Summary
Cmr2 is the largest and an essential subunit of a CRISPR RNA-Cas protein complex (the Cmr complex) that cleaves foreign RNA to protect prokaryotes from invading genetic elements. Cmr2 is thought to be the catalytic subunit of the effector complex because of its N-terminal HD nuclease domain. Here however, we report that the HD domain of Cmr2 is not required for cleavage by the complex in vitro. The 2.3Å crystal structure of Pyrococcus furiosus Cmr2 (lacking the HD domain) reveals two adenylyl cyclase-like and two α-helical domains. The adenylyl cyclase-like domains are arranged as in homodimeric adenylyl cyclases and bind ADP and divalent metals. However, mutagenesis studies show that the metal- and ADP-coordinating residues of Cmr2 are also not critical for cleavage by the complex. Our findings suggest that another component provides the catalytic function, and that the essential role by Cmr2 does not require the identified ADP- or metal-binding, or HD domains in vitro.
Introduction
Clustered Regularly Interspaced Short Palindromic Repeats (CRISPRs) confer adaptive immunity to prokaryotes in their defense against mobile genetic elements (Barrangou et al., 2007; Brouns et al., 2008; Garneau et al., 2010; Marraffini and Sontheimer, 2010b) and see general reviews (Deveau et al., 2010; Karginov and Hannon, 2010; Makarova et al., 2011b; Marraffini and Sontheimer, 2010a; Terns and Terns, 2011). CRISPR loci found in the genomes of prokaryotes contain short spacer elements derived from bacteriophages and conjugative plasmids, and occur in arrays of repeat-spacer-repeat sequences (Bolotin et al., 2005; Mojica et al., 2005; Pourcel et al., 2005). CRISPRs produce small RNAs that recognize homologous foreign nucleic acids. Adjacent to the CRISPRs are CRISPR-associated (cas) genes that encode enzymes and factors involved in all functional stages of CRISPR-mediated immunity (Haft et al., 2005; Makarova et al., 2011a; Makarova et al., 2006; Makarova et al., 2011b). There are multiple families of Cas proteins associated with different types of CRISPR repeats that comprise distinct CRISPR-Cas immune systems (Haft et al., 2005; Makarova et al., 2011a; Makarova et al., 2006; Makarova et al., 2011b).
The mechanism of CRISPR-based immunity is divided into three stages. In the adaptation stage, a short sequence derived from an invader is integrated into the CRISPR array as a spacer between two repeats (Barrangou et al., 2007; Garneau et al., 2010; Horvath et al., 2008). In the expression stage, the CRISPR array is transcribed into long precursor RNAs from a promoter found in a leader region preceding the array. The precursor RNAs are processed into CRISPR RNAs (crRNAs), which are loaded into effector complexes composed of Cas proteins (Brouns et al., 2008; Carte et al., 2008; Hale et al., 2008; Hale et al., 2009; Haurwitz et al., 2010). In the third stage, the effector complexes use the crRNA to guide the cleavage of invader DNA or RNA (Hale et al., 2009; Jore et al., 2011; Lintner et al., 2011; Semenova et al., 2011; Wiedenheft et al., 2011a; Wiedenheft et al., 2011b).
Insights into mechanisms of crRNA-mediated targeting of DNA by several CRISPR-Cas systems are beginning to emerge. Organisms that contain the Csn-type systems have been shown to cleave foreign DNA (Garneau et al., 2010), apparently through the activity of Csn1 (or Cas9) (Sapranauskas et al., 2011). CRISPR-guided DNA silencing is also mediated by the Csm-type Cas proteins, although the mechanism is unknown (Marraffini and Sontheimer, 2008, 2010b). DNA silencing by the Cse-type system appears to involve the nuclease-helicase protein Cas3. This protein has been shown to have single-stranded DNA endonuclease activity (Mulepati and Bailey, 2011; Sinkunas et al., 2011) or single-stranded DNA and RNA endo- and exonuclease activity (Beloglazova et al., 2011) as well as dsDNA unwinding activity (Beloglazova et al., 2011; Howard et al., 2011; Sinkunas et al., 2011). In E. coli, CRISPR interference requires both Cas3 and a protein complex called Cascade, which consists of five Cse-type proteins (CasABCDE) (Brouns et al., 2008). The Cascade complex was shown to bind the target strand of dsDNA through base-pairing and to displace the non-complementary strand (Jore et al., 2011). Cas3 is hypothesized to join the DNA-bound Cascade complex, and move along and cleave the target DNA (Jore et al., 2011; Sinkunas et al., 2011; Wiedenheft et al., 2011a; Wiedenheft et al., 2011b). Cas3 proteins are also associated with other types of CRISPR-Cas systems and may function similarly in those systems.
The Cmr (Cas RAMP module)-type effector complex recognizes and destroys complementary RNAs (Hale et al., 2011; Hale et al., 2009). This CRISPR-Cas effector complex has been found to cleave target RNAs in vivo, but has also been reconstituted and analyzed in vitro (Hale et al., 2011; Hale et al., 2009). The complexes characterized from Pyrococcus furiosus (Pfu) include six proteins, Cmr1-6 (Figure 1A), and crRNAs of 39 and 45 nucleotides (nts) (Hale et al., 2011; Hale et al., 2009). Each of the 6 Cmr proteins and at least one crRNA is essential for function. As is the case for some other CRISPR-Cas systems, crRNAs associated with Cmr complexes contain an 8-nt repeat sequence tag at the 5′ end upstream of an invader-targeting or guide element of variable sequence (Brouns et al., 2008; Lintner et al., 2011; Marraffini and Sontheimer, 2010b). The 5′ 8-nt repeat tag is generated by Cas6, which binds to the CRISPR locus transcript and cuts within each repeat 8 nt upstream of the guide element (Carte et al., 2008). The Cmr complex cleaves the target RNA 14 nucleotides from the 3′ end of the bound crRNA (Hale et al., 2009). Interestingly, although the cleavage reaction requires specific divalent metals, the complex cleaves on the 5′ side of the phosphodiester bond and produces a 3′, or 2′,3′ cyclic phosphate terminus and a 5′ OH terminus (Hale et al., 2009). These products are reminiscent of metal-independent catalysis, in which the 2′ OH of ribose is used as a nucleophile to attack the adjacent 3′ phosphate and break the RNA backbone (Yang, 2011). Little is known about how the Cmr complex assembles, interacts with the crRNA and substrate RNA, and finally facilitates RNA cleavage.
