Abstract
The enterococci are an ancient genus that evolved along with the tree of life. These intrinsically rugged bacteria are highly adapted members of the intestinal consortia of a range of hosts that spans the animal kingdom. Enterococci are also leading opportunistic hospital pathogens, causing infections that are often resistant to treatment with most antibiotics. Despite the importance of enterococci as hospital pathogens, the vast majority live outside of humans, and nearly all of their evolutionary history took place before the appearance of modern humans. Because hospital infections represent evolutionary end points, traits that exacerbate human infection are unlikely to have evolved for that purpose. However, clusters of traits have converged in specific lineages that are well adapted to colonize the antibiotic-perturbed gastrointestinal tracts of patients and that thrive in the hospital environment. Here we discuss these traits in an evolutionary context, as well as how comparative genomics is providing new insights into the evolution of the enterococci.
Keywords: Enterococcus, genomics, human-associated microbial flora
Introduction: Origins and Evolution of the Enterococci
Enterococci predate the split of humans from primates by aeons, but these bacteria were not specifically noticed until the end of the nineteenth century, when Thiercelin (117) and MacCallum & Hastings (66) separately described human enterococcal infections. After more than a hundred years, the experimental proof of Koch's postulates by the latter still stands as one of the finest studies in the field (66). Nearly all research on enterococci has been in the context of human infection, with comparatively little attention paid to their wide distribution or roles in nature. Yet it is only through the lens of their native biology—the result of evolution over millennia—that we are likely to understand their emergence as hospital pathogens (39, 129) capable of transmitting antibiotic resistances to other clinically important microbes (55, 64).
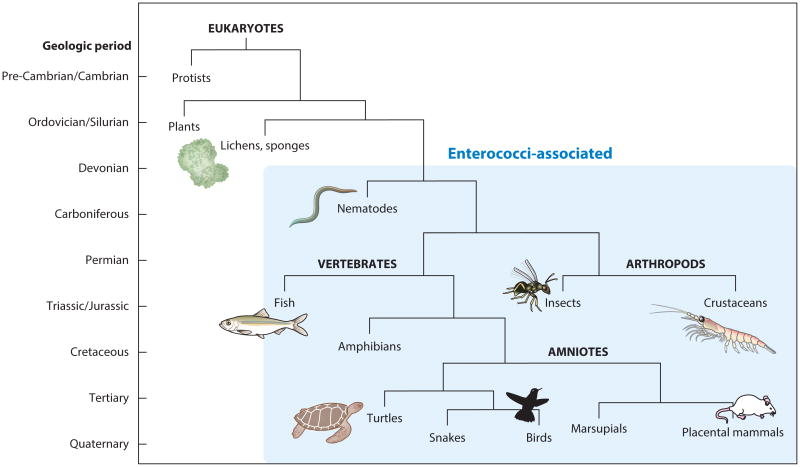
The genus Enterococcus consists of nonsporulating gram-positive bacteria that were initially grouped within the genus Streptococcus (65). The genus was officially named Enterococcus in 1984, based on sequence divergence of 16S rRNA (102). Enterococci inhabit the intestinal tracts of humans, animals, and insects (8, 20, 71, 72, 77) and the environments these organisms inhabit, including plants, soil, and water (40, 78). The most parsimonious explanation for their occurrence in this vast array of hosts is that they were core members of the intestinal microbiome of the last common ancestor of mammals, reptiles, birds, and insects, which existed in the early Devonian period, over 400 mya (39) (Figure 1).
Figure 1.
Simplified tree of eukaryotic life, with blue shading indicating animals with which enterococci have been found to be associated. The labels to the left of the tree show approximate geologic periods.
Given the broad distribution of enterococci among animals and in the environment, the fraction of enterococci that reside within humans is very small. Furthermore, as a core member and the most abundant gram-positive coccus in the human gastrointestinal (GI) tract (58, 124), the fraction of human-associated enterococci causing infection is infinitesimally small. It is therefore unlikely that any particular trait found in enterococci outside of hospitals evolved or was acquired because of its ability to exacerbate disease. Rather, native enterococcal factors that contribute to disease exist because of their contribution to bacterial survival and proliferation in one or more natural habitats. Ironically, we currently know almost nothing about these natural roles.
Enterococci are ubiquitous members of GI tract consortia. This may be because of their ability to survive harsh innate defense systems of host GI tract environments, including the grinder of one of the oldest and most abundant forms of animal life, nematodes (36). The ability to colonize the intestine of nematodes, where they are bathed in the lysates of other bacteria, would provide enterococci with a nutrient-rich habitat. This may have contributed to initial host adaptation, resulting in the streamlined genomes observed today. Enterococci are auxotrophic for many amino acids, vitamins, and micronutrients (84), and the genomes of commensal strains are only about 2.75 megabases (88). Enterococci are also able to survive and grow in a wide range of temperatures and pHs and in both hypertonic and hypotonic conditions (106, 107).
As a typical omnivorous mammal, humans shed the equivalent of their entire intestinal microbiota into the environment every 2–3 weeks (37, 130). With an average historic life span of 25–30 years (52), this corresponds to depositing more than 500 colon equivalents into the environment over a lifetime. Therefore, far less than 1% of human-derived enterococci actually reside within the human GI tract at any given time (or, looked at another way, a small fraction of the enterococcal life cycle is spent in the human, let alone causing disease). Enterococci are able to survive in sediments and soils, on the surface of aquatic and terrestrial vegetation, and in freshwater and marine water (10, 111), and they persist in these settings longer than most bacteria (110). Despite the auxotrophies of enterococci, some studies indicate that they are able to replicate in the environment (27, 134), possibly as the result of collaboration within a polymicrobic consortium. This survival ability results from their tolerance of UV irradiation (70), salt, starvation (43), and possibly predation by protozoa (44) and bacteriophages (93). This inherent ruggedness has no doubt contributed to their emergence in the harsh environment of the modern hospital.
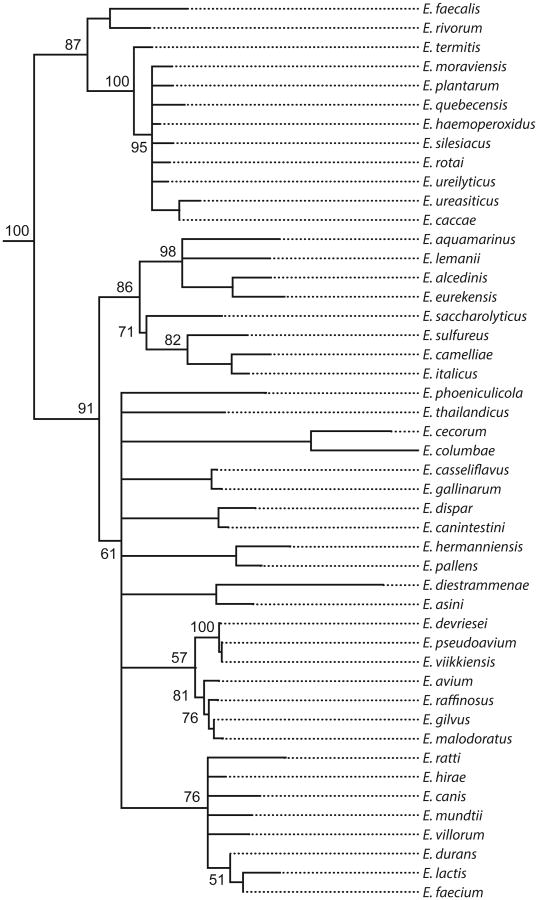
Nearly 50 species of enterococci have been identified, mostly in the genomic era (90) (Figure 2). Speciation occurs when populations become isolated and diverge over time (73). We currently know little about the natural distribution of enterococcal species or the ecological isolation that led to speciation. E. faecalis and E. faecium are the most common species isolated from the human GI tract, with the former being predominant (69, 85). They are also the enterococci most often associated with human infection (39). It is therefore surprising that these two species are at opposite ends of the phylogenetic tree (Figure 2). E. faecalis occurs in one of the oldest branches of the genus, whereas E. faecium arose more recently. However, their common occurrence in the human intestine and in hospital infection shows that they possess common features that allow them to inhabit similar ecologies. Given that both species share overlapping, if not identical, microecologies within humans, it is clear that the isolation required for speciation of E. faecium did not occur there.
Figure 2.
Phylogenetic tree of the genus Enterococcus. Available 16S rRNA gene sequences for each species were compiled using Geneious software (Biomatters, Ltd., San Francisco, California) with a neighbor-joining algorithm. The 16S rRNA sequence of Vagococcus lutrae was used as an outgroup. Bootstrap values are shown for nodes with more than two branches and were generated over 1,000 iterations.
What drives enterococcal speciation? The specific nature of an animal's intestinal tract (microbial diversity, diet, host defense and other secretions, body temperature, pH) selects for the outgrowth of favorably matched strains, resulting in host adaptation (22, 69). In intestinal tracts that are not ecologically isolated, such as the intestinal tracts of an insect and insectivore (where enterococci of predator and prey comingle in the GI tract of the predator), separate species may not evolve, because of horizontal movement of genes. However, enterococci within the intestinal tract of an herbivore may be sufficiently isolated from other members of the genus to permit speciation—the ultimate extension of host adaptation.
