 2009) in the paired end fashion using default parameters and setting the maximum insert size to 1000 (http://bio-bwa.sourceforge.net/bwa.shtml). Approximately 93% of the reads mapped for each strain which gave ~30× genome-wide coverage.
Candidate Gene Analysis
For the purposes of this study, candidate genes were defined as those found within the 90% Bayesian credible interval of a given behavioral QTL and whose expression was either cis-regulated (cis-eQTL) and/or contained a clear functional coding region variant. eQTL mapping using the LXS exon array data was performed in PhenoGen as described above. There was a nearly complete overlap between the strains used for the behavior testing and those used for eQTL mapping (see Venn diagram in supplemental table S1). Functional coding variants (SNPs and small indels) were called from the ILS/ISS sequences using the GATK UnifiedGenotyper (v. 2.4–9) enhanced by using previously identified variants in dbSNP build 137 (McKenna et al., 2010). The variants were filtered for highest quality and homozygosity using GATK Variant Quality Score Recalibrator (VQSR) and were annotated against Ensembl reference annotations.
Results
QTL Mapping
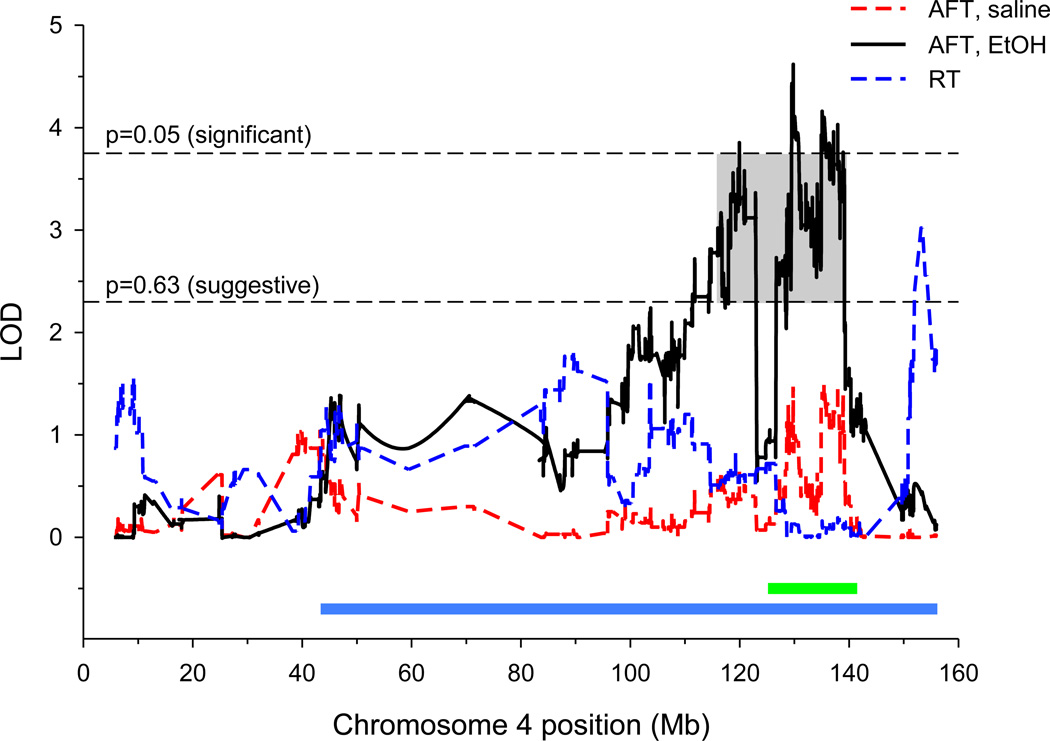
The results of QTL mapping for the 5 LORR-related phenotypes are shown in Table 1. Overall, nine suggestive (genome-wide p-value<0.63; Lander and Kruglyak, 1995) and one significant (genome-wide p-value<0.05) QTLs were mapped. The most noteworthy QTL is for AFT_Et located in the distal region of chromosome 4, shown in Figure 2 (black solid line), accounting for 2.6% of the genetic variance (VG). Also shown in Figure 2 is the map for AFT_sal which was well under the suggestive level. QTLs for drinking behavior in the mouse have also been mapped to the chromosome 4 region: a meta-analysis of unlimited access two-bottle choice drinking in BXD crosses (Belknap and Atkins, 2001) and drinking in the dark in the LXS RI panel (Saba et al., 2011) shown as a green and blue bar in the lower part of Figure 2, respectively. A suggestive QTL for RT also mapped to distal chromosome 4 (Figure 2). Although the 90% Bayesian credible interval overlapped with the AFT_Et peak (Table 1), it is clear from Figure 1 that the two loci are distinct.
Table 1.
LORR- and AFT-related QTLs mapped in the LXS RI strains.
| Trait | Chrom. | QTL Peak (Mb) | CI (Mb)1 | LOD2 | P value3 | %VG | Increaser Allele5 |
|---|---|---|---|---|---|---|---|
| ST_sal | 1 | 109.161 | 65.1 – 135.8 | 3.03 | 0.22 | ILS | |
| 8 | 28.627 | 17.4 – 126.9 | 2.59 | 0.44 | ISS | ||
| 12 | 34.972 | 16.3 – 81.7 | 2.68 | 0.39 | ILS | ||
| ST_Et | 1 | 80.637 | 41.7 – 135.8 | 3.10 | 0.17 | ILS | |
| 8 | 28.628 | 18.3 – 33.1 | 3.59 | 0.07 | ISS | ||
| 12 | 34.649 | 16.3 – 53.5 | 2.48 | 0.48 | ILS | ||
| 17 | 35.771 | 9.3 – 47.4 | 2.86 | 0.26 | ISS | ||
| AFT_sal | (none) | ||||||
| AFT_Et | 4 | 129.798 | 116.0 – 139.1 | 4.62 | 0.007 | 2.6 | ILS |
| 15 | 11.311 | 5.0 – 72.6 | 2.87 | 0.25 | ISS | ||
| RT | 4 | 153.199 | 6.1 – 156.1 | 3.02 | 0.17 | ISS |
90% Bayesian credible interval.
