Abstract
The GATA family proteins Gln3p and Gat1p mediate nitrogen catabolite repression (NCR)-sensitive transcription in Saccharomyces cerevisiae. When cells are cultured with a good nitrogen source (glutamine, ammonia), Gln3p and Gat1p are restricted to the cytoplasm, whereas with a poor nitrogen source (proline), they localize to the nucleus, bind to the GATA sequences of NCR-sensitive gene promoters, and activate transcription. The target of rapamycin-signaling cascade and Ure2p participate in regulating the cellular localization of Gln3p and Gat1p. Rapamycin, a Tor protein inhibitor, like growth with a poor nitrogen source, promotes nuclear localization of Gln3p and Gat1p. gln3Δ and ure2Δ mutants are partially resistant and hypersensitive to growth inhibition by rapamycin, respectively. We show that a vid30Δ is more rapamycin-sensitive than wild type but less so than a ure2Δ. VID30 expression is modestly NCR-sensitive, responsive to deletion of URE2, and greatly increases in low ammonia medium. Patterns of gene expression in a vid30Δ suggest that the Vid30p function shifts the balance of nitrogen metabolism toward the production of glutamate, especially when cells are grown in low ammonia. CAN1, DAL4, DAL5, MEP2, DAL1, DAL80, and GDH3 transcription is down-regulated by Vid30p function with proline as the nitrogen source. An effect, however, that could easily be indirect.
Saccharomyces cerevisiae has evolved to live in widely varying nutritional environments from rich YPD medium in a laboratory flask to poor, depleted soil. A significant contributor to the organism’s success is its ability to selectively use a variety of nitrogen sources. The basis of this selectivity is nitrogen catabolite repression (NCR).1 Expression of nearly all genes whose products participate in the uptake and catabolism of nitrogenous compounds are NCR-sensitive and dependent upon two GATA family transcription factors, Gln3p and Gat1p/Nil1p (see Refs. 1–4 for reviews of the field). When nitrogen availability limits growth, as occurs with poor nitrogen sources such as proline, Gln3p and/or Gat1p bind to the GATA sequences upstream of NCR-sensitive genes and activate their transcription (5–11). However, in nitrogen excess, the GATA sites are unoccupied because Gln3p and Gat1p are excluded from the nucleus (12–15). Ure2p, which binds to Gln3p and Gat1p (15–17), has long been known to be a negative regulator of NCR-sensitive gene expression and more recently Gln3p- and Gat1p-mediated transcription (12–20).
In a manner that appears to be similar to that which occurs when cells are provided with a poor nitrogen source, the macrolide antibiotic rapamycin participates in the regulation of Gln3p and Gat1p intracellular localization (12, 15, 17). Rapamycin specifically binds Fpr1p (peptidylprolyl isomerase), and the resulting complex binds to and inactivates Tor1p and Tor2p protein kinases (21, 22). Inactivation of Tor1p/Tor2p has been implicated in a broad range of cellular functions, including cell cycle progression (22–24), translation initiation (24), ribosome biosynthesis (13), autophagy (25, 26), amino acid permease stability (27), and microtubule assembly (28). More recently, genome-wide transcriptional analyses have shown that a substantial fraction of the total transcriptome changes following rapamycin addition to the medium (13, 14, 29).
A group of genes significantly affected in the transcriptome analyses were those whose expression is NCR-sensitive, i.e. Gln3p-/Gat1-dependent (13, 14, 29). Deletion of GLN3 results in partial resistance to rapamycin, while ure2Δ mutants are hypersensitive (13). The phosphorylation state of Gln3p/Gat1p and Ure2p correlates with these data. Gln3p, Gat1p, and Ure2p are hyperphosphorylated in cells provided with excess nitrogen and underphosphorylated when nitrogen is limiting or rapamycin is added to the medium (12–14, 17). Together these data suggest that the Tor proteins play a role in transduction of the signal that responds to the cell’s nitrogen supply.
A search of the literature for rapamycin-related molecules identified a protein about which little is known, the product of the YGL227w open reading frame, which has been given the temporary designations Vid30p and Tin1p.2 Since so little is known about this gene, we investigated its transcription profile and mutant phenotype. Here we show that VID30 expression possesses characteristics expected of an NCR-sensitive gene. Although a vid30Δ mutant exhibits alterations in NCR-sensitive gene expression, the data we collected are most consistent with the suggestion that Vid30p function shifts nitrogen metabolism toward the formation of glutamate, especially when cells are grown in limiting ammonia.
EXPERIMENTAL PROCEDURES
Strains and Media
S. cerevisiae strains used in this work are all derivatives of wild type BY4742 (Table I). Yeast were cultured at 30 °C in YNB-glucose medium (0.17% yeast nitrogen base without ammonium sulfate or amino acids (Difco), 2% glucose; quantities of amino acids (in micrograms) were provided to cover auxotrophies where necessary). Proline (0.1%), ammonium sulfate (0.05 or 7.6 mM [0.1%]), or glutamine (0.1%) were provided as sole nitrogen sources. For all experiments, cells were harvested at a density of A600 nm ~0.8 and used for total RNA extraction.
Table I.
Strains used in this research
| Strain | Genotype |
|---|---|
| S. cerevisiae | |
| BY4742 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 |
| BY14594 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 vid30Δ |
| BY11983 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 ure2Δ |
| BY10173 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 gln3Δ |
| E. coli | |
| DH5α | E. coli DH5α F′/endA1 hsdR17(rK–mK+) supE44 thi1 recA1 gyrA (Nalr) relA1 Δ(lacZYA-argF)U169 (m80lacZΔM15) |
The transcriptional effects of overexpressing VID30 were analyzed by growing transformants (pYES2 or pYES-V) of BY4742 in YNB-raffinose/galactose medium (1.5% raffinose + 0.5% galactose) with proline (0.1%) or glutamine (0.1%). Effects of VID30 overexpression on cell growth were determined by growing wild type transformants (pYES2 or pYES-V) to exponential phase at 30 °C in YNB-glucose (2%)-ammonium sulfate (0.1%) medium. Cells were harvested, washed, resuspended in sterile water, and streaked onto YNB plates containing either 2% glucose or 2% galactose and 0.1% proline, 0.1% glutamine, or 0.1% ammonium sulfate as sole carbon and nitrogen sources, respectively.