The lack of clade structure, or even substantial genetic drift among populations of E. faecalis (54, 88), might be explained by the observation that it is widely disseminated along the food chain (71, 77). It is tempting to speculate that this stems from genetic mixing between enterococci of predator and prey, possibly facilitated by the near-exclusive occurrence of highly efficient pheromone-responsive plasmid gene transfer systems in E. faecalis (30, 31). It was recently shown that large pieces of the chromosome could in fact be mobilized and transferred between E. faecalis strains by pheromone-responsive plasmids (68), possibly limiting divergence and speciation.
The relative ecological isolation of the gut flora of herbivores leads to two predictions: First, a main driver of Enterococcus speciation may have been the large radiation in mammalian herbivore species that accompanied the global expansion of C4 photosynthesizing plants eight to three million years ago (32). Second, more species divergence is likely to occur among the host-adapted enterococci of various herbivores. The occurrence in the phylogenetic tree of E. faecium as a modern species, along with considerable branching and divergence, suggests its genesis at a time of considerable host species radiation (Figure 2).
Evolution of Enterococcal Virulence Factors
Virulence is traditionally defined as the extent to which an organism can cause disease (11). It is often viewed as a means to an end—enhanced growth and dissemination to other hosts (57, 118). However, enterococcal infection in humans and close relatives with a long history of hygienic funerary rituals (95) is an evolutionary dead end. In other words, because there is little chance of bloodstream-to-bloodstream or heart-valve-to-heart-valve transmission of enterococci, there is no sustained advantage for them to proliferate at these common sites of infection (9). Although enterococci can cause infection in nonhospitalized patients, outside of the hospital setting this is rare and often results from an underlying predisposition. This appears to have been the case for the first Enterococcus-infected patient, described by MacCallum & Hastings (66). The patient had a prior history of rheumatic fever, which may have caused heart valve scarring and blood flow abnormalities, predisposing him to endocarditis. Almost 10 years later, he developed and succumbed to enterococcal endocarditis, perhaps related to a recent severe respiratory infection (66). Although in this classic case the patient was not infected in a hospital, most invasive enterococcal infections today occur within hospitals, in patients with underlying conditions and whose native flora has been destabilized by prior antibiotic use (39).
Not all enterococci are equal. The bulk of the multidrug-resistant enterococcal infection problem is caused by discrete lineages of E. faecalis and E. faecium, and we now know the principal characteristics of those lineages (54, 60, 74, 79, 100, 112). In the case of E. faecalis, hospital-adapted, multidrug-resistant lineages include strains of multiple-locus sequence type clonal clusters CC2, CC9, CC28, and CC40 (74, 100, 112). For E. faecium, most hospital-acquired infections are caused by CC17 and closely related strains (54, 60). What, then, makes these lineages different from commensal E. faecalis and E. faecium strains? In both species, a variety of auxiliary traits have converged on mobile elements, resulting in genomes that are significantly larger than those of commensal strains (61, 88). The hospital-adapted clonal lineages are also unique in their ability to be transmitted patient-to-patient within the hospital setting, being well suited to thrive in that environment (2).
Because invasive infection is an evolutionary end point, the hospital environment has selected for traits that make colonization and transmission more likely. As a result, the variable traits that have converged in hospital-adapted, multidrug-resistant enterococci include antibiotic resistances (61, 74, 112), an Esp-containing pathogenicity island (59, 61, 104, 112), and more complex integral cell wall carbohydrate operons (88, 112). In the prototype CC2 E. faecalis strain, V583, a protein termed aggregation substance (AS) is encoded by antibiotic-resistance plasmids, and additional alleles occur on the chromosome (91). AS is a prototypical example of an enterococcal virulence factor, in that it exacerbates various measures of infection (15, 33, 114). But because invasive enterococcal infection is an evolutionary end point, it is more likely that the presence of AS in hospital lineages of E. faecalis stems from its contribution to GI tract colonization, mainly through association with antibiotic-resistance plasmids [but also possibly because of its ability to increase binding to colonic epithelial cells (56, 86)]. AS is an LPXTG-containing surface protein that is an important component of pheromone-responsive plasmid transfer systems of E. faecalis (30, 31). Briefly, peptide pheromones are secreted by potential E. faecalis plasmid recipient cells, and these pheromones are sensed by E. faecalis cells harboring pheromone-responsive plasmids. Induction of AS expression promotes effective mating pair formation, a first step in plasmid transfer. During infection, AS increases vegetation size and severity of E. faecalis experimental endocarditis (14, 16) and promotes internalization of bacteria by cultured enterocytes (127). AS also contributes to biofilm formation, which results in bacterial adherence and persistence on tissues and medical devices (16). Its association with postsegregational killing systems on pheromone-responsive plasmids, which also often contain antibiotic-resistance genes, likely selects for the maintenance of AS within cells. These postsegregational killing systems promote retention of antibiotic-resistance plasmids, even in the absence of antibiotic selection (17, 41).
Another virulence trait that undoubtedly evolved for other purposes is the cytolysin of E. faecalis. The cytolysin is a broadly active bacteriocin that is effective against nearly all species of gram-positive bacteria (123). Originally described in the 1930s (119), the cytolysin is now known to belong to the large and diverse class of lantibiotic peptides (132). Yet even among lantibiotics, the cytolysin is unique in its stereochemistry (115), as well as in its ability to lyse erythrocytes and other eukaryotic cells, in addition to bacteria (19). The cytolysin contributes to enterococcal virulence in a wide array of infection models (14, 36, 46, 50) and is found on the pathogenicity island initially characterized in the E. faecalis CC2 strain MMH594 (104), or on pheromone-responsive, AS-expressing plasmids, such as pAD1 (45). Like AS, the cytolysin is found in both clinical and other E. faecalis strains (7, 47). The cytolysin probably evolved initially as a colonization factor, potentially in a nonhuman host, and likely allowed E. faecalis to access new ecological space via its bacteriocin activity (99). It is currently unknown whether the ability to lyse eukaryotic cells is an evolved function or simply a consequence of the broad antiprokaryotic activity of this molecule. Speculatively, the ability to lyse eukaryotic cells may have allowed for the killing of eukaryotic intestinal parasites in an insect or other primitive host, which would confer a selective advantage to bacterium and host, a formula for evolutionary success.
A third factor that has converged in multidrug-resistant, hospital-adapted enterococcal lineages of both E. faecalis and E. faecium, but that likely evolved in response to other selective forces, is the surface protein Esp (105). As noted above, Esp is encoded within collections of genes that have converged in pathogenic lineages on pathogenicity islands (105); and importantly, Esp occurs in the leading hospital lineages of both E. faecalis and E. faecium (105, 131). A version of Esp has also been identified in some strains of Staphylococcus aureus (75). In S. aureus and in enterococci, Esp promotes tissue attachment and biofilm formation (120, 121). This attachment contributes to persistence and survival at sites of infection and also likely plays a similar role at sites of commensal colonization in other hosts, although this has not been studied extensively.
The structure of the cell wall also appears to be enhanced in hospital-adapted lineages: E. faecalis strains belonging to CC2, CC9, and CC28 (but not CC40) express different versions of the capsular polysaccharide (42, 74, 112). This trait enhances E. faecalis resistance to phagocytosis (42) and has not been identified in E. faecium. Because bloodstream-to-bloodstream transmission of enterococci is unlikely, this trait most likely evolved to enhance general environmental resistance, or possibly resistance to predation by amoebas, bacteriophages, or other predators. Hospital strains of both E. faecalis and E. faecium do have more elaborate operons for production of the integral cell wall carbohydrate Epa (88, 112), which may confer additional environmental resistance.
The variable features that have converged in hospital-adapted lineages occur in the context of general features of enterococci that also make them more likely to cause infection than many other species. The many background traits that occur in most enterococcal strains and also contribute to the virulence of enterococci have been extensively examined and reviewed (49, 76, 79–81, 94, 97). Among other traits, these include the adhesin Ace (21, 82, 83, 109) and the extracellular metalloprotease gelatinase, both of which are expressed by E. faecalis (6, 67, 96). A main role of gelatinase in nature appears to be remodeling the E. faecalis surface, which can include modulating surface levels of Ace in mature communities, in response to accumulation of quorum signals (92).
Evolution of Antibiotic Resistance
Antibiotics and antibiotic resistance have ancient roots that may stretch as far back as the origins of life (23). Yet our ability to produce, purify, and release hundreds of millions of tons of antibiotics into the human ecosystem has fundamentally changed the human–proximal microbial landscape (24, 63, 48). Much of the enterococcal antibiotic-resistance problem stems from horizontally acquired traits (89). Enterococci are particularly well positioned to both acquire and serve as a depot for antibiotic-resistance factors for two reasons. First, enterococci exist in complex microbial ecosystems, in intimate contact with a large diversity of potential sources of genetic material (37). Second, because of their high level of intrinsic antibiotic resistance (49, 79–81), enterococci occur in environments that are substantially enriched for antibiotic-resistance elements.
