Abstract
The Saccharomyces cerevisiae allantoate/ureidosuccinate permease gene (DAL5) is often used as a reporter in studies of the Tor1/2 protein kinases which are specifically inhibited by the clinically important immunosuppressant and anti-neoplastic drug, rapamycin. To date, only a single type of cis-acting element has been shown to be required for DAL5 expression, two copies of the GATAA-containing UASNTR element that mediates nitrogen catabolite repression-sensitive transcription. UASNTR is the binding site for the transcriptional activator, Gln3 whose intracellular localization responds to the nitrogen supply, accumulating in the nuclei of cells provided with poor nitrogen sources and in the cytoplasm when excess nitrogen is available. Recent data raised the possibility that DAL5 might also be regulated by the retrograde system responsible for control of early TCA cycle gene expression, prompting us to investigate the structure of the DAL5 promoter in more detail. Here, we show that clearly one (UASB), and possibly two (UASA), additional cis-acting elements are required for full DAL5 expression. One of these elements (UASB) is in a region that is heavily protected from DNaseI digestion and functions in a highly synergistic manner with the two UASNTR elements. Cis-acting elements UASNTR-UASA and UASNTR-UASB are situated on the same face of the DNA two and one turn apart, respectively. We also found that decreased DAL5 expression in glutamate-grown cells, a characteristic shared with retrograde regulation, likely derives from decreased nuclear Gln3 levels that occur under these growth conditions rather than direct retrograde system control.
Keywords: DAL5, Gln3, Gat1, Nitrogen catabolite repression, Retrograde transcription, Rtg3, Allantoate permease, GATA-sequences
1. Introduction
Rapamycin has become a clinically important drug owing to its immunosuppressant and anti-neoplastic properties [1-5]. Its physiological action occurs as a result of binding to the prolyl isomerase, Fkbp12, and it is this complex that acts as a specific inhibitor of Tor protein kinases [6,7]. Inactivation of the Tor proteins, either by mutation or with rapamycin, elicits broad changes in cellular activities including: decreased translational initiation, eIF-4G instability, inhibition of cell cycle progression, aberrant actin cytoskeleton reorganization, decreased polymerase I and III activities, as well as increased autophagy, protein degradation and expression of retrograde and nitrogen catabolic genes [8-15]. Studies in Saccharomyces cerevisiae are contributing significantly to our understanding of the Tor1/2 proteins and how they regulate the above cellular processes, for example, by controlling intracellular localization of numerous transcription factors. This was first shown for S. cerevisiae transcription factors Gln3 and Msn2 [16-19]; a similar mode of control has more recently been suggested for retrograde transcription factors Rtg1/3[20,63].
Gln3, and Gat1, are C–X2–C–N17–20–C–X2–C zinc-finger proteins responsible for nitrogen responsive (nitrogen catabolite repression (NCR)-sensitive) gene expression in S. cerevisiae [21-23]. When cells are provided with good nitrogen sources (e.g., glutamine or ammonia in some strains), Gln3 localizes to the cytoplasm and genes encoding permeases and degradative enzymes for poor nitrogen sources (e.g., proline or allantoin) are not expressed [21-24]. In contrast, when nitrogen is limiting or only poor nitrogen sources are available, Gln3 accumulates in the nucleus and NCR-sensitive transcription increases [21-24].
Treating cells with rapamycin generates some of the same outcomes as growth in a poor nitrogen source, i.e., nuclear accumulation of Gln3 and increased NCR-sensitive transcription [16-19,24,25]. Not only does Gln3 accumulate in the nuclei of rapamycin-treated cells, but its electrophoretic mobility increases, leading to the conclusion that Tor1/2-mediated changes in Gln3 phosphorylation/dephosphorylation are responsible for its intracellular localization [16,19]. This conclusion has been recently questioned by the fact that changes in Gln3 phosphorylation, detected so far, often fail to correlate with Gln3 intracellular localization [24]: (i) Gln3 is dephosphorylated and accumulates in the nuclei of cells treated with rapamycin for 30 min. However, after 60 min of treatment, Gln3 is cytoplasmic even though it remains dephosphorylated; (ii) Gln3 exhibits the same phosphorylation profile with proline, ammonia, or glutamine as nitrogen source even though Gln3 is nuclear in proline-grown cells and cytoplasmic when glutamine or ammonia is provided.
One of the common reporter genes used in the study of Tor1/2 and the control of NCR-sensitive transcription is DAL5, encoding the allantoate/ureidosuccinate permease [21-23]. It was in this promoter that the GATA-containing cis-acting element (UASNTR) was discovered and found to be necessary and sufficient for NCR-sensitive transcription [26]. The DAL5 promoter contains nine UASNTR-homologous sequences, but only two of them can be demonstrated to be functionally significant (they are underlined in Fig. 1). Although no other cis-acting elements have been reported in the DAL5 promoter, the possibility that additional elements might exist was raised by the observation that DAL5 expression closely parallels that of the retrograde gene, CIT2.
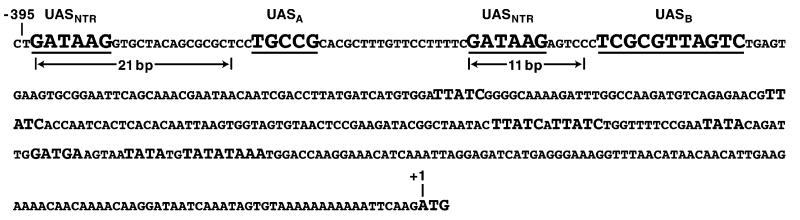
Fig. 1.
DNA sequence upstream of the DAL5 gene. The sequence begins at position −395. The four cis-acting elements discussed in this work are underlined. Two of them, UASNTRS, are the elements responsible for nearly all NCR-sensitive DAL5 expression. Also indicated in the figure are the distances of one (11-bp) and two (21-bp) helical turns of a B-DNA molecule with 10.5 bp per turn. UASNTR-homologous and TATA sequences appear in capital letters.
The retrograde genes encode early TCA cycle enzymes, citrate synthetase (CIT2), aconitase (ACO1), and isocitrate dehydrogenase (IDH1/2), that synthesize α-ketoglutarate needed to assimilate ammonia when cells are growing fermentatively in high glucose-ammonia medium [28-32]. In keeping with this function, retrograde gene expression correlates with intracellular levels of ammonia [27]. Growth with ammonia, or nitrogen sources degraded to ammonia, elicits high retrograde gene expression. Conversely, retrograde gene expression is low in cells grown with glutamate, or compounds degraded to glutamate [27,33]. Four regulatory proteins control retrograde gene expression: Rtg1, Rtg2, Rtg3 and Mks1. Rtg1/3 are responsible for transcriptional activation of the retrograde genes, Rtg2 is a positive regulator that interacts with Mks1, and Mks1 is a strong negative regulator of retrograde transcription[20,33-36].
To further understand DAL5 expression and its regulation, we investigated whether the DAL5 promoter contained additional cis-acting elements. Evidence is presented that clearly one (UASB), and possibly two (UASA), previously unidentified cis-acting elements, in addition to the two known UASNTRS upstream of DAL5, are required for full expression. Elements UASNTR–UASA and UASNTR–UASB, are situated on the same face of the DNA two and one turn apart, respectively. At least one of these elements, UASB, functions in a highly synergistic manner with UASNTR. Even though DAL5 (NCR-sensitive) and CIT2 (retrograde) expression profiles are identical with some nitrogen sources, we found no evidence to support the possibility that NCR-sensitive DAL5 and retrograde CIT2 expression are regulated in common. Rather, expression of DAL5 appears to be more NCR-sensitive than some of the other nitrogen catabolic genes, e.g., GAP1. We propose that this expression profile derives from: (i) GATA-elements contained in the DAL5 promoter binding the GATA-family transcription factors poorly, and (ii) the ability of UASB and UASA to increase the overall level of transcription, but only when they can function synergistically with UASNTR elements.
