Abstract
Objective
Gastric cancer (GC) remains difficult to cure due to heterogeneity in a clinical challenge and the molecular mechanisms underlying this disease are complex and not completely understood. Accumulating evidence suggests that microRNAs (miRNAs) play an important role in GC, but the role of specific-miRNAs involved in this disease remains elusive. We performed next generation sequencing (NGS) based whole-transcriptome profiling to discover GC-specific miRNAs, followed by functional validation of results.
Design
NGS-based miRNA profiles were generated in matched pairs of GCs and adjacent normal mucosa (NM). Quantitative RT-PCR validation of miR-29c expression was performed in 274 gastric tissues, which included 2 cohorts of matched GC and NM specimens. Functional validation of miR-29c and its gene targets was undertaken in cell lines, as well as K19-C2mE and K19-Wnt1/C2mE transgenic mice.
Results
NGS analysis revealed four GC-specific miRNAs. Among these, miR-29c expression was significantly decreased in GC vs. NM tissues (P<0.001). Ectopic expression of miR-29c mimics in GC cell lines resulted in reduced proliferation, adhesion, invasion, and migration. High miR-29c expression suppressed xenograft tumor growth in nude mice. Direct interaction between miR-29c and its newly discovered target, ITGB1, was identified in cell lines and transgenic mice. MiR-29c expression demonstrated a step-wise decrease in wild type-hyperplasia-dysplasia cascade, in transgenic mice models of GC.
Conclusions
MiR-29c acts as a tumor suppressor in GC by directly targeting ITGB1. Loss of miR-29c expression is an early event in the initiation of gastric carcinogenesis, and may serve as a diagnostic and therapeutic biomarker for patients with GC.
Keywords: Gastric cancer, next-generation sequencing, miR-29c, ITGB1, transgenic mice
INTRODUCTION
Gastric cancer (GC) is the second leading cause of cancer-related death worldwide.1 Although clinical outcome of GC has gradually improved through earlier diagnosis, surgical resection, and chemotherapy, 5-year survival rates of patients with GC are only 20-30%.2 GC is a biologically heterogeneous disease that evolves in the background of various genetic and epigenetic alterations. Therefore, it is essential to have a more comprehensive understanding of molecular variables that affect GC disease pathways, in order to develop appropriate approaches for its diagnosis and treatment.
Although a number of molecular drivers of GC have been described over the years,3 only very recently, microRNAs (miRNAs) have emerged as key players in the pathogenesis of this disease.4 MiRNAs are short, non-coding RNA molecules that regulate gene expression by directly binding to the 3’-UTR region of their target gene mRNA. MiRNAs have been found to regulate a variety of cellular processes such as cell proliferation, differentiation, invasion, migration, and epithelial-mesenchymal transition (EMT). Accumulating evidence indicates that miRNAs are frequently dysregulated in human cancers including GC,5-8 and that these non-coding RNAs play oncogenic or tumor-suppressive roles in cancer cells. Thus, establishment of a miRNA expression profiles is not only important for investigating the underlying functional mechanisms for a specific cancer, but a better knowledge of their expression patterns could reveal molecular signatures that can be developed as prognostic/predictive biomarkers as well.
Most previous miRNA profiling studies were conducted using miRNA expression microarray platforms. Using this technology, an aberrant miRNA expression signature in GC tissues has been described previously.9,10 More recently, the emergence of Next Generation Sequencing (NGS) platforms has revolutionized the field of genomic medicine, and has helped in the identification of comprehensive, previously unrecognized and specific DNA and RNA targets in human cancers. In comparison to microarray platforms, NGS-based sequencing technologies have several advantages, including massive parallel analysis of widely expressed miRNAs in the genome, quantification of absolute abundance of miRNAs, identification of miRNA sequence variations, and discovery of novel miRNAs. Although few recent studies have attempted transcriptomic profiling in GC, 11,12 to the best of our knowledge, none of the previous studies have used NGS-based platform for the discovery and validation of GC-specific miRNAs by analyzing matched tumor and non-cancerous gastric tissues.
In the present study, we performed comprehensive miRNA profiling using a NGS platform, identified several GC-specific miRNAs, and discovered miR-29c expression to be significantly downregulated in GC tissues vis-à-vis matched normal tissues. We systematically validated the tumor suppressive role of miRNA-29c in a series of experiments performed in cell lines, and transgenic mice models. Moreover, using cell lines and animal models, we identified that ITGB1 (integrin β1), is a novel, downstream gene target of miR-29c, which plays an important role in cell signaling, differentiation, migration, and apoptosis - all processes that are essential for the evolution and development of gastric carcinogenesis.
MATERIALS AND METHODS
Cell lines
Four human GC cell lines, SNU-601, SNU-668, AGS, MKN28 and one human cervical cancer cell line, HeLa were obtained from the Korean Cell Line Bank (Seoul, Korea), and were cultured and maintained in appropriate culture conditions.
Tissue specimens
This study utilized 286 tissue specimens including 143 matched pairs of GC and corresponding normal mucosa tissues (NM) from 3 different GC patient cohorts, as described in supplementary table 1. For NGS analysis, four matched pairs of frozen GCs and adjacent normal mucosa, and two additional NM specimens were obtained from Mie University Medical Hospital, Japan. For validation, 24 pairs of frozen GC and adjacent NM were obtained from Seoul National University Hospital, Korea. In addition, 113 pairs of formalin-fixed, paraffin-embedded (FFPE) GC tissues and matched corresponding normal gastric mucosa tissues from the Mie University Medical Hospital, Japan were analyzed. These studies were approved by the Institutional Review Boards (IRB) of all involved institutions, and written informed consent was obtained from all patients.
Discovery of miR-29c using Next-Generation Sequencing (NGS)
TruSeq miRNA libraries generated from GC and NM tissues were sequenced using an Illumina HiSeq 2000 sequencer with single end read length of 50 bases, following the manufacturer’s instructions. The miRNA sequencing results were also compared with small RNA-seq data sets from the NCBI Sequence Read Archive (GSE36968)11 and miRNA microarray data sets from the GEO database (GSE28700)13.
For the computational analysis of Illumina’s small RNA-seq data, raw sequencing reads were subjected to quality filters as described previously.14 Before alignment, raw reads were initially filtered for (1) quality, (2) presence of the 3’ adapter, to ensure a small RNA was ligated and sequenced completely, and (3) size of small RNA reads (17 to 27 nt). Alignment of reads was compared against human miRNA hairpin sequences in the miRBase v.19 using Novoalign V2.08.01 (www.novocraft.com) with the following parameters: -m -r All 1 -l 18 -t 30 -h 90 -o SAM, default options. After alignment, the reads were further separated into two categories of mapped reads vs. unmapped reads. For the mapped reads, we filtered out reads containing more than two mismatches.