Logarithm of the odds ratio.
Adjusted genome-wide p values.
Percent of the genetic variance accounted for by the QTL.
Indicates from which progenitor the allele came that causes the phenotype to increase in value.
Figure 2.
QTL maps of AFT and RT in the LXS RI strains. The dashed red line shows the QTL map for AFT_sal, the solid black line is for AFT_Et, and the dashed blue line is for RT. The horizontal black dashed lines show levels for significant (upper) or suggestive (lower) peaks. The grey box indicates the 90% Bayesian credible interval for the AFT_Et QTL. The green and blue horizontal lines at the bottom of the graph depict the location of previously mapped QTLs for drinking behavior in the BXD (green) and LXS (blue) RI panels (see text for more information).
Numerous suggestive QTLs were mapped for the behavioral phenotypes (Table 1). Note that the increaser allele (progenitor allele that causes the phenotype to increase) for two of the ST_sal peaks was ILS while the third was ISS. Since the ILS and ISS were selected for ST, it is unlikely that an ISS allele would be responsible for an increase in ST; it is more likely that the chromosome 8 peak, the smaller of the three, is a false positive. Three QTLs for ST_Et were in common with the ST_sal peaks which is not surprising since they show a significant genetic correlation (Radcliffe et al., 2013). The increaser allele for the ST_Et QTL on chromosome 17 is also ISS suggesting that it may also be a false positive.
Enrichment Analysis
LXS Exon Array Analysis
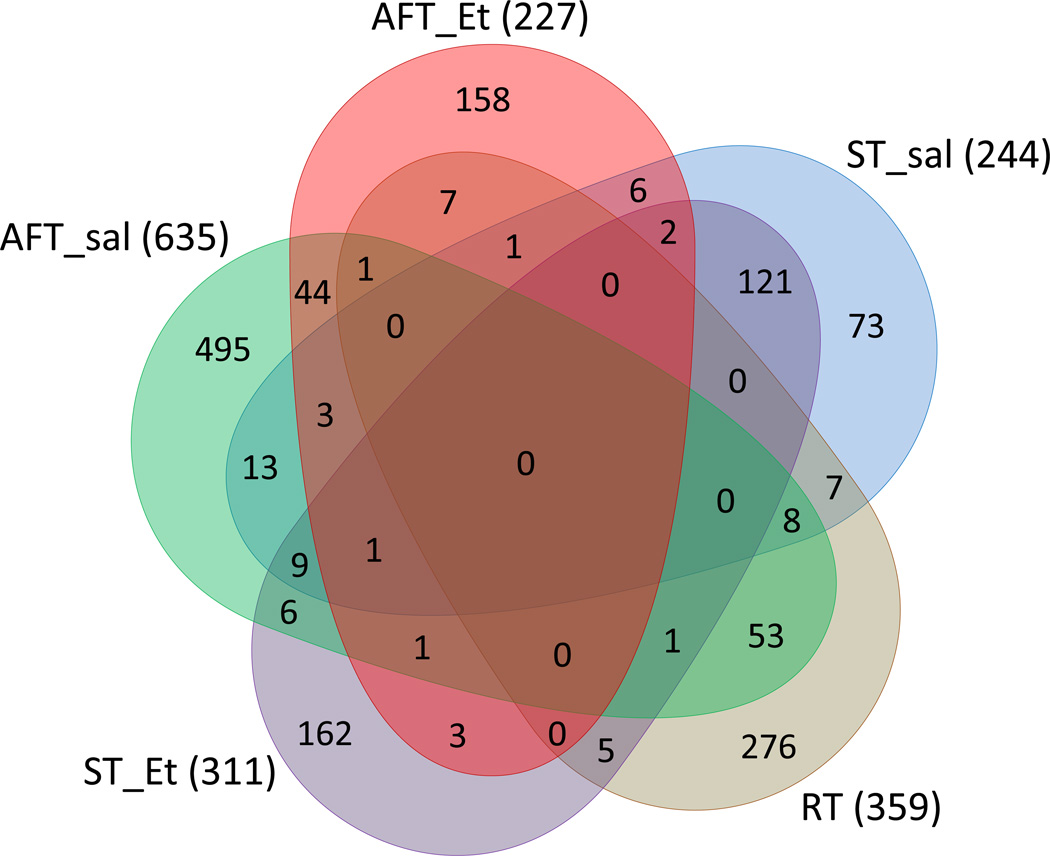
Genes assayed in the LXS were correlated to the behavioral traits under the hypothesis that they are related en masse to the phenotype and would therefore shed biological insight into the traits. The number of annotated genes in each list and the overlap between them is shown as a Venn diagram in figure 3. Note that AFT_sal has approximately twice as many genes as any of the others. Also note that, with the exception of ST_sal, the majority of genes in each list were unique; i.e., for ST_Et, AFT_sal, AFT_Et, and RT, the percent of unique genes were 52%, 78%, 70%, and 77%, respectively. Only 30% of the genes were unique to ST_sal; the majority (50%) was in common with ST_Et. The full lists of correlated genes can be found in supplemental table S1.
