SUMMARY
Islet amyloid polypeptide (IAPP) is a hormone cosecreted with insulin. IAPP proceeds through a series of conformational changes from random coil to β-sheet via transient α-helical intermediates. An unknown subset of these events are associated with seemingly disparate gains-of-function including catalysis of self-assembly, membrane penetration, loss of membrane integrity, mitochondrial localization and finally cytotoxicity, a central component of diabetic pathology. A series of small molecule, α-helical mimetics, oligopyridylamides, was previously shown to target the membrane bound α-helical oligomeric intermediates of IAPP. In this study, we develop an improved, microwave assisted synthesis of oligopyridylamides. A series of designed tripyridylamides demonstrate that lipid-catalyzed self-assembly of IAPP can be deliberately targeted. These molecules additionally affect IAPP induced leakage of synthetic liposomes and cellular toxicity in insulin secreting cells. The tripyridylamides inhibit these processes with identical rank orders of effectiveness. This indicates a common molecular basis for the disparate set of observed effects of IAPP.
Keywords: Amylin, amyloidogenesis, membrane protein, synthetic helical mimetics, membrane permeabilization, INS-1 cell toxicity
INTRODUCTION
Protein-protein interactions (PPIs) regulate numerous biological processes such as proliferation, differentiation, and apoptosis. Consequently, misregulation of these interactions contributes to diseases as diverse as cancer, HIV infection, and many forms of neurodegeneration. A significant proportion (∼40%) (Davis, Tsou, and Hamilton, 2007) of PPIs are mediated by α-helical structural motifs. The structure-based design of small molecules that can stabilize or disrupt helix-protein interactions with high specificity and efficacy is a central challenge in medicinal chemistry. Small molecules which can present functional groups on a pre-organized framework have been identified that disrupt the helix-protein recognition surfaces in interactions such as p53/hDM2 (Prabhakaran, Barnard, et al, 2013; Yin, Lee, et al, 2005), Bak/Bcl-xL (Ernst, Becerril, et al, 2003), and the six helix-bundle assembly of GP41 (Ernst, Kutzki, et al, 2002). Plainly, helix-protein interactions are an important and addressable category of biological target.
Helix-helix interactions can themselves be a component of pathological gains-of-function. Such interactions have been implicated as important to Alzheimer's, Parkinson's, and Type II diabetes (Davis, Tsou, and Hamilton, 2007). In diabetes, pathological self-assembly occurs in the 37 residue peptide, islet amyloid polypeptide (IAPP) (Figure 1). IAPP is a hormone, coexpressed, processed, packaged, and secreted with insulin by pancreatic β-cells. Its hormonal role is likely autocrine as IAPP knockout mice display a loss of precision in the pulsatile nature of insulin secretion (Braun, Ramracheya, and Rorsman, 2012). Importantly, rats transgenic for the human form of IAPP show a progression of prediabetic and diabetic symptoms suggesting that IAPP may also have a causal role in the etiology of this disease.
Figure 1.
Schematic of molecules used in this work. (A) Synthesis of the analogs of tripyridylamide. Reagents and conditions: (a) 33% HBr in AcOH, reflux, 24 h; (b) 2M NH3 in EtOH, 0°C for 30 min and then at r.t. for 24 h; (c) NaNO2, conc. H2SO4, 0 °C for 30 min and then at r.t. for 3 h; (d) Alkyl iodide, Ag2CO3, hexane (dry), 300 W, 150 °C, 12 min; (e) Pd(OAc)2, PPh3, DMF (dry), MeOH (dry), Et3N (dry), CO (g), 100 °C, 20 h; (f) LiOH, THF/H2O, r.t., 30 min; (g) Pd/C, H2 (g), THF, r.t., 4 h; (h) Compound 1-R, 2-chloro-1-methylpyridinium iodide, DCM (dry), Et3N (dry), reflux, 4 h; (i) Pd/C, H2 (g), THF, r.t., 4 h; (j) Compound 2-R, 2-chloro-1-methylpyridinium iodide, DCM (dry), Et3N (dry), reflux, 4 h; (k) DCM:TFA:TES (80:15:5, v/v), r.t., 3h. (B) Structural representations of the analogs of oligopyridylamides. (C) Primary sequence of human IAPP (hIAPP) used in the study. (D) A helical wheel representation of the helical sub-domain of IAPP. Blue and green represent positively charged and hydrophobic amino acid residues respectively.
Structurally, IAPP is predominantly disordered in aqueous solution with weak sampling of α-helical states across residues 5–22. Upon interaction with a phospholipid bilayer, IAPP binds, stabilizes this helical sub-domain (Apostolidou, Jayasinghe, and Langen, 2008; Williamson, Loria, and Miranker, 2009), and samples parallel and antiparallel helical assemblies prior to the conversion of IAPP to an amyloid form (Nath, Miranker, and Rhoades, 2011). Mutagenic analyses of IAPP have shown this helical region to be important to gains-of-toxic function in cell culture (Cao, Abedini, et al, 2013; Magzoub, and Miranker, 2012). Whether or not these states are themselves toxic, or are on/off the pathways to toxicity remains an open question. Regardless, these early interactions are important as they either directly lead to the formation of membrane active toxic oligomers, or serve as an off-pathway pool of protein that can modulate toxic assembly. Perturbation of these states by small molecules are thus desirable for both basic understanding of the disease and therapeutic lead development.
The structural basis of helix-helix interactions relevant to function can be characterized by a reduced description: namely that of the interactions of side chain residues on one helical face at positions i, i+3/i+4, and i+7 positions with corresponding positions of one or more other α-helices. The Hamilton lab has pioneered the development of synthetic small molecules that can mimic the placement of R-groups at comparable spatial positions (Cummings, and Hamilton, 2010). Thus, disruption using small molecule protein mimetics can be based on the premise of mimicking one half of a helix target interaction. An interaction might alternatively be stabilized allosterically by binding the solvent exposed component of an α-helix containing complex.
Oligopyridylamide based mimetics were previously found to act as antagonists of membrane catalyzed self-assembly of IAPP (Hebda, Saraogi, et al, 2009; Saraogi, Hebda, et al, 2010). Inhibition was suggested to have been achieved via charge complementarity in which the oligopyridylamide scaffold (Figure 1A), derivatized with carboxylate functionalities, interacts with basic side chain residues (R11, H18, K1, and N-terminus) presented on the IAPP α-helical surface. The site of contact was confirmed using NMR binding studies between a pentapyridylamide (IS-5) and IAPP. However, the study indicates that the use of carboxylates was an oversimplification of the binding interaction. For example, two positive charges, R11 and H18 are separated by two α-helical turns while our published molecules display three carboxylates over a corresponding distance. In this work, we have therefore conducted a structure activity analysis of the molecular nature of oligopyridylamide:IAPP interactions. We have improved the synthesis strategy for this class of molecules, and designed and synthesized analogs of tripyridylamide in which various functionalities are presented in-between flanking carboxylates. This systematic study enables us to gain insight into the molecular basis for interactions involved in self-assembly of membrane bound α-helical intermediates of IAPP.
RESULTS AND DISCUSSION
A convergent synthesis based approach was developed using a microwave-assisted protocol to facilitate the generation of a library of tripyridylamides. The bromo-substituted aromatic imidate was synthesized from 2, 6-dichloro-3-nitropyridine in three steps (Figure 1). This was then selectively O-alkylated using microwave-assistance in the presence of alkyl iodides and silver carbonate (Figure 1) (Singh, Cavalluzzo, et al, 2009). The bromo-substituted O-alkylated aromatic imidates were converted to methyl esters in the presence of Pd(OAc)2 under an atmosphere of CO(g) (450 psi, 75 °C) followed by saponification. Chain elongation was achieved using iterative amide coupling between aryl amines and acids using 2-chloro-1-methylpyridinium iodide (Mukaiyama’s reagent) and subsequent reduction of nitro groups (see synthesis and characterization in SI text). The acid labile tert-butyl esters were cleaved using TFA cocktail (DCM:TFA:TES, 80:15:5, v/v) in the last step to afford the series of tripyridylamides as free acids with varying degree of hydrophobicity on the central pyridine scaffold in nearly quantitative yields (Figure 1). In comparison to the earlier reported synthetic procedure for O-alkylation of bromo imidates (time = overnight and yield = 75%), microwave assisted protocol (time = 10 min. and yield ∼75–93% for various monomers, Table S1) enabled us to efficiently generate a library of analogs of O-alkylated bromo imidates. Additionally, the O-alkylation in tandem with carbonylation of bromo imidates makes it a powerful tool to generate libraries of oligopyridylamides.