For SOLiD small RNA re-analysis of the Profile#1,11 colorspace read alignments were performed using LifeScope Genomic Analysis Software V2.5 with the following parameters: workflow = small.rna, reference = hg19, smallRNA.genome.mapping = false, smallRNA.mirBase.mapping.scheme = 18.2.0. After alignment, the filtering procedures, normalization, log transformation, and differential analyses were performed like the Illumina datasets. After raw reads were mapped and filtered, an expression value (abundance) for each miRNA across all samples was calculated. For all differential expression analyses, the number of reads for a given miRNA was normalized by dividing the total number of mapped reads in that sample. This resulted in the percent normalized read count for each miRNA, which was then multiplied by 106 to scale the expression value to an appropriate range. Principal Component Analysis (PCA) was performed to identify possible outlier samples. Log transformation was applied to further scale the range of expression values. One-way ANOVA tests were performed between case and control groups in our cohort. For the Profile#1,11 tumors were compared by stage using normal tissues as control.
miRNA expression analysis
Expression of miR-29c was analyzed using TaqMan miRNA assays (Applied Biosystems, Foster City, CA). The average expression level of miR-29c was normalized against U6 and RNU44 as described previously.6
For in situ hybridization (ISH) analysis, five micrometer thick FFPE tissue sections were hybridized with the miR-29c probe (LNA-modified and 5`- and 3`-DIG-labeled oligonucleotide, Exiqon, Woburn, Massachusetts, USA) as described previously.6 Positive (U6 snRNA, Exiqon) and negative controls (scrambled miRNA control, Exiqon) were included in each hybridization procedure as described previously.6
Gene expression analysis
Total RNAs were reverse transcribed to cDNA and quantitative real-time PCR (qRT-PCR) was performed as previously described.6,15,16 Primer sequences are described in supplementary table 2.
Protein expression analysis
Proteins were isolated and western immunoblotting was performed using anti-ITGB1 (Abcam, Cambridge, UK), anti-α-tubulin (Sigma-Aldrich, St. Louis, MO, USA), and anti-β-actin (Cell Signaling Technology, Danvers, MA, USA) antibodies, as described previously.6,15
For immunocytochemistry, cells were fixed in 4% paraformaldehyde and incubated with 10% blocking buffer. Following blocking, cells were incubated with anti-ITGB1 (Abcam, Cambridge, UK) and Alexa Fluor 555 goat anti-rabbit IgG (H+L; Invitrogen, Carlsbad, CA, USA).
Transfection experiments using miR-29c mimic, miR-29c inhibitor, stable miR-29c expressing vector and ITGB1 siRNA
In order to transiently induce or inhibit miR-29c expression, hsa-miR-29c mimics (Applied Biosystems) or anti hsa-miR-29c inhibitor (Applied Biosystems) was used to transfect GC cells, as described previously.6 Verification of transfection efficiency was conducted using the Pre-miR miRNA Precursor Molecules Negative Control (Applied Biosystems) and Anti-miR miRNA Inhibitor Negative Control (Applied Biosystems), respectively.
To establish cell lines stably expressing miR-29c, a fragment containing the full-length coding region of miR-29c cDNA was amplified and cloned into the pcDNA3.1 vector.16 Plasmids were transfected into each cell line, and miRNA-expressing clones were selected as described previously.16 Primers for these steps are described in supplementary table 2. To suppress ITGB1 expression, cells were transfected with either ITGB1 siRNA (Bioneer, Korea) or control scrambled siRNA (Bioneer) using Lipofectamine-2000 (Invitrogen), following the manufacturer’s instructions.
Cell proliferation, adhesion, invasion, and wound healing assays
Cell proliferation was measured using Cell Counting Kit-8 (Dojindo Laboratories, Kumamoto, Japan) following manufacturer’s instructions. For the cell adhesion assay, 96-well plates were coated with fibronectin (10 μg/ml) at 4°C for 18 h and cells were allowed to adhere for 1.5 hours at 37°C. At the end of this time period, adherent cells were quantified using the Cell Counting Kit-8 (Dojindo Laboratories, Kumamoto, Japan) following the manufacturer’s instructions. Cell invasion and wound healing assays were performed as previously described.6
3’-UTR luciferase reporter assays
ITGB1 3’UTR was amplified from human cDNA using primers. The PCR product was cloned into pGL13UC as described previously.17 Primers are shown in supplementary table 2. Luciferase reporter vectors were transfected into the cells and luciferase activity was measured as described previously.6
Xenograft and transgenic mice models
To establish a tumor xenograft mice model, cancer cells stably expressing miR-29c were implanted into the flanks of six-week-old female athymic nude mice (Balb/c nu; Orient Bio Inc., Seoul, Korea).16 Tumor size was measured and the volume was calculated at specified time intervals.16
Gastritis and GC animal models were developed using K19-C2mE [Tg(Krt19-Ptgs2,Krt19-Ptges)8Tko] and Gan (K19-Wnt1/C2mE) [Tg(Krt19-Wnt1)2Maos/Tg(Krt19-Ptgs2,Krt19-Ptges)8Tko] transgenic mice, as described previously.18,19 All transgenic mice models were based on C57BL/6 mouse. Additional experimental details are provided in supplementary materials and methods.
All animal experiment protocols were approved by the Ethics Committees on Animal Experimentation of the Seoul National University and Kanazawa University.
Histology and immunostaining
Stomach tissues from mice were fixed in 4% paraformaldehyde, embedded and sectioned at 4 μm thickness. These sections were stained with H&E and anti-F4/80 (1:100, AbD Serotec, Oxford, United Kingdom), as described previously.18,19
Statistical analysis
Paired t-test, student t-test, and Kruskal-Wallis tests were used to analyze miRNA and gene expression. The Spearman’s correlation test was used to examine correlation between miRNA and target gene expression. Data are presented as mean±S.D. (standard deviation) and all statistical analyses were conducted using the Medcalc version 12.3 (Broekstraat, Belgium) and the GraphPad Prism version 5.0 (GraphPad Software, San Diego, CA).