Figure 3.
Venn diagram of genes whose RNA expression correlated to AFT-related behaviors among the LXS RI strains.
The correlation lists were evaluated for functional enrichment using DAVID’s Functional Annotation Clustering tool which assesses the shared gene membership among all represented terms and clusters them accordingly (Huang et al., 2009). Tables 2 (ST and RT) and 3 (AFT) list representative and significant GOTERM_BP_FAT terms (FDR≤0.10) from clusters with significant Enrichment Scores (≥1.3); also listed are six GOTERM_BP_FAT terms that were significant, but not found in significant clusters. The complete results can be found in supplemental table S1. Despite only modest overlap among the gene lists (Figure 3), terms related to cell adhesion, transcription, chromatin modification, morphogenesis/development, lipid biosynthesis, and protein metabolism/processing were in common among several of the behavioral traits.
Table 2.
Enrichment analysis of LXS exon array genes correlated to ST or RT.
| Annotation cluster #1 |
Representative Terms: ST_sal2 |
Annotation cluster #1 |
Representative Terms: ST_Et2 |
Annotation cluster #1 |
Representative Terms: RT2 |
|---|---|---|---|---|---|
| (No significant GOTERM_BP_FAT terms) | 1 (3.5) | Oxidation reduction (24) | 1 (3.9) |
Cell-cell adhesion (14) Homophilic cell adhesion (11) |
|
| 7 (1.5) | Lipid biosynthetic process (13) | 3 (2.0); 4 (2.0) |
Transcription (51) Positive regulation of transcription (20) |
||
| 22 (NS) | Heart morphogenesis (6) | 11 (1.4) | Striated muscle cell development (7) | ||
| 23 (NS) | Chromatin modification (12) | ||||
| 27 (NS) | Cellular component morphogenesis (17) |
Number in parentheses is the Enrichment Score for each cluster; the score of 1.3 is equivalent to non-log scale p of 0.05 (Huang et al., 2009). In one case, distinct clusters contained similar terms; these have been grouped here (see supplemental Table S-1). NS: Enrichment Score < 1.3.
Number in parentheses indicates the number of genes found within that term; terms are from GOTERM_BP_FAT (FDR≤0.10).
Table 3.
Enrichment analysis of LXS exon array genes correlated to AFT.
| Annotation cluster #1 |
Representative Terms: AFT_sal2 | Annotation cluster #1 |
Representative Terms: AFT_Et2 |
|---|---|---|---|
| 4 (2.5) |
Regulation of transcription (105) Transcription (94) |
4 (1.6) | Fatty acid metabolic process (8) |
| 5 (2.4) |
Chromatin modification (21) Chromosome organization (28) Chromatin organization (24) Covalent chromatin modification (10) |
5 (1.3); 10 (NS) | Mammary gland morphogenesis (5) |
| 8 (2.0) | Modification-dependent protein catabolic process (29) | 8 (NS) | Negative regulation of cell growth (5) |
| 9 (1.8) |
Cellular calcium ion homeostasis (29) Calcium ion homeostasis (10) |
9 (NS) | Phosphate metabolic process (20) |
| 11 (1.7); 26 (1.3) |
Forebrain development (15) Pallium development (8) Regulation of cell projection organization (8) |
||
| 14 (1.6) | Gland development (15) | ||
| 17 (1.6) | Transforming growth factor beta receptor signaling pathway (7) | ||
| 19 (1.5) | Cel-matrix adhesion (7) |
Number in parentheses is the Enrichment Score for each cluster; the score of 1.3 is equivalent to non-log scale p of 0.05 (Huang et al., 2009). In two cases, distinct clusters contained similar terms; these have been grouped here (see supplemental Table S-1). NS: Enrichment Score < 1.3.
Number in parentheses indicates the number of genes found within that term; terms are from GOTERM_BP_FAT.
ILS/ISS Alcohol Time Course Analysis
The goal of ILS/ISS microarray time course analysis was to identify genes that were alcohol-responsive in the progenitor strains that could potentially contribute to the pretreatment-mediated effect on AFT. A summary of the results is shown in Table 4. Note that only probe sets that showed significant pretreatment or strain-by-pretreatment interaction effects were retained for further analysis; however, a substantial number of these genes also showed a significant main effect of strain (Table 4). There was modest overlap in genes between the time points: 115 genes were in common between 2 and 8 hours and the single 24 hour gene was not found in either of the other two time points. In total, there were 1648 annotated genes derived from the complete time course analysis; the genes are listed in supplemental file S2. Enriched functional categories were identified from the time course gene list using the DAVID Functional Annotation Clustering tool as above (Table 5; supplemental file S2). Genes for all three time points were combined for the analysis.
Table 4.
Summary of time course two-way ANOVA
| Time point | Effect | # Probe sets | # Unique annotated genes |
|---|---|---|---|
| 2 hours | Pretreatment (FDR≤0.01) | 1142 | 822 |
| Interaction (FDR≤0.05) | 562 | 431 | |
| Strain (FDR≤0.01) | 405 | 291 | |
| 8 hours | Pretreatment (FDR≤0.01) | 610 | 544 |
| Interaction (FDR≤0.05) | 19 | 12 | |
| Strain (FDR≤0.01) | 169 | 153 | |
| 24 hours | Pretreatment (FDR≤0.01) | 1 | 1 |
| Interaction (FDR≤0.05) | 0 | 0 | |
| Strain (FDR≤0.01) | 1 | 1 | |
Table 5.