Whereas the ability of enterococci to readily pick up antibiotic resistances in a hospital setting seems obvious, there are few reasons to believe that this phenomenon is exclusive to modern hospitals. Given their wide distribution in nature, enterococci have had opportunities to acquire antibiotic resistances throughout their evolutionary history, and they likely have done so for hundreds of millions of years. Rotting plant matter is a main substrate on which many antibiotic-producing streptomycetes live, as well as a principal food source for many insects (103). Ingestion of antibiotic-producing/resistant streptomycetes, along with antibiotic-laden rotting plant matter, would give insect-resident enterococci access to the necessary genetic material, as well as a potential selective advantage, for acquiring resistance genes. Antibiotic resistance determinants identified in enterococci, such as aminoglycoside and vancomycin-resistance genes, bear obvious relationships to resistance operons from antibiotic-producing soil organisms (5, 26). Whereas in nature this is undoubtedly an exceedingly rare event, the release into the human-associated biosphere of millions of tons of antibiotics over the last 75 years provided opportunities for the outgrowth of potentially naturally occurring resistant strains, as well as strong selective pressure for spreading resistance determinants worldwide (24).
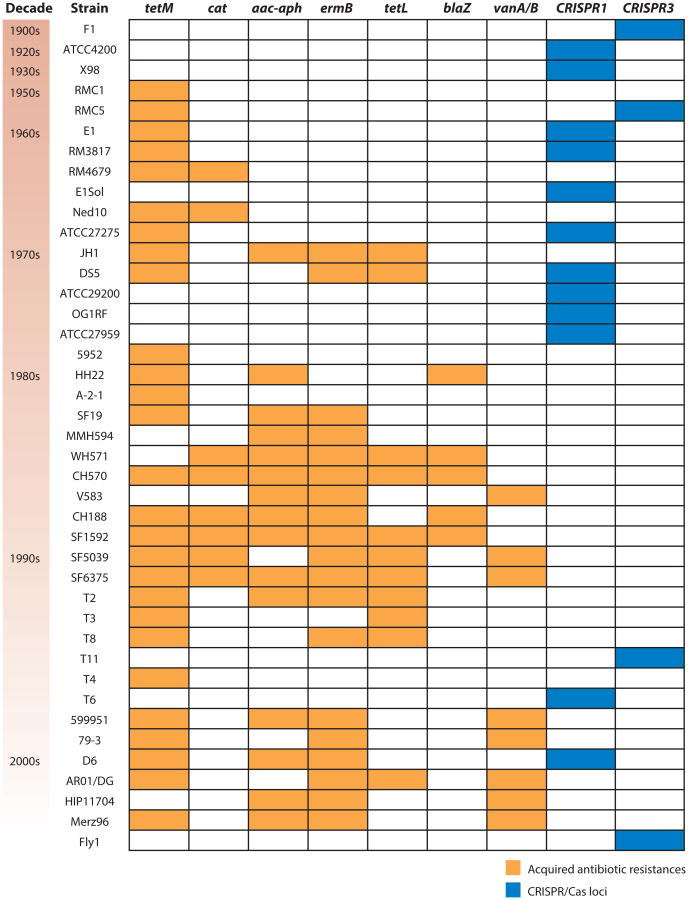
Isolates of E. faecalis that have ended up in strain collections, mainly from clinical infection sources, appear to have acquired resistance to tetracycline and chloramphenicol in the 1950s and 1960s, followed by gentamicin and erythromycin resistance in the 1970s, then ampicillin and vancomycin resistance in the 1980s and 1990s (Figure 3) (87). Interestingly, the occurrence of multiple acquired drug resistances in E. faecalis is inversely correlated with the presence of a functional clustered regularly interspaced short palindromic repeats (CRISPR)/Cas locus. Moreover, in several E. faecalis strains the CRISPR/Cas loci contain spacers derived from plasmids known to transmit antibiotic resistances (87). This suggests that the CRISPR/Cas system helps protect the E. faecalis genome from intrusion of mobile genetic elements, and that a defective CRISPR/Cas system might allow for acquisition of mobile elements and the antibiotic resistances that they encode. Speculatively, antibiotic selection for loss of CRISPR/Cas may also have made hospital-adapted strains of E. faecalis more receptive to other mobile elements, including the pathogenicity island and phages (87). The role played by the CRISPR/Cas system in E. faecium, where it occurs less frequently, is less clear (61, 87).
Figure 3.
Clustered regularly interspaced short palindromic repeats (CRISPR)/Cas loci and acquired antibiotic resistances in Enterococcus faecalis. Strains are ordered by date of isolation, from oldest to most recent, and approximate decades of isolation are shown to the left of the strain names. Presence of acquired antibiotic resistances is shown with orange shading, and CRISPR/Cas locus presence is shown in blue. Only strains containing an acquired antibiotic resistance or a CRISPR/Cas locus are included. Antibiotic resistances were profiled in Reference 74 and correspond to the following compounds: tetracycline (tetM and tetL), chloramphenicol (cat), gentamicin (aac-aph), erythromycin (ermB), ampicillin (blaZ), and vancomycin (vanA/B). Figure is adapted from Reference 87.
The recent emergence of vancomycin resistance in hospital-adapted enterococci provides an apt illustration of the importance of understanding their natural ecology and evolutionary history. Details of key events are missing, and competing interests shift the focus back and forth regarding possible interpretations of the available data. As a result, how and why vancomycin resistance emerged among the enterococci remain somewhat controversial. The bottom line is unambiguous—you get what you select for. Like Newton's third law, because of the tremendous diversity of life, for every action that affects an ecology, there is an equal and opposite reaction. The advent of the antibiotic era has therefore provided a figurative opportunity for Darwin to meet Newton (38).
The facts are these: It is indisputable that the glycopeptide avoparcin was widely used as an animal growth promoter in Europe and Australia (51, 125), and not the United States, prior to and throughout the emergence of vancomycin-resistant enterococci (VRE) in hospitals. Outbreaks of VRE occurred first in England (122) and France (62), nearly three years before similar outbreaks occurred in the United States (101). In France, the first VRE were E. faecium (62), whereas England saw a mixture of mostly E. faecium but also some E. faecalis (122). Not surprisingly, the vancomycin-resistance determinants were found to occur on mobile elements (62). At the time, there were no rigorous studies of the emergence of VRE in hospitals, nor of the environments to which patients or health-care workers had been exposed prior to hospitalization, so cause and effect were not experimentally proven. However, a logical proof of cause and effect can be convincingly advanced, as detailed further below.
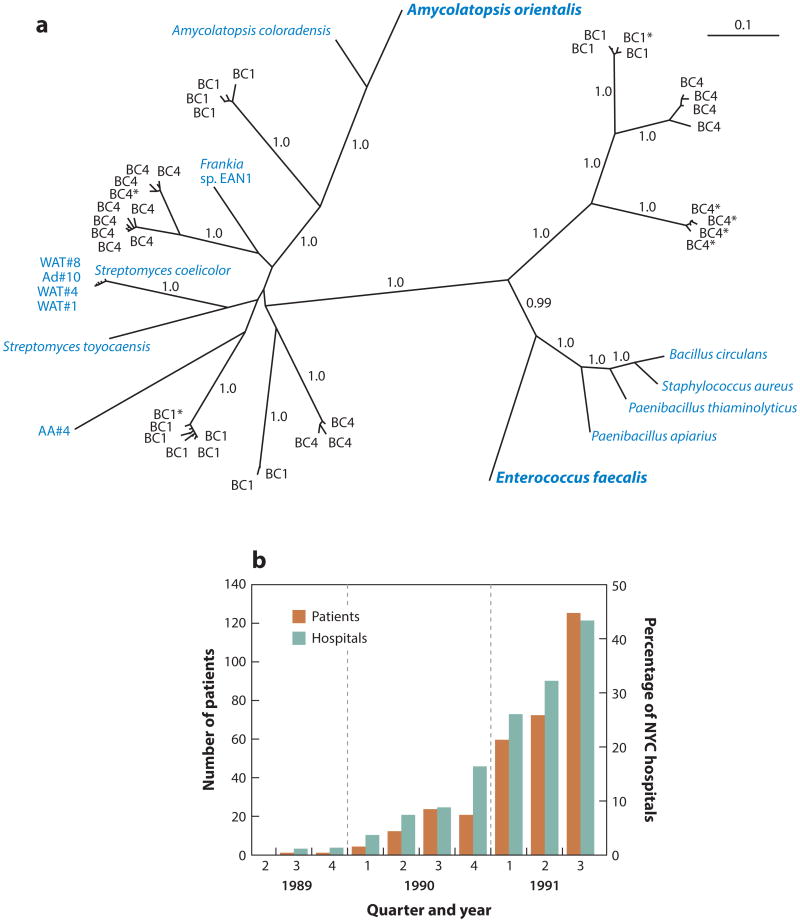
The appearance of VRE hospital strains stems from a series of reactions for which we have little information to accurately model. Furthermore, the spread of VRE involves yet other factors. For the first 30 years following the introduction of vancomycin, resistance in clinically important microbes was not reported. We currently know little of the rate-limiting step for vancomycin resistance to develop or the rates of each necessary reaction that occurred along the way. First, vancomycin resistance had to enter the enterococci in ecologies that overlap with hospitalized patients. The most common vancomycin-resistance determinants among hospital isolates are the VanA and VanB types (12). As shown in Figure 4a, the modern VanA operon has existed for at least 30,000 years, as have derivatives that show its evolution from the ultimate precursor in the Amycolatopsis orientalis vancomycin producer (25). Closely related operons have also been identified in the soil-dwelling-insect pathogen Bacillus (now Paenibacillus) papillae (98). Moreover, related vancomycin-resistance operons naturally occur in the genomes of E. gallinarum and E. casseliflavus (12, 88). This is important because E. casseliflavus was identified in 43.5% of insects [using culture methods (71)], indicating that both pathogenic and commensal enterococci of insects have had reasons to maintain vancomycin-resistance elements for aeons.