RESULTS
Next-generation sequencing (NGS) based discovery of miR-29c and validation of its expression pattern in GC tissues
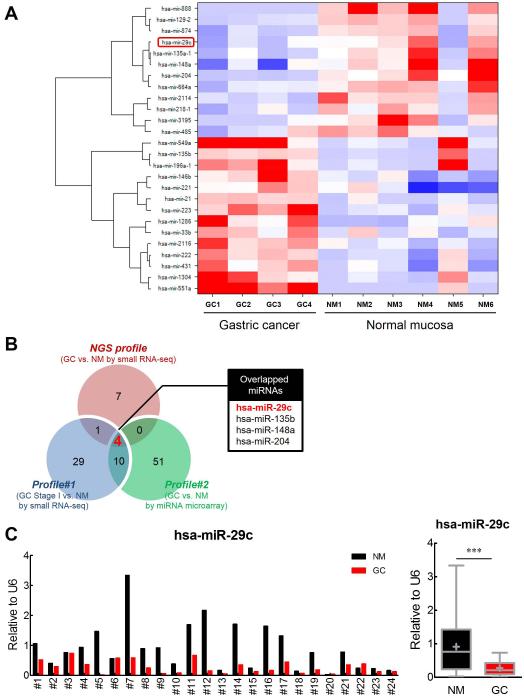
To discover miRNA transcriptomes that distinguish GC from non-cancerous or normal gastric tissues, NGS of small RNAs was performed on four pairs of frozen GCs and adjacent normal tissues, plus two additional normal gastric mucosal specimens. NGS generated a total 22×106 reads (supplementary table 3). For all samples, the number of filtered reads for a given miRNA was normalized to the total mapped reads by scaling the expression values through logarithmic transformation. After filtering the low quality reads and 3’ adapter sequences, a total of 3,555,838 effective miRNA reads in GC tissues and 6,076,342 miRNA reads in the corresponding adjacent normal gastric mucosa tissues were obtained (supplementary table 3). Next, using one-way ANOVA test, 26 miRNAs were found to be differentially expressed between GC and NM tissues (P<0.05 with >2 fold change; figure 1A and supplementary table 4).
Figure 1. Discovery and validation of miR-29c transcriptome in GC tissues by next-generation sequencing (NGS).
(A) Differential expression of 26 GC-specific miRNAs between paired gastric cancer (GC) and normal mucosa tissues (NM) in NGS analysis. (B) Identification of 4 shared miRNAs (miR-29c, miR-135b, miR-148a, and miR-204) between our data set (NGS profile) and two other data sets (Profiles #1 and 2). (C) Expression status of miR-29c in an independent validation cohort of 24 pairs of matching GC and NM tissues. ***p<0.0001, paired t-test
In order to further confirm the robustness of our GC-specific miRNA signature discovered by NGS, we compared our miRNA sequencing results (NGS profile) with two independent miRNA expression profiles that were generated from GCs using two different methods: a small RNA sequencing platform and a miRNA microarray. Kim et al. reported small RNA-seq data using the SOLiD sequencing platform in 19 gastric tumor samples and 6 non-cancerous gastric tissues (Profile#1; supplementary table 5).11 In addition, Tseng et al. generated miRNA expression profiles (Profile#2) using miRNA microarray in 22 paired tumor and non-tumor tissue specimens of GC patients.13 By comparing our NGS-based miRNA sequencing results with these two independent miRNA profiles, we identified 4 miRNAs that were shared between these datasets and were differentially expressed in GCs (miR-29c, miR-135b, miR-148a, and miR-204; figure 1B and supplementary table 6).
Previously, ultra-deep miRNA sequencing profiles of normal stomach tissues revealed that miR-29c was one of most highly expressed miRNAs in normal tissues.20 Since miR-29c was among the four miRNAs that was significantly downregulated in GCs vs. NMs, and was shared between our NGS profile and Profiles#1 and #2, we selected this miRNA for further validation and functional analyses in GC cell lines and animal models. In terms of validation, we quantified the expression of miR-29c in 24 pairs of frozen GCs and matched NMs, as well as in five GC cell lines. The qRT-PCR analyses revealed that miR-29c expression was significantly down-regulated in GC tissues (P=0.0003; figure 1C) and cell lines, compared to NM tissues (figure 2A). These results suggest that miR-29c is consistently downregulated in GCs, and may serve as tumor suppressor in this disease.
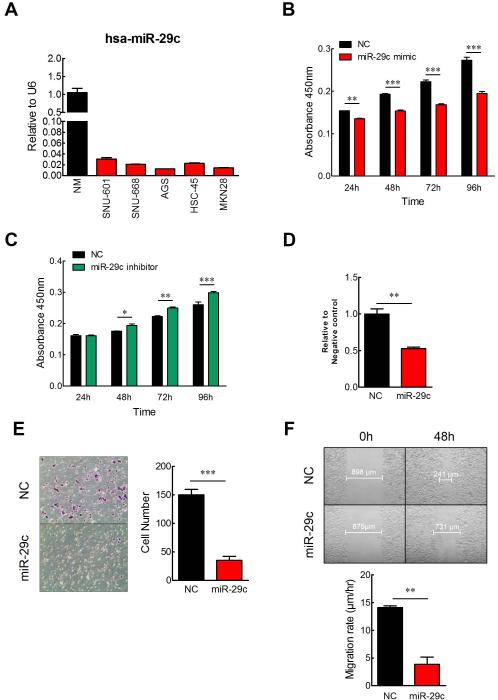
Figure 2. In vitro functional analysis of miR-29c.
(A) Expression of miR-29c in GC cell lines was analyzed by real-time TaqMan PCR. MTT cell proliferation assay following transfection with either (B) miR-29c mimic or (C) anti miR-29c inhibitor. (D) Cell adhesion assay, in which adherence ability of GC cell to fibronectin. (E) Cell invasion assays using Matrigel-coated transwell membrane. (F) Wound-healing assay. Cell monolayers were scratched with a pipette tip and images were taken 0 h and 48 h after wound formation. *p<0.05, **p<0.01, ***p<0.001, t test.
Restoration of miR-29c expression inhibits cell proliferation, adhesion, invasion, and migration in GC cells
To better understand the mechanistic role of miR-29c in gastric carcinogenesis, GC cell lines were transfected with either a miR-29c mimic or a miR-29c inhibitor. Since all five GC cell lines expressed miR-29c at very low levels, we selected SNU-601 for transfection experiments, as this cell line has been well characterized (figure 2A). Cell proliferation in miR-29c transfected cell lines was assayed by CCK-8 assay, each day, for up to 4 days. Restoration of miR-29c expression in GC cells resulted in decreased cell proliferation (figure 2B), whereas inhibition of miR-29c expression significantly increased GC cell proliferation compared to the negative controls (figure 2C).