Enrichment analysis of genes that were significant for pretreatment or strain-by-pretreatment interaction in the ILS/ISS time course. Genes for all three time points were included in the analysis.
| Annotation cluster #1 | Representative Terms (GOTERM_BP_FAT)1 |
|---|---|
| 2 (7.3); 4 (4.0); 18 (2.0); 53 (NS) |
Regulation of transcription (238) Positive regulation of transcription (65) Positive regulation of transcription from RNA polymerase II promoter (44) RNA processing (56) |
| 8 (2.8) |
Chromosome organization (51) Chromatin modification (31) |
| 10 (2.5) |
Protein catabolic process (61) Proteolysis involved in cellular protein catabolic process (59) |
| 12 (2.3); 17 (2.0) |
ncRNA processing (28) Ribosome biogenesis (18) |
| 13 (2.3) |
Regulation of apoptosis (60) Positive regulation of apoptosis (32) Induction of apoptosis (25) |
| 22 (1.8) |
Negative regulation of catalytic activity (17) Negative regulation of transferase (11) |
| 23 (1.8) |
Establishment of protein localization (71) Intracellular transport (50) |
| 25 (1.7) | Protein amino acid dephosphorylation (20) |
| 27 (1.6) | Small GTPase mediated signal transduction (34) |
| 37 (1.4) | Cerebral cortex development (9) |
Number in parentheses is the Enrichment Score for each cluster; score 1.3 is equivalent to non-log scale p of 0.05 (Huang et al., 2009). In two cases, distinct clusters contained similar terms; these have been grouped here (see supplemental Table S-2). NS: Enrichment Score < 1.3.
Number in parentheses indicates the number of genes found within that term; terms are from GOTERM_BP_FAT.
Candidate Gene Studies
The LXS exon array data were used in a genetical genomic analysis to identify cis-regulated candidates in the AFT_Et QTL which contained 716 annotated genes (Ensembl). A gene was considered to be an expression candidate if its expression showed a significant correlation to AFT_Et (p<0.05) and had a cis-eQTL (LOD>3.0, peak within 10 Mb of the gene). This resulted in 48 genes with LOD scores ranging from 3.0 to 27.3 and an average distance from QTL peak to gene of 1.1 Mb. The complete list can be found in supplemental file S3. Note that only 303 (42%) of the genes found within the QTL interval were on the exon array; the other 58% were removed through filtering or were not represented on the array.
Deep sequencing of the ILS and ISS detected 113,400 ILS/ISS variants within the 23 Mb QTL interval on chromosome 4 (Table 6). The majority were SNPs (92,001) with the remainder being indels (21,399) the largest of which was 50 nucleotides. A non-synonymous SNP or a coding region indel was found in 150 genes; 6 of the SNPs were nonsense. The complete list can be found in supplemental file S3.
Table 6.
Summary of ILS/ISS DNA variants (SNPs and small indels) in the chromosome 4 AFT_Et QTL (116–139 Mb).
| Total # annotated genes | 716 |
| Total # DNA variants | 113,400 |
| SNPs | 92,001 |
| ndels | 21,399 |
| Average distance between variants | 203 ± 4 bp |
| Indels in exons of annotated genes | 27 |
| Frameshifts | 10 |
| SNPs in exons of annotated genes | 1340 |
| Synonymous SNPs | 954 |
| Non-synonymous SNPs | 386 |
| Nonsense | 6 |
| Missense | 380 |
| Total # annotated genes with one or more variants in an exon | 247 |
| Total # annotated genes with one or more indels or non-synonymous SNPs in an exon | 150 |
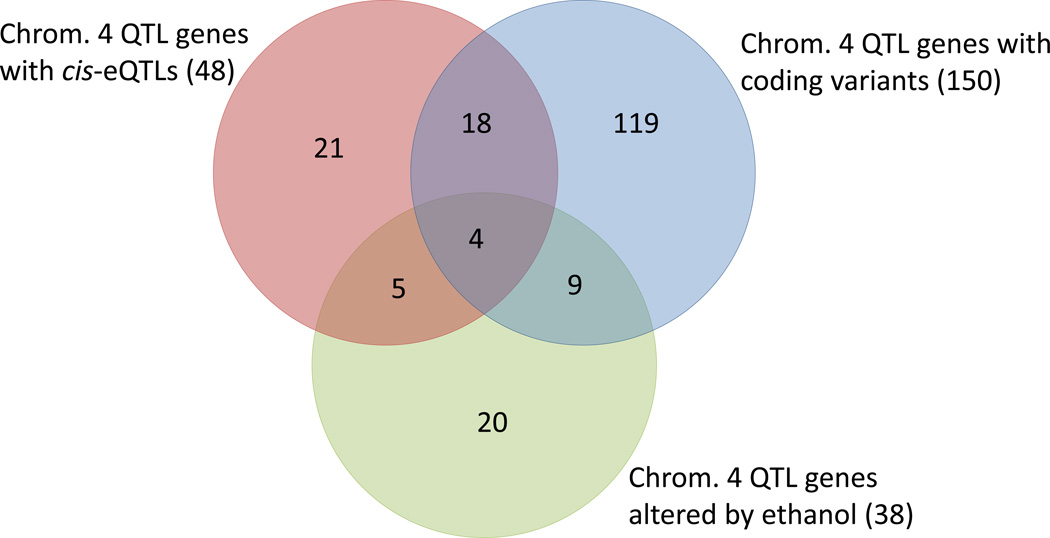
Many of the genes in the chromosome 4 QTL credible interval were found in more than one candidate category; i.e., they were in common among cis-eQTL regulated genes, genes affected by alcohol in the ILS/ISS time course study, or genes with a non-synonymous sequence variant. Their relationship is illustrated as a Venn diagram in Figure 4. The four genes that were common to all three groups were Map3k6, Ece1, Hp1bp3, and Mul1; genes in the other groups can be found in supplemental file S3.