The kinetic profile of compound effects on lipid-catalyzed and lipid free IAPP fibrillation was probed using an exogenous dye, Thioflavin T (ThT) as reporter (Wolfe, Calabrese, et al, 2010). ThT binds specifically to IAPP amyloid fibers without perturbing the kinetics of amyloid assembly. The kinetic profile for IAPP fibrillation includes a lag period, a growth phase, and a subsequent equilibrated phase in which the protein is predominantly in the amyloid fiber state (Figure 2A). Analyses in this work are restricted to the midpoint of the reaction (t50, Figure S1). The model membrane system used here is an equimolar mixture of dioleoylphosphatidylglycerol (DOPG) and dioleoylphosphatidylcholine (DOPC) extruded into 100 nm large unilamellar vesicles (LUV’s). The presence of a phospholipid surface catalyzes IAPP fibrillation and promotes α-helical membrane bound prefibrillar structures (Williamson, Loria, and Miranker, 2009).
Figure 2.
Effect of oligopyridylamides on the kinetics of IAPP amyloid assembly. (A) Time dependent ThT fluorescence-based kinetic profiles of lipid-catalyzed IAPP fibrillation alone (black), and in the presence of ADM-1 (blue), IS-5 (brown), or ADM-3 (red) at a ratio of 10:1 (ligand:IAPP). (Inset) A sigmoidal fit is used to determine the t50, time required to reach 50% fluorescence. (B) Summary of reaction t50 for the effect of analogs of tripyridylamide on the kinetics of lipid-free (black) and lipid-catalyzed (purple) self-assembly. (C) Dose dependent effect of ADM-3, ADM-1, ADM-7, and IS-5 on lipid-catalyzed fibrillation. (D) CD spectra of IAPP in the absence and presence of ADM-3 under lipid containing conditions. TEM images of lipid-catalyzed 10 µM IAPP after 1 h in the absence (E) and presence (F) of ADM-3. Lipid catalyzed conditions: [IAPP] = 10 µM, [ADM-X] = 100 µM, [ThT] = 5 µM, [DOPG:DOPC, 1:1, 100 nm] = 630 µM. Lipid free conditions: [IAPP] = 40 µM, [ADM-X] = 40 µM, [ThT] = 20 µM. For TEM, [IAPP] = 10 µM, [ADM-3] = 10 µM, [DOPG:DOPC, 1:1, 100 nm] = 630 µM. All error bars represent standard deviations from a minimum of three repeated measurements.
Inhibition of lipid-catalyzed IAPP fibrillation is markedly affected by the elimination of the central carboxylate of our previously published compound IS-3 (Saraogi, Hebda, et al, 2010). Lipid membrane catalysis accelerates IAPP fibrillation to a t50 of 0.92 ± 0.02 h. This is ∼18 times faster than the lipid-free condition (t50 = 17.1±1.2 h). IS-3, referred to here as ADM-1, possesses three carboxylate functionalities on its surface. ADM-1 inhibits IAPP fibrillation with a t50 that is 1.7 ± 0.04 fold higher than the control reaction (Figure 2A, 2B, and 2C), in agreement with our earlier work (t50 = 2.3 ± 0.04 times higher than control reaction) (Saraogi, Hebda, et al, 2010). ADM-3, a tripyridylamide with an ethyl substitution at position R, inhibits IAPP fibrillation with a t50 that is at least >10 fold higher than the control reaction (Figure 2). IS-5, a pentacarboxylate pyridylamide, delays IAPP fibrillation with a t50 that is 3.5 fold higher than the control reaction (IAPP only) at a stoichiometric ratio of 10:1 (IS-5:IAPP, Figure 2B and 2C), in aggrement with our previously published work (Saraogi, Hebda, et al, 2010). ADM-3 produced the same effect by delaying IAPP fibrillation with a t50 that is 3.7 fold higher than the control reaction (IAPP only) at a stoichiometric ratio of 1:1 (IS-5:IAPP, Figure 2C). Additionally, at higher concentrations (up to mM), IS-5’s inhibitory activity maximizes with a t50 which is 3.5 fold higher than the control reaction. In marked contrast, ADM-3 completely eliminates lipid catalyzed IAPP fibrillation. Plainly, trimeric ADM-3 is far better inhibitor of lipid catalyzed IAPP fibrillation than our previously demonstrated pentameric construct.
ADM-3 inhibited IAPP is non-fibrillar in spectroscopic and imaging assays. Dose dependent effects of ADM-3 on the t50 of fibrillation excludes the concern of intensity based artifacts in the ThT assay (Figure 2C). Moreover, both CD and transmission electron microscope (TEM) based results corroborate well with the ThT assay in terms of exhibiting inhibitory activity of ADM-3 on IAPP fibrillation. IAPP exhibits predominantly α-helical structure in the presence of lipid membrane (LUVs, DOPG:DOPC, 1:1, 100 nm) characterized by two minima at ∼208 and ∼222 nm in the CD spectrum (Figure 2D). The conformation of IAPP alone switches from α-helical to β-sheet in 1 h. In marked contrast, IAPP remains in an α-helical state in the presence of ADM-3 after 2 h at a stoichiometric ratio (Figure 2D). TEM showed IAPP fiber formation after 1 h (Figure 2E); however no fibers were observed in the presence of ADM-3 even after 2 h at an equimolar ratio (Figure 2F). As a control, denoted by ADM-3*, a mixture of monomeric building blocks for ADM-3 (Scheme S2) has no effect on lipid-catalyzed IAPP fibrillation indicating the importance of functionalities at suitable spatial distance and orientation. The tripyridylamides have no effect on ThT fluorescence intensity by themselves (Figure S2). Thus, the observation of inhibition by ADM-3 is robustly observed across several complementary assays.
The ethyl carbon of R occupies a site of specific importance to lipid-catalyzed IAPP assembly. This is evident by first comparing the inhibitory effect of ADM-3 and ADM-2 on IAPP fibrillation. The former inhibits with a t50 that is >10 fold higher than the control reaction (Figure 2B). In marked contrast, a single carbon loss to a methyl R group (ADM-2) is strongly detrimental. The effectiveness of ADM-2 is 2.9 ± 0.02 fold inhibitory relative to the control reaction. Substitution to a longer chain at R (ADM-4) preserves the ethyl carbon and effectiveness is also maintained. Additionally, a long aliphatic chain substitution at position R (ADM-5) is more inhibitory than ADM-2 (Figure 2B). Interestingly, inhibitory activity of ADM-5 is weakened relative to the ethyl (ADM-3) and butyl (ADM-4) substituted compounds. We conjecture this is due to self-association of the aliphatic chains which could decrease its effectiveness.
The introduction of branched substituents at ethyl carbon (ADM-6 and ADM-8) diminish the inhibitory activity towards IAPP fibrillation compared to ADM-3 (Figure 2B). This is reflected by the decrease in t50 of ADM-6 and ADM-8 containing reactions in comparison to ADM-3. Additionally, the introduction of branched substituents at the methyl position (ADM-7 to ADM-9) also produced similarly diminished effects on IAPP fibrillation. This suggests the formation of steric clash for branched substitution at methyl and ethyl positions of R. It is worth noting that while inhibitory activity is diminished compared to ADM-3, these molecules are still more potent inhibitors of IAPP fibrillation than ADM-1. The sensitivity of effectiveness to branching suggests a close packed interaction of tripyridylamides with IAPP.