Next, we analyzed the effect of ectopic miR-29c expression on cellular adhesion, invasion, and migration potential of SNU-601 cells. High miR-29c expression significantly reduced the adherence of GC cells to fibronectin, which is a major component of the extracellular matrix (P=0.0033; figure 2D). Similarly, overexpression of miR-29c significantly inhibited cell invasion into Matrigel-coated transwell membranes (P<0.001; figure 2E). In addition, high miR-29c expression significantly suppressed the ability of cells to migrate in a scratch wound healing assay (P<0.01; figure 2F). Collectively, these data suggest the tumor suppressive role of miR-29c in gastric carcinogenesis.
Overexpression of miR-29c reduces tumor growth in the xenograft nude mouse model, and therapeutic miR-29c delivery suppresses gastric tumorigenesis in nude mice
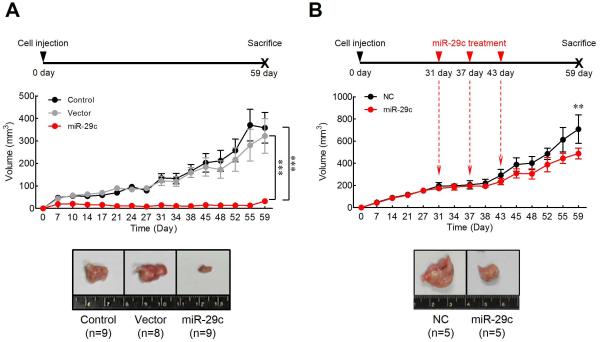
To ascertain the cellular mechanisms underlying miR-29c-mediated tumor suppression, we established a clone of SNU-601 cell line with stable overexpression of miR-29c, which was used for the xenograft nude mouse model experiments (figure 3A). The untreated parental SNU-601 cells, and a stable clone of SNU-601 cells bearing the empty vector were used as controls. The mice implanted with miR-29c overexpressing cells revealed a significantly slower tumor growth compared to animals injected with either controls at the end of eight weeks (miR-29c vs. control and vector, respectively; P<0.0001).
Figure 3. In vivo GC tumorigenesis analysis in the xenograft nude mouse models.
(A) Up-regulation of miR-29c expression inhibited tumor growth in the xenograft nude mouse model. Control, SNU-601 parental cell line; Vector, SNU-601 cell line transfected with empty vector; miR-29c, SNU-601 cell line transfected with miR-29c expression vector (B) Treatment of miR-29c suppressed xenograft nude mouse tumor. Either miR-29c mimic (miR-29c) or negative control (NC) were intratumorally injected using liposome at 31, 37, and 43 days after SNU-601 cells implantation. Tumor volumes represented as means ± SD. **p<0.01, ***p<0.001, two-way ANOVA test.
To further examine the therapeutic effect of miR-29c in gastric tumorigenesis, we used a liposome-based delivery system for the delivery of miR-29c into gastric tumors (figure 3B). To establish gastric tumors in vivo, SNU-601 cells were implanted in 6-week-old Balb/c nude mice. At 31, 37 and 43 days after implantation, miR-29c mimic or negative control constructs was injected intratumorally using liposome-based vehicle. Mice treated with miR-29c mimics demonstrated a significant suppression of tumor growth, which started as early as after third treatment (43 day) compared to controls (P<0.01). Taken together, these results demonstrate that miR-29c expression strongly suppresses gastric tumor growth in vivo. Moreover, our miR-29c liposome-mediated delivery data also highlight the therapeutic potential of this non-coding RNA in gastric neoplasia.
MiR-29c acquires tumor suppressor abilities by directly targeting ITGB1 in GC
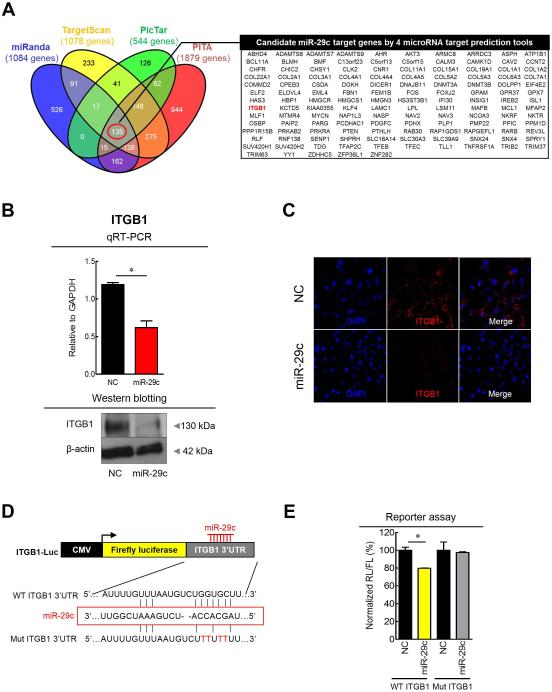
To gain further insight into the molecular mechanism(s) of miR-29c tumor suppressive activity, we sought out determine its gene targets in the gastric mucosa by interrogating the interaction between miR-29c and its target mRNA transcripts. Candidate targets were first determined using target prediction engine including miRanda, TargetScan, PicTar, and PITA (figure 4A). Although two miR-29c target genes were suggested previously: RCC221 and MCL122, but we identified a novel target gene, ITGB1 (integrinβ1), as it seemed more germane to this study as it has been shown to associate with prognosis and metastasis in patients with GC.23,24
Figure 4. Prediction and validation of miR-29c target gene in GC cell line.
(A) 135 target genes which have miR-29c seed sites were predicted via 4 different miRNA target prediction tools (miRanda, TargetScan, PicTar, and PITA). (B) Overexpression of miR-29c suppressed mRNA (qRT-PCR) and protein expression (Western blotting) of ITGB1. (C) Immunocytochemistry for ITGB1 showed low ITGB1 fluorescence intensity following miR-29c treatment. (D-E) Luciferase reporter assays revealed direct binding of miR-29c to the wild-type (WT), but not the mutant (Mut) sequences within the 3’UTR regions of TGB1. NC, transfection of negative control; miR-29c, transfection of miR-29c mimic; FL, firefly luciferase; RL, Renilla luciferase. *p<0.05, t test
To further confirm the functional interaction between miR-29c and ITGB1 generated by the target prediction algorithms, we performed a series of assays to determine the relationship between the ITGB1 and miR-29c in GC cell lines. Overexpression of miR-29c by transfecting miR-mimics in the SNU-601 cells resulted in significant reduction in ITGB1 mRNA transcription as well as protein expression by western blotting (figure 4B). Moreover, immunocytochemical analysis revealed decreased ITGB1 fluorescence intensity following transfection with miR-29c mimics (figure 4C). Since miRNAs are known to affect translation of gene transcripts, we performed luciferase reporter assays to determine whether miR-29c directly interacts with the ITGB1 3’UTR. GC cells were co-transfected with various combinations of pG13 luciferase reporter vectors. The plasmids were transfected with either empty luciferase vector, luciferase vector containing wild-type ITGB1-3’UTR, or luciferase vector containing mutant-type ITGB1-3’UTR and miR-29c mimic or a negative control. In cells transfected with the wild-type ITGB1-3’UTR and the miR-29c mimic, a significant decrease in luciferase activity was observed compared to wild-type ITGB1-3’UTR vector and negative controls (figure 4D-E). However, the luciferase activity of mutant-type ITGB1-3’UTR vector did not change following co-transfection with the miR-29c mimic, indicating miR-29c directly interacts with the 3’-UTR of ITGB1 to mediate its tumor suppressive function.