2. Materials and methods
2.1. Strains and media
The Saccharomyces cerevisiae strains used in this work are listed in Table 1. Strains were grown in media containing 0.17% yeast nitrogen base (Difco, without amino acids and ammonium sulfate) supplemented with 2% glucose and 0.1% of the indicated nitrogen source. GABA and proline are both nitrogen sources that support high NCR-sensitive gene expression. The main reason GABA was used in some experiments rather than proline was that GABA supports somewhat more robust growth than proline especially with gln3Δ strains. Where necessary, l-lysine (40 mgl −1), l-histidine (20 mgl −1), uracil (20 mgl −1), l-leucine (120 mgl −1), adenine (20 mgl −1), and l-tryptophan (20 mgl −1) were added to cover auxotrophies. Yeast strains were transformed using the lithium acetate method of Ito et al. [37]. Bacteria were transformed by the method of Tschumper and Carbon [38].
2.2. Construction of fusion plasmids
DAL5 promoter fragments (double-stranded synthetic oligonucleotides) were cloned into heterologous expression vectors. In addition to the DAL5 promoter region, SalI and EagI sites were synthesized onto the TATA-distal and proximal ends of each fragment, respectively, to permit asymmetric cloning into the heterologous CYC1 expression vector, pNG15, derived by deleting the CYC1 UAS elements from an in-frame CYC1-lacZ fusion gene [39]. This vector has been broadly used to identify and analyze UAS elements from the promoters of many genes.
Full-length wild-type and mutant DAL5-lacZ fusion plasmids (Figs. 4 and 10) were constructed using double-stranded 102 bp SalI-EcoRI fragments prepared from synthetic oligonucleotides. The SalI restriction site was synthesized onto the 5′ terminus of each fragment to permit cloning, whereas the EcoRI site at the 3′ terminus was present in the DAL5 promoter. The 102-bp SalI–EcoRI DNA fragment was gel-purified and ligated to the 2.6 kb EcoRI–SacI fragment derived from a DAL5-lacZ fusion plasmid (pRR30) [26]. This EcoRI–SacI fragment spans the in-frame DAL5-lacZ junction, the SalI site being in the DAL5 promoter and the SacI site in the lacZ gene. The ligation product was then digested with SacI to resolve polymers generated during ligation and the EcoRI–SacI fragment purified and ligated into 2 μm plasmid pLG669Z [40] digested with SalI and SacI. The 2 μm vector was used in this instance because, at the time the plasmids were constructed, it was the preferred vector for reasons of stable inheritance. The mutant oligonucleotides were identical to the wild-type alleles except at the mutated bases (these modifications are indicated with lower case letters). The structure of every plasmid insert including the restriction site used for cloning was verified by dideoxy sequencing. The DAL5 5′ deletion plasmids were constructed earlier [26]. Other techniques used in the cloning were standard and described in detail by Sambrook et al. [41].
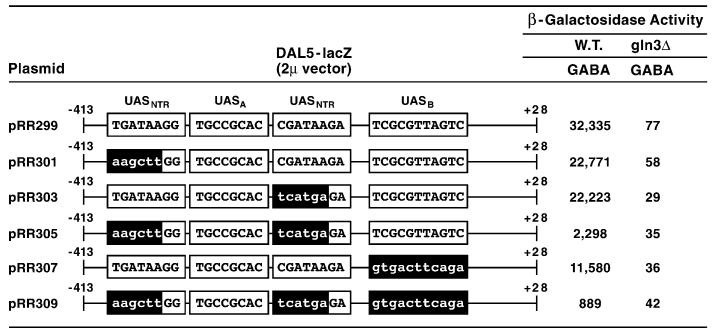
Fig. 4.
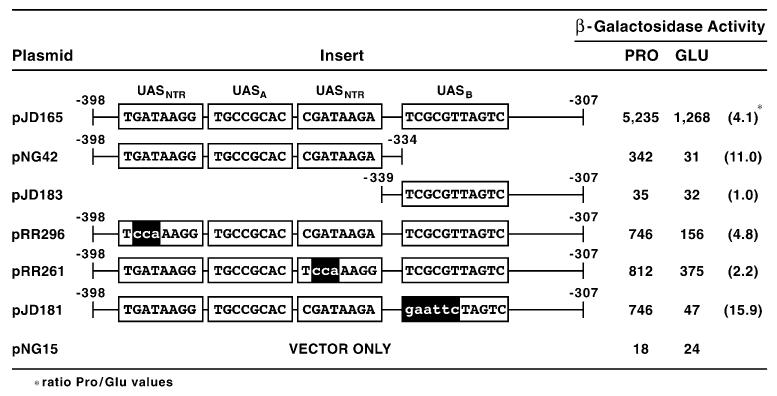
β-Galactosidase production supported by wild type and mutant in frame DAL5-lacZ fusion plasmid. Wild-type (TCY1) and gln3Δ (RR91) strains were transformed with the indicated DAL5-lacZ fusion plasmids cloned into 2 μm vector pLG669Z. Mutant sequences are indicated by lower-case letters. Transformants were then grown in minimal-γ-aminobutyrate (GABA) medium and assayed for β-galactosidase activity.
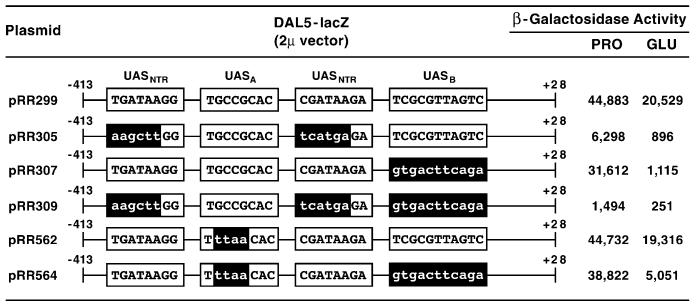
Fig. 10.
β-Galactosidase production supported by wild type and mutant in frame DAL5-lacZ. Wild type (TCY1) was transformed with the indicated fusion plasmids in vector pLG669Z. Mutant sequences are indicated by lower-case letters. Transformants were then grown in minimal proline (PRO) or glutamate (GLU) medium and assayed for β-galactosidase.
2.3. DNaseI footprinting
The procedures used for DNaseI footprinting were those described by Rai et al. [26]. Protein extracts were prepared as described by Pfeifer et al. [42] and Luche et al. [43]. The DNA probe was prepared by digesting DAL5 pRR29 [26] with EcoRI, radioactively labeling the product using the polynucleotide kinase reaction, followed by XhoI digestion to generate radioactive fragment (− 305 to − 411): TCGAGGAGCTATCATTTG CTGATAAGGTGCTACAGCGCGCTCCTGCCGCA CGCTTTGTTCCTTTTCGATAAGAGTCCCTCGCGTTAGTCTGAGTGAAGTGCGGAATT (sequences pertinent to the discussion are underlined).
2.4. Electrophoretic mobility shift assays
Electrophoretic mobility shift assays (EMSA)s were performed as described by Pfeifer et al. [42] and Luche et al. [44]. Wild-type and mutant DNA fragments used as radioactive probes were generated from oligonucleotides (−344 to −306) 5′-AAGAGTCCCTCGCGTTAGTCTGAGTGAAGTGCGGAATTC-3′,5′-GAATTCCGCACTTCACTCAGACTAACGCG AGGGACTCTT-3′, 5′-AAGAGTCCCgaattcTAGTCTGAGTGAAGTGCGGAATTC-3′, and 5′-GAATTC CGCACTTCACTCAGACTAgaattcGGGACTCTT-3′, respectively. Mutant substitutions are indicated in lower case letters. Wild-type and mutant competitor DNA fragments were generated from oligonucleotides (−362 to −312) 5′-GCTTTGTTCCTTTTCGATAAGAGTCCCTCGCGTTAGTCTGAGTGAAGTGCG-3′, 5′-CGCACTTCACTCAGACTAACGCGAGGGACTCTTATCGAAAAGGAACAAAGC-3′, 5′-GCTTTGTTCCTTTTCGATAAGAGTCCCTgtgacttcagaGAGTGAAGTGCG-3′, and 5′-CGCACTTCACTCtctgaagtcacAGGGACTCTTATCGAAAAGGAACAAAGC-3′, respectively. Highly sheared calf thymus DNA was used in all reaction mixtures as non-specific competitor DNA.
2.5. RNA preparation and hybridization
Total RNA was prepared by the methods of Rai et al. [44] and hybridization reactions carried out as described by Cox et al. [45].