To gain mechanistic insight into the role of phospholipid surface, kinetic assays were conducted in solution with no lipid (Figure 2B). Tripyridylamides in general were ineffective against IAPP fibrillation except ADM-5 which was a modest inhibitor of IAPP fibrillation at a stoichiometric ratio of 1:1 (tripyridylamide:IAPP). We have shown earlier that IS-5 acts as an agonist to IAPP fibrillation (lipid free, Figure 2B) (Hebda, Saraogi, et al, 2009; Saraogi, Hebda, et al, 2010). Here, ADM-3, which is 40% smaller and has 60% fewer anionic charges than IS-5, shows modest inhibition indicating that one or both of these changes are responsible for oligopyridylamide to act instead as agonist in lipid-free conditions. We earlier conjectured that IS-5 mimics the membrane surface and catalyzes IAPP fibrillation. Tripyridylamides on the other hand, have only small effects on IAPP fibrillation under lipid free conditions even at higher stoichiometric ratios (2:1, tripyridylamide:IAPP). This suggests that the mechanism is more nuanced than simply a competition with membrane surface. ADM-5 is an exception to this observation. The presence of self-associated ADM-5 (see below) may contribute to its capacity to act as an inhibitor even in the absence of a lipid-surface catalyst.
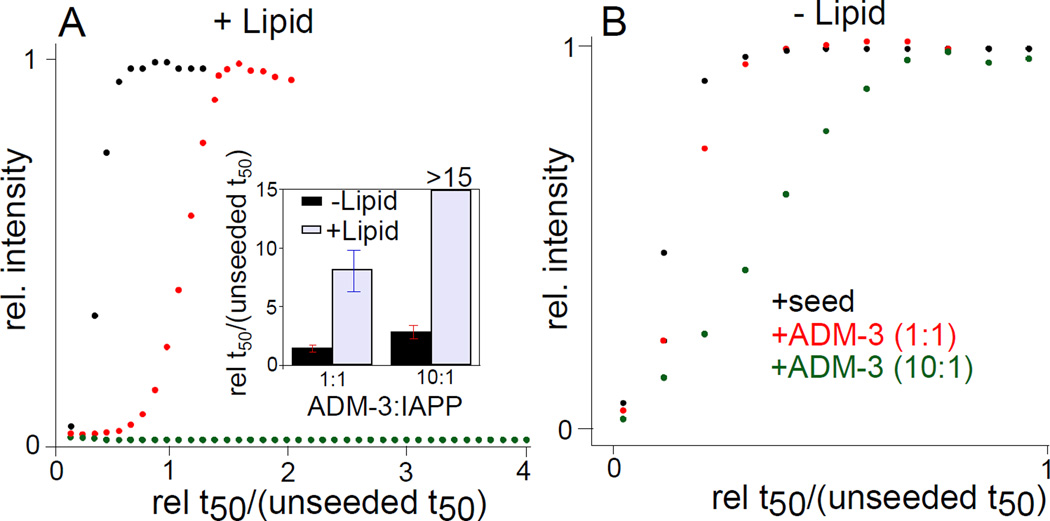
ADM-3 is an effective inhibitor of membrane associated elongation processes. Elongation was directly assessed by accelerating the IAPP self-assembly reaction using preformed fibers as seed. This bypasses primary nucleation processes, characterized by shortening of the lag phase (Figure 3). Here, lipid surface catalyzed IAPP fibrillation was accelerated to a t50 7.7 ± 0.3 fold faster by the addition of 1% seeds to a standard reaction (Figure 3A). Under this condition, the reaction is complete while still in the lag phase of the seed-free control. At an equimolar ratio, addition of ADM-3 slows down the elongation process to a t50 that is 8.1 ± 0.4 fold slower than the seeded reaction (Figure 3A). At a stoichiometric ratio of 10:1 (ADM-3:IAPP), IAPP amyloid self-assembly is wholly arrested. In marked contrast, the equivalent reactions performed in the absence of lipid give no inhibition at 1:1 and only modest inhibition of seeded assembly at 10:1, ADM-3:IAPP (Figure 3B). We conjecture that ADM-3 binds to α-helical intermediates of prefibrillar IAPP structures and attenuates their access to mature fibers. The greater sampling of α-helical states on the membrane surface facilitates the increase in apparent effectiveness of the helix targetted small molecules.
Figure 3.
Effect of ADM-3 on the kinetics of seeded IAPP fibrillation. Representative kinetic profiles of seeded reactions measured under lipid catalyzed (A) and lipid-free (B) conditions. Note, the x-axes are expressed as reaction time divided by the reaction t50 of an unseeded control. (Inset) Statistics of repeats of experiments shown in (A,B). For lipid catalyzed conditions: [IAPP] = 10 µM, [DOPG:DOPC, 1:1, 100 nm] = 630 µM, [seed] = 1% (w/w), ThT = 5 µM. For lipid free conditions: [IAPP] = 30 µM, [seed] = 5% (w/w), ThT = 15 µM.
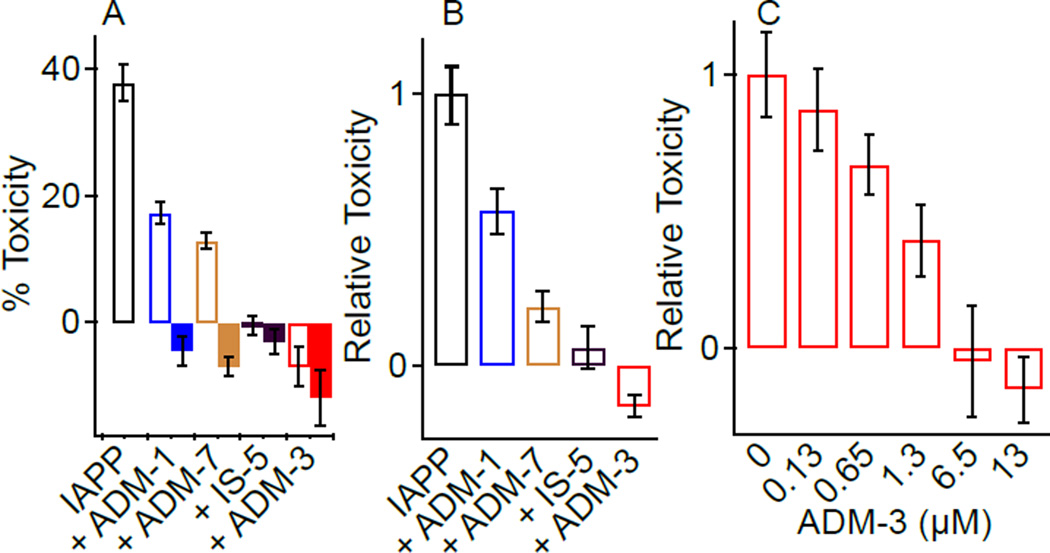
Lipid surface catalyzed amyloid formation serves as a downstream readout of events that may be associated with cell toxicity (Hebda, Magzoub, and Miranker, 2014). We therefore investigated the capacity of ADM-3 to affect cell toxicity and membrane permeation gains-of-function. ADM-3 rescues IAPP-mediated cytotoxicity (Figure 4). Toxicity is readily induced in an established model for pancreatic β-cells, INS-1 (Huang, Lin, et al, 2007). In the presence of 13 µM IAPP, the viability of INS-1 cells is reduced to 64 ± 3% (36 ± 3% toxicity) compared to addition of IAPP-free vehicle. Compounds alone are non-toxic under these conditions and do not interfere with the colorimetric assay (Figure S4). ADM-3, the most effective inhibitor of IAPP fibrillation, completely rescues INS-1 cell toxicity at a stoichiometric ratio of 1:1 (Figure 4A, B). ADM-1 (IS-3) and IS-5 rescued toxicity by 43 ± 9% (ADM-1:IAPP, 1:1) and 94 ± 7% (IS-5:IAPP, 1:1) respectively (Figure 4A, B). Dose dependent analysis of ADM-3 reveals it to function at sub-stoichiometric concentrations with 100% rescue evident at 1:0.5, IAPP:ADM-3. Clearly, the effectiveness of ADM-3 on lipid catalyzed fiber formation is reflected in its capacity to rescue INS-1 cells from IAPP induced toxicity.
Figure 4.