Since the functional role of the ITGB1 gene has not been previously studied in the pathogenesis of GC, we examined whether miR-29c regulation of ITGB1 translation actually influences the phenotype of gastric mucosa. To investigate the potential role of ITGB1 in GC pathogenesis, we performed ITGB1 knockdown in SNU-601 cells by transfecting them with either siRNA or the negative control. The knockdown effect by ITGB1 siRNA was confirmed by qRT-PCR as well as western blotting (figure 5A). Suppression of ITGB1 significantly reduced the ability of GC cells to adhere to fibronectin (figure 5B), and reduced ITGB1 expression lead to significant decrease in their invasive and migratory abilities (figure 5C-D).
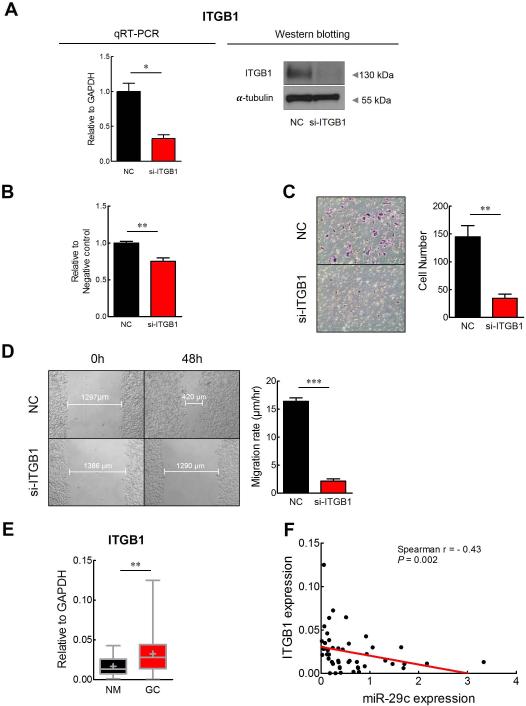
Figure 5. In vitro functional analysis and expression of ITGB1 in GC.
(A) Treatment of si-ITGB1 diminished mRNA (qRT-PCR) and protein expression (Western blotting) of ITGB1 in GC cell line. Down-regulation of ITGB1 by transfection with si-ITGB1 suppressed GC cell (B) adherence ability to fibronectin, (C) invasion ability into matrigel-coated transwell membrane, and (D) cell migration ability in a scratch wound healing assay. (E) Expression status of ITGB1 in human GC tissues (GC) and corresponding normal mucosa tissues (NM). GAPDH, glyceraldehyde-3-phosphate dehydrogenase. (F) Correlation analysis between miR-29c and target gene (ITGB1) expression in GC tissues. Red line represents linear regression line.*p<0.05, **p<0.01, ***p<0.001, t test.
Next, ITGB1 expression was measured in 24 pairs of GC and NM tissues using qRT-PCR. The expression of ITGB1 was significantly up-regulated in GC compared to corresponding NM tissues (P=0.0024; figure 5E). More interestingly, ITGB1 expression was significantly and inversely correlated with miR-29c expression in the same patient cohort (r =-0.43, P=0.002; figure 5F). Collectively, these findings highlight that miR-29c directly inhibits ITGB1 gene expression, which in turn prevents the growth proliferation program in the gastric cells.
Suppression of miR-29c is an early event in gastric carcinogenesis
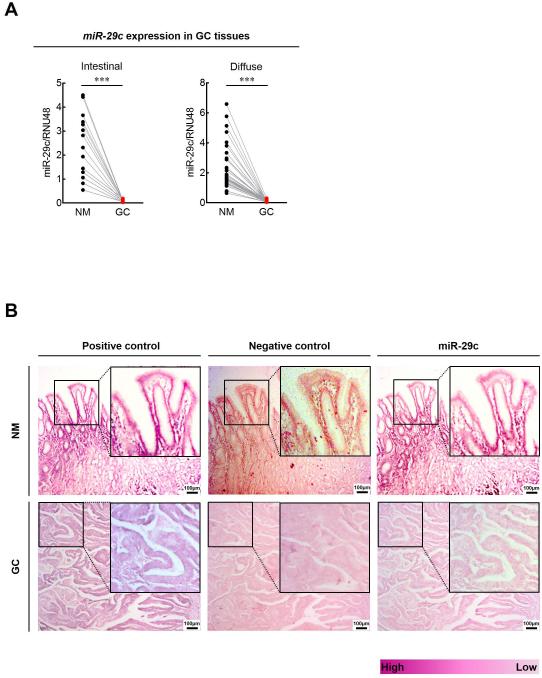
To investigate the clinical relevance of miR-29c expression in GC, we analyzed miR-29c expression in an independent cohort of 113 matched pairs of GC and corresponding NM tissues (the Clinical validation cohort). Consistent with our NGS validation cohort specimens, miR-29c expression was significantly lower in GC compared to NM (P<0.0001; figure 6A). These results were further confirmed by ISH studies, which also revealed marked suppression of miR-29c expression in GC compared to NM tissues (figure 6B).
Figure 6. Clinical relevance and in situ hybridization (ISH) expression of miR-29c in GC tissues.