To analyze the effects of rapamycin, vid30Δ, ure2Δ, gln3Δ, and wild type strains growing exponentially in YPD medium were harvested, washed, resuspended in sterile water, and streaked onto YPD or YPD + 50 ng/ml rapamycin solid media. The plates were incubated at 30 °C, 3 days for YPD and 5 days for YPD + rapamycin.
Northern Analysis
Total RNA was isolated as described previously (31). Poly(A)+ RNA was obtained using mRNA Isolation Kits (Roche Molecular Biochemicals) according to the manufacturer’s recommendations. Samples of poly(A)+ RNA were resolved on 1.4% agarose-formaldehyde gels and transferred to either GeneScreen Plus nylon membranes 66 (PerkinElmer Life Sciences) or positively charged nylon membranes (Roche Molecular Biochemicals). Double-stranded DNA probes (radioactively labeled using a Random Primed DNA Labeling Kit [Roche Molecular Biochemicals]) were synthesized by PCR using the oligonucleotides in Table II as primers. pC4 (loading and transfer standard) and pTSC317 (DAL80) were also used as probes (32, 33). Oligonucleotides used as probes for GDH1 and GDH3 (Table II) were end-labeled using polynucleotide kinase (Roche Molecular Biochemicals). Standard prehybridization, hybridization, and washing conditions were followed (31).
Table II.
Oligonucleotides used for PCR amplifications
| Target gene | Oligonucleotide |
|---|---|
| CAN1 | CAN15: 5′-ATGAGCCGGTCACAACCCTC-3′ |
| CAN13: 5′-GATGGAAGCGACCCAGAACT-3′ | |
| DAL1 | DAL15: 5′-TGGCGTTCGCGGGTTCAAAG-3′ |
| DAL13: 5′-TGCAACTTTAGTTTAACGTC-3′ | |
| DAL4 | DAL45: 5′-ATGGCTAACGACGCTCT-3′ |
| DAL43: 5′-TATGACACAATAGATGT-3′ | |
| DAL5 | DAL55: 5′-CAGTATTCATGGGTTACTTCC-3′ |
| DAL53: 5′-TAAGGTTCTCCAGCCTTTAAT-3′ | |
| GDH1a | GDH1-P: 5′-ACCTTACCGTCCTTAGTGTACTTCTTGGCATAGTCG-3′ |
| GDH2 | GDH25: 5′-ATAACAAAAATCGCGGTG-3′ |
| GDH23: 5′-TATCCTTACCATGCTCCA-3′ | |
| GDH3a | GDH3-P: 5′-GTGTTTGTATTTTTTTCCGTAGAGTACTCTTGTGCGGCCTGT-3′ |
| GLN1 | GLN15: 5′-ATGGCTGAAGCAAGCATC-3′ |
| GLN13: 5′-CTTACCGGCACCAACACC-3′ | |
| GLT1 | GLT15: 5′-TATTAGGTTGGAGAAACG-3′ |
| GLT13: 5′-AGTTCTAAAACGTTATCC-3′ | |
| HHF1 | H45: 5′-GGCCGGATCCATGTCCGGTAGAGGTAAAGG-3′ |
| H43: 5′-GCCGAATTCTTAACCACCGAAACCGTATAAGG-3′ | |
| MEP2 | MEP25: 5′-GTCTTACAATTTTACAGG-3′ |
| MEP23: 5′-TACCCAATTTGACCAACC-3′ | |
| VID30 | VID305: 5′-ATGTCTGAATATATGGATGA-3′ |
| VID303: 5′-TCACGTTCGGATAAAACGGG-3′ | |
| VKpn-5: 5′-AATTGGTACCAAATGTCTGAATATATGGAT-3′ | |
| VXho-3: 5′-TTGGCTCGAGAATGACTGATATCACATGGC-3′ |
Oligonucleotides were end-labeled and used as probes.
Plasmid Construction
Overexpression of VID30 was achieved by fusing it to the GAL1 promoter in multicopy pYES2 (Invitrogen Corporation, Carlsbad, CA). VID30 was amplified from S. cerevisiae genomic DNA using heat-stable DNA polymerase PWO, and primers VKpn-5 and VXho-3 (Table II), into which KpnI and XhoI sites had been engineered, respectively. The resulting PCR product was digested with KpnI and XhoI, yielding a 3,028 base pair fragment, which was cloned into the KpnI and XhoI sites of pYES2 to create pYES-V.