Figure 1.

The RNA silencing pathway mediated by the Cmr2-containing Cmr complex in Pyrococcus furiosus. (A) The Pyrococcus furiosus Cas protein locus #1. The processing endoribonuclease Cas6 is shown in grey and the Cmr proteins are shown in blue. Cmr2 is the largest of the six Cmr proteins and is a member of the Cas10 superfamily of CRISPR polymerase proteins. (B) Schematic of crRNA processing, assembly into the Cmr RNP complex and guided cleavage of a target RNA. The pre-crRNA transcript is initially processed by Cas6 within the repeat sequence to form intermediate crRNAs which contain 8 nt of the repeat at the 5′ end (called the tag; shown in black) and the remaining 22 nt of repeat at the 3′ end. These intermediate crRNAs are then processed by an unknown mechanism into mature crRNAs, which retain the 5′ tag sequence upstream of 31 or 37 nts of the spacer sequence. The mature crRNAs are loaded on the Cmr effector complex and guide the recognition and cleavage of target RNAs through base-pairing. The Cmr complex cleaves the target at the nucleotide opposite to that 14nt from the 3′ end of the crRNA. (C) In vitro cleavage activity of the reconstituted P. furiosus Cmr complex containing the full-length Cmr2 or an N-terminal domain truncated mutant, Cmr2dHD. The Cmr complex (1 μM) containing full-length Cmr2 (wt), no Cmr2 (-Cmr2) or Cmr2 lacking the N-terminal HD domain (Cmr2dHD) was incubated with either the 39 or 45 nucleotide crRNA, followed by addition of radiolabeled target RNA (indicated by an open arrow). – lanes contain no crRNA or proteins. The 39 nt crRNA guides cleavage that results in a 20 nt cleavage product while the 45 nt crRNA results in a 14 nt cleavage product (indicated by asterisks). crRNA/target RNA duplexes are indicated by closed arrows. See also Figure S1.
Cmr2 is the largest of the six proteins in the Cmr complex and a member of COG1353 (Haft et al., 2005; Makarova et al., 2002). It belongs to the greater Cas10 family of proteins (Makarova et al., 2011b). Many of the COG1353 proteins are predicted to have a permuted Histidine-Aspartate (HD)-superfamily hydrolase domain (predicted nuclease domain) at the N-terminus, a zinc ribbon domain, and a ferredoxin-like fold similar to the catalytic domain of many polymerases and nucleotide cyclases (Makarova et al., 2002). In addition, the ferredoxin-like fold in Cmr2 contains a degenerate GGDEF (GGDD) motif similar to that found in nucleotide cyclases (Anantharaman et al., 2010). The acidic residues of the GGDEF motif are known to bind divalent metal ions in these enzymes (Anantharaman et al., 2010; Wang et al., 2007). Cmr2 had been predicted to catalyze RNA/DNA polymerization for some aspect of CRISPR-Cas function (Anantharaman et al., 2010; Haft et al., 2005; Makarova et al., 2006), however, the only experimentally observed function of Cmr2 is its essential role in crRNA-mediated RNA cleavage by the Cmr1-6 complex ((Hale et al., 2009), Figure 1). Also based largely on its predicted functional domains, Cmr2 is considered the most likely Cmr complex subunit responsible for target RNA cleavage.
Here, we biochemically and structurally characterized Pyrococcus furiosus (Pf) Cmr2. We found that a truncated Cmr2 that lacks the N-terminal predicted HD domain (Cmr2dHD) is fully functional in RNA cleavage as part of the Cmr complex. The structure of Cmr2dHD in complex with adenosine diphosphate (ADP) contains a full and a partial adenylyl cyclase-like domain; together, the two domains form a nucleotide and divalent metal binding pocket. Cmr2dHD possesses some of the residues required for nucleotide cyclization in adenylyl cyclases. However, it does not exhibit detectable ATP hydrolysis activity in vitro. Furthermore, mutation of the conserved acidic residues that interact with bound metal ions or ADP does not affect the ability of the reconstituted Cmr complex containing Cmr2dHD to cleave target RNA. Implications of these findings in the biochemical mechanism of the Cmr-mediated RNA silencing reaction are discussed.
Results
The predicted HD domain of Cmr2 is not required for target RNA cleavage activity
Sequence alignment of Pf Cmr2 with other Cmr2 proteins using PSI-BLAST and COBALT (Papadopoulos and Agarwala, 2007) showed that the N-terminal 215 amino acids comprise a permuted HD hydrolase domain ((Makarova et al., 2002), Figure S1). To test the predicted function of Cmr2 and the HD domain in RNA cleavage, we constructed a truncated version of Cmr2, Cmr2dHD, in which the first 215 amino acids are removed. The Cmr1-6 complex was reconstituted with either the wild-type or Cmr2dHD protein and tested in a previously established activity assay (Hale et al., 2009). The complex containing Cmr2dHD showed similar cleavage activity to that containing the wild-type protein, producing the correctly sized 14- and 20-nt target RNA cleavage products when reconstituted with the 45- and 39-nt crRNA, respectively (Figure 1C). This result indicates that the putative HD domain is not required for RNA cleavage by the complex. Since Cmr2dHD expression is more robust than the wild-type Cmr2, all further experiments were performed using Cmr2dHD.
Overview and domain structures of Cmr2dHD
The crystal structure of Cmr2dHD without bound ADP was solved by single wavelength anomalous diffraction phasing methods using a L-Selenomethionine (SeMet)-substituted protein crystals and refined to 2.4 Å resolution (Table 1). The crystal of ADP-bound Cmr2dHD has the same space group and unit cell dimensions as those of the ADP-free crystal and its structure was refined to 2.3Å resolution (Table 1). Cmr2dHD was crystallized in an orthorhombic space group (P212121) with one protein in the asymmetric subunit without extensive interfaces among its symmetry mates (Table 1). The ADP-free and ADP-bound structures of Cmr2dHD do not differ in overall structure and the ADP-bound structure is used for subsequent discussions on general structural features of Cmr2dHD. Cmr2dHD structure shows a flat triangular arrangement of four structural domains (D1, D2, D3, D4) connected by loops (Figure 2A, 2B and 2E). Domains D1 and D3 are ferredoxin(-like) domains positioned side-by-side and each is followed by a small α-helical domain, D2 and D4, respectively.