(A) Expression status of miR-29c in pairs of matching GC and NM tissues. (left panel, intestinal type GC; right panel, diffuse type GC; ***p<0.001, t test) (B) In situ hybridization analysis of miR-29c in matching GC and NM tissues. (Positive control, U6 snRNA, purple color; Negative control, scrambled miRNA control, pink color; Inserts show higher magnifications)
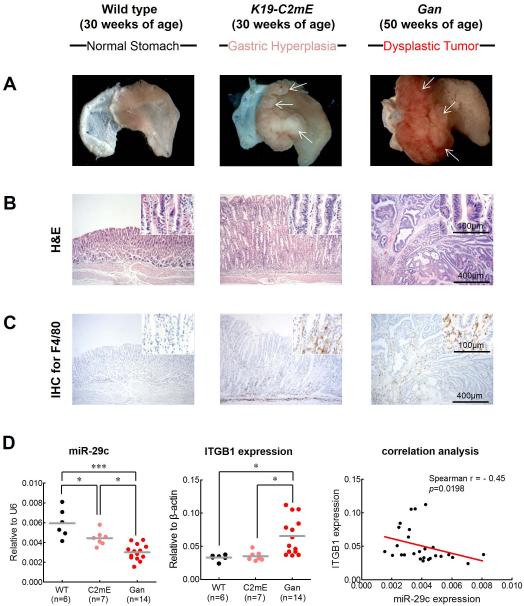
Based on the results to date, we hypothesized that miR-29c may affect the initiation of gastric carcinogenesis. To address this issue, we established two different transgenic animal models: a K19-C2mE mouse model for gastritis, and a Gan mouse model for GC. Previously, we demonstrated that K19-C2mE mice develop gastric hyperplasia in the stomach via induction of the COX-2/PGE2 pathway and Gan mice acquire inflammation-associated GC via induction of COX-2/PGE2 and Wnt signaling pathways.18,19 We found that the K19-C2mE mice developed mucosal hyperplasia in the proximal glandular stomach at 30 weeks of age, while Gan mice developed dysplastic gastric tumors that invaded the muscle layers in the stomach (figure 7A-B) at 50 weeks of age. Abundant activated macrophages are accumulated in the gastric mucosa of the transgenic mice, which promote proliferation of epithelial progenitor cells through Wnt signaling.25-27 Thus, to estimate gastric tumorigenesis process, we analyzed macrophage infiltration in these mice by IHC for F4/80, the best known marker for mature mouse macrophages28 (figure 7C). The wild-type mouse displayed rare macrophage infiltration in glandular stomach, whereas abundant macrophage infiltrates were observed in mucosal stroma of the K19-C2mE mice and in the Gan mice bearing gastric tumors.
Figure 7. Functional analysis of miR-29c in gastritis (K19-C2mE mice) and gastric tumor (Gan mice) transgenic mice models.
Gastritis in K19-C2mE mice and gastric tumors in K19-Wnt1/C2mE mice. (A) Macroscopic photographs and (B) H&E staining of the glandular stomach of the wild-type (left), K19-C2mE (middle), and Gan (right) mice. White arrows in middle panel of (A) indicate gastric hyperplasia lesions. White arrows in middle panel of (A) indicate gastric dysplastic gastric tumors. (C) Immunohistochemistry (IHC) staining for macrophages using F4/80 antibody in the wild-type mouse stomach (left), K19-C2mE (middle), and Gan (right) mice. (D) Expression status of miR-29c (left) and ITGB1 (middle), and correlation analysis (right) between miR-29c and target gene (ITGB1) expression in transgenic mice models. Red line represents linear regression line.*p<0.05, **p<0.01, ***p<0.001, one-way ANOVA test.
Finally, we analyzed the expression of miR-29c and its target gene, ITGB1, in these two transgenic mice models. Compared to the wild-type animals, both K19-C2mE and the Gan mice showed significant decrease in miR-29c expression (P<0.0001; figure 7D). Furthermore, miR-29c expression were significantly lower in the gastric tumors of the Gan mice compared to the K19-C2mE mice with gastric hyperplasia (P=0.0011). By contrast, ITGB1 expression was significantly increased in the Gan mice compared to the C2mE and wild-type animals, respectively. Moreover, ITGB1 expression was significantly and inversely correlated with miR-29c expression in both transgenic mice models (r=-0.45, P=0.0198). These results provide novel and important evidence for the functional role of miR-29c in gastric mucosa and suggest that loss of miR-29c expression in the precancerous state may facilitate initiation of gastric carcinogenesis.
DISCUSSION
In this study, we first performed state-of-the-art next generation sequencing in matched pairs of human GCs and normal mucosa to identify miR-29c as a GC-specific miRNA. Thereafter, using a series of in vitro and in vivo assays, we uncovered that miR-29c act as an important tumor suppressor in the normal gastric mucosa. Although decreased miR-29c expression has been reported previously in human cancers,21,22,29 ours is the first study that provides a novel and comprehensive insight into the functional role of miR-29c as it relates to the pathogenesis of GC. To the best of our knowledge, this is the first study that has: (i) used NGS to identify GC-specific miRNA signatures, (ii) utilized a series of cell culture and animal models to uncover the tumor suppressive role of miR-29c in gastric epithelium, (iii) provided promising evidence for the potential therapeutic use of miR-29c in GC, (iv) identified ITGB1 as a novel target gene of miR-29c, and (v) demonstrated that suppression of miR-29c in transgenic animal models prevented gastric tumor growth.
Although a few previous studies have attempted NGS for the development of genomic signatures in GC, the primary goal of our study was to focus on GC-specific miRNA signatures. Using the Illumina HiSeq 2000 platform, deep-sequencing was performed to generate whole transcriptome profiles of miRNAs in paired GC and corresponding NM tissues. To further ensure the reliability and reproducibility of our discovery data set and to narrow down the most promising candidate miRNAs, we compared our results with two other data sets that were generated using two different high-throughput profiling techniques (SOLiD sequencing platform and miRNA microarray platform) in independent GC cohorts. A total of four miRNAs (miR-29c, miR-135b, miR-148a and miR-204) were common between these three independent GC miRNA profiles. Among these, compared to the cancerous tissue, high levels of miR-29c expression were observed in normal gastric tissues, and these results were confirmed by ultra-deep sequencing. In additional data analysis using a different tool for miRNA deep sequencing analysis, miRNAkey,30 miR-29c was confirmed as a prominent candidate miRNA dysregulated in GC (supplementary figure 1). This consistent finding from these independent cohorts as well as different analysis tools was in part the rationale for selection and systematic exploration for the role of miR-29c in gastric neoplasia.