Figure 4.
(a) Unrooted Bayesian phylogeny of translated vanA sequences analyzed in Reference 25. Blue denotes strains with VanHAX clusters confirmed to confer resistance to vancomycin; sequences containing stop codons but homology throughout are noted with a single asterisk. The scale bar represents 0.1 substitutions per site. Panel is reproduced from Reference 25. (b) Number of patients and cumulative percentage of New York City (NYC) hospitals with vancomycin-resistant enterococci infection between 1989 and 1991. Panel is reproduced from Reference 35.
What is the effect of applying orally consumed avoparcin to animals? It clearly enriches the glycopeptide-resistance determinants in the ecology (1). This enrichment may first occur by the selective outgrowth of microbes associated with insects living near antibiotic-containing agricultural animal excreta (e.g., microbes akin to P. papillae or E. casseliflavus). An increase in the abundance of vancomycin-resistance operons in the vicinity of animal-adapted enterococcal strains, especially in the presence of selection, increases the probability of movement of resistance into animal-adapted enterococci. Few studies have systematically examined insects or other nonmammalian ecologies in agricultural areas treated with avoparcin. However, within years of the initial reports of VRE infections, VRE were found to be common among the commensal flora of Europeans and their farm animals (4, 28), whereas they are rare among the commensal flora of farm animals in the United States (18, 108, 116, 128). Moreover, following the ban on the use of avoparcin in agriculture, there have been statistically significant decreases in the abundance of VRE associated with farm animals in Denmark (3) and France (53), strongly suggesting direct cause and effect between agricultural use of avoparcin and VRE occurrence.
The first outbreak of VRE infection began in November 1986 in Dulwich Hospital, London (122). Over the course of roughly a year, 55 enterococcal isolates were obtained from patients who had been treated typically with vancomycin every five to seven days, along with a cephalosporin. The VRE that emerged included both E. faecium and E. faecalis, indicating that this outbreak was not entirely clonal and that the vancomycin resistance was likely mobile. Nearly all strains of both species were reported to be resistant to both vancomycin and teicoplanin, a hallmark of the VanA phenotype (12, 122). Most vancomycin-resistant E. faecium in this outbreak were also ampicillin resistant (122) and exhibited other resistances indicative of well-adapted hospital strains. Two vancomycin-resistant E. faecium strains nearly simultaneously isolated in a French hospital (62) were also resistant to ampicillin and other drugs, suggesting that vancomycin resistance had entered the hospital-adapted enterococci. Around 1986, then, the VanA operon entered hospital-adapted lineages of enterococci in geographical areas where avoparcin had been used.
The first cases of VRE in the United States occurred beginning in February 1987 at Barnes Hospital in Saint Louis, Missouri (101). Although this was only months after the European cases, there were important differences. First, the first US VRE isolates were E. faecalis. They were resistant to multiple antibiotics (101) and were found to be prototypical CC2 hospital strains (74). However, these isolates displayed a different phenotype, being vancomycin resistant but teichoplanin susceptible (101). A similar vancomycin-resistant E. faecalis that exhibited teichoplanin susceptibility was nearly simultaneously isolated in France (133), and this phenotype was termed VanB (12). As with hospital-endemic strains of E. faecium, CC2 strains of E. faecalis are mainly associated with hospital environments (74), which suggests that shortly after the movement of VanA into the enterococci, likely in Europe, VanB moved into E. faecalis, possibly in the United States or in Europe.
In the United States, VanB E. faecalis did not ultimately emerge as the main problem. Even now, only about 10% of E. faecalis infections in the United States are vancomycin resistant (39), whereas vancomycin-resistant E. faecium, mainly of the VanA phenotype, has become endemic. In the United States, the VanA outbreak appears to have started in New York City in 1989 (35). This outbreak looked very much like the one that began almost three years earlier in London. Because of the London outbreak, an active surveillance program was in place in New York City, and no VRE were detected throughout 1988. The first VRE strain was not detected until September 1989, and by October 1991, 361 patients at 38 hospitals in the New York City area were identified with VRE infections (Figure 4b). The vast majority of strains were E. faecium;ofthe E. faecalis strains, most were of the VanA phenotype, although a few VanB strains were noted. One patient had both E. faecium and E. faecalis VRE, and by Southern blot it appeared that vancomycin resistance in both species was mediated by related elements, suggesting horizontal transfer within the patient (35). All E. faecium strains were resistant to ampicillin and other antibiotics, emblematic of establishment of the VanA operon in the highly-hospital-adapted CC17 lineage. This lineage rapidly spread to become a leading cause of hospital-acquired VRE infection throughout the United States in the 1990s, before spreading globally in the 2000s (12).
The question, then, is why did the first cases of VRE occur in Europe but not become an endemic problem there until the 2000s, whereas VRE immediately took off in the United States? One likely reason is the level of vancomycin used in the United States. In the mid-1990s, physicians in the United States used about 12 times more vancomycin per capita in hospitals than did their colleagues in the Netherlands (125). Furthermore, even within the comparatively small geographic area of Europe, clinical practices vary by country. As a result, rates of VRE bloodstream infection in Luxembourg are on average 10 times those of surrounding countries (12), indicating that differences in clinical practices, not geographic isolation, are now the main drivers of VRE incidence.
There was great concern of further spread of vancomycin resistance to S. aureus, because vancomycin is an important last-line therapy for methicillin-resistant S. aureus (MRSA) infections. Surprisingly, it took over 15 years for vancomycin resistance to transfer from enterococci into S. aureus in clinical isolates (13, 55, 126). However, likely because of the fitness cost (34), each case of vancomycin-resistant S. aureus appears to have arisen de novo in each patient in which it was identified (55), and there are no well-documented cases of patient-to-patient spread. However, just as rare events gave rise to well-adapted VRE, many additional mutations may not be required in S. aureus to mitigate the current fitness costs.
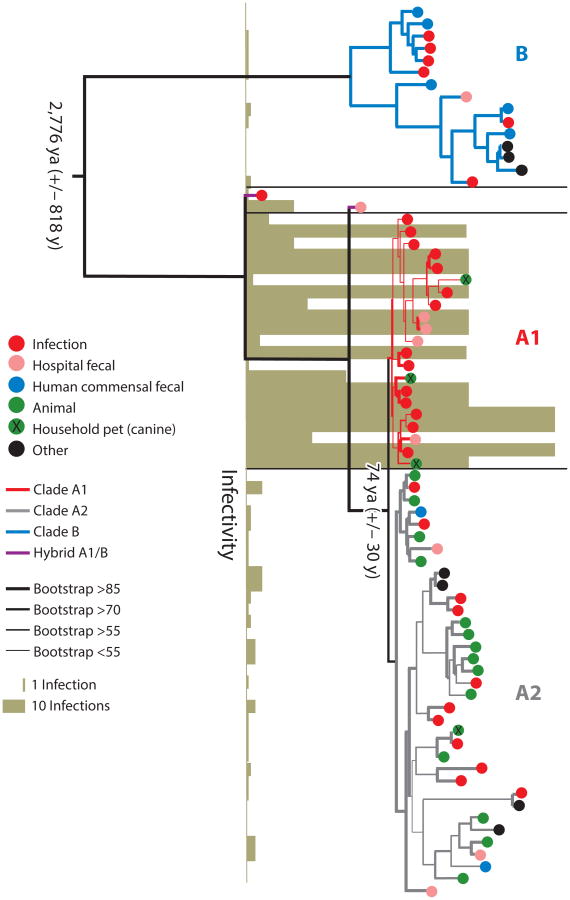
Recent comparative genomics studies have shown that the CC17 lineage of E. faecium is a hypermutator strain that arose from a clade mainly inhabiting the GI tracts of domestic animals roughly 75 years ago—exactly the time of the introduction of antibiotics into medical practice (Figure 5) (61). Interestingly, the main differences between the clade from which CC17 branched and human commensal microbes, in addition to the previously mentioned accumulation of antibiotic-resistance elements, pathogenicity islands, and other mobile elements, are differences in carbohydrate utilization pathways. Human commensal E. faecium strains appear to be well equipped to metabolize carbon derived from dietary sources. However, animal strains and the hospital-endemic CC17 lineage have traded many of these operons for pathways for metabolizing amino sugars of the type found on the surface of gut epithelial cells and in secretions such as mucin (54, 61, 135). In other words, human commensal strains appear to be exactly that—sharing at the table with the host. Animal and hospital E. faecium strains, however, appear to have evolved the ability to utilize host secretions and cell surface modifications as carbohydrate sources in a more parasitic relationship. The divergence between human and animal E. faecium strains appears to have started a little more than 3,000 years ago—a time of increasing urbanization of humans, increasing domestication of animals, and introduction of hygiene measures (61). Conceivably, forcing animals onto a restricted diet through domestication may have been an important driving force in the evolution of animal strains of E. faecium. Rather than deriving carbon from dietary sources that no longer existed because of dietary shifts imposed by domestication, they evolved to derive carbon from host secretions. Coincidentally, this would also be an asset in colonizing the GI tract of a hospitalized patient in the intensive care unit, where feeding may be by intravenous administration, with few sources of carbon other than host secretions in the GI tract. Experimental results are emerging that support this prospect (135).