Table 1. Crystallographic data collection and refinement statistics*.
| Data Collection Statistics | |||
|---|---|---|---|
| Cmr2dHD | Cmr2dHD-ADP-Ca | Cmr2dHD-ADP-Mn% | |
| Space group | P212121 | P212121 | P212121 |
| a | 70.6 | 73.2 | 73.0 |
| b | 80.2 | 85.0 | 85.6 |
| c | 143.9 | 140.1 | 140.8 |
| Resolution range (Å) | 50-2.4 (2.5-2.4) | 50-2.3 (2.34-2.30) | 50-3.6 (3.7-3.6) |
| No. of observed unique reflections | 29299 (1037) | 30172 (2687) | 18205 (685) |
| Redundancy | 6.7 (4.4) | 6.7 (5.1) | 3.5 (1.7) |
| Completeness (%) | 90.7 (65.7) | 96.9 (88.2) | 97.3 (71.4) |
| I/σ(I) | 27.6 (2.49) | 25.9 (2.28) | 30.5 (9.43) |
| Rsym(%) | 8.3 (55) | 7.4 (72) | 8.9 (49.0) |
| Refinement statistics | |||
| Resolution range (Å) | 42.7-2.4 (2.5-2.4) | 29.6-2.3 (2.37-2.30) | |
| Rwork(%) | 20.8 | 20.6 | |
| Rfree(%) | 26.4 | 26.1 | |
| Model information | |||
| No. of protein/ADP complexes | 1/0 | 1/1 | |
| No. of protein/ADP atoms/water | 4476/0/31 | 4476/26/34 | |
| No. of amino acids | 550 | 550 | |
| Root-mean-square deviations (rmsd) | |||
| Bond length (Å) | 0.014 | 0.018 | |
| Bond angle (°) | 1.093 | 1.508 | |
| Ramachandran plot of protein residues | |||
| Preferred regions (%) | 96.2 | 95.9 | |
| Allowed regions (%) | 3.8 | 4.1 | |
| Disallowed region (%) | 0.0 | 0.0 | |
Values in parentheses are for the last resolution shell.
This data set was only used for identification of bound metals and the structure was not further refined.
Figure 2.

Crystal structure of P. furiosus Cmr2dHD. (A) Overview and domain structure of Cmr2dHD. Cmr2dHD is a flat, triangular, four-domain protein. The N-terminal domain D1 (blue) contains a ferredoxin-like fold (which contains a P-loop) and a cysteine cluster. The D3 domain (orange) contains a ferredoxin fold that includes a conserved GGDD motif on a β-hairpin and a P-loop structure. Domains D2 (green) and D4 (red) are exclusively α-helical. (B) Topology of the Cmr2dHD structure with the same coloring scheme as panel A. Arrows represent β-sheets and cylinders represent α-helices. Dashed lines represent disordered regions. The P-loops of D1 and D3, the β-hairpin of D3 and the cysteine cluster of D1 are indicated. (C) Cmr2dHD contains two homologous ferredoxin folds (colored in blue and orange for the D1 and D3 domains respectively). The D1 ferredoxin-like fold lacks the β-hairpin but maintains the P-loop (purple). The D3 domain contains both the β-hairpin (yellow) and P-loop (purple). (D) D1 and D3 domains are homologous. The two domains have a root-means-square-deviation of 2.6 Å. (E) Electrostatic surface potential of Cmr2dHD bound with adenosine diphosphate (ADP). See also Figure S4.
The N-terminal domain (D1, residues 216-503) is the largest of the four domains and is composed of nine α-helices and three β-strands. The three β-strands (β1–β3) and two of the nine α-helices (α2 and α6) form a ferredoxin-like fold short of a β-strand (Figure 2C). (If the loop connecting α3 to α4 were replaced by a β-strand, D1 would be a bona fide ferredoxin fold.) Incorporated into D1 and below the ferredoxin-like fold, is a cysteine cluster that is formed by two CXXC motifs, one at the α10-α11 loop and one at the beginning of helix α11 (Figure 2A). The two pairs of cysteine residues (Cys448 and Cys481, Csy478 and Cys451) form a right- and left-handed disulfide bond, respectively (Figure S4). The CXXC motif of the four cysteine residues in Cmr2 is the basis for the predicted Zinc ribbon structure (Makarova et al, 2011b), however, the center of the cysteine cluster does not have space to accommodate a zinc ion and the cysteine cluster structure does not resemble the rubredoxin-like zinc ribbon (Wang et al, 2007).
Domain D3 (residues 593-764) is the most conserved domain of Cmr2dHD and contains a ferredoxin fold (Figure 2C). The ferredoxin fold of D3 is a typical βαββαβ structure comprised of β4–β7 and α18-α19 (Figure 2A, C). The ferredoxin fold is extended by secondary structure elements α20 to β9 leading to an extended 7-stranded central sheet sandwiched by 3 helices. Short loops connect secondary structures at the lower region of the D3 domain while most of the upper region loops are disordered (Figure 2A and 2B).
Surprisingly, despite limited sequence similarity (∼16% identity) between the D1 and D3 domains, their three-dimensional structures superimpose well including regions beyond the ferredoxin fold (Root-Means-Square-Deviation, or RMSD, is 2.6 Å for 102 Cα atoms) (Figure 2D). This finding suggests a domain duplication event followed by diversification.
Cmr2dHD resembles homodimeric adenylyl cyclases
Among available structures, Cmr2dHD has the highest similarity to nucleotide cyclases when aligned using the DALI multiple structural alignment server (http://ekhidna.biocenter.helsinki.fi/dali_server/). The best alignment was obtained with domain D3 of Cmr2dHD and a monomer of Mycobacterium tuberculosis (Mt) adenylyl cyclase (PDB 1YBU, (Sinha et al, 2005). Despite a 14% amino acid sequence identity between the two proteins, the alignment Z-score is 11.9 and the RMSD is 2.9 Å (166 aligned Cα atoms). The homologous domain D1 also resembles the adenylyl cyclase monomer, but to a lesser degree (Z-score 10.0, RMSD 2.1). Together, domains D1 and D3 have an overall resemblance to the dimeric form of Mt adenylyl cyclase (Figure 3A & 3C). Each Mt adenylyl cyclase monomer consists of a typical ferredoxin fold (βαββαβ) plus an extra αβαββ folding unit at the C-terminal end, leading to a continuous 7-stranded β-sheet sandwiched by three α-helices on one side and one α-helix on the other (Figure 3C) (Artymiuk et al, 1997; Sinha et al, 2005). Domain D3 has the same 7-stranded β-sheet and flanking helices with only a small difference in the C-terminal folding unit (Figure 3A). Domain D3 has an αββ while Mt adenylyl cyclase has an αβαββ element. Domain D1, on the other hand, lacks the entire C-terminal folding unit except for the α-helix, α9. Thus, due to the missing β-strand in its ferredoxin fold and the C-terminal folding unit, D1 contains a 3-stranded rather than a 7-stranded β-sheet sandwiched by helices (Figure 3A). In addition, the relative orientation of the ferredoxin fold in D1 with respect to that in D3 is significantly different from that between the two monomers of Mt adenylyl cyclase. As a result, the D1 and D3 domains form a more open interface than that between the two monomers of Mt adenylyl cyclase.
