Abstract
Reversible site-specific DNA inversion reactions are widely distributed in bacteria and their viruses. They control a range of biological reactions that most often involve alterations of molecules on the surface of cells or phage. These programmed DNA rearrangements usually occur at a low frequency, thereby preadapting a small subset of the population to a change in environmental conditions, or in the case of phages, an expanded host range. A dedicated recombinase, sometimes with the aid of additional regulatory or DNA architectural proteins, catalyzes the inversion of DNA. RecA or other components of the general recombination-repair machinery are not involved. This chapter discusses site-specific DNA inversion reactions mediated by the serine recombinase family of enzymes and focuses on the extensively studied serine DNA invertases that are stringently controlled by the Fis-bound enhancer regulatory system. The first section summarizes biological features and general properties of inversion reactions by the Fis/enhancer-dependent serine invertases and the recently described serine DNA invertases in Bacteroides. Mechanistic studies of reactions catalyzed by the Hin and Gin invertases are then discussed in more depth, particularly with regards to recent advances in our understanding of the function of the Fis/enhancer regulatory system, the assembly of the active recombination complex (invertasome) containing the Fis/enhancer, and the process of DNA strand exchange by rotation of synapsed subunit pairs within the invertasome. The role of DNA topological forces that function in concert with the Fis/enhancer controlling element in specifying the overwhelming bias for DNA inversion over deletion and intermolecular recombination is emphasized.
A. Introduction
Reversible site-specific inversions of DNA segments occur within the genomes of many bacteria and their phages. These reactions are catalyzed by a dedicated recombinase and do not employ the use of the general recombination-repair machinery. In some cases, additional host regulatory proteins also perform critical functions, and DNA superstructure can play a profound role. In general, site-specific DNA inversions occur at a low frequency and regulate gene expression by coupling alternative protein coding segments to a fixed promoter or by switching the orientation of a promoter with respect to coding region(s). In this manner a small subset of the population becomes pre-adapted to a potential change in the environment.
The two major classes of enzymes mediating site-specific recombination include a diverse family of tyrosine recombinases and a more conserved family of serine recombinase, as named from their catalytic active site residues (1). This chapter focuses on the Fis/enhancer-dependent DNA invertase subfamily of serine recombinases. The DNA invertases are closely related to resolvases, which primarily mediate deletions, and as a group these are often referred to as small serine recombinases. Serine recombinases are relatively easily identified by conserved signature motifs over the active site regions within their catalytic domains. As of the beginning of 2014 there were tens of thousands of entries in the sequence database that exhibit >30% identity over at least part of the catalytic domains of small serine recombinases like the Hin DNA invertase.
The chapter begins by reviewing the biology of well characterized Fis/enhancer-dependent DNA inversion reactions. A more detailed discussion on biological aspects of these reactions plus other site-specific DNA inversion systems is provided in Mobile DNA II Chapter 13 (2). The multiple DNA inversion reactions mediated by small serine recombinases in Bacteriodes, which were discovered after publication of the Mobile DNA II chapter, are also briefly discussed here.
The emphasis of most of the chapter is on recent developments concerning mechanistic studies of the Fis/enhancer-dependent inversion reactions, focusing on the Hin- and Gin-catalyzed reactions. Two defining features of these serine DNA invertase systems are discussed in detail. One is the requirement for the enhancer regulatory element that functions in cis, but in a largely distance- and orientation-independent manner with respect to the sites of recombination. The enhancer specifies two binding sites for the Fis protein, which was initially discovered because of its activity in the inversion reactions – hence its full name Factor for inversion stimulation. Fis is now known as a general nucleoid-associated protein that controls many different DNA reactions.