Oligopyridylamides effects IAPP-mediated toxicity. (A) Representative toxicity of 13 µM IAPP with and without the cointroduction of equimolar amounts of the indicated compound (open). Toxicity of 13 µM compound alone (no IAPP) is also shown (filled). Error bars reflect variability across 4–8 technical replicates within a single execution of the assay. (B) Statistics of relative toxicity from repeated independent execution of toxicity assays shown in (A). Data is expressed as toxicity in the presence of compound relative to toxicity of IAPP alone measured at the same time on the same batch of cells. (C) Rescue from 13 µM IAPP toxicity as a function of the indicated concentration of ADM.
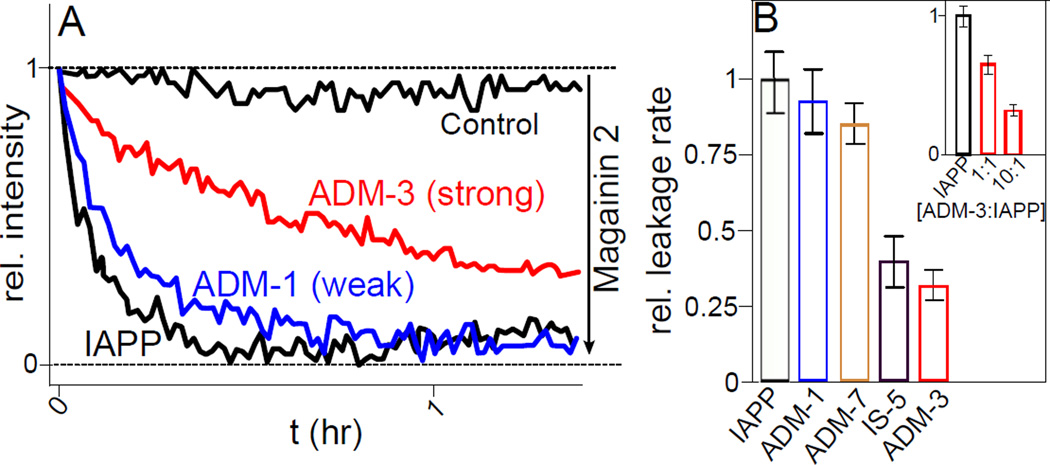
Membrane poration at the mitochondrial membrane is a potential contributor to IAPP mediated-cell toxicity. Hence, in vitro leakage may serve as a complementary assay for cell toxicity (Last, and Miranker, 2013). Liposome leakage kinetics were employed to assess the binding interaction between membrane bound α-helical intermediates of IAPP and tripyridylamides (Figure 5). Our assay uses 100 nm diameter, unilamellar extruded vesicles comprised of 100% DOPG. Fluorescein dextran at 70 kDa was encapsulated during extrusion. This size dextran has previously been shown not to escape from IAPP permeabilized vesicles. The kinetics of leakage was probed by quenching of fluorescence intensity using DPX (p-xylene-bis-pyridinium bromide, ∼400 Da). This results in well defined, single exponential profiles in which 100% leakage is observed. The leakage rate constant for 4 µM IAPP was 9.3 ± 1.1×10−4 s−1 which is within the range of our published report (Figure 5) (Last, Rhoades, and Miranker, 2011).
Figure 5.
Attenuation of IAPP mediated liposome leakage. (A) Representative assessment of leakage competance performed using fluorescein dextran encapsulated liposomes. At t=0, 200 µM of DOPG LUVs are exposed to the soluble quencher DPX only (control), or with addition of 4 µM IAPP (black), or with addition of 10:1 of the indicated compound:IAPP (colored). The fully leaked response and scale is established with 20 µM magainin 2. (B) Statistics from repeat analysis of single exponential fits to leakage data shown in (A). Data is expressed relative to IAPP only control (black). (inset) Dose dependence of ADM-3.
Small molecules that affect fiber formation and toxicity, also affect the leakage competence of IAPP. The rate constant for liposome leakage induced by IAPP in the presence of ADM-3 (Figure 5A) was decreased by a factor of 1.6 ± 0.3 and 3.2 ± 0.4 at a stoichiometric ratio of 1:1 (ADM-3:IAPP) and 10:1 respectively (inset, Figure 5B). On the other hand, at 10:1 (ligand:IAPP), ADM-1/IS-3 and IS-5 delay the kinetics of IAPP mediated leakage by a factor of 1.1 ± 0.1 and 2.2 ± 0.4 respectively (Figure 5B). To assess the completeness of the leakage reaction, magainin 2, an antimicrobial peptide from Xenopus, was added to the wells after 90 min. A significant decrease in the fluorescence intensity is apparent for the solutions without IAPP. In contrast, no significant changes in fluorescence are observed for IAPP containing samples indicative of the complete permeabilization of fluorescein encapsulated liposomes. As a further control, fluorescein encapsulated liposomes were assayed with oligopyridylamides only. No leakage was observed (Figure S3). Plainly, the helical targetted oligopyridyl amides have the capacity to diminish membrane permeation.
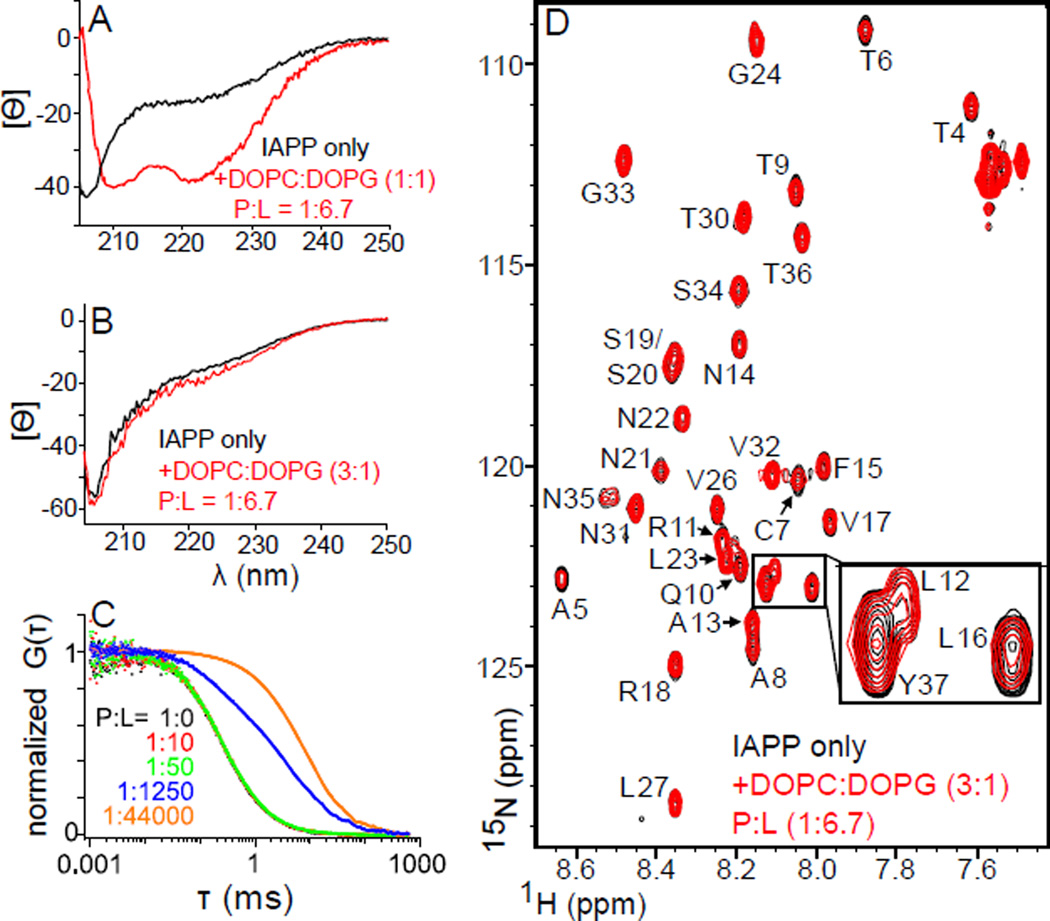
A common precursor underpins poration, fiber formation and toxicity. ADM-3 and three other molecules are shown here to inhibit IAPP induced membrane permeabilization as well as lipid-catalyzed IAPP amyloid assembly. Moreover, these molecules are capable of rescuing cytotoxicity induced in INS-1 cells. Importantly, the rank order for molecules affecting all three processes is the same: ADM-3 > IS-5 > ADM-7 > ADM-1. This suggests a common, membrane bound helix conformation underpins all three gains-of-function. Our observed correlation is at odds with a recently published report (Cao, Abedini, et al, 2013) which suggests that α-helical structure is not required for membrane leakage, and that membrane leakage is not directly related to toxicity. We matched the conditions at or near the P:L ratio used in that work, 1:6.7. In response to the addition of liposomes, we observe no change in CD signal (Figure 6B and Figure S5A and Figure S5B), no change in the linear diffusion (Figure 6C), and no change in NMR chemical shifts (Figure 6C and Figure S5C). This suggests that it is the absence of membrane binding which results in a lack of a change in secondary structure. Observed leakage in that work is measured at two time points, 10 min and 2 days. Reported observables are significant, but often fractional (i.e. <100%) and mainly unchanged over the time period. For example, 30 µM human IAPP shows ∼30% leakage at 10 min and ∼60% after 2 days. The suggests the presence of a dead-time event. Our assay gives full, single exponential profiles from 0 to 100% leaked. The assay used by Cao et al is likely reporting on a leakage process that is not the same as ours. Our own efforts using mutagenesis suggest that membrane-bound α-helical sub-states are associated with several solution and cellular gains-of-function including localizing to mitochondria (Magzoub, and Miranker, 2012). Such work does not implicate α-helical states directly, but underpins the idea that manipulation of the helical assemblies can affect gains-of-function. Such manipulation includes direct interaction with helical sub-domains, or binding to one or more intermediates downstream from the helical states. Toxic oligomers that are neither amyloid fiber, nor monomeric precursor have been detected, for example, by immunochemical methods (Kayed, Head, et al, 2003), and crystallography (Laganowsky, Liu, et al, 2012). The small molecules synthesized here are designed to interact with the α-helical subdomain. As a result, we assert that our observations support the view that leakage competent α-helical oligomeric states provide either the conditions from which toxic species arise, or are directly responsible for IAPP mediated toxicity.