Figure 3.

Structure of Cmr2dHD bound with adenosine diphosphate (ADP) and Ca2+, and structure comparison with Mycobacterium tuberculosis adenylyl cyclase (Mt Ac). (A) Cmr2dHD binds to ADP (stick model) at its interface between D1 and D3 domain. Cyan spheres represent bound Ca2+ ions and red spheres represent water oxygen atoms. (B) Close-up view of ligand binding site of Cmr2dHD. ADP interacts with the P-loops (purple) of the D1 domain and the bound Ca2+ atoms interact with the β-hairpin (yellow) and P-loop (purple) of domain D3. (C) Crystal structure of the AB dimer (colored in blue and orange respectively) of the Mycobacterium tuberculosis (Mt) Rv1900c adenylyl cyclase bound with α, β-methyleneadenosine 5′-triphosphate (AMPCPP) ((Sinha et al., 2005), 1YBU). The Mt adenylyl cyclase dimer is displayed with its monomer A (orange) in the same orientation as D3 domain of Cmr2dHD. (D) Close-up view of ligand binding site of Mt adenyly cyclase. AMPCPP interacts primarily with the β-hairpin (yellow) and P-loop (purple) of monomer A in Mt adenylyl cyclase.
A β-hairpin in Mt adenylyl cyclase contains the degenerate GGDEF motif (GDGF) that is known to facilitate catalysis of phosphate cyclization (Figure 3D, Sinha et al, 2005). Adjacent to the β-hairpin is the P-loop structure with residues that participate in binding to the nucleotide (Sinha et al, 2005; Tesmer et al, 2000) (Figure 3D). The same architecture is observed in the D3 (but not in the D1) domain of Cmr2dHD (Figure 3B). The D3 β-hairpin contains a GGDD sequence motif (Gly671-Asp674) and is positioned next to the P-loop (residues Gly601 to Gly605) (Figure 3B and 3D, Figure 5A). In the D1 domain, the region corresponding to the GGDEF β-hairpin is replaced by an insertion of two α-helices (α3 and α4), although the P-loop is maintained (Figure 3B). The similarity that we found between the D3 domain of Cmr2 and Mt adenylyl cyclase monomer suggested that Cmr2dHD may bind nucleotides and metal ions.
Figure 5.

Differences in interaction of the conserved residues of Cmr2dHD and Mt adenylyl cyclase with nucleotide and metal ions. (A) Multiple sequence alignment of the most conserved portion of selected Cmr2 proteins across archaea and bacteria. Secondary structures are colored in the same scheme as in Figure 2 and are shown above the alignment (cylinders for α-helices, arrows for β-strands and lines for loops). Levels of conservation in amino acids are indicated by shades of blue with darker blue corresponding to greater conservation. The locations of the P-loop and β-hairpins are indicated. Residues that are mutated in Figure 6 or S3 are indicated by asterisks. (B) Detailed interactions at the ligand binding site of Pf Cmr2dHD. Residues within 3.5 Å of the bound ligands are displayed using the same color scheme as in Figure 2. Cyan spheres represent bound Ca2+ ions and red spheres represent water oxygen atoms. (C) Detailed interactions of Mt adenylyl cyclase active site. Residues are similarly identified and the purple sphere represents bound Mn2+. See also Figures S2, S3 and S5.
Cmr2dHD binds nucleotides and metal ions
To address how Cmr2dHD binds ADP, we compared ADP-free and ADP-bound structures. No major conformational changes were observed between the unbound and nucleotide-bound Cmr2dHD except for a loop-to-helix transition near the P-loop of D1 domain (α1, residues 229-234). This region is unstructured but forms a short helix upon ADP binding (Figure 4). Interestingly, although ADP is localized to the expected binding site near the GGDD β-hairpin and the P-loop on the D3 domain, it is bound in a different orientation in Cmr2dHD than in Mt adenylyl cyclase (Figure 3B and 3D; Figure 5B and 5C) (Sinha et al., 2005). In Mt adenylyl cyclase, an ATP analog, α,β-methyleneadenosine 5′-triphosphate (AMPCPP), inserts into a binding pocket formed by hydrophobic residues of monomer B and stacks on the peptide plane of Gly345 and Asp346 of monomer A (Figure 5C) (Sinha et al, 2005). The bound ADP in Cmr2dHD is also stabilized by residues from both domains D1 and D3. However, the details of the interaction and the location of the bound ATP are different between the two structures. The strictly conserved Asp674 of the GGDD motif of Cmr2dHD interacts with ADP by contacting its 2′ ribose hydroxyl group (Figure 5A and 5B). The adenine ring of ADP is sandwiched by Tyr669 of D3 β-hairpin, and Val229 and Ile233 of α1 in D1. It is clear that the loop-to-helix transition of α1 is critical for Val229 and Ile233 to stabilize the bound ADP. Ser250 and Ser246 of α2 make hydrogen bond contacts to the N6 and N1 atoms of the ADP. The phosphate backbone is stabilized by bound metal ions and water molecules that further interact with protein residues (Figure 3A and 5B). As a result, the ADP is closer to the P-loop of the D1 domain of Cmr2dHD than AMPCPP is to that of the Mt adenylyl cyclase monomer B (Figure 3B & 3D). The observed interactions suggest Cmr2 is specific for the adenine base. This is supported by the fact that Cmr2dHD crystals soaked with GTP, UTP or CTP did not yield electron densities corresponding to these nucleotides (data not shown).
Figure 4.