Figure 5.
RAxML (113) single-nucleotide-polymorphism-based tree based on concatenated alignments of DNA sequences of 1,344 single-copy core genes in 73 Enterococcus faecium genomes. Bootstrapping was performed with 1,000 replicates. The origins of the strains and the dates for the split between the clades, estimated by BEAST analysis (29), are indicated. The infectivity score reflects the number of strains of a particular sequence type, in the MLST database, isolated from infection. Three clades are highlighted: B in blue, A1 in red, and A2 in grey. Figure is reproduced from Reference 61.
Conclusion
Since the discovery of the enterococci most attention has been paid to the anomalous biology of a small number of lineages that are associated with human infection. Relatively few studies have aimed to contribute to an understanding of the natural history and ecology of the enterococci. Understanding the forces that have shaped the evolution of the enterococci over aeons will enable us to learn how these resilient bacteria have adapted to the unique conditions of the hospital environment in the antibiotic era.
Acknowledgments
The authors thank the members of the Gilmore lab, and in particular François Lebreton, for helpful discussions throughout the preparation of this review. Production of this work was made possible by support from Public Health Service Grants AI083214, AI108710, and AI072360. D.V.T. is supported by Grant EY007145 from the National Institutes of Health.
Glossary
- Devonian period
a geologic period spanning approximately 420–360 mya
- Coccus
a spherical bacterium
- Hypertonic
an environment with a higher concentration of solutes outside the cell than inside the cell, which causes water to flow out of the cell by osmosis
- Hypotonic
an environment with a higher concentration of solutes inside the cell than outside the cell, which causes water to flow into the cell by osmosis
- Protozoa
single-celled eukaryotic organisms
- Bacteriophages
viruses that infect and replicate within bacteria
- Phylogenetic tree
a branching diagram that shows the evolutionary relationships between species, based on differences in their genetic characteristics
- Endocarditis
infection caused by inflammation of the heart lining and heart valves
- Streptomycetes
actinobacteria found predominantly in soil and decaying vegetation that produce the majority of clinically useful antibiotics of natural origin
- Avoparcin
a glycopeptide antibiotic used in agriculture that shares chemical similarity with vancomycin
Footnotes
Disclosure Statement: The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
Contributor Information
Daria Van Tyne, Email: vantyne@fas.harvard.edu.
Michael S. Gilmore, Email: michael_gilmore@meei.harvard.edu.
Literature Cited
- 1.Acar J, Casewell M, Freeman J, Friis C, Goossens H. Avoparcin and virginiamycin as animal growth promoters: a plea for science in decision-making. Clin Microbiol Infect. 2000;6:477–82. doi: 10.1046/j.1469-0691.2000.00128.x. [DOI] [PubMed] [Google Scholar]
- 2.Arias CA, Murray BE. The rise of the Enterococcus: beyond vancomycin resistance. Nat Rev Microbiol. 2012;10:266–78. doi: 10.1038/nrmicro2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bager F, Aarestrup FM, Madsen M, Wegener HC. Glycopeptide resistance in Enterococcus faecium from broilers and pigs following discontinued use of avoparcin. Microb Drug Resist. 1999;5:53–56. doi: 10.1089/mdr.1999.5.53. [DOI] [PubMed] [Google Scholar]
- 4.Bates J. Epidemiology of vancomycin-resistant enterococci in the community and the relevance of farm animals to human infection. J Hosp Infect. 1997;37:89–101. doi: 10.1016/s0195-6701(97)90179-1. [DOI] [PubMed] [Google Scholar]
- 5.Benveniste R, Davies J. Aminoglycoside antibiotic-inactivating enzymes in actinomycetes similar to those present in clinical isolates of antibiotic-resistant bacteria. Proc Natl Acad Sci USA. 1973;70:2276–80. doi: 10.1073/pnas.70.8.2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bleiweis AS, Zimmerman LN. Properties of proteinase from Streptococcus faecalis var. Liquefaciens. J Bacteriol. 1964;88:653–59. doi: 10.1128/jb.88.3.653-659.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brock TD, Peacher B, Pierson D. Survey of the bacteriocines of enterococci. J Bacteriol. 1963;86:702–7. doi: 10.1128/jb.86.4.702-707.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Broderick NA, Raffa KF, Goodman RM, Handelsman J. Census of the bacterial community of the gypsy moth larval midgut by using culturing and culture-independent methods. Appl Environ Microbiol. 2004;70:293–300. doi: 10.1128/AEM.70.1.293-300.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown SP, Cornforth DM, Mideo N. Evolution of virulence in opportunistic pathogens: generalism, plasticity, and control. Trends Microbiol. 2012;20:336–42. doi: 10.1016/j.tim.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Byappanahalli MN, Nevers MB, Korajkic A, Staley ZR, Harwood VJ. Enterococci in the environment. Microbiol Mol Biol Rev. 2012;76:685–706. doi: 10.1128/MMBR.00023-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Casadevall A, Pirofski L. Host-pathogen interactions: the attributes of virulence. J Infect Dis. 2001;184:337–44. doi: 10.1086/322044. [DOI] [PubMed] [Google Scholar]
- 12.Cattoir V, Leclercq R. Twenty-five years of shared life with vancomycin-resistant enterococci: Is it time to divorce? J Antimicrob Chemother. 2013;68:731–42. doi: 10.1093/jac/dks469. [DOI] [PubMed] [Google Scholar]
- 13.Chang S, Sievert DM, Hageman JC, Boulton ML, Tenover FC, et al. Infection with vancomycin-resistant Staphylococcus aureus containing the vanA resistance gene. N Engl J Med. 2003;348:1342–47. doi: 10.1056/NEJMoa025025. [DOI] [PubMed] [Google Scholar]
- 14.Chow JW, Thal LA, Perri MB, Vazquez JA, Donabedian SM, et al. Plasmid-associated hemolysin and aggregation substance production contribute to virulence in experimental enterococcal endocarditis. Antimicrob Agents Chemother. 1993;37:2474–77. doi: 10.1128/aac.37.11.2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chuang ON, Schlievert PM, Wells CL, Manias DA, Tripp TJ, Dunny GM. Multiple functional domains of Enterococcus faecalis aggregation substance Asc10 contribute to endocarditis virulence. Infect Immun. 2009;77:539–48. doi: 10.1128/IAI.01034-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chuang-Smith ON, Wells CL, Henry-Stanley MJ, Dunny GM. Acceleration of Enterococcus faecalis biofilm formation by aggregation substance expression in an ex vivo model of cardiac valve colonization. PLoS ONE. 2010;5:e15798. doi: 10.1371/journal.pone.0015798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clewell DB, Weaver KE. Sex pheromones and plasmid transfer in Enterococcus faecalis. Plasmid. 1989;21:175–84. doi: 10.1016/0147-619x(89)90041-3. [DOI] [PubMed] [Google Scholar]
- 18.Coque TM, Tomayko JF, Ricke SC, Okhyusen PC, Murray BE. Vancomycin-resistant enterococci from nosocomial, community, and animal sources in the United States. Antimicrob Agents Chemother. 1996;40:2605–9. doi: 10.1128/aac.40.11.2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cox CR, Coburn PS, Gilmore MS. Enterococcal cytolysin: a novel two component peptide system that serves as a bacterial defense against eukaryotic and prokaryotic cells. Curr Protein Pept Sci. 2005;6:77–84. doi: 10.2174/1389203053027557. [DOI] [PubMed] [Google Scholar]
- 20.Cox CR, Gilmore MS. Native microbial colonization of Drosophila melanogaster and its use as a model of Enterococcus faecalis pathogenesis. Infect Immun. 2007;75:1565–76. doi: 10.1128/IAI.01496-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Creti R, Imperi M, Bertuccini L, Fabretti F, Orefici G, et al. Survey for virulence determinants among Enterococcus faecalis isolated from different sources. J Med Microbiol. 2004;53:13–20. doi: 10.1099/jmm.0.05353-0. [DOI] [PubMed] [Google Scholar]
- 22.David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2013;505:559–63. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davies J. What are antibiotics? Archaic functions for modern activities. Mol Microbiol. 1990;4:1227–32. doi: 10.1111/j.1365-2958.1990.tb00701.x. [DOI] [PubMed] [Google Scholar]
- 24.Davies J, Davies D. Origins and evolution of antibiotic resistance. Microbiol Mol Biol Rev. 2010;74:417–33. doi: 10.1128/MMBR.00016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.D'Costa VM, King CE, Kalan L, Morar M, Sung WW, et al. Antibiotic resistance is ancient. Nature. 2011;477:457–61. doi: 10.1038/nature10388. [DOI] [PubMed] [Google Scholar]