2.6. β-Galactosidase assays
β-Galactosidase assays were performed by the method of Guarente and Mason [46], and activity expressed in units defined by Miller [47]. At least duplicate assays for each of two independent transformants were performed for each value reported. The precision of our β-galactosidase assays has been characterized in detail [48]. There is a 10–15% day to day variation in the absolute values observed, but the patterns of the data were invariant. Plasmids (2 μm-based) used in some experiments supported far greater β-galactosidase production than others (ARS1-based); the latter being constructed much earlier [26]. The potential effects of differences in reporter gene plasmid copy number were evaluated earlier for NCR-sensitive gene promoter analyses and found not to adversely affect the analyses. The only instances in which copy number does alter interpretation of the data are when the copy number or expression of the GLN3, GAT1, and URE2 genes is altered [49].
3. Results
3.1. A novel cis-acting element in the DAL5 promoter
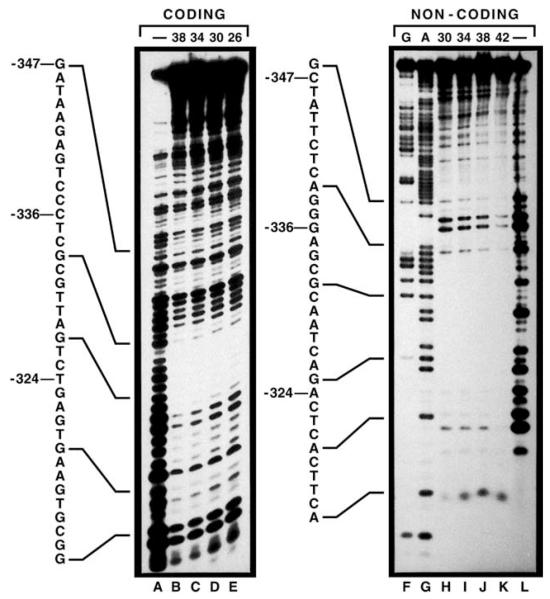
The GATA sequences responsible for most DAL5 transcription are at positions −393 to −388 and −347 to −342 (Fig. 1, underlined), with the 5′ GATA sequence situated in a region previously shown to be protected from DNaseI digestion [26]. As expected, we found the 3′ GATA required for DAL5 expression (−347 to −342) was also in a protected region (Fig. 2, right panel), although this footprint was weaker than the one covering the 5′ GATA (compare Fig. 2 of this work with Fig. 5, [26]). In addition, we observed a large and strongly protected region downstream of the 3′ GATA (−336 to −324, left panel; and −339 to −320, right panel). There also appeared to be a protected region 5′ of −347 of the non-coding but not the coding strand. These data suggested one or more proteins, in addition to Gln3/Gat1, might bind to the DAL5 upstream region.
Fig. 2.
DNaseI protection of sequences required for DAL5 gene expression. Lanes F and G contain the separated products of the Maxam–Gilbert G and A+G reactions. Lanes A–E and H–L contain the digestion products of the 106-bp XhoI EcoRI fragment following incubation without added extract (lanes)- or with increasing amounts of wild-type (strain M970) extract (concentrations in micrograms, numbers at the top of each panel) and limiting amounts of DNasel.
Fig. 5.
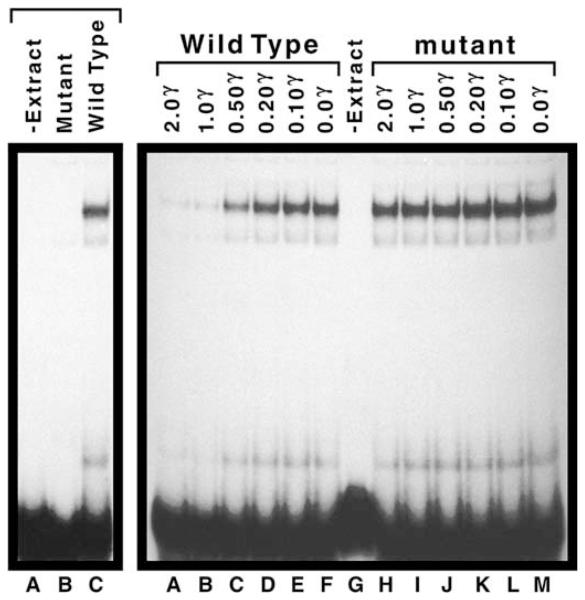
Electrophoretic-mobility shift assay employing DNA probes derived from the DAL5 promoter region. Left panel, lane A and right panel, lane G contained no protein extract (-Extract). All remaining lanes contained 30 μg of extract prepared from strain W303-1A. The radioactive DNA probes as well as the wild-type (right panel, lanes A–F) and mutant (right panel, lanes H–M) competitor DNA fragments were prepared as described in Section 2. They were added to the EMSA reaction mixtures in the amounts (in μg) indicated. Identities of the two weak complexes are not known.
3.2. DAL5 sequences protected from DNaseI digestion are required for DAL5 expression
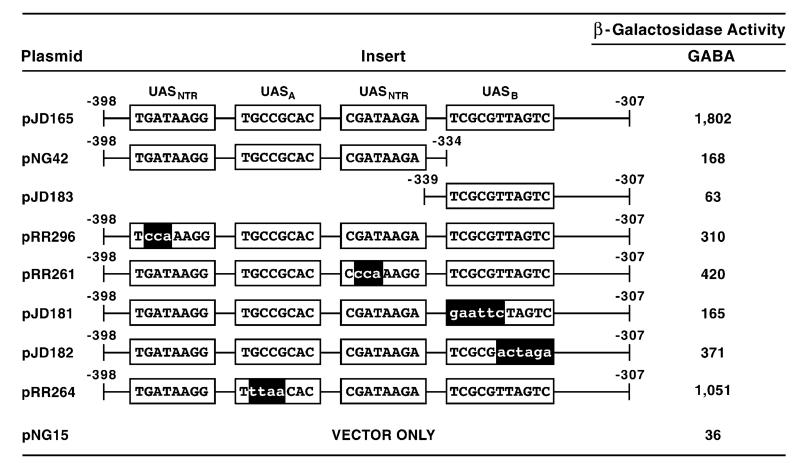
To determine whether sequences −339 to −320 were required for DAL5 expression, we analyzed wild-type and mutant DNA fragments covering this region (−398 to −307) using a CYC1 heterologous expression vector system. The first two mutant DNA fragments were chosen such that the functional UASNTR elements were contained on one fragment (pNG42) and the region beneath the footprint was contained on the other (pJD183). The parent fragment (pJD165) supported substantial β-galactosidase production in cells provided with a relatively poor nitrogen source, γ-aminobutyrate (GABA) (Fig. 3). Expression decreased substantially when this fragment was divided into two sub-fragments. The first sub-fragment (pNG42) supported low reporter gene expression, while values for the second sub-fragment (pJD183) were barely above background (pNG15) (Fig. 3). These data indicated that the two previously identified GATA elements supported low-level expression that was synergistically enhanced (tenfold) by a second element, which could not function alone. We designated this newly identified cis-acting element UASB.
Fig. 3.
β-Galactosidase production supported by wild-type and mutant plasmids containing synthetic DAL5 fragments covering positions −398 to −307. The synthesized DNA fragments were cloned into CYC1 (Ars1) heterologous expression vector pNG15. pJD165, pNG42 and pJD183 contain native DAL5 sequences, whereas their derivatives contain the substitution mutations shown with lower-case letters. The transformation recipient was wild-type strain TCY1 which was grown in minimal γ-aminobutyrate (GABA) medium prior to assay for β-galactosidase activity.
To insure the above results did not derive aberrantly from the large deletions we made in the parent fragment, we successively inactivated each of the UASNTRS (pRR296 and pRR261) and UASB of pJD165 using substitution mutations (pJD181 and pJD182) (Fig. 3). Either GATA element alone (pRR296 and pRR261), when present with UASB, supports greater β-galactosidase production than the two GATA elements together in the absence of the latter sequence (pNG42 and pJD181) (Fig. 3). Further, mutating the 5′ half of the footprinted region (pJD181) resulted in less than half as much reporter gene expression than when the 3′ half was mutated (pJD182), suggesting the most important sequence for function is likely TCGCG along with one or more of the bases TTAGTC.