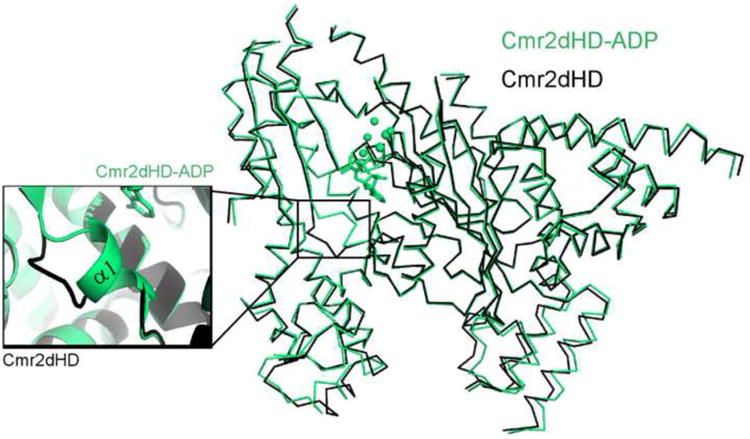
Overlay of the ADP-bound (green) and unbound (black) structures of Cmr2dHD. Conformational change is restricted to widening of the distance between the lower part of the D1 and D2 domains and the formation of the α1 helix at the nucleotide binding site (inset).
As in adenylyl cyclases, the GGDD motif of Cmr2dHD also binds divalent ions (Figure 5B). Metal binding sites were first suggested by high levels of Fourier difference densities and later confirmed by anomalous difference densities obtained from Mn2+ soaked crystals (Figure S2). Weaker difference density around the metal sites was also observed that could be attributed to coordinated water molecules. Since the best crystals were grown in the presence of 0.2 M Ca2+, we placed Ca2+ at the binding sites identified from the Mn2+-soaked crystals. The two Ca2+ ions are separated by 3.5 Å and are coordinated to protein, nucleotide ligands and ordered water molecules (Figure 5B; Figure S2). Although the precise coordination geometry cannot be established at the resolution of the structure, both Ca2+ ions appear to have an octahedral coordination geometry (Figure 5B). The B site calcium is coordinated to Asp673 of the GGDD motif, Asp600 and the amide of Gly601 of the P-loop, and two or possibly three ordered water molecules (Figure 5B). Unlike adenylyl cyclases where metal A coordinates with the pro-R oxygen of the α-phosphate, which was postulated to facilitate the nucleophilic attack of the 3′-hydroxyl group on the α-phosphate (Figure 5C), the calcium ion at the A metal site is coordinated to an oxygen of the β-phosphate of ADP, three aspartates (Asp673 and Asp674 from the GGDD motif and Asp600 from the P-loop), and two ordered water molecules (Figure 5B). The structural difference in nucleotide binding observed between Cmr2dHD and Mt adenylyl cyclase is consistent with the fact that Cmr2dHD (alone or in the context of the Cmr complex) does not release detectable phosphate or pyrophosphate when incubated with γ 32P-ATP or αββ32P-ATP (Figure S5).
Metal-coordinating aspartates of Cmr2 are not required for target RNA cleavage activity
The Asp cluster that coordinates both metals in Cmr2dHD is highly conserved (Figure 5A, (Makarova et al, 2002)). We hypothesized that the metal-binding site could be involved in phosphodiester bond cleavage as is observed in metal-dependent ribonucleases (Yang, 2011). Mutation of the catalytic metal-coordinating Asp residues in the RNase H family of nucleases abolished their RNA cleavage activity (Glavan et al, 2006; Nowotny et al, 2005). We mutated the Asp residues and tested crRNA-guided RNA cleavage activity of the Cmr complex containing the Cmr2dHD mutants. To our surprise, mutation of Asp600, Asp602, Asp673, or Asp673 and Asp674, did not abolish the cleavage activity of the Cmr complex (reconstituted with either the 39-nt and the 45-nt crRNA) (Figure 6 and Table S1). We further mutated other highly conserved residues that may or may not be involved in nucleotide binding. Again, mutants of Ser246, Ser250, Tyr669, Asp460 and His762 (Figure 6), or Glu475 and Lys739, or Lys744 (Figure S3) all maintained a significant level of catalytic activity. We thus conclude that the nucleotide and metal binding residues of Cmr2 are not required for Cmr complex-mediated target RNA cleavage.
Figure 6.
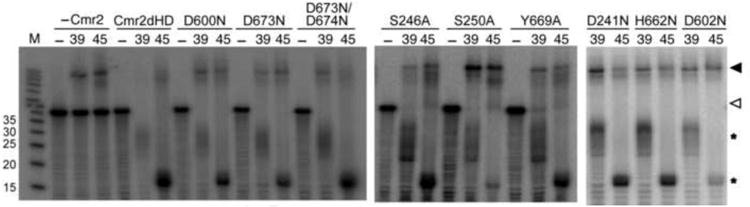
Mutational analysis of the nucleotide and metal-binding residues of Cmr2dHD. ReconstitutedCmr complexes (Cmr1-6, 1 μ M) containing no Cmr2 (-Cmr2), Cmr2dHD or mutants thereof (asindicated) were incubated with crRNAs (0.1 μ M) of 39 nt or 45 nt (as indicated) for 30 minutesbefore the addition of γ 32P-labeled target RNA (labeled by an open arrow). The reactionmixtures were incubated for 1 hour and analyzed on 15% denaturing polyacrylamide gel. – lanescontain no crRNA or proteins. Cleavage products are indicated by asterisks. Target RNA/crRNA duplexes are indicated by a closed arrow. See also Table S1.
Discussion
The Cmr2 protein from Pyrococcus furiosus is a large, multidomain protein whose sequence contains features reminiscent of DNA polymerases and nucleotide cyclases, which originally led to its classification as a CRISPR-Cas system polymerase (Makarova et al, 2002; Makarova et al, 2011a; Makarova et al, 2006). The crystal structure of a truncated but functional Cmr2 protein, Cmr2dHD, has revealed that the resemblance of Cmr2dHD to DNA polymerases is limited to a βαββαβ fold and secondary elements for binding divalent metals and nucleotides (Figure 2). Cmr2 more closely resembles homodimeric adenylyl cyclases in terms of protein fold, ligand binding sites, and overall architecture (Figures 3, 5). In addition, a Cys-cluster and two α-helical domains surround the adenylyl cyclase-like core in Cmr2.