RESULTS
VID30 Expression Is NCR-sensitive and Rapamycin-responsive
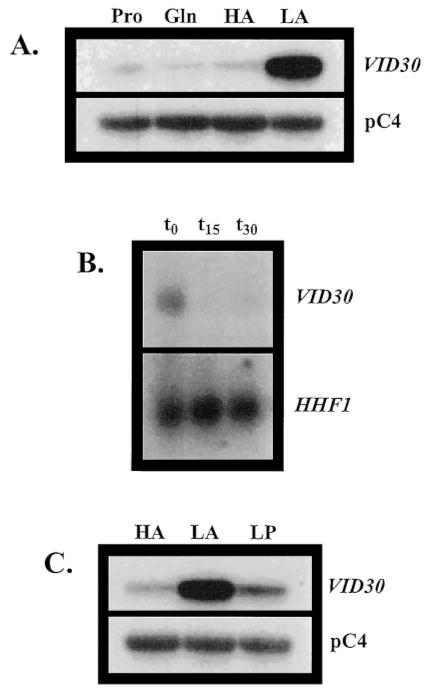
To ascertain whether Vid30p played a part in the rapamycin cascade that participates in the regulation of NCR-sensitive gene expression, we compared the effects of the TOR protein inhibitor, rapamycin, on wild type and mutant cells. Growth of ure2Δ strains are hypersensitive to rapamycin, while gln3Δ mutants are more resistant than wild type (13, 17). A vid30Δ mutant is slightly less hypersensitive than a ure2Δ strain (Fig. 1), implying a relationship potentially exits between the TOR signaling cascade and Vid30p function. This prompted us to determine whether VID30 expression was NCR-sensitive by assaying steady-state VID30 mRNA derived from cells cultured in YNB-proline (nitrogen-derepressive) and YNB-glutamine (nitrogen-repressive) media. VID30 expression is modestly NCR-sensitive (Fig. 2A); this result can be even more clearly observed when the autoradiogram is highly overexposed. The NCR sensitivity of VID30 expression and rapamycin sensitivity of a vid30Δ mutant predicted a response of VID30 expression to deletion of URE2 or addition of rapamycin to the medium. In agreement with these expectations, VID30 expression increases following addition of 200 ng/ml rapamycin to nitrogen-rich medium, but less dramatically so than occurs with the control gene, DAL5 (Fig. 2B). VID30 expression in cells provided with high ammonia or glutamine (excess nitrogen) also increases when URE2 is deleted (Fig. 2C). It is significant that the magnitude of NCR sensitivity, rapamycin-mediated induction, and ure2Δ-generated derepression of VID30 expression are quantitatively similar. Together, these data argue that VID30 expression is modestly NCR-sensitive.
Fig. 1. Deletion of VID30 confers hypersensitivity to rapamycin.
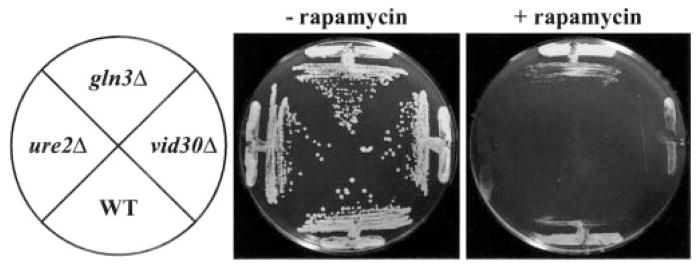
Wild type (WT; BY4742), gln3Δ (BY10173), ure2Δ (BY11983), and vid30Δ (BY14594) strains grown on rich medium containing (+Rap) or devoid (−Rap) of rapamycin (50 ng/ml). Plates were incubated at 30 °C for three (−Rap) and 5 days (+Rap), respectively.
Fig. 2. VID30 expression is NCR-sensitive and induced by the TOR protein inhibitor rapamycin.
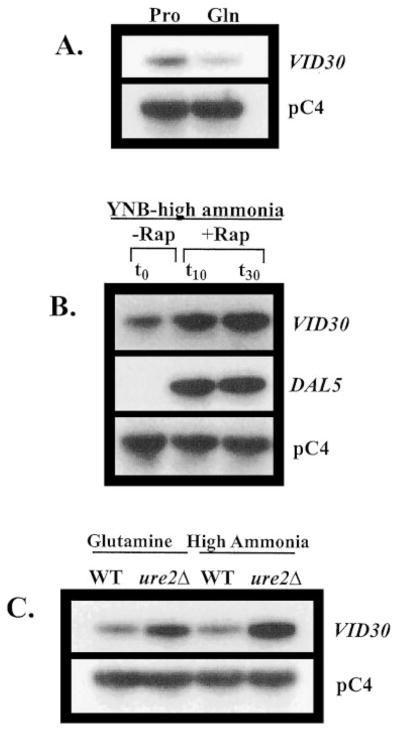
A, Northern blot analysis of poly(A)+ RNA (1 μg/lane) from BY4742 grown in YNB-0.1% proline (Pro) and YNB-0.1% glutamine (Gln). B, Northern blot analysis of poly(A)+ RNA (5 μg/lane) from BY4742 grown in YNB-0.1% ammonium sulfate to late log phase (t0) and then 10 min (t10) and 30 min after (t30) addition of rapamycin to a final concentration of 200 ng/ml to the growth medium. C, poly(A)+ RNA (2.5 μg/lane) from strains BY4742 (WT) and BY11983 (ure2Δ) grown in either YNB-0.1% glutamine or YNB-0.1% ammonium sulfate. pC4 was used as the loading standard. WT, wild type.
Vid30p Negatively Regulates Expression of Multiple Genes Associated with Nitrogen Catabolism
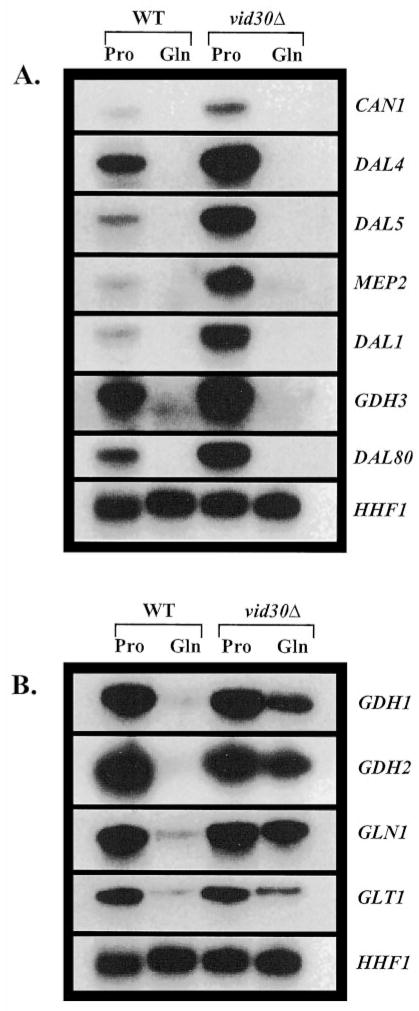
The similar hypersensitivities of vid30Δ and ure2Δ mutants to rapamycin prompted us to investigate whether Vid30p, like Ure2p, might be a negative regulator of NCR-sensitive gene expression. We analyzed expression of representative NCR-sensitive genes in wild type and vid30Δ strains. Included in the analysis were genes encoding (i) permeases: MEP2 (low capacity ammonia), DAL4 (allantoin), DAL5 (allantoate), and CAN1 (basic amino acid); (ii) enzymes for the interconversion of ammonia, glutamate, and glutamine: GDH1 (NADP-glutamate dehydrogenase), GDH3 (NADP-glutamate dehydrogenase), GDH2 (NAD-glutamate dehydrogenase), GLN1 (glutamine synthetase), and GLT1 (glutamate synthase (GOGAT, glutamine amide: 2-oxoglutarate aminotransferase)); (iii) an enzyme participating in catabolism of allantoin: DAL1 (allantoinase); and (iv) the GATA-specific transcriptional repressor: DAL80.