3.3. Functional analysis of DAL5 cis-acting elements in a native promoter
Although promoter fragment analysis in heterologous expression vector systems is useful in the study of transcriptional regulation, the data obtained do not always reflect the native condition. Therefore, we constructed DAL5-lacZ fusion plasmids in which the sequences discussed above were mutated in a full length wild type DAL5 promoter. β-Galactosidase production decreased about one third when either of the two GATA elements were mutated (Fig. 4, pRR301 and pRR303). When both GATA elements were mutated (pRR305), reporter gene expression decreased almost fifteenfold. However, the observed expression was still fully Gln3-dependent, suggesting one or more of the unmutated GATA sequences situated elsewhere upstream of DAL5 (Fig. 1) were able to support 5–10% of the wild-type expression level. This is consistent with earlier experiments showing that the GATA homologous sequence at −135 to −130 supports low-level β-galactosidase production in the CYC1 heterologous expression system [50].
Mutating UASB decreased promoter function two-thirds compared to wild type (Fig. 4, pRR307), and when all three cis-acting elements were mutated (pRR309), β-galactosidase production decreased over 35-fold. The fact that reporter gene expression supported by pRR299 is far more than the sum of that supported by pRR305 and pRR307, again indicates that UASB functions synergistically with the two GATA elements. Moreover, this putative element appears capable of functioning, albeit poorly, with one or more of the remaining seven GATA-homologous sequences upstream of DAL5 as evidenced by data obtained with pRR305 and pRR309. On this occasion, data observed with the intact promoter parallel those derived from experiments with isolated DNA fragments.
3.4. Assay of protein binding to wild type and mutant DNA fragments covering −344 to −306
The above evidence suggested one or more proteins required for DAL5 expression bound to UASB. To test this suggestion more directly, we performed an electrophoretic-mobility shift assay (EMSA) using wild-type and mutant DNA fragments. As shown in Fig. 5, when a radioactive 39-bp wild-type DNA fragment or one mutated as in pJD181 (Fig. 3) were incubated with crude cell extract, a strong, extract-dependent shift in mobility of the wild-type DNA fragment occurred (left panel, lanes A and C). This did not occur with the mutant fragment (left panel, lane B). Supporting this observation, a non-radioactive 51-bp wild-type DNA fragment containing UASB was an effective competitor in this assay (right panel, lanes A–G), whereas a similar DNA fragment mutated as in pRR307 (Fig. 4) was not (right panel, lanes G–M). Together these data support the suggestion that one or more proteins, required for DAL5 expression, bind to UASB.
3.5. Decreased DAL5 expression with glutamate as a nitrogen source
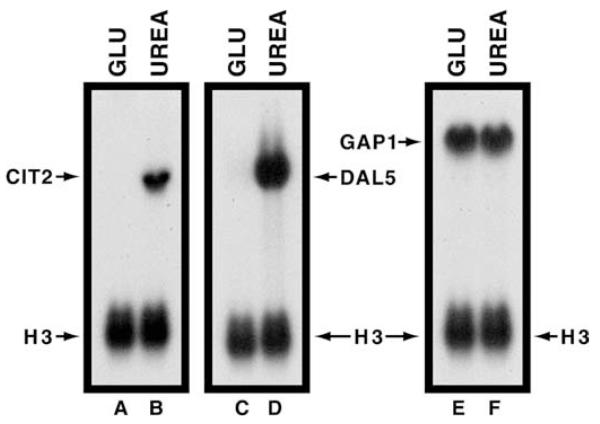
The discovery of an additional DAL5 cis-acting element, unrelated to NCR-sensitive UASNTRS, provided an opportunity of explaining a puzzling characteristic of DAL5 expression. DAL5 exhibits the same expression profile as the retrograde reporter gene, CIT2, i.e., expression is high when urea is provided as sole nitrogen source and low with glutamate (Fig. 6, lanes A–D). This stands in sharp contrast with some other NCR-sensitive genes, such as GAP1, which are expressed more or less equivalently in cells provided with glutamate versus urea (Fig. 6, lanes E, F). The similarity of CIT2 and DAL5 expression profiles at first raised the possibility that retrograde regulatory proteins might also control DAL5. However, this was eliminated by the demonstration that DAL5 expression was high in rtg2 mutant cells provided with ammonia as sole nitrogen source, whereas CIT2 was not, indicating that DAL5 expression occurred independently of retrograde transcription [27]. The reason that DAL5 is expressed in ammonia-grown rtg2 mutant but not in wild-type cells is that the mutant, lacking the ability to produce the retrograde enzymes, cannot synthesize the α-ketoglutarate required for ammonia assimilation [27]. Hence, under these conditions, ammonia is a poor nitrogen source and DAL5 expression is not repressed as in the wild type. Further, in contrast with CIT2 expression, DAL5 was not expressed in the mks1 mutant with glutamate or ammonia as nitrogen source, indicating Mks1 did not play a role in DAL5 expression [33]. Together, these data argued against the possibility that loss of DAL5 expression in glutamate-grown cells derives from retrograde control.
Fig. 6.
Northern-blot analyses of steady-state CIT2, DAL5 and GAP1 mRNA levels in wild-type (M970) cells provided with glutamate (GLU) or urea as sole nitrogen source. A histone H3 (HHT1) DNA probe was used to assess loading precision and transfer efficiencies.
Since low DAL5 expression with glutamate as nitrogen source did not derive from retrograde control, our next objective was to locate the region(s) of its promoter that were influenced by glutamate as nitrogen source. Low expression with glutamate as nitrogen source is by no means a universal characteristic of NCR-sensitive gene expression, which decreased the likelihood that glutamate specifically influenced GATA-mediated regulation per se. This reasoning, coupled with the newly identified UASB element, prompted us to query whether it might account for decreased DAL5 expression in glutamate medium. We addressed this possibility by analyzing wild-type and mutant DNA fragments in proline vs. glutamate-grown cells (Fig. 7). In only two instances did β-galactosidase production decrease more than five-fold in glutamate vs. proline medium, those with pNG42 and pJD181 (Fig. 7). This occurred in the latter case even though pJD181 supported approximately the same amount of lacZ expression as pRR296 and pRR261 in proline-grown cells (Fig. 7). Correlating with the expression data just mentioned, pNG42 and pJD181 are equivalent in terms of function. In the latter plasmid, UASB was inactivated by substitution mutations, whereas in the former it was destroyed by deletion. It is interesting to note that mutating either one of the two GATA elements on pJD165 (pRR296, pRR261) did not decrease lacZ expression with glutamate as nitrogen source nearly as much as did loss of UASB (pJD181). These data argued that UASB played a significant role in the diminished DAL5 expression that occurs in minimal glutamate medium.
Fig. 7.
β-Galactosidase production supported by wild-type and mutant plasmids containing synthetic DAL5 fragments covering positions −398 to −307. The synthesized DNA fragments were cloned into heterologous expression vector pNG15. pJD165, pNG42 and pJD183 contain native DAL5 sequences, whereas their derivatives contain the substitution mutations shown with lower-case letters. The transformation recipient was strain TCY1 which was grown in minimal proline (PRO) or glutamate (GLU) medium prior to assay for β-galactosidase. Figures in parentheses are the quotient of the proline values divided by the glutamate values to yield fold repression.