Figure 6.
Characterization of IAPP membrane binding under alternate conditions. CD spectra of 60 µM human IAPP (A) or rat IAPP (B) in the absence (black) and presence (red) of liposomes at the indicated stoichiometry of DOPC:DOPG and with P:L=1:6.7 (IAPP:lipid). (C) Normalized fluorescence autocorrelation curves for 100 nM rat IAPPRG. Shown are protein alone (black) and in the presence of DOPC:DOPG (3:1) liposomes at the indicated P:L. Shifts of curves to the right are indicative of binding to the liposomes. Note, red and black are obscured by the green points. (D) NMR (15N HSQC) of 50 µM rat IAPP in the absence (black) or presence (red) of lipid bilayer at 1:6.7 (IAPP:lipid), and DOPC:DOPG at 3:1. (inset) A three fold magnification of resonances from the α-helical subdomain (L12, L16), and the C-terminus (Y37). Elipticity in (A) and (B) are in units of deg.cm2.dmol−1.res−1. Buffer conditions (A–C): 20 mM Tris·HCl, 100 mM NaCl, pH 7.4. (D) As in (A–C), but at pH 6.5 here, and pH 7.5 in supplement (Figure S5).
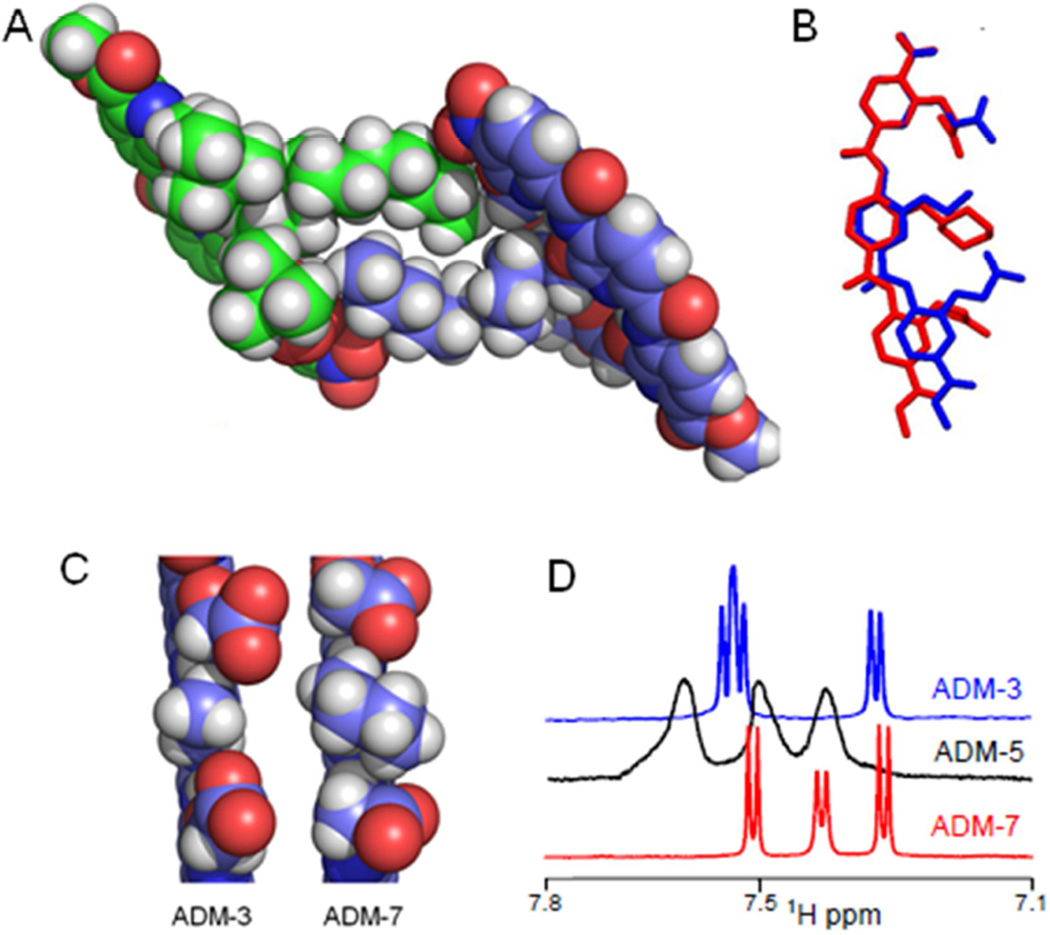
The crystal structure of tert-butyl analogs of ADM-5 and ADM-7 were determined (Figure 7). The structures adopt a rod-like elongated conformation with a stable curved backbone stabilized by the bifurcated hydrogen bonding (Figure 7). The crystal packing of ADM-5 shows self-assembly possibly assisted by the aliphatic chain (Figure 7A). This possibility was confirmed under our lipid-free, kinetic assay conditions by 1D, 1H NMR. The 1H peaks in spectra of ADM-5 were broad (Figures 7D, Figure S5D). In marked contrast, splitting of proton peaks in ADM-3 and ADM-7 were readily resolved. We conjecture that the dynamic self-assembly of ADM-5 (Figure 7D) attenuated the monomer’s accessibility to inhibit IAPP fibrillation and reduced its inhibitory activity in comparison to ADM-3 and ADM-4. A model of ADM-3 can be derived from ADM-5 by deletion of atoms (Figure 7B). ADM-3 presents the side chains on one face which is the potential complementary recognition surface for the membrane bound helical structure of IAPP (Figure 7B, blue). ADM-7, where cyclohexane is substituted at position R, effects the positioning of adjacent side chains probably due to the steric hindrance (Figure 7B and 7C). We speculate that the range of the conformations accessible for COOH functional groups are limited by the steric clash offered by the central bulky group (here cyclohexane) which might be contributing to their lower inhibitory activity towards IAPP fibrillation. In summary, a series of analogs of tripyridylamide was designed and synthesized with varying functionalities presented on its surface. The selection process was sensitive to formation of an optimal complementary surface to side chain residues of the α-helical domain of IAPP. The inhibitory activity of tripyridylamides is strongly sensitive to the selection of the functionality at the R position on the central pyridyl moiety. Indeed, structure based design enabled us to create a trimeric construct that wholly eclipsed the molar activity of our previously published pentapyridyl amide (Hebda, Saraogi, et al, 2009; Saraogi, Hebda, et al, 2010). The study lays a foundation for designing putative small molecules inhibitors of membrane bound α-helical oligomeric intermediates which have been suggested as relevant not only to type 2 diabetes, but also Alzheimer's and Parkinson's diseases.