Deletion of VID30 increases CAN1, DAL4, DAL5, MEP2, DAL1, GDH3, and DAL80 mRNA levels 2–10-fold in YNB-proline-cultured cells (Fig. 3A). However, the NCR sensitivity of these genes is not affected, i.e. there is no alteration of expression when a vid30Δ mutant is growing in glucose-glutamine medium (Fig. 3A). At this point, we cannot rigorously distinguish whether the effects observed when a vid30Δ mutant is grown in glucose-proline medium are direct or indirect.
Fig. 3. Expression of genes associated with nitrogen metabolism in wild type and vid30Δ strains provided with a repressive (glutamine) or derepressive (proline) nitrogen source.
Northern blot analysis of poly(A)+ RNA (1 μg/lane) wild type (BY4742) and vid30Δ (BY14594) strains grown in either YNB-0.1% proline (Pro) or YNB-0.1% glutamine (Gln). Probes are indicated on the sides of the panels.
In contrast to typical NCR-sensitive genes, the expression profiles of genes associated with the interconversion of ammonia, glutamate, and glutamine (GDH1, GDH2, GLN1, and GLT1) are quite different in two respects (Fig. 3B): (i) these genes are expressed more or less equivalently when wild type and vid30Δ strains are grown in YNB-proline medium, and (ii) their expression is markedly increased when a vid30Δ mutant growing in YNB-glutamine medium is compared with wild type; we estimate the increase to be in the range of 2–10-fold (Fig. 3B). These results argue that Vid30p negatively regulates CAN1, DAL4, DAL5, MEP2, DAL1, GDH3, and DAL80 when cells are growing with proline as the nitrogen source and GDH1, GDH2, GLN1, and GLT1 when cells are provided with glutamine. However, deletion of VID30 does not restore expression of the second group of genes in glucose-glutamine grown cells to the level seen in the glucose-proline medium. They are more highly expressed in proline medium regardless of whether or not Vid30p is present (Fig. 3B), indicating that negative regulation by Vid30p is shared, in a formal sense, by at least one other protein.
Low Ammonia-mediated VID30 Expression
Although VID30 expression is modestly NCR-sensitive, more striking is the dramatic increase in VID30 expression that occurs when cells are grown in medium containing low ammonia (0.05 mM) (Fig. 4A). VID30 expression in low ammonia medium is more than 10-fold greater than with proline as nitrogen source (Fig. 4A). This high level of VID30 expression is lost within 15 min after the ammonia concentration in the culture medium is increased to 0.1% (Fig. 4B).
Fig. 4. VID30 expression is enhanced by low ammonia.
A, Northern blot analysis of poly(A)+ RNA (1 μg/lane) from BY4742 grown in YNB-0.1% proline (Pro), YNB-0.1% glutamine (Gln), YNB-0.1% ammonium sulfate (HA), or YNB-0.05 mM ammonium sulfate (LA). B, BY4742 was grown in YNB-0.05 mM ammonium sulfate. Total RNA (30 μg/lane) were extracted from BY4742 cells grown to late log phase in YNB-0.05 mM ammonium sulfate (t0), and then 15 min after (t15) and 30 min after (t30) addition of ammonium sulfate to a final concentration of 0.1% to the growth medium. C, Northern blot analysis of poly(A)+ RNA (1 μg/lane) from BY4742 grown in YNB-0.1% ammonium sulfate (HA), YNB-0.05 mM ammonium sulfate (LA), and YNB-0.1 mM proline (LP). HHF1 and pC4 were used as loading standards.
The VID30 expression we observe in low ammonia medium may conceivably be derived in two ways: (i) a response of VID30 expression to the low concentration of ammonia or (ii) nitrogen starvation per se. To distinguish these possibilities, we divided a culture into two portions, one being transferred to low ammonia and the other a similarly low concentration of proline. As shown in Fig. 4C, the response was observed only in low ammonia medium, arguing against starvation as the driving force behind increased VID30 expression.