A second defining feature is the strict specificity for promoting intramolecular inversions over deletions or intermolecular recombination reactions. This directionality of recombination is mediated by the Fis/enhancer element in concert with DNA supercoiling. Thus, a major focus of research in the field has been to elucidate the steps involved in formation of the active recombination complex, called an invertasome. The Fis/enhancer element functions as a molecular scaffold in the assembly of the invertasome, whereby inactive DNA invertase dimers are remodeled into an active tetramer that breaks, exchanges, and ligates the DNA strands into the inverted orientation.
Research on DNA invertases and resolvases has elucidated our current understanding of the subunit rotation reaction that mediates the exchange of DNA strands by the serine recombinases. Although the translocation of subunits after double strand DNA cleavages would seem like a perilous reaction because of the potential for chromosome breaks, recent evidence provides a compelling case for this unique mechanism of DNA recombination (see also chapters by Rice and Stark (3, 4)).
At the end of the chapter, recent progress on identifying specific residues on serine DNA invertases that control key conformational transitions during the reaction is discussed. Mutations in these residues often enable efficient recombination without the requirement for the Fis/enhancer element and thus without control of directionality.
B. Biology and basic features of serine DNA invertase reactions
B.1 Hin-catalyzed site-specific DNA inversion controlling flagellar phase variation in Salmonella
Antigenic variation, whereby clonally-derived populations of Salmonella are converted to an alternate antigenic form, was first described by Andrewes in 1922 (5). Subsequent genetic studies on phase variation in Salmonella by the laboratories of Stocker, Lederberg, and Iino showed that the variable H antigen was the result of alternate expression of two unlinked flagellin genes originally called H1 and H2, but renamed fliC and fljB, respectively (6–9). Stocker, and more recently Kutsukake et al., measured switching rates between the two flagellins in rich media cultures to be in the range of 10−4 to 10−5 per cell per generation (9, 10). Gillen and Hughes reported 2- to 20-fold higher switching rates using chromosomal lac reporters (11). All of these studies have shown a 2.5- to 30-fold bias in favor of switching from FljB to FliC expression over the reverse direction, but the molecular basis for this difference remains unknown. The low rate of switching is consistent with the postulated role of flagellar phase variation in escaping a host immune response. A clonally-derived population of Salmonella will express primarily one flagellin type, enabling the few members of the population that have switched to be insensitive to antibody generated by the host against the dominant flagellin.
The flagellar phase variation system is present in many but not all of the thousands of serovars of Salmonella enterica subsp. enteric, including the intensively studied serovar Typhimurium (Salmonella Typhimurium). Studies have shown that Salmonella Typhimurium cells expressing either flagellin are equally efficient at invading mouse intestinal epithelial cells and colonizing Peyers patches, but strains genetically locked into expressing only FliC are more virulent than FljB phase-locked strains (12). The FliC phase-locked cells were found to be more efficient at colonizing the spleen during later stages of infection. Moreover, infecting wild-type bacteria that were initially in the FljB phase tended to have switched to the FliC phase when spleens were analyzed 2 weeks post infections, whereas infecting FliC cells remained in the FliC phase.
Molecular analyses in the latter 1970s by Silverman, Zieg, and Simon first demonstrated that the “H-controlling region” adjacent to the H2/fljB flagellin gene in the Salmonella Typhimurium chromosome contains an inverting segment of DNA. Restriction mapping, together with the formation of hybridization bubbles between DNA molecules cloned from strains expressing the different flagellin forms, provided the physical proof for inversion of a ~1 kb DNA segment (13, 14). The region can be transferred into E. coli where it also undergoes inversion. Genetic and sequence analyses showed that the hin (H inversion) gene, coding for a 190 amino acid residue protein that is responsible for the inversion reaction, was contained within the 993 bp invertible segment (Fig. 1A) (15–17). The segment is bounded by two imperfectly palindromic 26 bp recombination sites hixL and hixR (Fig. 1A and 2) and contains a σ28-dependent promoter that initiates transcription 28 bp upstream of hixR (18). In the fljB ON orientation, transcription from this promoter reads through hixR, and translation of the FljB flagellin begins just 5 bases after the end of the hixR sequence. hixR has diverged from recombination site sequences of other Fis/enhancer-dependent DNA inversion systems (Fig. 2), probably because the locus must also encode the fljB ribosome binding site. The fljA gene (originally called rH1, repressor of H1), whose protein product is responsible for preventing synthesis of the alternative flagellin FliC, is co-transcribed with and begins 68 nt downstream of fljB (Fig. 1A)(19). FljA binds to the fliC mRNA to both inhibit its translation and promote its degradation (20–22). In the opposite or fljBA OFF orientation, the absence of fljA transcription enables the FliC flagellin, which is encoded elsewhere on the chromosome, to be expressed. The closest gene, iroB, which is annotated as a glycosyl transferase family protein, begins about 650 bp from the σ28-dependent promoter in the fljBA OFF orientation.