Figure 7.
Atomic structures of tripyridylamides. (A) A crystallographic dimer of the tert-butyl ester analog of ADM-5. (B) Overlay of crystal structure models of ADM-3 and ADM-7. ADM-3 is derived from the crystal structure of tert-butyl ester ADM-5, with tert-butyl ester and excess R group atoms removed (blue). Crystals were not obtainable for ADM-3. ADM-7 is also shown with tert-butyl ester groups removed. Alignment was performed using only atoms from one of the pyridyl rings. (C) Side-on, space filled views of the structures shown in (B). (D) Comparison of 1D 1H NMR spectra of the aromatic region (full spectra in Figure S5D) of ADM-3, ADM-5, and ADM-7 under lipid-free, kinetic assay conditions (see Figure 2).
SIGNIFICANCE
Self-assembly of disease specific proteins into ordered fibrillar aggregates is a hallmark of many clinical conditions. Islet amyloid polypeptide (IAPP), a 37 residue peptide hormone cosecreted with insulin, proceeds through a series of conformations starting with weakly helical states in solution to membrane bound α-helical forms that eventually lead to β-sheet rich, filamentous aggregates. One diabetes relevant possibility is that some subset of this dynamic and heterogeneous ensemble of membrane bound states are directly or indirectly responsible for the several pathological gains-of-function including, membrane translocation, membrane poration, mitochondrial localization, and cytotoxicity. A SAR study, using a novel small molecule template, has been employed to gain insight. The template allows linking of the membrane bound IAPP ensemble to amyloidogenesis, membrane leakage, and cytotoxicity.
EXPERIMENTAL PROCEDURES
Materials
Unless otherwise indicated, all IAPP used in this work is the WT, human sequence. Human islet amyloid polypeptide (IAPP) was synthesized by t-Boc methods and purified by Genscript (Piscataway, NJ) and Elim Biopharmaceuticals (Hayward, CA). Uniformly labelled with 15N, wild-type rat IAPP was prepared recombinantly as described previously (Williamson, and Miranker, 2007). Thioflavin T (ThT) was purchased from Acros Organics (Fair Lawn, NJ). Lipids [dioleoylphosphatidylglycerol (DOPG) and dioleoylphosphatidylcholine (DOPC)] were purchased from Avanti Polar Lipids, Inc. (Alabaster, AL). The 96-well plates (black, w/flat bottom) were bought from Greiner Bio-One (Monroe, NC). All of the chemicals were purchased from commercial suppliers and used without further purification. Silica plates (w/UV254, aluminum backed, 200 micron) and silica gel (standard grade, particle size = 40–63 micron, 230 × 400 mesh) for flash column chromatography were purchased from Sorbent Technologies (Atlanta, GA). Dry solvents were purchased from Sigma Aldrich (St. Louis, MI) or VWR (Bridgeport, NJ). 2,6-dichloro-3-nitropyridine, alkyl iodides, triethylamine (dry), 2-chloro-1-methylpyridinium iodide, tert-butyl bromoacetate, trifluoroacetic acid (TFA), and triethylsilane (TES) were purchased from Sigma Aldrich (St. Louis, MI). The microwave assisted synthetic steps were performed in 5 mL vials (Biotage, Charlotte, NC) using a Biotage Initiatior Classic Synthesizer (Charlotte, NC).
Preparation of IAPP
IAPP (∼2 mg) was solubilized in 7 M guanidinium hydrochloride. The solution was filtered (0.2 micron) and transferred to Silica C-18 spin column, 50–450 µL loading, 30–300 µg capacity (The Nest Group, Inc. Southborough, MA), washed twice with water (400 µL each) followed by 10% acetonitrile in water, 0.1% formic acid (v/v) and then eluted into 200 µL of 50% acetonitrile in water, 0.1% formic acid (v/v). The concentration of IAPP (oxidized form) was calculated using absorbance measurements at 280 nm (ε = 1400 M−1cm−1). The IAPP solution was divided into several aliquots (20–50 µL, 1–2 mM), lyophilized, and stored as a white solid at −80 °C. Fresh stock solution of IAPP was prepared in water for each experiment.
Preparation of rat IAPPRG
IAPP was fluorescently labeled on its amino terminus with Rhodamine Green succinimidylester by adding a 3.5 molar excess to the peptide in a pH 7.4, 100mM sodium bicarbonate solution and incubating for 4 hours at room temperature. Free dye was separated from labeled peptide using a C-18 reverse phase analytical HPLC column (Grace Vydac, Columbia, MD). Fractions containing labeled IAPP, IAPPRG, were pooled, lyophilized and stored at −80°C. For measurements, samples were solubilized with a 20 mM Tris pH 7.4, 100mM NaCl buffer.
Preparation of Unilameller Vesicles
LUVs were prepared from mixture of DOPG:DOPC at ratios specified in the text. The solution of DOPG and DOPC in chloroform (20 mg/ml each) was first mixed and then dried with argon for 1 h followed by drying on lyophilizer for 4 h. The mixture was hydrated in 100 mM KCl, 50 mM sodium phosphate, and pH 7.4 for 20 minutes. A 20 mg/mL solution of lipid in buffer was passed 21 times through 100 nm diameter filters. (Whatman, GE Healthcare, Piscataway, NJ). The phospholipid content of the final material was also confirmed by measurement of total phosphorous (Chen, Toribara, and Warner, 1956).
Kinetic assay
Unless otherwise stated, kinetic reactions were conducted in buffer containing liposome (630 µM, DOPG:DOPC, 1:1, size = 100 nm) and 20 µM ThT in a black 96-well plate. This was followed by addition of the small molecule dissolved in DMSO (final DMSO conc.: 0.5%, v/v). Fiber formation was initiated by addition of IAPP stock solution (final conc.:10 µM). Final volume in each well was 200 µL. The kinetics of IAPP fibrillation was monitored by ThT fluorescence (Ex 450 nm and Em 485 nm) using a FluoDia T70 fluorescence plate reader (Photon Technology International, Edison, NJ). The data were blank subtracted and renormalized to the maximum intensity of reactions containing only IAPP.
Reaction profiles were fit using the built in sigmoidal fit on Origin 5.0 separately for each run. This was used to extract separate t50 (time required to reach 50% change in fluorescence) from which average and standard deviation were calculated. All presented fits were normalized for clarity. Kinetic assays under lipid free condition were repeated in the same way as mentioned above except there is no lipid present. Error bars shown in this work represent standard deviations (SD) from the mean of 3 or more repeat experiments.
Electron Microscopy
IAPP (10 µM) was incubated in buffer (100 mM KCl, 50 mM sodium phosphate, pH 7.4) under lipid catalyzed amyloid formation conditions (DOPG:DOPC, 1:1, 630 µM, d = 100 nm) both in the absence and presence of ADM-3. Aliquots of these samples were then applied on glow-discharged (25 mA and 30 sec.) carbon-coated 300-mesh copper grids for 1 min. and then dried. Grids were then negatively stained for 1 min. with uranyl acetate (2%, w/v) and dried. Micrographs of grids were examined on a Phillips Tecnai 12 transmission electron microscope at 120 kV accelerating voltages. All conclusions drawn from images in this work include at least one repeat in which the sample identity was withheld from the investigator preparing and analyzing images.
Circular Dichroism
The stock solution of IAPP (1–2 mM) in water was diluted to 25 µM IAPP in 100 mM KCl and 50 mM sodium phosphate at pH 7.4 and premixed with LUV’s (1200 µM, DOPG:DOPC, 1:1, 100 nm) before measuring the spectra. CD Spectra were collected at 0.5 nm intervals from 200 to 260 nm with 10 s averaging time with an average of 4 repeats. The CD spectra in the presence of ADM-3 were recorded using a similar method as described above except in the presence of ADM-3 at a stoichiometric ratio of 1:1 (ADM-3:IAPP). The CD spectra recorded for 60 µM IAPP in Fig. 6 (main text) and Fig. S5 were conducted in 100 mM NaCl and 20 mM Tris.HCl at pH 7.4 in the absence and presence of LUV’s (400 µM, DOPG:DOPC, 1:3, 100 nm). IAPP was preincubated with LUV’s for 10–15 min. before recording the spectra.