3.6. A second previously unrecognized cis-acting element upstream of DAL5
Although the above data point to UASB being responsible for DAL5 expression in glutamate medium, it was subject to a caveat. The full DAL5 upstream region had not been analyzed for potential effects of growth with glutamate as nitrogen source. Therefore, we used an existing set of DAL5-lacZ nested 5′ deletion plasmids to investigate this question [26]. As shown in Fig. 8, β-galactosidase production supported by full length DAL5-lacZ pRR26 was threefold less with glutamate than with proline as nitrogen source, and as with all NCR-responsive genes in these strains, was highly repressed (24-fold) by growth with ammonia. Note that the glutamate effect here was smaller than observed in the Northern blots. Besides possible differences deriving from the fact that interpretation of data from heterologous plasmid-borne β-galactosidase production are subject to other limitations than those from steady-state mRNA measurements, the strain used here is less NCR-sensitive than the one used for the Northern blot experiments [27]. Deleting sequences −414 to −370 (pRR41) decreased overall β-galactosidase production and rendered it approximately twofold more sensitive to growth with both glutamate and ammonia as nitrogen source (Fig. 8, values in parentheses). Decreased lacZ expression with pRR41 was not surprising since one of the two important GATA elements responsible for DAL5 expression is at −393 and −388. Further deletion to −353 (pRR42) did not diminish the activity with proline as nitrogen source, but increased NCR-sensitivity two additional fold (relative to pRR41) with both glutamate and ammonia (values in parentheses). Finally, deletion of the second of the two important DAL5 GATA elements (pRR44) eliminated most remaining β-galactosidase production. The fact that reporter gene expression supported by pRR41 and pRR42 diminished in parallel (comparing proline to glutamate to ammonia levels) was consistent with the suggestion that DAL5 was just more NCR-sensitive than other genes. The data further argued that sequences between −370 and −353 were necessary for full lacZ expression with increasingly repressive nitrogen sources, glutamate and ammonia, but not proline, even though this region did not contain GATA elements. DAL5 region −370 to −353 did contain, however, an inverted repeat, TGCCGCA, which we hypothesized might be associated with the phenotype of pRR42. We tested this hypothesis by measuring the effects of mutating the GCCGC sequence of pJD165 and found lacZ expression decreased by nearly half (Fig. 3, pRR264). This suggested that the GCCGC sequence, which we designated UASA, was required for full lacZ expression.
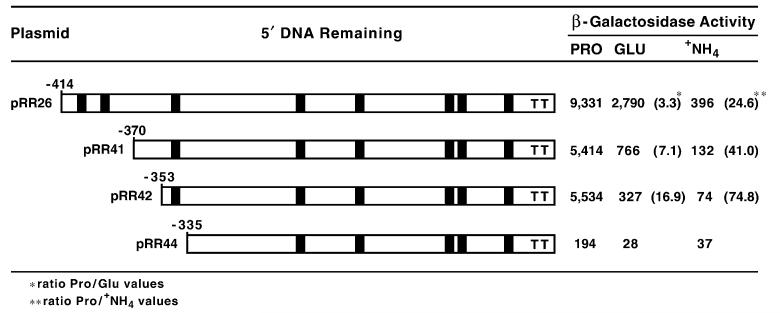
Fig. 8.
β-Galactosidase production supported by nested 5′-deletions of the DAL5 upstream region fused in frame to the lacZ reporter gene. The deletion plasmids were from the work of Rai et al. [26]. The deletion plasmids were transformed into wild-type strain TCY5, and the transformants grown in minimal medium containing proline (PRO), glutamate (GLU) or ammonia (+NH4) as sole nitrogen source prior to assay for β-galactosidase. Values in parentheses are the quotient of the proline values divided by the glutamate (*) or ammonia (**) values to yield fold repression. Filled boxes indicate the positions of UASNTR-homologous sequences. Ts indicate the relative positions of TATA elements.
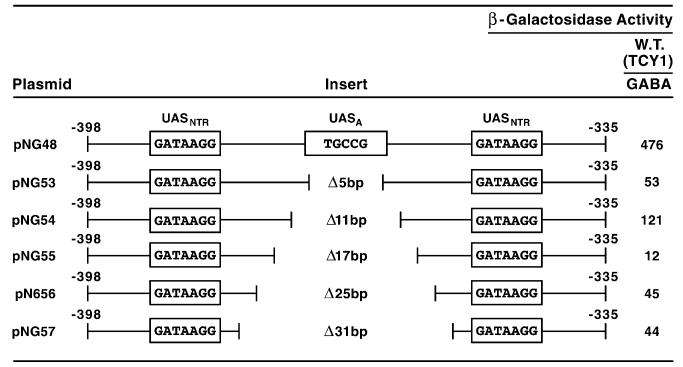
Additional indirect evidence supporting the physiological importance of UASA derives from plasmids designed to test whether the two important DAL5 GATA elements had to be on the same side of the DNA molecule in order to function. To test this question, deletions of 5, 11, 17, 25 and 31 bps were introduced into DAL5 parental fragment −398 to −335 (Fig. 9, pNG48); this was the fragment originally used for the saturation mutagenesis that identified the important bases of UASNTR [50]. The first deletion (pNG53, −370 to −366), which positioned the GATA elements on opposite sides of the DNA molecule, decreased reporter gene expression ninefold to near background. Deleting an additional 6 bp (pNG54), which returned the GATA sites to the same side of the DNA molecule, increased lacZ expression 2.5-fold. All further deletions reduced lacZ expression to near background levels (Fig. 9, pNG55-57).
Fig. 9.
β-Galactosidase production supported by plasmids containing synthetic DAL5 fragments covering positions −398 to −335. The synthesized DNA fragments were cloned into heterologous expression vector pNG15, and are the same ones used earlier to identify the bases required for NCR-sensitive gene expression [26]. The extent of the deletions carried in the plasmids is indicated in the figure. DNA was deleted in a symmetrical manner beginning with the first deletion of the 5 bases, TGCCG at positions −370 to −366. The transformation recipient was wild-type RH218 which was grown in minimal γ-aminobutyrate (GABA) medium prior to enzyme assay.
Although these data suggested the two GATA elements functioned better when on the same side of the DNA molecule, one aspect of the data was disappointing, i.e., lacZ expression supported by pNG53 was quite low. This was unexpected because Gln3, in contrast to Dal80, can bind to DAL3 promoter fragments containing single GATA elements [51]. Further, we expected β-galactosidase values to be more like those observed with the wild type in cells containing pNG54. Data with pRR264 (Fig. 3) suggested a way to rectify these results. It demonstrated that the bases we deleted in pNG53-57 not only changed the topography of the GATA elements with respect to one another, but also serendipitously removed a cis-acting element (UASA) whose presence increases transcription twofold.
A final test of the possibility that UASA participates in DAL5 expression was to assess the effects of mutations in UASA (pRR264) in a full-length wild-type DAL5 promoter. As shown in Fig. 10, the GGCG to TTAA mutation did not affect β-galactosidase supported by pRR562 relative to wild-type pRR299. This argues that, unlike the case in which only UASA and two UASNTR elements are present (Fig. 9, pNG48 vs. pNG53), UASA is not crucial for gene expression when a full complement of DAL5 promoter elements is present. In contrast, when both UASA and UASB were mutated, there were demonstrable differences between the single and double mutants (pRR562 vs. pRR564 and pRR307 vs. pRR564): (i) with proline as nitrogen source, pRR564 supported 20% more lacZ expression than pRR307 (Fig. 10); (ii) with glutamate, lacZ expression increased over fourfold (Fig. 10). A priori, these are the characteristics expected when a negative element is mutated. There are known instances, however, when a similar phenotype has been observed upon mutating a positive cis-acting element [52]. Although the evidence for UASA is not nearly as convincing as in the case of UASB, it does support the contention that the inverted repeat at −369 to −364 may play a role in DAL5 expression.
3.7. Identity of the proteins binding to UASA and UASB
Knowledge of the proteins that bind to UASA and UASB would greatly increase our understanding of DAL5 transcription. Therefore, we compared the UASA and UASB sequences with those of known DNA-binding proteins. One protein was identified as a candidate for binding to UASB, but deletion of its cognate gene did not produce a phenotype in DAL5 expression, arguing against its binding to the DAL5 promoter. Therefore, the identities of at least two transcription factors that participate in DAL5 transcription remain unknown.
4. Discussion
Data presented in this work clearly identify one (UASB), and possibly two (UASA), previously unrecognized cis-acting elements which, together with the two major DAL5 GATA-containing UASNTR elements, are required for high-level DAL5 expression. These newly identified elements and the ability of UASB to function synergistically with UASNTR has significantly refined our view of how Gln3-mediated, NCR-sensitive transcription occurs. The 3′ most of the newly discovered elements, UASB, is situated beneath a strong DNaseI footprint, while only marginal protection of UASA can be demonstrated. The degree to which UASA and UASB are protected from DNaseI digestion correlates with the strength of their influence on DAL5 expression. As shown in Fig. 1, the distance from the beginning of the 5′ UASNTR to that of UASA is 23 bp and from the beginning of the 3′ UASNTR to UASB is 12 bp. The mammalian GATA sequences that bind transcription activator GATA-1 are located in the major groove of the DNA [53]. If, by analogy, either of the two functional DAL5 GATA elements analyzed in this work is arbitrarily placed in a major groove of the DNA molecule, then the cis-acting element adjacent to it is also in the major groove on the same face of the DNA: UASNTR – 2 turns – UASA and UASNTR – 1 turn – UASB.