Figure 1.
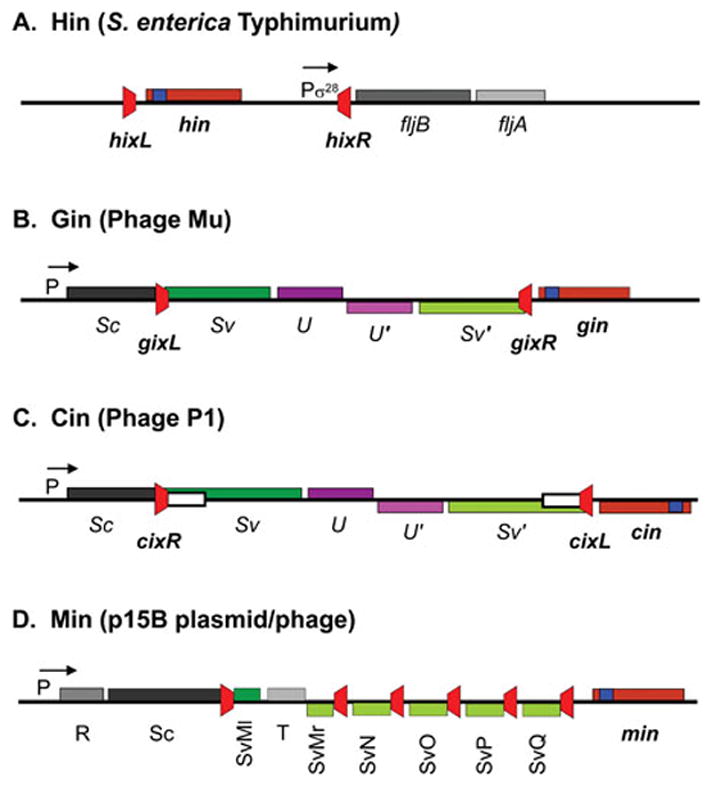
Genetic organization of Fis/enhancer-dependent DNA inversion systems. A. The Hin system controlling flagellin synthesis in Salmonella enterica Typhimurium. fljB codes for one of the alternatively expressed flagellins, and fljA is a repressor of the fliC flagellin gene located elsewhere on the chromosome. B and C. The Gin and Cin systems from phages Mu and P1, which control phage host range. Sv and Sv′ gene segments are alternatively fused in-frame to the Sc gene segment. The S and U genes encode tail fiber proteins. D. The complex Min locus from the p15B plasmid. The different Sv gene segments are alternatively fused to the Sc gene segment. In each case the recombination sites are colored red, and the positions of the DNA invertases (maroon) with their associated recombinational enhancer segments (blue) are denoted. P designates a promoter with the S. Typhimurium fliC and fljA genes being transcribed by the sigma 28 form of RNA polymerase.
Figure 2.

Fis/enhancer-dependent serine DNA invertase recombination sites. DNA sequences of recombination sites from the systems depicted in Fig. 1 are listed along with the consensus sequence at the top. Sequences matching the consensus are highlighted in cyan.