Figure 4.
Venn diagram of genes that were found within the 23 Mb 90% Bayesian credible interval of the significant chromosome 4 AFT_Et QTL and were either cis-regulated in the LXS RI panel, had a non-synonymous coding variant in the ILS compared to the ISS, or were alcohol-responsive in the ILS and/or ISS.
Discussion
The significant chromosome 4 QTL for AFT_Et is a promising region for harboring genes that are pleiotropic for AFT and drinking behavior. This locus replicates QTLs in the same region of chromosome 4 for DID in the LXS (Saba et al., 2011) and for two-bottle preference drinking in 5 independent studies using various crosses derived from C57BL/6 and DBA/2 mice (Belknap and Atkins, 2001). Additionally, a number of linkage and GWAS studies of drinking- and alcoholism-related traits have implicated the region on human chromosome 1p that is syntenic to the mouse chromosome 4 locus (e.g., Edwards et al., 2012, Uhl et al., 2008). Interestingly, the current locus appears to be specific for AFT in the alcohol pretreatment group further emphasizing that the change in AFT that results from previous experience with alcohol may be a more important predictor of drinking behavior than initial AFT (Radcliffe et al., 2013).
The chromosome 4 QTL region is gene- and variant-dense: 176 of the 716 annotated genes in this region could be considered high priority “candidates”. Even this large list is undoubtedly incomplete because there are probably additional cis-eQTLs or other types of DNA variants (copy number variants, inversions, large indels, etc.) that we were unable to identify using the current methods, not to mention the possibility of the less well understood effects of synonymous SNPs (Hunt et al., 2009). It is certainly possible that a single gene in this region is responsible for the QTL, but identification of a single “QT-gene” for any given QTL has been the exception rather than the rule (Flint et al., 2005). We thus propose that multiple genes in the region, perhaps displaying a variety of genetic mechanisms, are responsible for the QTL. These hypothetical genes may reach across many of the functional categories identified in the enrichment analyses. For example, 23 of the candidates are related to transcription, 4 relate to chromatin and chromatin modification, 15 are in protein phosphorylation pathways, 8 are related to proteolysis, 9 are related to lipid biosysnthesis, and all of the other functional categories are represented by at least one candidate. This result may be indicating that the QTL is being driven by several small clusters of functionally related genes which, if true, perhaps provides some hope for the successful dissection of this QTL and also more generally for any QTL: the effect of any individual gene among many may not be tractable, but the effect of a handful of functionally-related gene clusters may be.
Only two of the nine suggestive QTLs have been mapped for LORR in previous studies: the chromosome 1 QTL for ST_sal and ST_Et and the chromosome 15 AFT_Et QTL (Bennett et al., 2006, Bennett et al., 2002). This low level of validation may be a result of differences in the LORR test employed here compared to that used in the earlier mapping studies. It also is possible that the pretreatment caused enough of a change in the behavior to account for the difference. Of course these QTLs were mapped at only the suggestive level and, although the current study employed more RI strains than is typical, the statistical power to detect QTLs was still limited.
While all of the behavioral correlation gene lists except for ST_sal revealed at least one significantly enriched functional group, AFT_sal by far had the largest number of correlated genes and enriched functional categories. This may be due in part to the fact that the exon array data were generated from untreated mice suggesting that the correlated genes might be more related to an underlying predisposition towards a certain level of AFT rather than to a dynamic transcriptomic response that might be seen in the alcohol pretreatment group. As such, it is interesting that many of the enriched terms tend to point towards general cellular and molecular regulation; e.g., transcription and mRNA metabolism, proteolysis, chromatin modification, and protein phosphorylation. The latter two categories are of particular interest since both have been shown to interact with alcohol and modulate alcohol responses (Nielsen et al., 2012, Trudell et al., 2014). There is a substantial literature demonstrating that alcohol stimulates or suppresses protein phosphorylation-related activity and genetic deletion studies indicate that at least several protein kinases mediate a wide variety of responses to alcohol (Trudell et al., 2014). Epigenetics has become increasingly recognized as an important mechanism contributing to drug dependence (Robison and Nestler, 2011). In particular, alcohol has been shown to modify epigenetic markers (Finegersh and Homanics, 2014, Pandey et al., 2008) and gene networks related to chromatin modification have been identified in the context of alcohol drinking in the mouse (Wolstenholme et al., 2011) and in the brains of human alcoholics (Ponomarev et al., 2012). Inhibition of histone deacetylase (HDAC) activity led to increased drinking in mice (Wolstenholme et al., 2011) and reduced withdrawal-induced anxiety-like behavior in rats (Pandey et al., 2008). Of particular relevance here, HDAC inhibition blocked alcohol-induced sensitization (Legastelois et al., 2013) and also blocked the development of rapid tolerance to the anxiolytic effects of alcohol (Sakharkar et al., 2012). Considering these findings, it is notable that there were 11 genes in the chromosome 4 QTL region that have known associations with chromatin remodeling, although only 4 would be considered clear candidates.