Ours is the first study that used a series of in vitro and in vivo experiments to provide a direct experimental evidence for the functional role of miR-29c as a tumor suppressor in gastric neoplasia. In addition, we identified ITGB1 as a novel miR-29c-target gene, and confirmed its direct interaction with the miR-29c. In terms of miR-29c target genes, RCC221 and MCL122 have been previously reported, and we were also able to confirm a direct interaction of the two target genes (RCC2 and MCL1) with miR-29c using luciferase reporter assays (supplementary figure 2). The expression status of these two genes did not have a direct correlation with miR-29c expression in GC tissues (supplementary figure 3), while high levels of ITGB1 expression have been shown to associate with poor prognosis and recurrence in patients with GC.23,24 In addition, previous studies have shown the interaction of ITGB1 with cytokines, growth factors and extracellular matrix (ECM) proteins.31,32 Overexpression of ITGB1 has been found in various epithelial malignancies including breast cancer and glioblastoma, during invasion, angiogenesis and metastasis.33-35 Intriguingly, our mechanistic and functional data permit us to better appreciate the functional role of ITGB1 in human cancers, its expression positively regulated GC cell adhesion, invasion and migration. Furthermore, our observation for an inverse correlation between miR-29c expression and ITGB1 expression in GC tissues fills this important void in literature for the missing experimental evidence for the function of miR-29c and ITGB1 in gastric pathogenesis.
Another interesting aspect of our study is that we firstly demonstrated the tumor initiator role of miR-29c using transgenic mice models of gastric hyperplasia and carcinogenesis. Our data revealed that while miR-29c was significantly down-regulated in a step-wise manner in wild type- hyperplasia- dysplasia cascade, the expression of its target gene, ITGB1, was conversely up-regulated in these animal models of gastric carcinogenesis. Collectively, our findings provide a novel mechanistic insight that was previously unrecognized, and highlight that suppression of miR-29c is an early phenomenon in gastric carcinogenesis and may trigger the initiation of GC.
Our study also highlighted the potential clinical application of miR-29c in the diagnosis and treatment of patients with GC. Although miR-29c expression status was not associated with prognosis and metastasis in our patient cohort, suppression of miR-29c was an early event in gastric carcinogenesis, suggesting the possibility for the application of miR-29c as a diagnostic biomarker for this malignancy. In regard to its therapeutic potential, we noted that treatment of miR-29c mimics markedly reduced tumor volume in GC xenograft mouse model. In addition, a recent study showed restoration of miR-29c expression by treatment of celecoxib, a selective COX-2 inhibitor, in human GC cell lines.22 In this context, our observation that miR-29c was significantly suppressed in gastritis and gastric tumor mice models through genetic induction of COX-2/mPGES-1 and Wnt/PGE2 pathways, suggests the importance of miR-29c as a potential therapeutic target for COX-2 meditated gastric carcinogenesis.
In conclusion, this study provides discovery and validation of GC-specific miRNA transcriptome profiles generated by NGS. We identified that miR-29c is a potent tumor suppressor in the stomach, and its growth inhibitory effects are in part, mediated through its downstream target gene, ITGB1. Using cell culture and animal models, functional characterization for the role of miR-29c reveals that loss of its expression is an early event in gastric carcinogenesis.
Supplementary Material
SIGNIFICANCE OF THIS STUDY.
What is already known about this subject?
Gastric cancer (GC) is a biologically heterogeneous disease accompanying various genetic and epigenetic alterations.
Several microRNAs (miRNAs) are frequently dysregulated in human cancers.
Most previous miRNA profiling studies were conducted using miRNA expression microarray platforms; however, conventional hybridization-based microarray methodologies have several limitations including lack of sensitivity, narrow dynamic range, and non-specific hybridization.
What are the new findings?
This is the first study that performed next generation sequencing (NGS) based whole-transcriptome profiling to identify GC-specific miRNA signatures.
We discovered the most promising GC-specific miRNAs, miR-29c, by comparison three different high-throughput miRNAs profiling generated from independent GC cohorts.
We elucidate a direct experimental evidence for the tumor suppressor role of miR-29c by regulating target gene (ITGB1) in GC through a series of in vitro and in vivo experiments.
We firstly demonstrated that suppression of miR-29c is an early event in gastric carcinogenesis using transgenic mouse models for gastritis (K19-C2mE) and GC (K19-Wnt1/C2mE).
How might it impact on clinical practice in the foreseeable future?
Our study provide GC-specific miRNAs signature generated by state-of-the-art NGS technique. Moreover, we highlight a potent tumor suppressor role of miR-29c through its downstream target gene, ITGB1, which may be an essential event in gastric carcinogenesis. Taken together, these results underscore the potential early diagnostic and therapeutic biomarker for patients with GC.
ACKNOWLEDGEMENTS
This study was supported by A3 Foresight Program from National Research Foundation of Korea. We thank Dr. Margaret M, Hinshelwood for her skillful editing and revision of the manuscript.