Vid30p Acts as a Positive Regulator in Low Ammonia
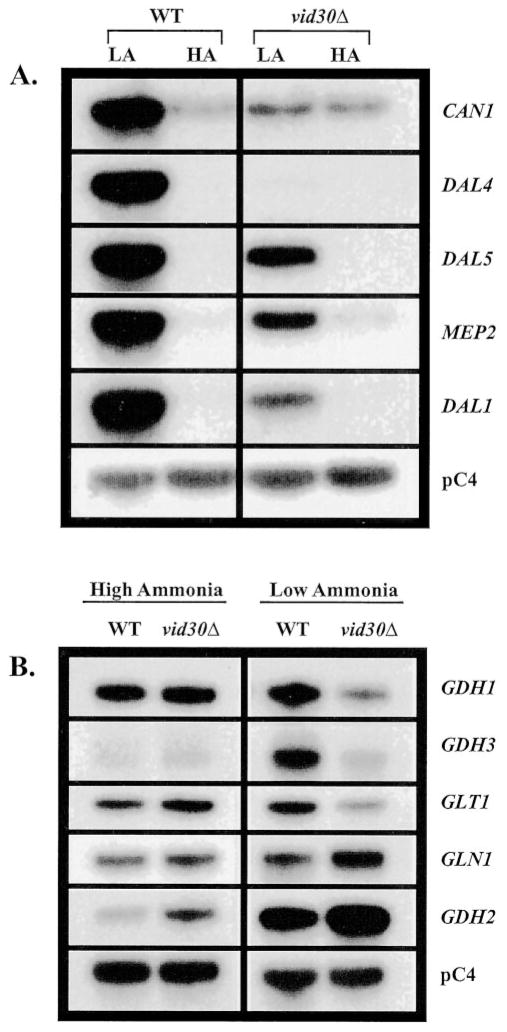
High level VID30 expression in low ammonia (0.05 mM) medium, and its loss when ammonia concentrations are high (7.6 mM (0.1%)), prompted us to investigate Vid30p regulation of other nitrogen metabolism-related genes under similar experimental conditions. We analyzed expression of the two groups of genes described in Fig. 3 in wild type and vid30Δ strains. For CAN1, DAL4, DAL5, MEP2, and DAL1, the fold increase observed when comparing expression in low versus high ammonia-grown cells, is smaller in the vid30Δ mutant than the wild type (Fig. 5A). This result derives from the fact that deletion of VID30 decreases expression of these genes in low ammonia medium with no demonstrable effect in high ammonia medium. These results suggest that Vid30p acts as a positive regulator of CAN1, DAL4, DAL5, MEP2, and DAL1 gene expression in low ammonia. Taken together, these data and those in Fig. 3 suggest that Vid30p functions as a negative regulator with proline as the nitrogen source and as a positive regulator with low ammonia.
Fig. 5. Expression of genes associated with nitrogen metabolism in wild type and vid30Δ strains grown in high and low ammonia.
Northern blot analysis of poly(A)+ RNA (1 μg/lane) from wild type (BY4742) and vid30Δ (BY14594) strains grown in YNB-0.1% ammonium sulfate (HA) or YNB-0.05 mM ammonium sulfate (LA).
With exception of GDH2, deletion of VID30 does not effect the expression of ammonia-glutamate-glutamine interconversion genes when cells are growing in high ammonia medium (Fig. 5B). The steady-state level of GDH2 mRNA modestly increases in the vid30Δ. In addition, there is no detectable expression of GDH3, which is not surprising given its high NCR sensitivity and the fact that high ammonia is a repressive nitrogen source. On the other hand, in low ammonia medium, there is less expression of the GDH1, GDH3, and GLT1 genes in vid30Δ than wild type. GDH2 and maybe GLN1 are the exceptions, i.e. their expression increases rather than decreases when the vid30Δ mutant is grown in low ammonia (Fig. 5B). Therefore, Vid30p behaves as a positive regulator for all of the interconversion genes in low ammonia-grown cells with the exception of GDH2 and GLN1, where control is modestly negative.
Overexpression of VID30 Inhibits Cell Growth and Alters Nitrogen-regulated Gene Expression
The NCR-sensitive, rapamycin-responsive expression of VID30 prompted us to examine the effect of VID30 overexpression on cell growth in different nitrogen conditions. VID30 was fused to the GAL1 promoter in pYES2, allowing VID30 expression to be induced independently of the nitrogen source in the medium. Wild type strain BY4742, transformed with vector pYES2, grows similarly on YNB-glucose or YNB-galactose medium containing 0.1% ammonium sulfate or glutamine as the sole nitrogen source (Fig. 6). The transformants grow more slowly, however, on YNB-galactose-proline medium. Cells transformed with GAL1-VID30 pYES-V grow like the control cells on YNB-glucose, irrespective of the nitrogen source supplied. However, growth on all three nitrogen sources is inhibited when VID30 is overexpressed (galactose as a carbon source) (Fig. 6). These results indicate that controlled expression of VID30 is essential for optimal cell growth.
Fig. 6. Overexpression of VID30 negatively affects cell growth.
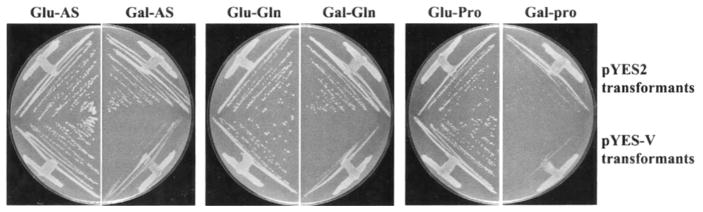
Growth of BY4742 transformed with pYES-V that overexpresses VID30; pYES2 was used as a negative control. Transformants were grown on YNB-2% glucose and YNB-2% galactose media containing either 0.1% ammonium sulfate (Glu-AS and Gal-AS), 0.1% glutamine (Glu-Gln and Gal-Gln) or 0.1% proline (Glu-Pro and Gal-Pro) as sole nitrogen sources. Ammonium sulfate- and glutamine-containing plates were incubated for 2 days and proline-containing plates for 4 days at 30 °C.
Next, we examined the effect of VID30 overexpression (Fig. 7, top panels) on the transcription of nitrogen-associated genes in cells provided with different nitrogen sources. If anything, Vid30p exhibits characteristics of a very modest negative regulator of NCR-sensitive gene expression in cells growing with proline as nitrogen source (Fig. 7A). There is somewhat more expression of CAN1, DAL4, DAL5, and MEP2 observed in the wild type strain transformed with pYES2 than with pYES-V; DAL4 expression is clearly most responsive. Two genes, DAL1 and DAL80, did not respond to VID30 overexpression (Fig. 7A). Overexpression of VID30 did not detectably alter expression of any of these genes in cells grown under repressive conditions (with glutamine as the nitrogen source) (Fig. 7A). These results support the suggestions that: (i) Vid30p negatively regulates NCR-sensitive gene expression in cells grown with proline as nitrogen source, and (ii) Vid30p has no effect on the transcription of these genes in cells grown under repressive nitrogen conditions. These conclusions are consistent with those derived from the data in Fig. 3A, where, by deletion, Vid30p was shown to be a negative regulator of CAN1, DAL4, DAL5, MEP2, DAL1, GDH3, and DAL80 gene expression in proline-grown cells.