D3 is the most conserved domain of Cmr2dHD. It contains a conserved GGDD β-hairpin and an adjacent P-loop that closely resemble those of cyclases (Mou et al, 2009; Sinha et al, 2005). The less conserved D1 domain also has a cyclase-like fold but lacks the equivalent β-hairpin. The interface of the two domains forms a metal and nucleotide binding site analogous to that of Mt adenylyladenylyl cyclase (Figures 3 and 5). Two divalent metal ions bound at the β-hairpin/P-loop are coordinated by three phylogenetically conserved aspartate residues (Asp600, Asp673, and Asp674) at this site (Figure 5). The bound ADP forms multiple hydrogen bond and hydrophobic interactions with highly conserved residues from both the β-hairpin/P-loop of D3 and the P-loop of D1. Despite their overall resemblance, important differences in domain orientation, metal coordination, and nucleotide location are observed between Cmr2dHD and Mt adenylyl cyclase. The interface between D1 and D3 of Cmr2dHD is more open than that between the two subunits of the Mt adenylyl cyclase. The metal A of Cmr2dHD is coordinated to the β-phosphate rather the α-phosphate pro-R oxygen as in Mt adenylyl cyclase. Thus, it is unlikely that Cmr2 supports catalysis of cyclization as Mt adenylyl cyclase does.
We discovered in this work that key conserved features of Cmr2 are not required for cleavage of target RNAs by the Cmr complex. The HD domain was hypothesized to serve as a nuclease domain (Makarova et al, 2002; Makarova et al, 2011a). Its removal from Cmr2, however, did not disrupt crRNA-guided RNA nuclease activity of the Cmr complex (Figure 1C). Furthermore, mutations of either nucleotide- or metal-binding aspartate residues also did not eliminate RNA cleavage activity. The HD domain and nucleotide and metal binding sites of Cmr2 are conserved and it was therefore somewhat surprising that mutations did not significantly impair the ability of Cmr2 to assemble into catalytically active Cmr complexes capable of cleaving target RNAs in vitro (Figure 6). Conceivably, these domains of Cmr2 are critical for a function that we did not assay, for example, a regulatory (perhaps efficiency or signaling) role in Cmr activity in vivo. These possibilities await confirmation from further biochemical and structural data.
Our findings suggest that Cmr2 is unlikely the catalytic subunit of the Cmr complex, and refocus the question of the identity of the catalytic subunit. Full activity of the Cmr complex requires five additional Cmr proteins (Hale et al., 2009). Four of these proteins (Cmr1, Cmr3, Cmr4, and Cmr6) belong to the greater RAMP family of Cas proteins (Haft et al., 2005; Makarova et al., 2002). Several RAMP proteins have been recently determined to function as metal-independent riboendonucleases (Brouns et al., 2008; Carte et al., 2008; Gesner et al., 2011; Haurwitz et al., 2010; Wang et al., 2011) and our findings suggest that that the catalytic activity of the Cmr effector complex resides in one or more of the RAMP proteins of the complex.
Experimental Procedures
Protein preparation
In order to construct the Cmr2dHD truncation, PCR was performed from the previously described PF1129-containing pET200D-TOPO plasmid (Hale et al., 2009) using 5′ CACCATGGTTAAGGATCCCACTTTGCTCAGG 3′ as a forward primer, and 5′ GTCATGCTAGCCATGTCATGGAAACACCTCCGCGTCCG 3′ as the reverse primer. PCR was performed using the Expand High Fidelity PCR system (Roche) as recommended. The resulting PCR product was cloned into the pET200D-TOPO plasmid using the manufacturer's protocol (Invitrogen). Resulting plasmids were sequenced to confirm the truncation. Point mutations were made using Quikchange (Agilent). The proteins were expressed in Escherichia coli BL21 RIPL cells (Agilent technologies, Santa Clara, CA). Cells were resuspended in lysis buffer (25 mM sodium phosphate at pH 7.5, 1.0 M NaCl, 10% (v/v) glycerol, 5.0 mM β-mercaptoethanol, and 0.2 mM phenylmethylsulfonyl fluoride) and disrupted by sonication. The cell debris was cleared by centrifugation. The supernatant was loaded onto a Ni-NTA column equilibrated with the lysis buffer supplemented with 5 mM imidazole. The column was washed with the lysis buffer containing 25 mM imidazole and the protein was eluted by increasing imidazole to 350 mM. Fractions containing the protein were pooled and loaded onto a Superdex 200 (Hiload 26/60, GE Healthcare) size-exclusion chromatography column that had been equilibrated with 20 mM Tris-HCl (pH 7.4), 500 mM NaCl, 5% (v/v) glycerol, and 5 mM β-mercaptoethanol. Fractions corresponding to the purified protein were pooled and concentrated.
Crystallization of native and L-Selenomethionine-labeled Cmr2dHD
Native and L-Selenomethionine (SeMet)-labeled Cmr2dHD were crystallized at 30°C by vapor diffusion method using hanging drops. Equal volumes of protein and well solution (0.1 mM HEPES sodium at pH 7.4, 0.2 M calcium chloride dihydrate, 28% (v/v) polyethylene glycol 400) were combined into 2.4 μl droplets on cover slips. Crystals formed in 6-7 days and had a hexagonal prism shape with typical dimensions of 0.4 mm × 1 mm × 0.4 mm.
Data collection and structure determination
Crystals were cryo-protected in a buffer containing 0.1 mM HEPES sodium (pH 7.4), 0.2 M calcium chloride dihydrate, 15% (v/v) polyethylene glycol 400, 1.0 M sodium chloride prior to being mounted on goniometer head. For the ADP-bound structure, 2 mM ADP was added to the cryo protecting solution. For the Mn2+-bound structure, native crystals were soaked in the cryo solution containing 0.2 M Mn2+ instead of Ca2+. X-ray diffraction data were collected at the Southeast Regional Collaborative Access Team (SER-CAT) beamline 22ID or 22BM. Data were indexed, integrated, and scaled using the HKL2000 software package (Otwinowski and Minor, 1997). The space group of the crystals was determined to be P212121 with one protein in the asymmetric unit and the cell dimensions are listed in Table 1. Phases for the Cmr2dHD structure were determined from a highly redundant single-wavelength anomalous dispersion (SAD) data set at the anomalous peak of selenium from a SeMet-labeled crystal. Structure determination, iterative model building and structure refinement were done using the PHENIX (Adams et al., 2010) and COOT programs (Emsley and Cowtan, 2004). The refinement statistics are shown in Table 1. The structure of the ADP-bound Cmr2dHD was obtained by molecular replacement using the ADP-free Cmr2dHD structure as the search probe.