Many of the functional categories identified in the ILS/ISS alcohol time course study were found in one or more of the genetic correlation groups, including regulation of transcription, chromatin modification, proteolysis, protein phosphorylation/dephosphorylation activity, and brain development. The finding of common functions between the two experiments, despite being completely independent, using completely different experimental designs, and using tissue from whole brain vs. striatum, suggests that these particular categories may be important in the behaviors. Moreover, many of these same categories also have been identified by others in the context of alcoholism using microarray analysis with a variety of species and experimental approaches, including from the brains of human alcoholics (Farris and Miles, 2013, Liu et al., 2006, Mulligan et al., 2006, Vanderlinden et al., 2013).
There are several important limitations and caveats to the current study. First, the microarrays did not contain all genes or gene transcripts that are expressed in brain and that may be of relevance. Second, there are many genomic features that are either unannotated and/or were not examined; e.g. non-coding RNAs and structural variants. Finally, while there is some consistency between the current results and those of others, the generality of these findings should be considered. The mechanism of AFT for a hypnotic dose may differ for lower doses, although there is probably at least some overlap, and to what extent hypnotic sensitivity or high-dose AFT relates to Schuckit’s LR hypothesis is unclear (Crabbe et al., 2010). Also the current results must be viewed in the context of an animal model – the LXS – that is only a fairly narrow genetic representation of the mouse.
In conclusion, we have mapped QTLs for LORR-related traits in the LXS RI panel, including AFT, and have used genomics-oriented approaches to identify candidate genes and candidate functions to investigate the genetic and molecular basis of these behaviors. The AFT_Et chromosome 4 QTL is especially interesting because drinking-related behaviors have been mapped in the mouse and in humans at this same locus. This QTL is thus consistent with our previous observation of a significant genetic correlation between AFT_Et and DID further highlighting our working hypothesis that it may be the interaction between AFT and initial exposures to alcohol that is a critical determinant in AUD risk (Radcliffe et al., 2013). Based on the results of the candidate gene studies, we postulate that the chromosome 4 QTL contains genes that are pleiotropic for AFT_Et and drinking behavior and that multiple genes each with relatively small, but not necessarily equal effects are responsible for the QTL. Two independent microarray experiments pointed towards a kind of broad cellular plasticity as mediating the genetic effects on ST, AFT, and RT. Future studies will include finer mapping of the chromosome 4 AFT_Et QTL and expression profiling of the LXS with and without alcohol to further refine the list of candidates. Network-based approaches such as Weighted Gene Co-expression Network Analysis (WGCNA) can be applied to continue to explore gene networks and biological pathways that mediate AFT. It is our hope that we can more clearly delineate the relationship between AFT and drinking behavior with the ultimate goal of developing more effective treatments for AUDs.
Supplementary Material
Acknowledgements
The authors would like to thank the UCD Genomics and microarray core for their help with the microarray processing and DNA sequencing and also the reviewers of this article for their helpful comments and suggestions. This research was funded from NIH grants R01-AA016957-04 (RAR), U01-AA-013517-13 (LMS), and R24-AA013162-13 (BT), and from a Sloan Foundation Fellowship (RD).
References
- Affymetrix. Exon array background Correction. Affymetrix Whitepaper. 2005 Available at: http://www.affymetrix.com/support/technical/whitepapers/exon_background_correction_whitepaper.pdf. [Google Scholar]
- Belknap JK. Effect of within-strain sample size on QTL detection and mapping using recombinant inbred mouse strains. Behav Genet. 1998;28:29–38. doi: 10.1023/a:1021404714631. [DOI] [PubMed] [Google Scholar]
- Belknap JK, Atkins AL. The replicability of QTLs for murine alcohol preference drinking behavior across eight independent studies. Mammalian Genome. 2001;12:893–899. doi: 10.1007/s00335-001-2074-2. [DOI] [PubMed] [Google Scholar]
- Bennett B, Beeson M, Gordon L, Johnson TE. Reciprocal congenics defining individual quantitative trait Loci for sedative/hypnotic sensitivity to ethanol. Alcohol Clin Exp Res. 2002;26:149–157. [PubMed] [Google Scholar]
- Bennett B, Carosone-Link P. Replication of small effect quantitative trait loci for behavioral traits facilitated by estimation of effect size from independent cohorts. Genes Brain Behav. 2006;5:404–412. doi: 10.1111/j.1601-183X.2005.00174.x. [DOI] [PubMed] [Google Scholar]
- Bennett B, Carosone-Link P, Zahniser NR, Johnson TE. Confirmation and fine mapping of ethanol sensitivity quantitative trait loci, and candidate gene testing in the LXS recombinant inbred mice. J Pharmacol Exp Ther. 2006;319:299–307. doi: 10.1124/jpet.106.103572. [DOI] [PubMed] [Google Scholar]