Fig. 7. Overexpression of VID30 affects the expression of genes associated with nitrogen metabolism.
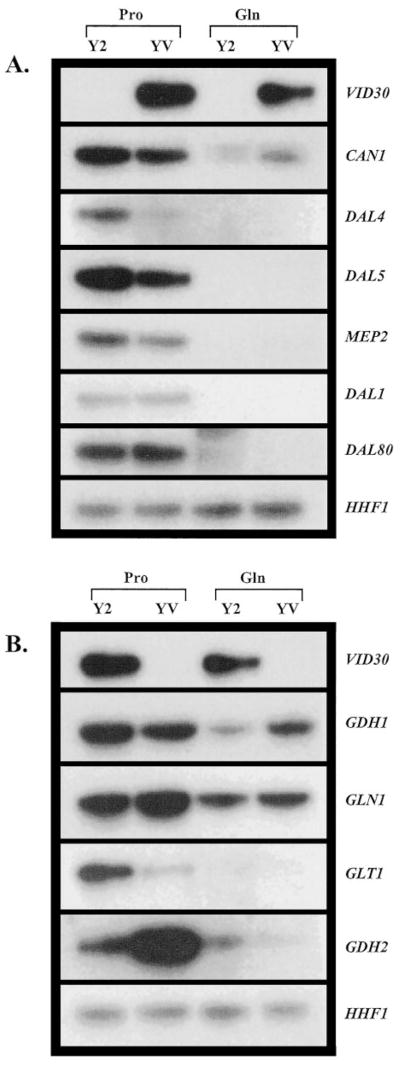
Northern blot analysis of poly(A)+ RNA (1 μg/lane) from transformants of BY4742 containing pYES2 (Y2) or pYES-V (YV) grown in YNB-0.1% proline (Pro) and YNB-0.1% glutamine (Gln) with 1.5% raffinose + 0.5% galactose as the carbon source.
The expression profiles of the ammonia-glutamate-glutamine interconversion genes were more complex. In YNB-proline-grown cells, GDH1 expression slightly decreases, while the decrease in GLT1 expression is dramatic; GLN1 expression slightly increases (Fig. 7B). In contrast, GDH2 expression increases rather markedly in proline grown cells overexpressing VID30 (Fig. 7B). Expression of GDH1 modestly increases, that of GDH2 decreases slightly, and that of GLN1 and GLT1 is not affected by VID30 overexpression with glutamine as the nitrogen source (Fig. 7B).
DISCUSSION
Data presented in this work demonstrate that Vid30p is a participant in the complex regulatory network controlling nitrogen metabolism in S. cerevisiae. VID30 is among the genes whose expression is NCR-sensitive, regulated by Ure2p, and induced by rapamycin. The strength of regulation, however, is only modest to moderate when compared with that of DAL5 or DAL80 under similar circumstances. The more dramatic regulation of VID30 expression occurs in response to ammonia. In low ammonia medium, VID30 expression is remarkably high and rapidly decreases when the ammonia concentration in the medium is increased (Fig. 8). That VID30 expression is enhanced in response to low ammonia rather than starvation is supported by the observation that it does not occur in similarly low concentrations of proline.
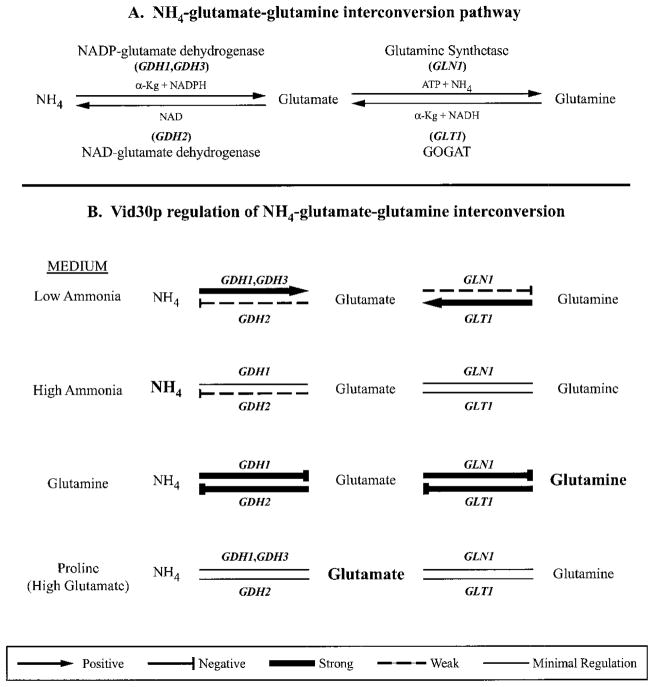
Fig. 8. Working model of Vid30p regulation of nitrogen metabolism in S. cerevisiae.
Arrows and bars indicate that Vid30p function is positively or negatively regulating expression of the indicated genes. For examples, Vid30p positively regulates GDH1 and GDH3 expression and negatively regulates GDH2 expression when cells are cultured in low ammonia.
The effects of Vid30p on the expression of genes associated with nitrogen metabolism are quite complex. With proline as the nitrogen source, Vid30p negatively regulates the nitrogen catabolic genes, CAN1, DAL4, DAL5, MEP2, DAL1, DAL80, and GDH3, but does not affect expression of GDH1, GLN1, or GLT1. There is a small positive effect on GDH2. With glutamine as the nitrogen source, the direction and strength of regulation is reversed. There is little if any effect of Vid30p on the regulation of CAN1, DAL4, DAL5, MEP2, DAL1, DAL80, and GDH3. In contrast, Vid30p is a significant negative regulator of GDH1, GDH2, GLN1, and GLT1 expression. The most obvious difference between these two sets of genes is that the latter are associated with the interconversion of ammonia, glutamate, and glutamine.