The newly discovered UASA and UASB elements appear to function synergistically with UASNTR GATA elements. Although necessary and sufficient for NCR-sensitive expression [54], DAL5 UASNTR elements support only low-level transcription by themselves. High-level transcription requires, in particular, UASB as well. In this respect, DAL5 joins several genes in which UASNTR elements have been shown to function synergistically with other non-GATA cis-acting elements: (i) protein(s) binding to a cis-acting element containing the sequence TTTGTTT in GLN1 [55,56]; (ii) Put3 in PUT1 [56,57]; (iii) Rap1 and Abf1 in CAR1 and CAR2 [48,58-60]; and (iv) Dal82 in DAL7 [39]. Further, ectopically placing a Dal82 binding site near a mutated GATA-element, which has lost 80% of its ability to support NCR-sensitive gene expression, suppresses the effects of the cis-acting mutation [35]. The latter observations were interpreted as suggesting Gln3 directly or indirectly interacts with Dal82 to increase the stability of Gln3 binding to the mutated GATA element. By this reasoning, we hypothesize proteins binding to UASA and UASB also directly or indirectly increase the stability of Gln3/Gat1 binding to their DAL5 UASNTR elements. Increased stability of Gln3 binding in turn generates increased gene expression. The mechanism of synergistic interaction, however, is unknown.
The above discussion offers a plausible explanation for the observed increase in NCR-sensitivity of DAL5 expression following inactivation of UASB (for example, Fig. 7, pJD165, pJD181 and Fig. 8, pRR41, pRR42 values in parentheses). In general, when cells are provided with a poor nitrogen source, e.g., proline, most Gln3 accumulates in the nucleus. Increased Gln3 levels facilitate binding to its target UASNTR elements which in turn increases transcription. On the other hand, with repressive nitrogen sources, e.g., glutamine, Gln3 localizes to the cytoplasm and hence its nuclear concentration is low as is transcription. With glutamate, a nitrogen source that elicits intermediate NCR [27,61], nuclear Gln3 accumulation is intermediate (K.H. Cox, J.J. Tate and T.G. Cooper, unpublished observations). If a gene’s UASNTR elements support strong association with Gln3 (e.g., GAP1), the effect of a modest to moderate decrease in nuclear Gln3 levels would not be expected to produce a great decrease in transcription. However, if the UASNTR elements associate poorly with Gln3 to begin with, any decrease in nuclear Gln3 concentration would concomitantly decrease transcription as seen in DAL5. With respect to UASB, high nuclear levels of Gln3 in proline-grown cells would tend to increase association between Gln3 and DAL5 UASNTRS, hence minimizing the effects of inactivating UASA/B. On the other hand, growing cells in glutamate lowers intranuclear Gln3 which results in Gln3’s interaction with the UASNTR elements becoming more labile. In this instance, inactivating UASA/B would further destabilize the Gln3-UASNTR interactions, lowering transcription even more and producing the increased NCR-sensitivity seen in Figs. 7 and 8. It will be possible to test this explanation when proteins binding to UASNTR, UASA and UASB can be purified and DNA binding studies performed in vitro.
An alternative explanation for low DAL5 expression in glutamate-grown cells is that UASA and UASB are negatively regulated by glutamate. We do not favor this interpretation for two reasons: (i) it hypothesizes glutamate potentially functions through two different cis-acting elements; and (ii) it does not explain parallel increased NCR-sensitivity seen with pRR41 and pRR42 in glutamate and ammonia-grown cells (Fig. 8).
Our interpretation of the above data may also offer insight into how DAL5 can economically meet the conflicting requirements that derive from encoding the permease that transports a precursor of biosynthetic reactions (ureidosuccinate) and a substrate of nitrogen catabolism (allantoate) [62]. The biosynthetic function of Dal5 requires constant low level permease activity – just enough to take advantage of any ureidosuccinate available in the environment, thereby alleviating the need for the cell to synthesize it. The catabolic function of Dal5 (a component of the allantoin degradative pathway), on the other hand, requires the ability to produce a broad range of permease activities depending upon environmental conditions. In excess nitrogen, Dal5 is unnecessary for catabolic purposes and should be restricted to low levels. In limiting nitrogen, on the other hand, high-level Dal5 activity is needed if the cell is to exploit allantoate as a nitrogen source. The DAL5 promoter is structured to meet these varying demands. In formal terms, the situation is analogous to that of preamplifier and amplifier circuits. A preamplifier circuit is capable of only low-level output, but one that is highly regulated. The DAL5 UASNTR elements possess just such characteristics, i.e., they are significantly more responsive to NCR than those of many other genes in this regulon, but on their own, function relatively poorly. An amplifier circuit is unable to regulate a signal, but can greatly amplify a weak regulated input signal. The DAL5 UASB element possesses these characteristics, i.e., it has no demonstrable regulated UAS activity on its own, but greatly amplifies UASNTRS ability to activate transcription when it is active. Therefore, except in conditions of nitrogen limitation, DAL5 expression is low, but always on. When nitrogen limitation does become significant, the DAL5 UASNTRS are able to function more effectively due to increased nuclear levels of Gln3, with the result being greatly amplified by the action of proteins binding to UASB.
Finally, early comparisons of allantoin pathway promoters led to the suggestion that two types existed: (i) those, like DAL5 and DAL3, that function constitutively (are not inducible) but are highly NCR-sensitive; and (ii) those, like DAL4, DAL7, DUR1 2 and DUR3, that are NCR-sensitive but also inducible with the inducer being the last unique compound in the pathway, allophanate. Present data, along with that cited above from GLN1, PUT1, CAR1 2 and DAL7, suggest only one NCR-sensitive promoter organization exists, i.e., one in which UASNTR elements act synergistically with a variety of other transcription factor binding sites. What is different about the two originally proposed promoter types are the characteristics of the non-UASNTR elements. For the inducible genes, the element that functions synergistically with UASNTR is inducer-dependent (e.g., UASI/Dal82 binding site, UASPRO/Put3 binding site, UASARG/Arg81-83), whereas for the constitutive genes, this synergistic element is inducer-independent (UASB in DAL5, TTTGTTT in GLN1 and Abf1/Rap1 binding sites in CAR1 and CAR2). This view of NCR-sensitive promoters per se and allantoin pathway promoters in particular is more biochemically unified than the one proposed earlier.
Taken together, the observations presented here provide a more complete view of the S. cerevisiae DAL5 promoter, a more unified view of the synergistic interaction of UASNTR elements with unrelated inducer-responsive or constitutive cis-acting elements and suggest that diminished DAL5 expression with glutamate as nitrogen source derives not from retrograde regulation but rather from the fact that poor UASNTR elements are more sensitive to nuclear Gln3 levels and hence NCR. The presence of DAL5 UASA and UASB also raise the possibility that the gene’s expression may be regulated beyond the effects of NCR.
Table 1.
Saccharomyces cerevisiae strains used in this work
| RH218 | MATa, trp1, CUP1, gal2, SUC2, Mal− |
| TCY1 | MATα, lys2, ura3 |
| TCY5 | MATα, lys2, ura3, trp1::hisG |
| W303-1A |
MATa, ade2-1, can1-100, his3-11 15, leu2-3 112, trp1-1, ura3-1 |
| RR91 | MATα, lys2, ura3, gln3::hisG |
| M970 | MATa, lys5/Matα, lys2 |
Acknowledgements
We thank Tim Higgins for preparing the artwork, and the UT Yeast Group for suggestions to improve the manuscript. This work was supported by NIH Grant GM-35642.