- Bennett B, Saba LM, Hornbaker CK, Kechris KJ, Hoffman P, Tabakoff B. Genetical genomic analysis of complex phenotypes using the PhenoGen website. Behav Genet. 2011;41:625–628. doi: 10.1007/s10519-010-9427-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman KW, Wu H, Sen S, Churchill GA. R/qtl: QTL mapping in experimental crosses. Bioinformatics. 2003;19:889–890. doi: 10.1093/bioinformatics/btg112. [DOI] [PubMed] [Google Scholar]
- Carlborg O, De Koning DJ, Manly KF, Chesler E, Williams RW, Haley CS. Methodological aspects of the genetic dissection of gene expression. Bioinformatics. 2005;21:2383–2393. doi: 10.1093/bioinformatics/bti241. [DOI] [PubMed] [Google Scholar]
- Churchill GA, Doerge RW. Empirical threshold values for quantitative trait mapping. Genetics. 1994;138:963–971. doi: 10.1093/genetics/138.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabbe JC, Bell RL, Ehlers CL. Human and laboratory rodent low response to alcohol: is better consilience possible? Addict Biol. 2010;15:125–144. doi: 10.1111/j.1369-1600.2009.00191.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cromer JR, Cromer JA, Maruff P, Snyder PJ. Perception of alcohol intoxication shows acute tolerance while executive functions remain impaired. Exp Clin Psychopharmacol. 2010;18:329–339. doi: 10.1037/a0019591. [DOI] [PubMed] [Google Scholar]
- Dick DM, Bierut LJ. The genetics of alcohol dependence. Current psychiatry reports. 2006;8:151–157. doi: 10.1007/s11920-006-0015-1. [DOI] [PubMed] [Google Scholar]
- Edwards AC, Aliev F, Bierut LJ, Bucholz KK, Edenberg H, Hesselbrock V, Kramer J, Kuperman S, Nurnberger JI, Jr, Schuckit MA, Porjesz B, Dick DM. Genome-wide association study of comorbid depressive syndrome and alcohol dependence. Psychiatric genetics. 2012;22:31–41. doi: 10.1097/YPG.0b013e32834acd07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erwin VG, Deitrich RA. Genetic selection and characterization of mouse lines for acute functional tolerance to ethanol. J Pharmacol Exp Ther. 1996;279:1310–1317. [PubMed] [Google Scholar]
- Farris SP, Miles MF. Fyn-dependent gene networks in acute ethanol sensitivity. PLoS One. 2013;8:e82435. doi: 10.1371/journal.pone.0082435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finegersh A, Homanics GE. Acute ethanol alters multiple histone modifications at model gene promoters in the cerebral cortex. Alcohol Clin Exp Res. 2014;38:1865–1873. doi: 10.1111/acer.12465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint J, Valdar W, Shifman S, Mott R. Strategies for mapping and cloning quantitative trait genes in rodents. Nat Rev Genet. 2005;6:271–286. doi: 10.1038/nrg1576. [DOI] [PubMed] [Google Scholar]
- Heath AC, Madden PA, Bucholz KK, Dinwiddie SH, Slutske WS, Bierut LJ, Rohrbaugh JW, Statham DJ, Dunne MP, Whitfield JB, Martin NG. Genetic differences in alcohol sensitivity and the inheritance of alcoholism risk. Psychological medicine. 1999;29:1069–1081. doi: 10.1017/s0033291799008909. [DOI] [PubMed] [Google Scholar]
- Hu W, Saba L, Kechris K, Bhave SV, Hoffman PL, Tabakoff B. Genomic insights into acute alcohol tolerance. J Pharmacol Exp Ther. 2008;326:792–800. doi: 10.1124/jpet.108.137521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Hubbell E, Liu WM, Mei R. Robust estimators for expression analysis. Bioinformatics. 2002;18:1585–1592. doi: 10.1093/bioinformatics/18.12.1585. [DOI] [PubMed] [Google Scholar]
- Hunt R, Sauna ZE, Ambudkar SV, Gottesman MM, Kimchi-Sarfaty C. Silent (synonymous) SNPs: should we care about them? Methods Mol Biol. 2009;578:23–39. doi: 10.1007/978-1-60327-411-1_2. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics (Oxford, England) 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- Lander E, Kruglyak L. Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet. 1995;11:241–247. doi: 10.1038/ng1195-241. [DOI] [PubMed] [Google Scholar]
- Legastelois R, Botia B, Naassila M. Blockade of ethanol-induced behavioral sensitization by sodium butyrate: descriptive analysis of gene regulations in the striatum. Alcohol Clin Exp Res. 2013;37:1143–1153. doi: 10.1111/acer.12088. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Lewohl JM, Harris RA, Iyer VR, Dodd PR, Randall PK, Mayfield RD. Patterns of gene expression in the frontal cortex discriminate alcoholic from nonalcoholic individuals. Neuropsychopharmacology. 2006;31:1574–1582. doi: 10.1038/sj.npp.1300947. [DOI] [PubMed] [Google Scholar]
- Lundquist F. The determination of ethyl alcohol in blood and tissue. Methods Biochem Anal. 1959;7:217–251. [Google Scholar]
- McClearn GE, Kakihana R. In: Selective breeding for ethanol sensitivity: Short-Sleep and Long-Sleep mice, in Development of Animal Models as Pharmacogenetic Tools, Development of Animal Models as Pharmacogenetic Tools. Deitrich RA, Erwin VG, editors. Washington, D.C.: National Institute of Alcohol Abuse and Alcoholism; 1981. pp. 147–159. [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellanby E. Alcohol: its absorption into and disappearance from the blood under different conditions. 1919 [Google Scholar]