Similar partitioning of these nitrogen metabolic genes is observed when the influence of Vid30p on their expression profiles was assayed in high and low ammonia. Since high ammonia is a strongly repressive condition, little if any expression was observed for CAN1, DAL4, DAL5, MEP2, DAL1, DAL80, and GDH3, and the presence or absence of Vid30p does not affect it. GDH1, GLN1, and GLT1 were expressed in high ammonia, but again Vid30p did not affect their expression. Only for GDH2 does Vid30p behave as a negative regulator. On low ammonia, Vid30p is a positive regulator of all genes except GLN1 and GDH2, where it behaves as a negative regulator. Data obtained with cells overexpressing VID30 are consistent with the patterns of regulation just summarized.
From these data, we conclude that Vid30p function most significantly influences the control of ammonia, glutamate, and glutamine interconversion (Fig. 8), functioning to keep metabolism directed toward glutamate formation, especially when limiting ammonia is provided to the cells. The following reasoning supports this suggestion. The GDH1, GDH2, GLN1, and GLT1 gene products can be functionally divided into two groups: (i) enzymes that, respectively, catalyze conversion of ammonia and glutamine to glutamate, GDH1, GDH3, and GLT1 and (ii) those that, respectively, catalyze conversion of glutamate to ammonia and glutamine, GDH2 and GLN1. GDH1, GDH3, and GLT1 expression decreases markedly when a vid30Δ is grown in low ammonia, whereas GDH2 and GLN1 expression modestly increases. In other words, Vid30p modestly represses expression of genes whose products decrease the glutamate pool (GDH2, GLN1) and markedly activates expression of those whose products increase it (GDH1, GDH3, GLT1). These responses shift the interconversion reactions toward production of glutamate and away from ammonia and glutamine.
When cells are grown with high ammonia, VID30 expression itself is strongly decreased, thereby decreasing its ability to serve as a regulator. Therefore, it is not too surprising that about the only thing Vid30p does in high ammonia is to repress expression of GDH2, whose product decreases the glutamate pool. The NCR-sensitive GDH3 gene is not expressed under these conditions. In high ammonia, as in low, Vid30p function appears to be shifting the balance of metabolism toward glutamate. GLT1, GLN1, and GDH1 expression is not demonstrably affected in high ammonia, while GDH2 and GLN1 do not greatly respond to a vid30Δ with low ammonia. This seeming contradiction, however, is more apparent than real. Recall that GLT1 and GLN1 can function in concert to form glutamate from ammonia. In this regard, it is pertinent to mention that the Km for glutamine of the S. cerevisiae GOGAT (encoded by GLT1) is reported to be 0.29 mM (34). Such high concentrations of glutamine are much more easily achieved in high rather than low ammonia medium. By this reasoning, GOGAT functions along with the GDH1 product to produce glutamate when environmental ammonia is high and when GDH3, whose product is responsible for one of the three routes to glutamate production, is largely not expressed due to its NCR sensitivity. If this suggestion is correct, GOGAT may be able to function quite differently in yeast than in bacteria where it, in collaboration with glutamine synthetase, is reported to be the primary route of ammonia assimilation (30). In summation, when ammonia is the nitrogen source, Vid30p function shifts nitrogen metabolism toward glutamate production in the face of many biosynthetic reactions that use glutamate to produce nitrogenous macromolecules.
The end products of nitrogen catabolic pathways associated with most poor nitrogen sources such as proline or allantoin are glutamate or ammonia, respectively, produced at a growth-limiting rate. Consistent with these physiological conditions, Vid30p function exhibits characteristics of a negative regulator for the expression of typical NCR-sensitive genes whose expression we measured when cells are provided with proline as the sole nitrogen source. It also is not particularly surprising that the ammonia-glutamate-glutamine interconversion genes are not highly regulated in proline medium, because the cell’s glutamate requirement will be fulfilled more readily than either that for glutamine or ammonia. This derives from the fact that glutamate is the primary end product of proline catabolism. The situation is quite different when the nitrogen source is glutamine. In this case, the need for ammonia is much smaller, and its supply is much greater. Furthermore, there is a good supply of glutamine and glutamate as well. Therefore, Vid30p function partially represses expression of all of the interconversion genes with GLT1 being the least down-regulated when glutamine is provided as the sole nitrogen source. Vid30p function doesn’t appear to repress GDH3 expression, but in this instance, Vid30p-mediated repression is not necessary, because GDH3 expression is already highly repressed due to its NCR sensitivity. Although the data provide new insights into the nature of potential Vid30p functions, the mechanistic details of how these functions are accomplished remain to be elucidated, as do the identities of direct Vid30p targets.
Acknowledgments
We thank the University of British Columbia Wine Research Center and the University of Tennessee Yeast Groups for suggested improvements to the manuscript and Michael Robertson and Russ Morris of the University of British Columbia Media Group for preparing the artwork.
Footnotes
This work was supported by Natural Sciences and Engineering Research Council of Canada Grant 217271-99 (to H. J. J. v. V.) and National Institutes of Health Grant GM-35642-14 (to T. G. C.).
The abbreviation used is: NCR, nitrogen catabolite repression.
J. M. Cherry, C. Ball, K. Dolinski, S. Dwight, M. Harris, J. C. Matese, G. Sherlock, G. Binkley, H. Jin, S. Weng, and D. Botstein, Saccharomyces Genome Data Base (genome-www.stanford.edu/Saccharomyces/cgi-bin/SGD/locus.pl?locus=VID30).