References
- [1].Schreiber SL. Immunophilin-sensitive protein phosphatase action in cell signaling pathways. Cell. 1992;70:365–368. doi: 10.1016/0092-8674(92)90158-9. [DOI] [PubMed] [Google Scholar]
- [2].Sigal NH, Dumont FJ. Cyclosporin A, FK-506, and rapamycin: pharmacologic probes of lymphocyte signal transduction. Annu. Rev. Immunol. 1992;10:519–560. doi: 10.1146/annurev.iy.10.040192.002511. [DOI] [PubMed] [Google Scholar]
- [3].Guba M, von Breitenbuch P, Steinbauer M, Koehl G, Flegel S, Hornung M, Bruns CJ, Zuelke C, Farkas S, Anthuber M, Jauch KW, Geissler EK. Rapamycin inhibits primary and metastatic tumor growth by antiangiogenesis: involvement of vascular endothelial growth factor. Nat. Med. 2002;8:128–135. doi: 10.1038/nm0202-128. [DOI] [PubMed] [Google Scholar]
- [4].Hidalgo M, Rowinsky EK. The rapamycin-sensitive signal transduction pathway as a target for cancer therapy. Oncogene. 2000;19:6680–6686. doi: 10.1038/sj.onc.1204091. [DOI] [PubMed] [Google Scholar]
- [5].Hosoi H, Dilling MB, Liu LN, Danks MK, Shikata T, Sekulic A, Abraham RT, Lawrence JC, Jr., Houghton PJ. Studies on the mechanism of resistance to rapamycin in human cancer cells. Mol. Pharmacol. 1998;54:815–824. doi: 10.1124/mol.54.5.815. [DOI] [PubMed] [Google Scholar]
- [6].Schreiber SL. Chemistry and biology of the immunophilins and their immunosuppressive ligands. Science. 1991;251:283–287. doi: 10.1126/science.1702904. [DOI] [PubMed] [Google Scholar]
- [7].Heitman J, Movva NR, Hall MN. Proline isomerases at the crossroads of protein folding, signal transduction, and immunosuppression. New. Biol. 1992;4:448–460. [PubMed] [Google Scholar]
- [8].Barbet NC, Schneider U, Helliwell SB, Stansfield I, Tuite MF, Hall MN. TOR controls translation initiation and early G1 progression in yeast. Mol. Biol. Cell. 1996;7:25–42. doi: 10.1091/mbc.7.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Dennis PB, Fumagalli S, Thomas G. Target of rapamycin (TOR), balancing the opposing forces of protein synthesis and degradation. Curr. Opin. Genet. Dev. 1999;9:49–54. doi: 10.1016/s0959-437x(99)80007-0. [DOI] [PubMed] [Google Scholar]
- [10].Schmelzle T, Hall MN. TOR, a central controller of cell growth. Cell. 2000;103:253–262. doi: 10.1016/s0092-8674(00)00117-3. [DOI] [PubMed] [Google Scholar]
- [11].Cyert MS. Regulation of nuclear localization during signaling. J. Biol. Chem. 2001;276:20805–20808. doi: 10.1074/jbc.R100012200. [DOI] [PubMed] [Google Scholar]
- [12].Raught B, Gingras AC, Sonenberg N. The target of rapamycin (TOR) proteins. Proc. Natl. Acad. Sci. USA. 2001;98:7037–7044. doi: 10.1073/pnas.121145898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Rohde J, Heitman J, Cardenas ME. The TOR kinases link nutrient sensing to cell growth. J. Biol. Chem. 2001;276:9583–9586. doi: 10.1074/jbc.R000034200. [DOI] [PubMed] [Google Scholar]
- [14].Crespo JL, Hall MN. Elucidating TOR signaling and rapamycin action, lessons from Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 2002;66:559–579. doi: 10.1128/MMBR.66.4.579-591.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Thomas G, Sabatini DM. In: Current Topics Microbiology and Immunolology. Hall MN, editor. Vol. 279. Springer; Berlin: 2003. [Google Scholar]
- [16].Beck T, Hall MN. The TOR signalling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature. 1999;402:689–692. doi: 10.1038/45287. [DOI] [PubMed] [Google Scholar]
- [17].Cardenas ME, Cutler NS, Lorenz MC, Di Como CJ, Heitman J. The TOR signaling cascade regulates gene expression in response to nutrients. Genes Dev. 1999;13:3271–3279. doi: 10.1101/gad.13.24.3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Hardwick JS, Kuruvilla FG, Tong JK, Shamji AF, Schreiber SL. Rapamycin-modulated transcription defines the subset of nutrient-sensitive signaling pathways directly controlled by the Tor proteins. Proc. Natl. Acad. Sci. USA. 1999;96:14866–14870. doi: 10.1073/pnas.96.26.14866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Bertram PG, Choi JH, Carvalho J, Ai W, Zeng C, Chan TF, Zheng XF. Tripartite regulation of Gln3p by TOR, Ure2p, and phosphatases. J. Biol. Chem. 2000;275:35727–35733. doi: 10.1074/jbc.M004235200. [DOI] [PubMed] [Google Scholar]
- [20].Komeili A, Wedaman KP, OÕShea EK, Powers T. Mechanism of metabolic control: target of rapamycin signaling links nitrogen quality to the activity of the Rtg1 and Rtg3 transcription factors. J. Cell. Biol. 2000;151:863–878. doi: 10.1083/jcb.151.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Hofman-Bang J. Nitrogen catabolite repression in Saccharomyces cerevisiae. Mol. Biotechnol. 1999;12:35–73. doi: 10.1385/MB:12:1:35. [DOI] [PubMed] [Google Scholar]
- [22].Cooper TG. Transmitting the signal of excess nitrogen in Saccharomyces cerevisiae from the Tor proteins to the GATA factors: connecting the dots. FEMS Microbiol. Rev. 2002;26:223–238. doi: 10.1111/j.1574-6976.2002.tb00612.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Cooper TG. Topics in Current Genetics. In: Winderickx J, Taylor PM, editors. Nutrient-induced Responses. 2004. pp. 225–257. Chapter 7. [Google Scholar]
- [24].Cox KH, Kulkarni A, Tate JJ, Cooper TG. Gln3 phosphorylation and intracellular localization in nutrient limitation and starvation differ from that generated by rapamycin-inhibition of Tor1/2 in Saccharomyces cerevisiae. J. Biol. Chem. 2004;279:10270–10278. doi: 10.1074/jbc.M312023200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Cox KH, Tate JJ, Cooper TG. Cytoplasmic compartmentation of Gln3 during nitrogen catabolite repression and the mechanism of its nuclear localization during carbon starvation in Saccharomyces cerevisiae. J. Biol. Chem. 2002;277:37559–37566. doi: 10.1074/jbc.M204879200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Rai R, Genbauffe FS, Sumrada RA, Cooper TG. Identification of sequences responsible for transcriptional activation of the allantoate permease gene in Saccharomyces cerevisiae. Mol. Cell. Biol. 1989;9:602–608. doi: 10.1128/mcb.9.2.602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Tate JJ, Cooper TG. Tor1/2 regulation of retrograde gene expression in Saccharomyces cerevisiae derives indirectly as a consequence of alterations in ammonia metabolism. J. Biol. Chem. 2003;278:36924–36933. doi: 10.1074/jbc.M301829200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Liao XS, Small WC, Srere PA, Butow RA. Intramitochondrial functions regulate nonmitochondrial citrate synthase (CIT2) expression in Saccharomyces cerevisiae. Mol. Cell. Biol. 1991;11:38–46. doi: 10.1128/mcb.11.1.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Liao X, Butow RA. RTG1 and RTG2: two yeast genes required for a novel path of communication from mitochondria to the nucleus. Cell. 1993;72:61–71. doi: 10.1016/0092-8674(93)90050-z. [DOI] [PubMed] [Google Scholar]
- [30].Chelstowska A, Butow RA. RTG genes in yeast that function in communication between mitochondria and the nucleus are also required for expression of genes encoding peroxisomal proteins. J. Biol. Chem. 1995;270:18141–18146. doi: 10.1074/jbc.270.30.18141. [DOI] [PubMed] [Google Scholar]
- [31].Velot C, Haviernik P, Lauquin GJ. The Saccharomyces cerevisiae RTG2 gene is a regulator of aconitase expression under catabolite repression conditions. Genetics. 1996;144:893–903. doi: 10.1093/genetics/144.3.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Zhegchang L, Butow RA. A transcriptional switch in the expression of yeast tricarboxylic acid cycle genes in response to a reduction or loss of respiratory function. Mol. Cell. Biol. 1999;19:6720–6728. doi: 10.1128/mcb.19.10.6720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Tate JJ, Cox KH, Rai R, Cooper TG. Mks1p is required for negative regulation of retrograde gene expression in Saccharomyces cerevisiae but does not affect nitrogen catabolite repression-sensitive gene expression. J. Biol. Chem. 2002;277:20477–20482. doi: 10.1074/jbc.M200962200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Dilova I, Chen CY, Powers T. Mks1 in concert with TOR signaling negatively regulates RTG target gene expression in S. cerevisiae. Curr. Biol. 2002;12:389–395. doi: 10.1016/s0960-9822(02)00677-2. [DOI] [PubMed] [Google Scholar]