RNA-cleavage reactions
Purified Cmr2 wild-type, Cmr2dHD, or a Cmr2dHD mutant and each of Cmr1, Cmr3, Cmr4, Cmr5, and Cmr6 were combined in equal molar amounts to a final concentration of 50 μM in 20 mM Tris-HCl (pH 7.4), 500 mM NaCl, 5% (v/v) glycerol, and 5 mM β-mercaptoethanol. The reconstituted complex was diluted to 1 μM when being incubated with either the 39-mer or the 45-mer Pf7.01 crRNA (Hale et al, 2009) at 0.1 μM for 30 min at 70°C in 20 mM sodium cacodylate (pH 6.2), 20 mM MgCl2 in the presence of 1 unit of SUPERase-In RNAse inhibitor. RNA cleavage reaction was initiated by adding 0.016 μM 5′-radiolabeled target RNA and the reaction was incubated for 1 hour at 70°C. The reaction was quenched by the addition of 96% formamide dye. The cleavage products were resolved on a 15% polyacrylamide, 7 M urea gel and visualized by phosphorimaging.
Supplementary Material
Acknowledgments
This work was supported by National Science Foundation grant MCB0817638 to H.L., American Heart Association 11PRE7090000 to A. C. and R01 GM54682 to M.T. and R.T. We thank Paul Griffith (UGA) for technical assistance in generating the Cmr2 HD domain mutation. X-ray diffraction data were collected from the Southeast Regional Collaborative Access Team (SER-CAT) 22-ID beamline at the Advanced Photon Source, Argonne National Laboratory.
Supporting institutions for APS beamlines may be found at http://necat.chem.cornell.edu/ and www.ser-cat.org/members.html. Use of the Advanced Photon Source was supported by the U. S. Department of Energy, Office of a Science, Office of Basic Energy Sciences, under Contract No. W-31-109-Eng-38.
Footnotes
Accession Numbers: Coordinates and structure factors have been deposited in the Protein Data Bank with accession number 3UR3 for the Cmr2 structure and 3UNG for the ADP-bound Cmr2 structure.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anantharaman V, Iyer LM, Aravind L. Presence of a classical RRM-fold palm domain in Thg1-type 3′- 5′nucleic acid polymerases and the origin of the GGDEF and CRISPR polymerase domains. Biol Direct. 2010;5:43. doi: 10.1186/1745-6150-5-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artymiuk PJ, Poirrette AR, Rice DW, Willett P. A polymerase I palm in adenylyl cyclase? Nature. 1997;388:33–34. doi: 10.1038/40310. [DOI] [PubMed] [Google Scholar]
- Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- Beloglazova N, Petit P, Flick R, Brown G, Savchenko A, Yakunin AF. Structure and activity of the Cas3 HD nuclease MJ0384, an effector enzyme of the CRISPR interference. EMBO J. 2011;30:4616–4627. doi: 10.1038/emboj.2011.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolotin A, Quinquis B, Sorokin A, Ehrlich SD. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology. 2005;151:2551–2561. doi: 10.1099/mic.0.28048-0. [DOI] [PubMed] [Google Scholar]
- Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, Dickman MJ, Makarova KS, Koonin EV, van der Oost J. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carte J, Wang R, Li H, Terns RM, Terns MP. Cas6 is an endoribonuclease that generates guide RNAs for invader defense in prokaryotes. Gene Dev. 2008;22:3489–3496. doi: 10.1101/gad.1742908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deveau H, Garneau JE, Moineau S. CRISPR/Cas system and its role in phage-bacteria interactions. Annu Rev Microbiol. 2010;64:475–493. doi: 10.1146/annurev.micro.112408.134123. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Garneau JE, Dupuis ME, Villion M, Romero DA, Barrangou R, Boyaval P, Fremaux C, Horvath P, Magadan AH, Moineau S. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468:67–71. doi: 10.1038/nature09523. [DOI] [PubMed] [Google Scholar]
- Gesner EM, Schellenberg MJ, Garside EL, George MM, Macmillan AM. Recognition and maturation of effector RNAs in a CRISPR interference pathway. Nat Struct Mol Biol. 2011;18:688–692. doi: 10.1038/nsmb.2042. [DOI] [PubMed] [Google Scholar]
- Haft DH, Selengut J, Mongodin EF, Nelson KE. A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Comput Biol. 2005;1:e60. doi: 10.1371/journal.pcbi.0010060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale C, Kleppe K, Terns RM, Terns MP. Prokaryotic silencing (psi)RNAs in Pyrococcus furiosus. RNA. 2008;14:2572–2579. doi: 10.1261/rna.1246808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale CR, Majumdar S, Elmore J, Pfister NT, Compton MM, Olson S, Resch AM, Glover CVC, Graveley BR, Terns RM, et al. Essential features and rational design of CRISPR RNAs that function with the Cas RAMP module complex to cleave RNAs. Mol Cell. 2011 doi: 10.1016/j.molcel.2011.10.023. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale CR, Zhao P, Olson S, Duff MO, Graveley BR, Wells L, Terns RM, Terns MP. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell. 2009;139:945–956. doi: 10.1016/j.cell.2009.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haurwitz RE, Jinek M, Wiedenheft B, Zhou K, Doudna JA. Sequence- and structure-specific RNA processing by a CRISPR endonuclease. Science. 2010;329:1355–1358. doi: 10.1126/science.1192272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath P, Romero DA, Coute-Monvoisin AC, Richards M, Deveau H, Moineau S, Boyaval P, Fremaux C, Barrangou R. Diversity, activity, and evolution of CRISPR loci in Streptococcus thermophilus. J Bacteriol. 2008;190:1401–1412. doi: 10.1128/JB.01415-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard JA, Delmas S, Ivancic-Bace I, Bolt EL. Helicase dissociation and annealing of RNA-DNA hybrids by Escherichia coli Cas3 protein. Biochem J. 2011;439:85–95. doi: 10.1042/BJ20110901. [DOI] [PubMed] [Google Scholar]
- Jore MM, Lundgren M, van Duijn E, Bultema JB, Westra ER, Waghmare SP, Wiedenheft B, Pul U, Wurm R, Wagner R, et al. Structural basis for CRISPR RNA- guided DNA recognition by Cascade. Nat Struct Mol Biol. 2011;18:529–536. doi: 10.1038/nsmb.2019. [DOI] [PubMed] [Google Scholar]