References
- 1.Cooper TG. In: The Molecular Biology of the Yeast Saccharomyces: Metabolism and Gene Expression. Strathern JN, Jones EW, Broach J, editors. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1982. pp. 39–99. [Google Scholar]
- 2.Magasanik B. In: The Molecular and Cellular Biology of the Yeast Saccharomyces: Gene Expression. Jones EW, Pringle JR, Broach JR, editors. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1992. pp. 283–318. [Google Scholar]
- 3.Cooper TG. In: Mycota III. Marzluf G, Bambrl R, editors. Springer-Verlag; Berlin: 1994. pp. 139–169. [Google Scholar]
- 4.ter Schure EG, van Riel NAW, Verrips CT. FEMS Microbiol Rev. 2000;24:67–83. doi: 10.1111/j.1574-6976.2000.tb00533.x. [DOI] [PubMed] [Google Scholar]
- 5.Cooper TG, Ferguson D, Rai R, Bysani N. J Bacteriol. 1990;172:1014–1018. doi: 10.1128/jb.172.2.1014-1018.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Daugherty JR, Rai R, El Berry HM, Cooper TG. J Bacteriol. 1993;175:64–73. doi: 10.1128/jb.175.1.64-73.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blinder D, Magasanik B. J Bacteriol. 1995;177:4190–4193. doi: 10.1128/jb.177.14.4190-4193.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cunningham TS, Svetlov V, Rai R, Cooper TG. J Bacteriol. 1995;178:3470–3479. doi: 10.1128/jb.178.12.3470-3479.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stanbrough M, Magasanik B. J Bacteriol. 1995;177:94–102. doi: 10.1128/jb.177.1.94-102.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coffman JA, Rai R, Cunningham T, Svetlov V, Cooper TG. Mol Cell Biol. 1996;16:847–858. doi: 10.1128/mcb.16.3.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stanbrough M, Magasanik B. J Bacteriol. 1996;178:2465–2468. doi: 10.1128/jb.178.8.2465-2468.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beck T, Hall MN. Nature. 1999;402:689–692. doi: 10.1038/45287. [DOI] [PubMed] [Google Scholar]
- 13.Cardenas ME, Cutler NS, Lorenz MC, Di Como CJ, Heitman J. Genes Dev. 1999;13:3271–3279. doi: 10.1101/gad.13.24.3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hardwick JS, Kuruvilla FG, Tong JK, Shamji AF, Schreiber SL. Proc Natl Acad Sci U S A. 1999;96:14866–14870. doi: 10.1073/pnas.96.26.14866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cunningham TS, Andhare R, Cooper TG. J Biol Chem. 2000;19:14408–14414. doi: 10.1074/jbc.275.19.14408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blinder D, Coschigano PW, Magasanik B. J Bacteriol. 1996;178:4734–4736. doi: 10.1128/jb.178.15.4734-4736.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bertram P, Choi JH, Carvalho J, Ai W, Zeng C, Chan TF, Zheng XFZ. J Biol Chem. 2000;275:35727–35733. doi: 10.1074/jbc.M004235200. [DOI] [PubMed] [Google Scholar]
- 18.Coffman JA, El Berry HM, Cooper TG. J Bacteriol. 1994;176:7476–7483. doi: 10.1128/jb.176.24.7476-7483.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Coschigano PW, Magasanik B. Mol Cell Biol. 1991;11:4455–4465. doi: 10.1128/mcb.11.9.4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Courchesne WE, Magasanik B. J Bacteriol. 1988;170:708–713. doi: 10.1128/jb.170.2.708-713.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cafferkey R, Young PR, McLaughlin MM, Bergsma DJ, Koltin Y, Sathe GM, Faucette L, Eng WK, Johnson RK, Livi GP. Mol Cell Biol. 1993;10:6012–6023. doi: 10.1128/mcb.13.10.6012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kunz J, Henriquez R, Schneider U, Deuter-Reinhard M, Movva NR, Hall MN. Cell. 1993;73:585–596. doi: 10.1016/0092-8674(93)90144-f. [DOI] [PubMed] [Google Scholar]
- 23.Heitman J, Movva NR, Hall MN. Science. 1991;253:905–909. doi: 10.1126/science.1715094. [DOI] [PubMed] [Google Scholar]
- 24.Barbet NC, Schneider U, Helliwell SB, Stansfield I, Tuite MF, Hall MN. Mol Biol Cell. 1996;7:25–42. doi: 10.1091/mbc.7.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Noda T, Ohsumi Y. J Biol Chem. 1998;273:3963–3966. doi: 10.1074/jbc.273.7.3963. [DOI] [PubMed] [Google Scholar]
- 26.Chan TF, Bertram PG, Ai W, Zheng XF. J Biol Chem. 2000;276:6463–6467. doi: 10.1074/jbc.M008162200. [DOI] [PubMed] [Google Scholar]
- 27.Schmidt A, Beck T, Koller A, Kunz J, Hall MN. EMBO J. 1998;17:6924–6931. doi: 10.1093/emboj/17.23.6924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Choi JH, Adames NR, Chan TF, Zeng C, Cooper JA, Zheng XF. Curr Biol. 2000;10:861–864. doi: 10.1016/s0960-9822(00)00599-6. [DOI] [PubMed] [Google Scholar]
- 29.Shamji AF, Kuruvilla FG, Schreiber SL. Curr Biol. 2000;10:1574–1581. doi: 10.1016/s0960-9822(00)00866-6. [DOI] [PubMed] [Google Scholar]
- 30.Reitzer LJ, Magasanik B. In: Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology. Neidhardt FC, Ingraham JL, Low KB, Magasanik B, Schaechter M, Umbarger HE, editors. ASM Press; Washington, D. C: 1987. pp. 301–320. [Google Scholar]
- 31.Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K, editors. Current Protocols in Molecular Biology. John Wiley & Sons, Inc; New York: 1995. pp. 1–27. [Google Scholar]
- 32.Law DTS, Segall J. Mol Cell Biol. 1988;8:912–922. doi: 10.1128/mcb.8.2.912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cunningham TS, Cooper TG. Mol Cell Biol. 1991;11:6205–6215. doi: 10.1128/mcb.11.12.6205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cogoni C, Valenzuela L, Gonzalez-Halphen D, Olivera H, Macino G, Ballario P, Gonzalez A. J Bacteriol. 1995;177:792–798. doi: 10.1128/jb.177.3.792-798.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]