NMR spectroscopy
50 µM 15N-labeled rat IAPP lipid-free NMR samples were prepared in either 20 mM Tris, 100 mM NaCl, 10% D2O, pH 6.5; or 20 mM Tris, 100 mM NaCl, 10% D2O, pH 7.4. 50 µM 15N-labeled rat IAPP NMR samples with liposomes were prepared under matched buffer conditions described above. All data were obtained on an Agilent 800 MHz spectrometer equipped with a triple resonance HCN cold-probe. All experiments were performed using an inbuilt 15N-HSQC pulse sequence using matched parameters. The mixing time for the 15N-HSQC NMR was 175 msec. The amide cross peaks of amino acid residues in HSQC NMR spectra were previously assigned (Williamson and Miranker, 2007). Data processing was conducted using nmrPipe (Delaglio, Grzesiek, et al, 1995) and plotted using Sparky (Goddard, and Kneller).
Liposome leakage assay
The kinetics of liposome leakage induced by IAPP was monitored on QuantaMaster C-61 fluorescence spectrometer (Photon Technology International, Edison, NJ) at 25 °C. The excitation and emission were observed at 480 nm and 526 nm respectively with 3 nm slit widths. The stock solution of IAPP used in the measurements was kept in water at a concentration of 1–2 mM. The stock solution of water soluble quencher, DPX (p-xylene-bis-pyridinium bromide) was prepared in buffer (100 mM KCl, 50 mM NaPi, pH 7.4) at a concentration of 100 mM. The small molecules used in leakage assay were dissolved in DMSO with a concentration range 10–20 mM. The final concentration of dye encapsulating liposome, human IAPP, and small molecules were 200, 4, and 40 µM respectively. The final buffer concentration was corrected using higher concentration of buffer to keep the osmolality balanced. The final concentration of DMSO used in the assay was less than 0.4%. The control sample contained everything except IAPP. To determine the leakage rate constant, first the fluorescence was normalized and corrected by subtracting it from normalized fluorescence of control reaction. The corrected fluorescence was fit using equation given below (using Matlab). All the experiments were performed in triplicate and the error values presented are standard deviations.
where b is the leakage rate constant.
Fluorescence correlation spectroscopy
FCS measurements were made on a lab-built instrument using a 488 nm DPSS laser for excitation (Coherent, Santa Clara, CA), as described previously (Sevcsik, Trexler, et al, 2011). Laser powers between 5.0–5.3 µW were measured prior to the laser entering the microscope. The fluorescence emission was collected through an objective and spectrally separated from the laser excitation using a Z488RDC long pass dichroic and an HQ600/200M bandpass filter. The resulting fluorescence emission was focused onto the aperture of a 50 µm diameter optical fiber coupled to an avalanche photodiode. The autocorrelation curves were created by a digital correlator (Flex03LQ-12, Correlator.com).
For measurements, 100 nM IAPPRG and liposomes were mixed by pipette in glass-bottomed 8-well chambers (Nunc, Rochester, NY) passivated with polylsine conjugated polyethylene glycol solution and incubated 5 minutes prior to measurement (Middleton, and Rhoades, 2010). For each lipid concentration, 30 traces of 10-seconds duration were measured.
Cytotoxicity assay
Rat insulinoma INS-1 cells (832/13, Dr. Gary W. Cline, Department of Internal Medicine, Yale University) were cultured at 37°C and 5% CO2 in phenol red free RPMI 1640 media supplemented with 10% fetal bovine serum, 1% penicillin/streptomycin (all from Life Technologies, Carlsbad, CA), and 2% INS-1 stock solution (0.5 M HEPES, 100 mM L-glutamine, 100 mM sodium pyruvate, and 2.5 mM β-mercaptoethanol). Cells were passaged upon reaching ∼95% confluence (0.25% Trypsin-EDTA, Life Technologies), propagated, and/or used in experiments. Cells used in experiments were pelleted, and cells were resuspended in fresh media with no Trypsin-EDTA.
Cell viability was measured using the Cell-Titer Blue (CTB) fluorescence-based assay. CTB reagent (Promega, Madison, WI) comprises nonfluorescent resazurin, which is metabolically reduced to fluorescent resorufin in living cells. Cells were plated at a density of 20000 cells/well (500 µL/well) in 24-well plates (BD Biosciences, San Diego, CA). After culturing for 48 h, media was replaced with fresh media containing hIAPP and small molecules pre-mixed at the desired concentration. Cells were incubated at 37°C and 5% CO2 with peptide and small molecules for an additional 48 h. After the incubation period, CTB reagent (100 µL) was added to each well and incubated at 37°C and 5% CO2 for 3 – 3.5 h. Fluorescence of the resorufin product was measured on a FluoDia T70 fluorescence plate reader (Photon Technology International, Birmingham, NJ). All solutions included 0.13% DMSO and 0.65% H2O to account for the addition of peptide and small molecule vehicle to sample wells. Wells that included vehicle but not peptide or small molecule served as the negative control (100% viable), and wells containing 10% DMSO were the positive control (0% viable). Percent toxicity was calculated using the following equation:
Each independent variable is the average fluorescence of 4 technical replicates from the negative control (<N>), positive control (<P>), and 2, 3, 4, or 8 technical replicates for samples (<S>, see manuscript).
NMR spectroscopy (small molecules)
The NMR spectra of small molecules (both 1H NMR and 13C NMR) were recorded on 400, 500, and 600 MHz Agilent spectrometers. The deuterated solvents used for all the compounds (except ADM-1-ADM-9) was CDCl3. For compounds ADM-1 to ADM-9, (CD3)2SO was used. Chemical shifts in NMR spectra are recorded in ppm and are calibrated against the residual solvent signals of CDCl3 [δ (H) = 7.26], (CD3)2SO (δ = 2.50). Splitting patterns which were difficult to interpret, recorded as multiplet (m) or broad (b). MestReNova (version 8.1.0) was used to integrate and process the NMR spectra.
Mol. Wt. determination (small molecules)
Mass spectra were obtained using either MALDI-TOF Voyager DE Pro (Yale, CBIC center) or University of Illinois Mass Spec. Facility. High-resolution electrospray ionization mass spectra were obtained using the Waters Synapt G2-Si ESI MS mass spectrometer. For MALDI-TOF, trans-2-[3-(4-tert-Butylphenyl)-2-methyl-2-propenylidene] malononitryile from Sigma Aldrich (St. Louis, MI) was used as the matrix. The MALDI-TOF spectra were collected using a N2 laser (337 nm wavelength), with an average of 500 shots per sample with a delayed time of 100 ns between each shot. Elemental analyses of tripyridylamides (ADM-2 to ADM-9) were carried out using PerkinElmer 2400 Series II CHNS/O Elemental Analyzer.
Purification of small molecules using flash column chromatography
The small molecules (except ADM-2 to ADM-9 and series 3) were purified using flash column chromatography. The solvent mixtures used for TLC and column chromatography were hexane/ethylacetate (80:20, v/v) and (60:40, v/v) for monopyridyls and oligopyridyls respectively. The purification of small molecules was visualized using UV lamp.
HPLC purification
HPLC purification of small molecules was carried out on Varian ProStar using VYDAC reverse-phase columns (4.6 × 100 mm, 1 mL/min, analytical; 10 × 100 mm, 3 mL/min, semiprep.). The mobile phase was composed of A: 5% ACN, 95% H2O, 0.1% TFA(v/v) and B: 95% ACN, 5% H2O, and 0.08% TFA (v/v). The solvent gradient used for compound purification was: 0–100% solvent B in solvent A for 10 min at a flow rate of 3.0 mL/min followed by 100% solvent B for 25 min (1 mL/min for analytical). The solvent elution was monitored at 220 nm.
Supplementary Material
HIGHLIGHTS.
A series of derivitizable oligopyridylamides is designed and synthesized.
Oligopyridylamides target α-helical structures of islet amyloid polypeptide (IAPP).