- Karginov FV, Hannon GJ. The CRISPR system: small RNA-guided defense in bacteria and archaea. Mol cell. 2010;37:7–19. doi: 10.1016/j.molcel.2009.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lintner NG, Kerou M, Brumfield SK, Graham S, Liu H, Naismith JH, Sdano M, Peng N, She Q, Copie V, et al. Structural and functional characterization of an archaeal clustered regularly interspaced short palindromic repeat (CRISPR)-associated complex for antiviral defense (CASCADE) J Biol Chem. 2011;286:21643–21656. doi: 10.1074/jbc.M111.238485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Aravind L, Grishin NV, Rogozin IB, Koonin EV. A DNA repair system specific for thermophilic Archaea and bacteria predicted by genomic context analysis. Nucleic Acids Res. 2002;30:482–496. doi: 10.1093/nar/30.2.482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Aravind L, Wolf YI, Koonin EV. Unification of Cas protein families and a simple scenario for the origin and evolution of CRISPR-Cas systems. Biol Direct. 2011a;6:38. doi: 10.1186/1745-6150-6-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Grishin NV, Shabalina SA, Wolf YI, Koonin EV. A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol Direct. 2006;1:7. doi: 10.1186/1745-6150-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Haft DH, Barrangou R, Brouns SJ, Charpentier E, Horvath P, Moineau S, Mojica FJ, Wolf YI, Yakunin AF, et al. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol. 2011b;9:467–477. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraffini LA, Sontheimer EJ. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science. 2008;322:1843–1845. doi: 10.1126/science.1165771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraffini LA, Sontheimer EJ. CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nat Rev Genet. 2010a;11:181–190. doi: 10.1038/nrg2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraffini LA, Sontheimer EJ. Self versus non-self discrimination during CRISPR RNA-directed immunity. Nature. 2010b;463:568–571. doi: 10.1038/nature08703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mojica FJ, Diez-Villasenor C, Garcia-Martinez J, Soria E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J Mol Evol. 2005;60:174–182. doi: 10.1007/s00239-004-0046-3. [DOI] [PubMed] [Google Scholar]
- Mou TC, Masada N, Cooper DM, Sprang SR. Structural basis for inhibition of mammalian adenylyl cyclase by calcium. Biochemistry. 2009;48:3387–3397. doi: 10.1021/bi802122k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulepati S, Bailey S. Structural and Biochemical Analysis of Nuclease Domain of Clustered Regularly Interspaced Short Palindromic Repeat (CRISPR)-associated Protein 3 (Cas3) J Biol Chem. 2011;286:31896–31903. doi: 10.1074/jbc.M111.270017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Method Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Papadopoulos JS, Agarwala R. COBALT: constraint-based alignment tool for multiple protein sequences. Bioinformatics. 2007;23:1073–1079. doi: 10.1093/bioinformatics/btm076. [DOI] [PubMed] [Google Scholar]
- Pourcel C, Salvignol G, Vergnaud G. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology. 2005;151:653–663. doi: 10.1099/mic.0.27437-0. [DOI] [PubMed] [Google Scholar]
- Sapranauskas R, Gasiunas G, Fremaux C, Barrangou R, Horvath P, Siksnys V. The Streptococcus thermophilus CRISPR/Cas system provides immunity in Escherichia coli. Nucleic Acids Research. 2011 doi: 10.1093/nar/gkr606. in press. Epub ahead of print retrieved November 23, 2011, http://nar.oxfordjournals.org/content/early/2011/08/03/nar.gkr606.long. [DOI] [PMC free article] [PubMed]
- Semenova E, Jore MM, Datsenko KA, Semenova A, Westra ER, Wanner B, van der Oost J, Brouns SJ, Severinov K. Interference by clustered regularly interspaced short palindromic repeat (CRISPR) RNA is governed by a seed sequence. Proc Natl Acad Sci U S A. 2011;108:10098–10103. doi: 10.1073/pnas.1104144108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha SC, Wetterer M, Sprang SR, Schultz JE, Linder JU. Origin of asymmetry in adenylyl cyclases: structures of Mycobacterium tuberculosis Rv1900c. EMBO J. 2005;24:663–673. doi: 10.1038/sj.emboj.7600573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinkunas T, Gasiunas G, Fremaux C, Barrangou R, Horvath P, Siksnys V. Cas3 is a single-stranded DNA nuclease and ATP-dependent helicase in the CRISPR/Cas immune system. EMBO J. 2011;30:1335–1342. doi: 10.1038/emboj.2011.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terns MP, Terns RM. CRISPR-based adaptive immune systems. Curr Opin Microbiol. 2011;14:321–327. doi: 10.1016/j.mib.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesmer JJ, Dessauer CW, Sunahara RK, Murray LD, Johnson RA, Gilman AG, Sprang SR. Molecular basis for P-site inhibition of adenylyl cyclase. Biochemistry. 2000;39:14464–14471. doi: 10.1021/bi0015562. [DOI] [PubMed] [Google Scholar]
- Wang R, Preamplume G, Terns MP, Terns RM, Li H. Interaction of the Cas6 riboendonuclease with CRISPR RNAs: recognition and cleavage. Structure. 2011;19:257–264. doi: 10.1016/j.str.2010.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Lee HS, Sugar FJ, Jenney FE, Jr, Adams MW, Prestegard JH. PF0610, a novel winged helix-turn-helix variant possessing a rubredoxin-like Zn ribbon motif from the hyperthermophilic archaeon, Pyrococcus furiosus. Biochemistry. 2007;46:752–761. doi: 10.1021/bi061870h. [DOI] [PubMed] [Google Scholar]
- Wiedenheft B, Lander GC, Zhou K, Jore MM, Brouns SJ, van der Oost J, Doudna JA, Nogales E. Structures of the RNA-guided surveillance complex from a bacterial immune system. Nature. 2011a;477:486–489. doi: 10.1038/nature10402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedenheft B, van Duijn E, Bultema JB, Waghmare SP, Zhou K, Barendregt A, Westphal W, Heck AJ, Boekema EJ, Dickman MJ, et al. RNA-guided complex from a bacterial immune system enhances target recognition through seed sequence interactions. Proc Natl Acad Sci U S A. 2011b;108:10092–10097. doi: 10.1073/pnas.1102716108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W. Nucleases: diversity of structure, function and mechanism. Q Rev Biophys. 2011;44:1–93. doi: 10.1017/S0033583510000181. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