- 26.D'Costa VM, McGrann KM, Hughes DW, Wright GD. Sampling the antibiotic resistome. Science. 2006;311:374–77. doi: 10.1126/science.1120800. [DOI] [PubMed] [Google Scholar]
- 27.Desmarais TR, Solo-Gabriele HM, Palmer CJ. Influence of soil on fecal indicator organisms in a tidally influenced subtropical environment. Appl Environ Microbiol. 2002;68:1165–72. doi: 10.1128/AEM.68.3.1165-1172.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Devriese LA, Ieven M, Goossens H, Vandamme P, Pot B, et al. Presence of vancomycin-resistant enterococci in farm and pet animals. Antimicrob Agents Chemother. 1996;40:2285–87. doi: 10.1128/aac.40.10.2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 2012;29:1969–73. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dunny GM. Genetic functions and cell-cell interactions in the pheromone-inducible plasmid transfer system of Enterococcus faecalis. Mol Microbiol. 1990;4:689–96. doi: 10.1111/j.1365-2958.1990.tb00639.x. [DOI] [PubMed] [Google Scholar]
- 31.Dunny GM, Leonard BA, Hedberg PJ. Pheromone-inducible conjugation in Enterococcus faecalis: interbacterial and host-parasite chemical communication. J Bacteriol. 1995;177:871–76. doi: 10.1128/jb.177.4.871-876.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Edwards EJ, Osborne CP, Stromberg CA, Smith SA, Bond WJ, et al. The origins of C4 grasslands: integrating evolutionary and ecosystem science. Science. 2010;328:587–91. doi: 10.1126/science.1177216. [DOI] [PubMed] [Google Scholar]
- 33.Fisher K, Phillips C. The ecology, epidemiology and virulence of Enterococcus. Microbiology. 2009;155:1749–57. doi: 10.1099/mic.0.026385-0. [DOI] [PubMed] [Google Scholar]
- 34.Foucault ML, Courvalin P, Grillot-Courvalin C. Fitness cost of VanA-type vancomycin resistance in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2009;53:2354–59. doi: 10.1128/AAC.01702-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Frieden TR, Munsiff SS, Low DE, Willey BM, Williams G, et al. Emergence of vancomycin-resistant enterococci in New York City. Lancet. 1993;342:76–79. doi: 10.1016/0140-6736(93)91285-t. [DOI] [PubMed] [Google Scholar]
- 36.Garsin DA, Sifri CD, Mylonakis E, Qin X, Singh KV, et al. A simple model host for identifying gram-positive virulence factors. Proc Natl Acad Sci USA. 2001;98:10892–97. doi: 10.1073/pnas.191378698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gill SR, Pop M, Deboy RT, Eckburg PB, Turnbaugh PJ, et al. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355–59. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gilmore MS. The molecular basis of antibiotic resistance: where Newton meets Darwin. Int J Med Microbiol. 2002;292:65. doi: 10.1016/s1438-4221(04)70085-5. [DOI] [PubMed] [Google Scholar]
- 39.Gilmore MS, Lebreton F, van Schaik W. Genomic transition of enterococci from gut commensals to leading causes of multidrug-resistant hospital infection in the antibiotic era. Curr Opin Microbiol. 2013;16:10–16. doi: 10.1016/j.mib.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Giraffa G. Functionality of enterococci in dairy products. Int J Food Microbiol. 2003;88:215–22. doi: 10.1016/s0168-1605(03)00183-1. [DOI] [PubMed] [Google Scholar]
- 41.Greenfield TJ, Ehli E, Kirshenmann T, Franch T, Gerdes K, Weaver KE. The antisense RNA of the par locus of pAD1 regulates the expression of a 33-amino-acid toxic peptide by an unusual mechanism. Mol Microbiol. 2000;37:652–60. doi: 10.1046/j.1365-2958.2000.02035.x. [DOI] [PubMed] [Google Scholar]
- 42.Hancock LE, Gilmore MS. The capsular polysaccharide of Enterococcus faecalis and its relationship to other polysaccharides in the cell wall. Proc Natl Acad Sci USA. 2002;99:1574–79. doi: 10.1073/pnas.032448299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hartke A, Giard JC, Laplace JM, Auffray Y. Survival of Enterococcus faecalis in an oligotrophic microcosm: changes in morphology, development of general stress resistance, and analysis of protein synthesis. Appl Environ Microbiol. 1998;64:4238–45. doi: 10.1128/aem.64.11.4238-4245.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hartke A, Lemarinier S, Pichereau V, Auffray Y. Survival of Enterococcus faecalis in seawater microcosms is limited in the presence of bacterivorous zooflagellates. Curr Microbiol. 2002;44:329–35. doi: 10.1007/s00284-001-0018-4. [DOI] [PubMed] [Google Scholar]
- 45.Ike Y, Clewell DB. Genetic analysis of the pAD1 pheromone response in Streptococcus faecalis, using transposon Tn917 as an insertional mutagen. J Bacteriol. 1984;158:777–83. doi: 10.1128/jb.158.3.777-783.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ike Y, Hashimoto H, Clewell DB. Hemolysin of Streptococcus faecalis subspecies zymogenes contributes to virulence in mice. Infect Immun. 1984;45:528–30. doi: 10.1128/iai.45.2.528-530.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ike Y, Hashimoto H, Clewell DB. High incidence of hemolysin production by Enterococcus (Streptococcus) faecalis strains associated with human parenteral infections. J Clin Microbiol. 1987;25:1524–28. doi: 10.1128/jcm.25.8.1524-1528.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Inst. Med. Antibiotic Resistance: Implications for Global Health and Novel Intervention Strategies: Workshop Summary. Washington, DC: Natl. Acad; 2010. [PubMed] [Google Scholar]
- 49.Jett BD, Huycke MM, Gilmore MS. Virulence of enterococci. Clin Microbiol Rev. 1994;7:462–78. doi: 10.1128/cmr.7.4.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jett BD, Jensen HG, Nordquist RE, Gilmore MS. Contribution of the pAD1-encoded cytolysin to the severity of experimental Enterococcus faecalis endophthalmitis. Infect Immun. 1992;60:2445–52. doi: 10.1128/iai.60.6.2445-2452.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jt. Expert Advis. Comm. Antibiot. Resist. (JETACAR) The Use of Antibiotics in Food-Producing Animals: Antibiotic-Resistant Bacteria in Animals and Humans. Canberra, Aust.: Commonw. Aust; 1999. [Google Scholar]
- 52.Kaplan H, Hill K, Lancaster J, Hurtado AM. A theory of human life history evolution: diet, intelligence, and longevity. Evol Anthropol. 2000;9:156–85. [Google Scholar]
- 53.Kempf I, Hellard G, Perrin-Guyomard A, Gicquel-Bruneau M, Sanders P, Leclercq R. Prevalence of high-level vancomycin-resistant enterococci in French broilers and pigs. Int J Antimicrob Agents. 2008;32:463–64. doi: 10.1016/j.ijantimicag.2008.04.029. [DOI] [PubMed] [Google Scholar]
- 54.Kim EB, Marco ML. Nonclinical and clinical Enterococcus faecium strains, but not Enterococcus faecalis strains, have distinct structural and functional genomic features. Appl Environ Microbiol. 2014;80:154–65. doi: 10.1128/AEM.03108-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kos VN, Desjardins CA, Griggs A, Cerqueira G, van Tonder A, et al. Comparative genomics of vancomycin-resistant Staphylococcus aureus strains and their positions within the clade most commonly associated with methicillin-resistant S. aureus hospital-acquired infection in the United States. mBio. 2012;3:e00112. doi: 10.1128/mBio.00112-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kreft B, Marre R, Schramm U, Wirth R. Aggregation substance of Enterococcus faecalis mediates adhesion to cultured renal tubular cells. Infect Immun. 1992;60:25–30. doi: 10.1128/iai.60.1.25-30.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kubinak JL, Potts WK. Host resistance influences patterns of experimental viral adaptation and virulence evolution. Virulence. 2013;4:410–18. doi: 10.4161/viru.24724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lawley TD, Walker AW. Intestinal colonization resistance. Immunology. 2013;138:1–11. doi: 10.1111/j.1365-2567.2012.03616.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Leavis H, Top J, Shankar N, Borgen K, Bonten M, et al. A novel putative enterococcal pathogenicity island linked to the esp virulence gene of Enterococcus faecium and associated with epidemicity. J Bacteriol. 2004;186:672–82. doi: 10.1128/JB.186.3.672-682.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Leavis HL, Bonten MJ, Willems RJ. Identification of high-risk enterococcal clonal complexes: global dispersion and antibiotic resistance. Curr Opin Microbiol. 2006;9:454–60. doi: 10.1016/j.mib.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 61.Lebreton F, van Schaik W, McGuire AM, Godfrey P, Griggs A, et al. Emergence of epidemic multidrug-resistant Enterococcus faecium from animal and commensal strains. mBio. 2013;4:e00534. doi: 10.1128/mBio.00534-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Leclercq R, Derlot E, Duval J, Courvalin P. Plasmid-mediated resistance to vancomycin and teicoplanin in Enterococcus faecium. N Engl J Med. 1988;319:157–61. doi: 10.1056/NEJM198807213190307. [DOI] [PubMed] [Google Scholar]