- [35].Sekito T, Liu Z, Thornton J, Butow RA. RTG-dependent mitochondria-to-nucleus signaling is regulated by MKS1 and is linked to formation of yeast prion [URE3] Mol. Biol. Cell. 2002;13:795–804. doi: 10.1091/mbc.01-09-0473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Liu Z, Sekito T, Spirek M, Thornton J, Butow RA. Retrograde signaling is regulated by the dynamic interaction between Rtg2p and Mks1p. Mol. Cell. 2003;12:401–411. doi: 10.1016/s1097-2765(03)00285-5. [DOI] [PubMed] [Google Scholar]
- [37].Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Tschumper G, Carbon J. Sequence of a yeast DNA fragment containing a chromosomal replicator and the TRP1 gene. Gene. 1980;10:157–166. doi: 10.1016/0378-1119(80)90133-x. [DOI] [PubMed] [Google Scholar]
- [39].van Vuuren HJ, Daugherty JR, Rai R, Cooper TG. Upstream induction sequence, the cis-acting element required for response to the allantoin pathway inducer and enhancement of operation of the nitrogen-regulated upstream activation sequence in Saccharomyces cerevisiae. J. Bacteriol. 1991;173:7186–7195. doi: 10.1128/jb.173.22.7186-7195.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Guarente L, Ptashne M. Fusion of Escherichia coli lacZ to the cytochrome C gene of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 1981;78:2199–2203. doi: 10.1073/pnas.78.4.2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning A Laboratory Manual. 2nd edn. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1989. [Google Scholar]
- [42].Pfeifer K, Arcangioli B, Guarente L. Yeast HAP1 activator competes with the factor RC2 for binding to the upstream activation site UAS1 of the CYC1 gene. Cell. 1987;49:9–18. doi: 10.1016/0092-8674(87)90750-1. [DOI] [PubMed] [Google Scholar]
- [43].Luche RM, Sumrada R, Cooper TG. Mol. Cell. Biol. 1990;10:3884–3895. doi: 10.1128/mcb.10.8.3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Rai R, Tate JJ, Cooper TG. Ure2, a prion precursor with homology to glutathione S-transferase, protects Saccharomyces cerevisiae cells from heavy metal ion and oxidant toxicity. J. Biol. Chem. 2003;278:12826–12833. doi: 10.1074/jbc.M212186200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Cox KH, Pinchak AB, Cooper TG. Genome-wide transcriptional analysis in S. cerevisiae by mini-array membrane hybridization. Yeast. 1999;15:703–713. doi: 10.1002/(SICI)1097-0061(19990615)15:8<703::AID-YEA413>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- [46].Guarente L, Mason T. Heme regulates transcription of the CYC1 gene of S. cerevisiae via an upstream activation site. Cell. 1983;32:1279–1286. doi: 10.1016/0092-8674(83)90309-4. [DOI] [PubMed] [Google Scholar]
- [47].Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1972. p. 403. [Google Scholar]
- [48].Smart WC, Coffman JA, Cooper TG. Combinatorial regulation of the Saccharomyces cerevisiae CAR1 (arginase) promoter in response to multiple environmental signals. Mol. Cell. Biol. 1996;16:5876–5887. doi: 10.1128/mcb.16.10.5876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Cunningham TS, Andhare R, Cooper TG. Nitrogen catabolite repression of DAL80 expression depends on the relative levels of Gat1p and Ure2p production in Saccharomyces cerevisiae. J. Biol. Chem. 2000;275:14408–14414. doi: 10.1074/jbc.275.19.14408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Bysani N, Daugherty JR, Cooper TG. Saturation mutagenesis of the UASNTR (GATAA) responsible for nitrogen catabolite repression-sensitive transcriptional activation of the allantoin pathway genes in Saccharomyces cerevisiae. J. Bacteriol. 1991;173:4977–4982. doi: 10.1128/jb.173.16.4977-4982.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Cunningham TS, Svetlov VV, Rai R, Smart W, Cooper TG. G1n3p is capable of binding to UASNTR elements and activating transcription in Saccharomyces cerevisiae. J. Bacteriol. 1996;178:3470–3479. doi: 10.1128/jb.178.12.3470-3479.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Kovari L, Sumrada R, Kovari I, Cooper TG. Multiple positive and negative cis-acting elements mediate induced arginase (CAR1) gene expression in Saccharomyces cerevisiae. Mol. Cell. Biol. 1990;10:5087–5097. doi: 10.1128/mcb.10.10.5087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Omichinski JG, Clore GM, Schaad O, Felsenfeld G, Trainor C, Appella E, Stahl SJ, Gronenborn AM. NMR structure of a specific DNA complex of Zn-containing DNA binding domain of GATA-1. Science. 1993;261:438–446. doi: 10.1126/science.8332909. [DOI] [PubMed] [Google Scholar]
- [54].Cooper TG, Rai R, Yoo HS. Requirement of upstream activation sequences for nitrogen catabolite repression of the allantoin system genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 1989;9:5440–5444. doi: 10.1128/mcb.9.12.5440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Minehart PL, Magasanik B. Sequence of the GLN1 gene of Saccharomyces cerevisiae: role of the upstream region inregulation of glutamine synthetase expression. J. Bacteriol. 1992;174:1828–1836. doi: 10.1128/jb.174.6.1828-1836.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Rai R, Daugherty JR, Cooper TG. UASNTR functioning in combination with other UAS elements underlies exceptional patterns of nitrogen regulation in Saccharomyces cerevisiae. Yeast. 1995;11:247–260. doi: 10.1002/yea.320110307. [DOI] [PubMed] [Google Scholar]
- [57].Huang HL, Brandriss MC. The regulator of the yeast proline utilization pathway is differentially phosphorylated in response to the quality of the nitrogen source. Mol. Cell. Biol. 2000;20:892–899. doi: 10.1128/mcb.20.3.892-899.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Kovari LZ, Cooper TG. Participation of ABF-1 protein in expression of the Saccharomyces cerevisiae CAR1 gene. J. Bacteriol. 1991;173:6332–6338. doi: 10.1128/jb.173.20.6332-6338.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Kovari LZ, Kovari I, Cooper TG. Participation of RAP1 protein in expression of the Saccharomyces cerevisiae arginase (CAR1) gene. J. Bacteriol. 1993;175:941–951. doi: 10.1128/jb.175.4.941-951.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Park HD, Scott S, Rai R, Dorrington R, Cooper TG. Synergistic operation of the CAR2 (Ornithine transaminase) promoter elements in Saccharomyces cerevisiae. J. Bacteriol. 1999;181:7052–7064. doi: 10.1128/jb.181.22.7052-7064.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Bossinger J, Cooper T. Possible failure of NADP-glutamate dehydrogenase to participate directly in nitrogen repression of the allantoin degradative enzymes in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 1975;66:889–892. doi: 10.1016/0006-291x(75)90723-8. [DOI] [PubMed] [Google Scholar]
- [62].Turoscy V, Cooper TG. Ureidosuccinate is transported by the allantoate transport system in Saccharomyces cerevisiae. J. Bacteriol. 1987;169:2598–2600. doi: 10.1128/jb.169.6.2598-2600.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Sekito T, Thornton J, Butow RA. Mitochondria-to-nuclear signalilng is regulated by the subcellular localization of the transcription factors Rtg1p and Rtg3p. Mol Biol Cell. 2000;11:2103–2115. doi: 10.1091/mbc.11.6.2103. [DOI] [PMC free article] [PubMed] [Google Scholar]