Membrane-bound α-helical oligomers of IAPP are associated with diabetes pathology.
The study suggests a common precursor underpins several IAPP functions.
ACKNOWLEDGMENTS
This research was supported, in part, by NIH GM094693 to ADM and an American Diabetes Association mentor-based postdoctoral fellowship to DES. We are thankful to the labs of Prof. Niley Hazari and Prof. David Spiegel (Chemistry Department, Yale University) for providing instruments and space to perform carbonylation and microwave based reactions en route to the synthesis of oligopyridylamides .
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL INFORMATION
Supplemental Information includes Experimental procedures for synthesis of oligopyridylamides, 126 figures, 2 schemes, and 1 table.
REFERENCE
- Apostolidou M, Jayasinghe SA, Langen R. Structure of α-Helical Membrane-bound Human Islet Amyloid Polypeptide and Its Implications for Membrane-mediated Misfolding. J. Biol. Chem. 2008;283:17205–17210. doi: 10.1074/jbc.M801383200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun M, Ramracheya R, Rorsman P. Autocrine regulation of insulin secretion. Diabetes Obes. Metab. 2012;14:143–151. doi: 10.1111/j.1463-1326.2012.01642.x. [DOI] [PubMed] [Google Scholar]
- Cao P, Abedini A, Wang H, Tu L, Zhang X, Schmidt AM, Raleigh DP. Islet amyloid polypeptide toxicity and membrane interactions. Proc. Natl. Acad. Sci. U S A. 2013;110:19279–19284. doi: 10.1073/pnas.1305517110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen PS, Toribara TY, Warner H. Microdetermination of Phosphorus. Anal. Chem. 1956;28:1756–1758. [Google Scholar]
- Cummings CG, Hamilton AD. Disrupting protein-protein interactions with non-peptidic, small molecule α-helix mimetics. Curr. Opin. Chem. Biol. 2010;14:341–346. doi: 10.1016/j.cbpa.2010.04.001. [DOI] [PubMed] [Google Scholar]
- Davis JM, Tsou LK, Hamilton AD. Synthetic non-peptide mimetics of small alpha]-helices. Chem. Soc. Rev. 2007;36:326–334. doi: 10.1039/b608043j. [DOI] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister G, Zhu G, Pfeifer J, Bax A. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Ernst JT, Becerril J, Park HS, Yin H, Hamilton AD. Design and Application of an α-Helix-Mimetic Scaffold Based on an Oligoamide-Foldamer Strategy: Antagonism of the Bak BH3/Bcl-xL Complex. Angew. Chem. Int. Ed. 2003;42:535–539. doi: 10.1002/anie.200390154. [DOI] [PubMed] [Google Scholar]
- Ernst JT, Kutzki O, Debnath AK, Jiang S, Lu H, Hamilton AD. Design of a Protein Surface Antagonist Based on α-Helix Mimicry: Inhibition of gp41 Assembly and Viral Fusion. Angew. Chem. Int. Ed. 2002;41:278–281. doi: 10.1002/1521-3773(20020118)41:2<278::aid-anie278>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- Goddard TD, Kneller DG. Sparky 3. San Francisco: University of California; [Google Scholar]
- Hebda JA, Magzoub M, Miranker AD. Small molecule screening in context: Lipid-catalyzed amyloid formation. Protein Sci. 2014;24:1341–1348. doi: 10.1002/pro.2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebda JA, Saraogi I, Magzoub M, Hamilton AD, Miranker AD. A Peptidomimetic Approach to Targeting Pre-amyloidogenic States in Type II Diabetes. Chem. Biol. 2009;16:943–950. doi: 10.1016/j.chembiol.2009.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Lin C, Haataja L, Gurlo T, Butler AE, Rizza RA, Butler PC. High Expression Rates of Human Islet Amyloid Polypeptide Induce Endoplasmic Reticulum Stress-Mediated β-Cell Apoptosis, a Characteristic of Humans With Type 2 but Not Type 1 Diabetes. Diabetes. 2007;56:2016–2027. doi: 10.2337/db07-0197. [DOI] [PubMed] [Google Scholar]
- Kayed R, Head E, Thompson JL, McIntire TM, Milton SC, Cotman CW, Glabe CG. Common Structure of Soluble Amyloid Oligomers Implies Common Mechanism of Pathogenesis. Science. 2003;300:486–489. doi: 10.1126/science.1079469. [DOI] [PubMed] [Google Scholar]
- Laganowsky A, Liu C, Sawaya MR, Whitelegge JP, Park J, Zhao M, Pensalfini A, Soriaga AB, Landau M, Teng PK, et al. Atomic View of a Toxic Amyloid Small Oligomer. Science. 2012;335:1228–1231. doi: 10.1126/science.1213151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Last NB, Miranker AD. Common mechanism unites membrane poration by amyloid and antimicrobial peptides. Proc. Natl. Acad. Sci. U S A. 2013;110:6382–6387. doi: 10.1073/pnas.1219059110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Last NB, Rhoades E, Miranker AD. Islet amyloid polypeptide demonstrates a persistent capacity to disrupt membrane integrity. Proc. Natl. Acad. Sci. U S A. 2011;108:9460–9465. doi: 10.1073/pnas.1102356108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magzoub M, Miranker AD. Concentration-dependent transitions govern the subcellular localization of islet amyloid polypeptide. FASEB J. 2012;26:1228–1238. doi: 10.1096/fj.11-194613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton ER, Rhoades E. Effects of Curvature and Composition on α-Synuclein Binding to Lipid Vesicles. Biophys. J. 2010;99:2279–2288. doi: 10.1016/j.bpj.2010.07.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nath A, Miranker AD, Rhoades E. A Membrane-Bound Antiparallel Dimer of Rat Islet Amyloid Polypeptide. Angew. Chem. Int. Ed. 2011;50:10859–10862. doi: 10.1002/anie.201102887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabhakaran P, Barnard A, Murphy NS, Kilner CA, Edwards TA, Wilson AJ. Aromatic Oligoamide Foldamers with a "Wet Edge" as Inhibitors of the α-Helix-Mediated p53/hDM2 Protein-Protein Interaction. Eur. J. Org. Chem. 2013;2013:3504–3512. [Google Scholar]
- Saraogi I, Hebda J, Becerril J, Estroff L, Miranker A, Hamilton A. Synthetic α-Helix Mimetics as Agonists and Antagonists of Islet Amyloid Polypeptide Aggregation. Angew. Chem. Int. Ed. 2010;49:736–739. doi: 10.1002/anie.200901694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevcsik E, Trexler AJ, Dunn JM, Rhoades E. Allostery in a Disordered Protein: Oxidative Modifications to α-Synuclein Act Distally To Regulate Membrane Binding. J. Am. Chem. Soc. 2011;133:7152–7158. doi: 10.1021/ja2009554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh BK, Cavalluzzo C, Maeyerc MD, Debyserd Z, Parmar VS, Eycken EV. Mirowave-Assisted Silver (I)-Mediated Selective O-Alkylation of Aromatic Imidate Systems. Synthesis. 2009;16:2725–2728. [Google Scholar]
- Williamson JA, Loria JP, Miranker AD. Helix Stabilization Precedes Aqueous and Bilayer-Catalyzed Fiber Formation in Islet Amyloid Polypeptide. J. Mol. Biol. 2009;393:383–396. doi: 10.1016/j.jmb.2009.07.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson JA, Miranker AD. Direct detection of transient alpha-helical states in islet amyloid polypeptide. Protein Sci. 2007;16:110–117. doi: 10.1110/ps.062486907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe LS, Calabrese MF, Nath A, Blaho DV, Miranker AD, Xiong Y. Protein-induced photophysical changes to the amyloid indicator dye thioflavin T. Proc. Natl. Acad. Sci. USA. 2010;107:16863–16868. doi: 10.1073/pnas.1002867107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H, Lee G, Park HS, Payne GA, Rodriguez JM, Sebti SM, Hamilton AD. Terphenyl-Based Helical Mimetics That Disrupt the p53/HDM2 Interaction. Angew. Chem. Int. Ed. 2005;117:2764–2767. doi: 10.1002/anie.200462316. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.