- Mulligan MK, Ponomarev I, Hitzemann RJ, Belknap JK, Tabakoff B, Harris RA, Crabbe JC, Blednov YA, Grahame NJ, Phillips TJ, Finn DA, Hoffman PL, Iyer VR, Koob GF, Bergeson SE. Toward understanding the genetics of alcohol drinking through transcriptome meta-analysis. Proc Natl Acad Sci U S A. 2006;103:6368–6373. doi: 10.1073/pnas.0510188103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen DA, Utrankar A, Reyes JA, Simons DD, Kosten TR. Epigenetics of drug abuse: predisposition or response. Pharmacogenomics. 2012;13:1149–1160. doi: 10.2217/pgs.12.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey SC, Ugale R, Zhang H, Tang L, Prakash A. Brain chromatin remodeling: a novel mechanism of alcoholism. J Neurosci. 2008;28:3729–3737. doi: 10.1523/JNEUROSCI.5731-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponomarev I, Crabbe JC. A novel method to assess initial sensitivity and acute functional tolerance to hypnotic effects of ethanol. J Pharmacol Exp Ther. 2002;302:257–263. doi: 10.1124/jpet.302.1.257. [DOI] [PubMed] [Google Scholar]
- Ponomarev I, Crabbe JC. Characterization of acute functional tolerance to the hypnotic effects of ethanol in mice. Alcohol Clin Exp Res. 2004;28:991–997. doi: 10.1097/01.alc.0000131978.79857.5e. [DOI] [PubMed] [Google Scholar]
- Ponomarev I, Wang S, Zhang L, Harris RA, Mayfield RD. Gene coexpression networks in human brain identify epigenetic modifications in alcohol dependence. J Neurosci. 2012;32:1884–1897. doi: 10.1523/JNEUROSCI.3136-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radcliffe RA, Floyd KL, Drahnak JA, Deitrich RA. Genetic dissociation between ethanol sensitivity and rapid tolerance in mouse and rat strains selectively bred for differential ethanol sensitivity. Alcohol Clin Exp Res. 2005;29:1580–1589. doi: 10.1097/01.alc.0000179208.05882.1f. [DOI] [PubMed] [Google Scholar]
- Radcliffe RA, Floyd KL, Lee MJ. Rapid ethanol tolerance mediated by adaptations in acute tolerance in inbred mouse strains. Pharmacol Biochem Behav. 2006a;84:524–534. doi: 10.1016/j.pbb.2006.06.018. [DOI] [PubMed] [Google Scholar]
- Radcliffe RA, Larson C, Bennett B. Genetic studies of acute tolerance, rapid tolerance, and drinking in the dark in the LXS recombinant inbred strains. Alcohol Clin Exp Res. 2013;37:2019–2028. doi: 10.1111/acer.12188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radcliffe RA, Lee MJ, Williams RW. Prediction of cis-QTLs in a pair of inbred mouse strains with the use of expression and haplotype data from public databases. Mamm Genome. 2006b;17:629–642. doi: 10.1007/s00335-005-0178-9. [DOI] [PubMed] [Google Scholar]
- Rhodes JS, Best K, Belknap JK, Finn DA, Crabbe JC. Evaluation of a simple model of ethanol drinking to intoxication in C57BL/6J mice. Physiol. Behav. 2005;84:53–63. doi: 10.1016/j.physbeh.2004.10.007. [DOI] [PubMed] [Google Scholar]
- Robison AJ, Nestler EJ. Transcriptional and epigenetic mechanisms of addiction. Nature reviews. Neuroscience. 2011;12:623–637. doi: 10.1038/nrn3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saba LM, Bennett B, Hoffman PL, Barcomb K, Ishii T, Kechris K, Tabakoff B. A systems genetic analysis of alcohol drinking by mice, rats and men: influence of brain GABAergic transmission. Neuropharmacology. 2011;60:1269–1280. doi: 10.1016/j.neuropharm.2010.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakharkar AJ, Zhang H, Tang L, Shi G, Pandey SC. Histone deacetylases (HDAC)-induced histone modifications in the amygdala: a role in rapid tolerance to the anxiolytic effects of ethanol. Alcohol Clin Exp Res. 2012;36:61–71. doi: 10.1111/j.1530-0277.2011.01581.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuckit MA. Self-rating of alcohol intoxication by young men with and without family histories of alcoholism. Journal of studies on alcohol. 1980;41:242–249. doi: 10.15288/jsa.1980.41.242. [DOI] [PubMed] [Google Scholar]
- Schuckit MA. Low level of response to alcohol as a predictor of future alcoholism. The American journal of psychiatry. 1994;151:184–189. doi: 10.1176/ajp.151.2.184. [DOI] [PubMed] [Google Scholar]
- Trudell JR, Messing RO, Mayfield J, Harris RA. Alcohol dependence: molecular and behavioral evidence. Trends Pharmacol Sci. 2014;35:317–323. doi: 10.1016/j.tips.2014.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhl GR, Drgon T, Johnson C, Li CY, Contoreggi C, Hess J, Naiman D, Liu QR. Molecular genetics of addiction and related heritable phenotypes: genome-wide association approaches identify "connectivity constellation" and drug target genes with pleiotropic effects. Ann N Y Acad Sci. 2008;1141:318–381. doi: 10.1196/annals.1441.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderlinden LA, Saba LM, Kechris K, Miles MF, Hoffman PL, Tabakoff B. Whole brain and brain regional coexpression network interactions associated with predisposition to alcohol consumption. PLoS One. 2013;8:e68878. doi: 10.1371/journal.pone.0068878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter NAR, McWeeney SK, Peters ST, Belknap JK, Hitzemann R, Buck KJ. SNPs matter: impact on detection of differential expression. Nat Methods. 2007;4:679–680. doi: 10.1038/nmeth0907-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RW, Bennett B, Lu L, Gu J, DeFries JC, Carosone-Link PJ, Rikke BA, Belknap JK, Johnson TE. Genetic structure of the LXS panel of recombinant inbred mouse strains: a powerful resource for complex trait analysis. Mamm Genome. 2004;15:637–647. doi: 10.1007/s00335-004-2380-6. [DOI] [PubMed] [Google Scholar]
- Wolstenholme JT, Warner JA, Capparuccini MI, Archer KJ, Shelton KL, Miles MF. Genomic analysis of individual differences in ethanol drinking: evidence for non-genetic factors in C57BL/6 mice. PLoS One. 2011;6:e21100. doi: 10.1371/journal.pone.0021100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.