Footnotes
Conflict of Interest: None of the authors have any conflicts to disclose
REFERENCES
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA: a cancer journal for clinicians. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 2.Hartgrink HH, Jansen EP, van Grieken NC, et al. Gastric cancer. Lancet. 2009;374:477–90. doi: 10.1016/S0140-6736(09)60617-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liang H, Kim YH. Identifying molecular drivers of gastric cancer through next-generation sequencing. Cancer letters. 2013;340:241–6. doi: 10.1016/j.canlet.2012.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thiel A, Ristimaki A. Gastric cancer: basic aspects. Helicobacter. 2012;17(Suppl 1):26–9. doi: 10.1111/j.1523-5378.2012.00979.x. [DOI] [PubMed] [Google Scholar]
- 5.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nature reviews. Cancer. 2006;6:857–66. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 6.Hur K, Toiyama Y, Takahashi M, et al. MicroRNA-200c modulates epithelial-to-mesenchymal transition (EMT) in human colorectal cancer metastasis. Gut. 2013;62:1315–26. doi: 10.1136/gutjnl-2011-301846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim YK, Yu J, Han TS, et al. Functional links between clustered microRNAs: suppression of cell-cycle inhibitors by microRNA clusters in gastric cancer. Nucleic acids research. 2009;37:1672–81. doi: 10.1093/nar/gkp002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Toiyama Y, Takahashi M, Hur K, et al. Serum miR-21 as a diagnostic and prognostic biomarker in colorectal cancer. Journal of the National Cancer Institute. 2013;105:849–59. doi: 10.1093/jnci/djt101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ueda T, Volinia S, Okumura H, et al. Relation between microRNA expression and progression and prognosis of gastric cancer: a microRNA expression analysis. The lancet oncology. 2010;11:136–46. doi: 10.1016/S1470-2045(09)70343-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Volinia S, Calin GA, Liu CG, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:2257–61. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim YH, Liang H, Liu X, et al. AMPKalpha modulation in cancer progression: multilayer integrative analysis of the whole transcriptome in Asian gastric cancer. Cancer research. 2012;72:2512–21. doi: 10.1158/0008-5472.CAN-11-3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li SC, Liao YL, Ho MR, et al. miRNA arm selection and isomiR distribution in gastric cancer. BMC genomics. 2012;13(Suppl 1):S13. doi: 10.1186/1471-2164-13-S1-S13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tseng CW, Lin CC, Chen CN, et al. Integrative network analysis reveals active microRNAs and their functions in gastric cancer. BMC systems biology. 2011;5:99. doi: 10.1186/1752-0509-5-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fehniger TA, Wylie T, Germino E, et al. Next-generation sequencing identifies the natural killer cell microRNA transcriptome. Genome research. 2010;20:1590–604. doi: 10.1101/gr.107995.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hur K, Cejas P, Feliu J, et al. Hypomethylation of long interspersed nuclear element-1 (LINE-1) leads to activation of proto-oncogenes in human colorectal cancer metastasis. Gut. 2014;63:635–46. doi: 10.1136/gutjnl-2012-304219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hur K, Han TS, Jung EJ, et al. Up-regulated expression of sulfatases (SULF1 and SULF2) as prognostic and metastasis predictive markers in human gastric cancer. The Journal of pathology. 2012;228:88–98. doi: 10.1002/path.4055. [DOI] [PubMed] [Google Scholar]
- 17.Park SY, Lee JH, Ha M, et al. miR-29 miRNAs activate p53 by targeting p85 alpha and CDC42. Nature structural & molecular biology. 2009;16:23–9. doi: 10.1038/nsmb.1533. [DOI] [PubMed] [Google Scholar]
- 18.Oshima H, Matsunaga A, Fujimura T, et al. Carcinogenesis in mouse stomach by simultaneous activation of the Wnt signaling and prostaglandin E2 pathway. Gastroenterology. 2006;131:1086–95. doi: 10.1053/j.gastro.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 19.Oshima H, Oshima M, Inaba K, et al. Hyperplastic gastric tumors induced by activated macrophages in COX-2/mPGES-1 transgenic mice. The EMBO journal. 2004;23:1669–78. doi: 10.1038/sj.emboj.7600170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ribeiro-dos-Santos A, Khayat AS, Silva A, et al. Ultra-deep sequencing reveals the microRNA expression pattern of the human stomach. PloS one. 2010;5:e13205. doi: 10.1371/journal.pone.0013205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matsuo M, Nakada C, Tsukamoto Y, et al. MiR-29c is downregulated in gastric carcinomas and regulates cell proliferation by targeting RCC2. Molecular cancer. 2013;12:15. doi: 10.1186/1476-4598-12-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saito Y, Suzuki H, Imaeda H, et al. The tumor suppressor microRNA-29c is downregulated and restored by celecoxib in human gastric cancer cells. International journal of cancer. Journal international du cancer. 2013;132:1751–60. doi: 10.1002/ijc.27862. [DOI] [PubMed] [Google Scholar]
- 23.Xu ZY, Chen JS, Shu YQ. Gene expression profile towards the prediction of patient survival of gastric cancer. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie. 2010;64:133–9. doi: 10.1016/j.biopha.2009.06.021. [DOI] [PubMed] [Google Scholar]
- 24.Zhao ZS, Li L, Wang HJ, et al. Expression and prognostic significance of CEACAM6, ITGB1, and CYR61 in peripheral blood of patients with gastric cancer. Journal of surgical oncology. 2011;104:525–9. doi: 10.1002/jso.21984. [DOI] [PubMed] [Google Scholar]
- 25.Oguma K, Oshima H, Aoki M, et al. Activated macrophages promote Wnt signalling through tumour necrosis factor-alpha in gastric tumour cells. The EMBO journal. 2008;27:1671–81. doi: 10.1038/emboj.2008.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oshima H, Hioki K, Popivanova BK, et al. Prostaglandin E(2) signaling and bacterial infection recruit tumor-promoting macrophages to mouse gastric tumors. Gastroenterology. 2011;140:596–607. doi: 10.1053/j.gastro.2010.11.007. e7. [DOI] [PubMed] [Google Scholar]
- 27.Pull SL, Doherty JM, Mills JC, et al. Activated macrophages are an adaptive element of the colonic epithelial progenitor niche necessary for regenerative responses to injury. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:99–104. doi: 10.1073/pnas.0405979102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leenen PJ, de Bruijn MF, Voerman JS, et al. Markers of mouse macrophage development detected by monoclonal antibodies. Journal of immunological methods. 1994;174:5–19. doi: 10.1016/0022-1759(94)90005-1. [DOI] [PubMed] [Google Scholar]
- 29.Bae HJ, Noh JH, Kim JK, et al. MicroRNA-29c functions as a tumor suppressor by direct targeting oncogenic SIRT1 in hepatocellular carcinoma. Oncogene. 2013 doi: 10.1038/onc.2013.216. [DOI] [PubMed] [Google Scholar]
- 30.Ronen R, Gan I, Modai S, et al. miRNAkey: a software for microRNA deep sequencing analysis. Bioinformatics. 2010;26:2615–6. doi: 10.1093/bioinformatics/btq493. [DOI] [PubMed] [Google Scholar]
- 31.Sastry SK, Burridge K. Focal adhesions: a nexus for intracellular signaling and cytoskeletal dynamics. Experimental cell research. 2000;261:25–36. doi: 10.1006/excr.2000.5043. [DOI] [PubMed] [Google Scholar]
- 32.van der Flier A, Sonnenberg A. Function and interactions of integrins. Cell and tissue research. 2001;305:285–98. doi: 10.1007/s004410100417. [DOI] [PubMed] [Google Scholar]
- 33.Foubert P, Varner JA. Integrins in tumor angiogenesis and lymphangiogenesis. Methods in molecular biology. 2012;757:471–86. doi: 10.1007/978-1-61779-166-6_27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gotzmann J, Mikula M, Eger A, et al. Molecular aspects of epithelial cell plasticity: implications for local tumor invasion and metastasis. Mutation research. 2004;566:9–20. doi: 10.1016/s1383-5742(03)00033-4. [DOI] [PubMed] [Google Scholar]
- 35.Tchaicha JH, Reyes SB, Shin J, et al. Glioblastoma angiogenesis and tumor cell invasiveness are differentially regulated by beta8 integrin. Cancer research. 2011;71:6371–81. doi: 10.1158/0008-5472.CAN-11-0991. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.