- 63.Levy SB. The Antibiotic Paradox: How the Misuse of Antibiotics Destroys Their Curative Power. Cambridge, MA: Perseus; 2002. [Google Scholar]
- 64.Levy SB, Marshall B. Antibacterial resistance worldwide: causes, challenges and responses. Nat Med. 2004;10:S122–29. doi: 10.1038/nm1145. [DOI] [PubMed] [Google Scholar]
- 65.Ludwig W, Seewaldt E, Kilpper-Balz R, Schleifer KH, Magrum L, et al. The phylogenetic position of Streptococcus and Enterococcus. J Gen Microbiol. 1985;131:543–51. doi: 10.1099/00221287-131-3-543. [DOI] [PubMed] [Google Scholar]
- 66.MacCallum WG, Hastings TW. A case of acute endocarditis caused by Micrococcus zymogenes (Nov. spec.), with a description of the microorganism. J Exp Med. 1899;4:521–34. doi: 10.1084/jem.4.5-6.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Makinen PL, Clewell DB, An F, Makinen KK. Purification and substrate specificity of a strongly hydrophobic extracellular metalloendopeptidase (“gelatinase”) from Streptococcus faecalis (strain 0G1-10) J Biol Chem. 1989;264:3325–34. [PubMed] [Google Scholar]
- 68.Manson JM, Hancock LE, Gilmore MS. Mechanism of chromosomal transfer of Enterococcus faecalis pathogenicity island, capsule, antimicrobial resistance, and other traits. Proc Natl Acad Sci USA. 2010;107:12269–74. doi: 10.1073/pnas.1000139107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Manson JM, Rauch M, Gilmore MS. The commensal microbiology of the gastrointestinal tract. Adv Exp Med Biol. 2008;635:15–28. doi: 10.1007/978-0-387-09550-9_2. [DOI] [PubMed] [Google Scholar]
- 70.Maraccini PA, Ferguson DM, Boehm AB. Diurnal variation in Enterococcus species composition in polluted ocean water and a potential role for the enterococcal carotenoid in protection against photoin-activation. Appl Environ Microbiol. 2012;78:305–10. doi: 10.1128/AEM.06821-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Martin JD, Mundt JO. Enterococci in insects. Appl Microbiol. 1972;24:575–80. doi: 10.1128/am.24.4.575-580.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mason KL, Stepien TA, Blum JE, Holt JF, Labbe NH, et al. From commensal to pathogen: translocation of Enterococcus faecalis from the midgut to the hemocoel of Manduca sexta. mBio. 2011;2:e00065–11. doi: 10.1128/mBio.00065-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mayr E. Ecological factors in speciation. Evolution. 1947;1:263–88. [Google Scholar]
- 74.McBride SM, Fischetti VA, Leblanc DJ, Moellering RC, Jr, Gilmore MS. Genetic diversity among Enterococcus faecalis. PLoS ONE. 2007;2:e582. doi: 10.1371/journal.pone.0000582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.McGavin MH, Krajewska-Pietrasik D, Rydén C, Höök M. Identification of a Staphylococcus aureus extracellular matrix-binding protein with broad specificity. Infect Immun. 1993;61:2479–85. doi: 10.1128/iai.61.6.2479-2485.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Michaux C, Martini C, Shioya K, Ahmed Lecheheb S, Budin-Verneuil A, et al. CspR, a cold shock RNA-binding protein involved in the long-term survival and the virulence of Enterococcus faecalis. J Bacteriol. 2012;194:6900–8. doi: 10.1128/JB.01673-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mundt JO. Occurrence of enterococci in animals in a wild environment. Appl Microbiol. 1963;11:136–40. doi: 10.1128/am.11.2.136-140.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mundt JO. Occurrence of enterococci on plants in a wild environment. Appl Microbiol. 1963;11:141–44. doi: 10.1128/am.11.2.141-144.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mundy LM, Sahm DF, Gilmore M. Relationships between enterococcal virulence and antimicrobial resistance. Clin Microbiol Rev. 2000;13:513–22. doi: 10.1128/cmr.13.4.513-522.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Murray BE. The life and times of the enterococcus. Clin Microbiol Rev. 1990;3:46–65. doi: 10.1128/cmr.3.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Murray BE. Diversity among multidrug-resistant enterococci. Emerg Infect Dis. 1998;4:37–47. doi: 10.3201/eid0401.980106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nallapareddy SR, Qin X, Weinstock GM, Hook M, Murray BE. Enterococcus faecalis adhesin, Ace, mediates attachment to extracellular matrix proteins collagen type IV and laminin as well as collagen type I. Infect Immun. 2000;68:5218–24. doi: 10.1128/iai.68.9.5218-5224.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nallapareddy SR, Singh KV, Duh RW, Weinstock GM, Murray BE. Diversity of ace, a gene encoding a microbial surface component recognizing adhesive matrix molecules, from different strains of Enterococcus faecalis and evidence for production of Ace during human infections. Infect Immun. 2000;68:5210–7. doi: 10.1128/iai.68.9.5210-5217.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Niven CF, Sherman JM. Nutrition of the enterococci. J Bacteriol. 1944;47:335–42. doi: 10.1128/jb.47.4.335-342.1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Noble CJ. Carriage of group D streptococci in the human bowel. J Clin Pathol. 1978;31:1182–86. doi: 10.1136/jcp.31.12.1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Olmsted SB, Dunny GM, Erlandsen SL, Wells CL. A plasmid-encoded surface protein on Enterococcus faecalis augments its internalization by cultured intestinal epithelial cells. J Infect Dis. 1994;170:1549–56. doi: 10.1093/infdis/170.6.1549. [DOI] [PubMed] [Google Scholar]
- 87.Palmer KL, Gilmore MS. Multidrug-resistant enterococci lack CRISPR-cas. mBio. 2010;1:e00227–10. doi: 10.1128/mBio.00227-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Palmer KL, Godfrey P, Griggs A, Kos VN, Zucker J, et al. Comparative genomics of enterococci: variation in Enterococcus faecalis, clade structure in E. faecium, and defining characteristics of E. gallinarum and E. casseliflavus. mBio. 2012;3:e00318–11. doi: 10.1128/mBio.00318-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Palmer KL, Kos VN, Gilmore MS. Horizontal gene transfer and the genomics of enterococcal antibiotic resistance. Curr Opin Microbiol. 2010;13:632–39. doi: 10.1016/j.mib.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Parte AC. LPSN—list of prokaryotic names with standing in nomenclature. Nucleic Acids Res. 2014;42:D613–16. doi: 10.1093/nar/gkt1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Paulsen IT, Banerjei L, Myers GS, Nelson KE, Seshadri R, et al. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science. 2003;299:2071–74. doi: 10.1126/science.1080613. [DOI] [PubMed] [Google Scholar]
- 92.Pinkston KL, Gao P, Diaz-Garcia D, Sillanpää J, Nallapareddy SR, et al. The Fsr quorum-sensing system of Enterococcus faecalis modulates surface display of the collagen-binding MSCRAMM Ace through regulation of gelE. J Bacteriol. 2011;193:4317–25. doi: 10.1128/JB.05026-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Purnell SE, Ebdon JE, Taylor HD. Bacteriophage lysis of Enterococcus host strains: a tool for microbial source tracking? Environ Sci Technol. 2011;45:10699–705. doi: 10.1021/es202141x. [DOI] [PubMed] [Google Scholar]
- 94.Rana NF, Sauvageot N, Laplace JM, Bao Y, Nes I, et al. Redox balance via lactate dehydrogenase is important for multiple stress resistance and virulence in Enterococcus faecalis. Infect Immun. 2013;81:2662–68. doi: 10.1128/IAI.01299-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rendu W, Beauval C, Crevecoeur I, Bayle P, Balzeau A, et al. Evidence supporting an intentional Neandertal burial at La Chapelle-aux-Saints. Proc Natl Acad Sci USA. 2013;111:81–86. doi: 10.1073/pnas.1316780110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Rich RL, Kreikemeyer B, Owens RT, LaBrenz S, Narayana SV, et al. Ace is a collagen-binding MSCRAMM from Enterococcus faecalis. J Biol Chem. 1999;274:26939–45. doi: 10.1074/jbc.274.38.26939. [DOI] [PubMed] [Google Scholar]
- 97.Rigottier-Gois L, Alberti A, Houel A, Taly JF, Palcy P, et al. Large-scale screening of a targeted Enterococcus faecalis mutant library identifies envelope fitness factors. PLoS ONE. 2011;6:e29023. doi: 10.1371/journal.pone.0029023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rippere K, Patel R, Uhl JR, Piper KE, Steckelberg JM, et al. DNA sequence resembling vanA and vanB in the vancomycin-resistant biopesticide Bacillus popilliae. J Infect Dis. 1998;178:584–88. doi: 10.1086/517480. [DOI] [PubMed] [Google Scholar]
- 99.Roux A, Payne SM, Gilmore MS. Microbial telesensing: probing the environment for friends, foes, and food. Cell Host Microbe. 2009;6:115–24. doi: 10.1016/j.chom.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ruiz-Garbajosa P, Bonten MJ, Robinson DA, Top J, Nallapareddy SR, et al. Multilocus sequence typing scheme for Enterococcus faecalis reveals hospital-adapted genetic complexes in a background of high rates of recombination. J Clin Microbiol. 2006;44:2220–28. doi: 10.1128/JCM.02596-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sahm DF, Kissinger J, Gilmore MS, Murray PR, Mulder R, et al. In vitro susceptibility studies of vancomycin-resistant Enterococcus faecalis. Antimicrob Agents Chemother. 1989;33:1588–91. doi: 10.1128/aac.33.9.1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Schleifer KH, Kilpper-Balz R. Transfer of Streptococcus faecalis and Streptococcus faecium to the genus Enterococcus nom. rev. as Enterococcus faecalis comb. nov. and Enterococcus faecium comb. nov. Int J Syst Bacteriol. 1984;34:31–34. [Google Scholar]
- 103.Seipke RF, Kaltenpoth M, Hutchings MI. Streptomyces as symbionts: an emerging and widespread theme? FEMS Microbiol Rev. 2012;36:862–76. doi: 10.1111/j.1574-6976.2011.00313.x. [DOI] [PubMed] [Google Scholar]
- 104.Shankar N, Baghdayan AS, Gilmore MS. Modulation of virulence within a pathogenicity island in vancomycin-resistant Enterococcus faecalis. Nature. 2002;417:746–50. doi: 10.1038/nature00802. [DOI] [PubMed] [Google Scholar]
- 105.Shankar V, Baghdayan AS, Huycke MM, Lindahl G, Gilmore MS. Infection-derived Enterococcus faecalis strains are enriched in esp, a gene encoding a novel surface protein. Infect Immun. 1999;67:193–200. doi: 10.1128/iai.67.1.193-200.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sherman JM. The streptococci. Bacteriol Rev. 1937;1:3–97. doi: 10.1128/br.1.1.3-97.1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sherman JM. The enterococci and related streptococci. J Bacteriol. 1938;35:81–93. doi: 10.1128/jb.35.2.81-93.1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Silverman J, Thal LA, Perri MB, Bostic G, Zervos MJ. Epidemiologic evaluation of antimicrobial resistance in community-acquired enterococci. J Clin Microbiol. 1998;36:830–32. doi: 10.1128/jcm.36.3.830-832.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Singh KV, Nallapareddy SR, Sillanpää J, Murray BE. Importance of the collagen adhesin Ace in pathogenesis and protection against Enterococcus faecalis experimental endocarditis. PLoS Pathog. 2010;6:e1000716. doi: 10.1371/journal.ppat.1000716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sinton LW, Braithwaite RR, Hall CH, Mackenzie ML. Survival of indicator and pathogenic bacteria in bovine feces on pasture. Appl Environ Microbiol. 2007;73:7917–25. doi: 10.1128/AEM.01620-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Sinton LW, Hall CH, Lynch PA, Davies-Colley RJ. Sunlight inactivation of fecal indicator bacteria and bacteriophages from waste stabilization pond effluent in fresh and saline waters. Appl Environ Microbiol. 2002;68:1122–31. doi: 10.1128/AEM.68.3.1122-1131.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Solheim M, Brekke MC, Snipen LG, Willems RJ, Nes IF, Brede DA. Comparative genomic analysis reveals significant enrichment of mobile genetic elements and genes encoding surface structure-proteins in hospital-associated clonal complex 2 Enterococcus faecalis. BMC Microbiol. 2011;11:3. doi: 10.1186/1471-2180-11-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–90. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 114.Sussmuth SD, Muscholl-Silberhorn A, Wirth R, Susa M, Marre R, Rozdzinski E. Aggregation substance promotes adherence, phagocytosis, and intracellular survival of Enterococcus faecalis within human macrophages and suppresses respiratory burst. Infect Immun. 2000;68:4900–6. doi: 10.1128/iai.68.9.4900-4906.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Tang W, van der Donk WA. The sequence of the enterococcal cytolysin imparts unusual lanthio-nine stereochemistry. Nat Chem Biol. 2013;9:157–59. doi: 10.1038/nchembio.1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Thal LA, Chow JW, Mahayni R, Bonilla H, Perri MB, et al. Characterization of antimicrobial resistance in enterococci of animal origin. Antimicrob Agents Chemother. 1995;39:2112–15. doi: 10.1128/aac.39.9.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Thiercelin ME. Sur un diplocoque saprophyte de l'intestin susceptible de devenir pathogen. C R Soc Biol. 1899;5:269–71. [Google Scholar]
- 118.Thrall PH, Burdon JJ. Evolution of virulence in a plant host-pathogen metapopulation. Science. 2003;299:1735–37. doi: 10.1126/science.1080070. [DOI] [PubMed] [Google Scholar]
- 119.Todd EW. A comparative serological study of streptolysins derived from human and from animal infections, with notes on pneumococcal hemolysin, tetanolysin, and staphylococcus toxin. J Pathol Bacteriol. 1934;39:299–321. [Google Scholar]
- 120.Toledo-Arana A, Valle J, Solano C, Arrizubieta MJ, Cucarella C, et al. The enterococcal surface protein, Esp, is involved in Enterococcus faecalis biofilm formation. Appl Environ Microbiol. 2001;67:4538–45. doi: 10.1128/AEM.67.10.4538-4545.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Tung H, Guss B, Hellman U, Persson L, Rubin K, Ryden C. A bone sialoprotein-binding protein from Staphylococcus aureus: a member of the staphylococcal Sdr family. Biochem J. 2000;345(Part 3):611–19. [PMC free article] [PubMed] [Google Scholar]
- 122.Uttley AH, Collins CH, Naidoo J, George RC. Vancomycin-resistant enterococci. Lancet. 1988;331:57–58. doi: 10.1016/s0140-6736(88)91037-9. [DOI] [PubMed] [Google Scholar]
- 123.Van Tyne D, Martin MJ, Gilmore MS. Structure, function, and biology of the Enterococcus faecalis cytolysin. Toxins. 2013;5:895–911. doi: 10.3390/toxins5050895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Vollaard EJ, Clasener HA. Colonization resistance. Antimicrob Agents Chemother. 1994;38:409–14. doi: 10.1128/aac.38.3.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Wegener HC. Historical yearly usage of glycopeptides for animals and humans: the American-European paradox revisited. Antimicrob Agents Chemother. 1998;42:3049. doi: 10.1128/aac.42.11.3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Weigel LM, Clewell DB, Gill SR, Clark NC, McDougal LK, et al. Genetic analysis of a high-level vancomycin-resistant isolate of Staphylococcus aureus. Science. 2003;302:1569–71. doi: 10.1126/science.1090956. [DOI] [PubMed] [Google Scholar]
- 127.Wells CL, Moore EA, Hoag JA, Hirt H, Dunny GM, Erlandsen SL. Inducible expression of Enterococcus faecalis aggregation substance surface protein facilitates bacterial internalization by cultured enterocytes. Infect Immun. 2000;68:7190–94. doi: 10.1128/iai.68.12.7190-7194.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Welton LA, Thal LA, Perri MB, Donabedian S, McMahon J, et al. Antimicrobial resistance in enterococci isolated from Turkey flocks fed virginiamycin. Antimicrob Agents Chemother. 1998;42:705–8. doi: 10.1128/aac.42.3.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Werner G, Coque TM, Hammerum AM, Hope R, Hryniewicz W, et al. Emergence and spread of vancomycin resistance among enterococci in Europe. Euro Surveill. 2008;13:19046. [PubMed] [Google Scholar]
- 130.Whitman WB, Coleman DC, Wiebe WJ. Prokaryotes: the unseen majority. Proc Natl Acad Sci USA. 1998;95:6578–83. doi: 10.1073/pnas.95.12.6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Willems RJ, Homan W, Top J, van Santen-Verheuvel M, Tribe D, et al. Variant esp gene as a marker of a distinct genetic lineage of vancomycin-resistant Enterococcus faecium spreading in hospitals. Lancet. 2001;357:853–55. doi: 10.1016/S0140-6736(00)04205-7. [DOI] [PubMed] [Google Scholar]
- 132.Willey JM, van der Donk WA. Lantibiotics: peptides of diverse structure and function. Annu Rev Microbiol. 2007;61:477–501. doi: 10.1146/annurev.micro.61.080706.093501. [DOI] [PubMed] [Google Scholar]
- 133.Williamson R, al-Obeid S, Shlaes JH, Goldstein FW, Shlaes DM. Inducible resistance to vancomycin in Enterococcus faecium D366. J Infect Dis. 1989;159:1095–104. doi: 10.1093/infdis/159.6.1095. [DOI] [PubMed] [Google Scholar]
- 134.Yamahara KM, Walters SP, Boehm AB. Growth of enterococci in unaltered, unseeded beach sands subjected to tidal wetting. Appl Environ Microbiol. 2009;75:1517–24. doi: 10.1128/AEM.02278-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Zhang X, Top J, de Been M, Bierschenk D, Rogers M, et al. Identification of a genetic determinant in clinical Enterococcus faecium strains that contributes to intestinal colonization during antibiotic treatment. J Infect Dis. 2013;207:1780–86. doi: 10.1093/infdis/jit076. [DOI] [PubMed] [Google Scholar]