A purified in vitro system supporting Hin-catalyzed DNA inversion was reported in 1986 (23). In addition to the Hin recombinase, two host factors were found to be required for efficient inversion. Factor I was identified as the nucleoid-associated protein HU, and Factor II was renamed Fis (Factor for inversion stimulation) and later also classified as a nucleoid protein (24–26). Along with the hixL and hixR recombination sites, a third cis-acting sequence was found to be required (27). In the Hin inversion system, this 69 bp recombinational enhancer locus is encoded within the amino-terminal coding segment of Hin (residues 8–30) and thus located within the invertible segment about 100 bp from the center of the hix site (Fig. 1A). However, it can function effectively from 10 bp closer to over 4 kb from the closest hix site, both inside and outside of the invertible segment and in either orientation (27, 28). The enhancer contains two binding sites for the Fis protein (29). Very low rates of inversion can occur on substrates lacking an enhancer when Fis is present, but inversion is nearly undetectable without Fis (23, 25). DNA supercoiling is absolutely essential for the inversion reaction (23, 30). The functions of these components in the inversion reaction will be discussed in detail below.
The primary determinant that is believed to be responsible for limiting the frequency of inversion in Salmonella (<10−4 per cell per generation) is the very low synthesis of Hin because ectopic increases in Hin synthesis result in coordinate increases in DNA inversion rates (31). LacZ reporter experiments have provided evidence for transcription initiating in the 74 bp segment between the hix recombination site and start of the hin gene (R. Johnson, unpublished). Experiments designed to test whether Hin binding to hix or to a low affinity Hin binding site between hix and the start of the hin gene have not provided evidence for regulation of hin transcription under laboratory culturing conditions. Likewise, there is no evidence that Fis regulates hin expression. Translation initiation signals do not appear to limit Hin protein synthesis. Fis levels, which are very low under slow growth and late exponential-stationary phase conditions (18, 32), and DNA supercoiling densities influence inversion rates (33). Supercoiling densities in Salmonella Typhimurium are reported to be high in the gut of infected mice but decrease within macrophages, which is a major route used to enter the blood stream to cause systemic infection (34).
B.2 Fis/enhancer-dependent serine invertase systems controlling phage host range
A large number of DNA inversion systems that are associated with phage or phage-like genetic elements are closely related to Hin. Among these, the Gin- and Cin-catalyzed inversion reactions from phages Mu and P1, respectively, are the best characterized (Fig. 1B and C). Gin catalyzes inversion of a 3015 bp G segment within the Mu genome, and Cin catalyzes inversion of a ~ 4.2 kb segment within the P1 genome (35, 36). Both of these invertible segments contain genes encoding alternative tail fiber proteins that enable the phage to adsorb to different bacterial hosts, thereby increasing the phage host range (37, 38). In the case of phage Mu, a constant N-terminal coding segment of the S gene, which is encoded immediately adjacent to the invertible segment, is linked to its two different C-terminal coding segments, Sv or Sv′, depending on the orientation (Fig. 1B) (39–43). The U/U′ genes are also alternatively expressed depending on the orientation. The gene encoding Gin is located immediately outside of the invertible segment on the side opposite to the Sc gene fragment (44). The organization of the phage P1 C-inversion region is analogous to the Mu system with one important difference: the cin gene is oriented oppositely to the gin gene relative to the invertible segments (Fig. 1C) (36, 45, 46). Fis, together with recombinational enhancer sequences that are located within the N-terminal end of the respective recombinase genes, are required for efficient DNA inversion in both systems (26, 47–51). Thus, whereas the hin enhancer is located within the invertible segment beginning about 100 bp from hixL/R, the gin enhancer is located outside of the invertible segment beginning about 90 bp from gixR and the cin enhancer is located outside of the invertible segment beginning about 500 bp from cixL. The different organizations provided an initial clue that the enhancer sequences could function at variable positions and orientations relative to the recombination sites, which was also experimentally confirmed for the phage systems (47, 48, 52).
Like Hin, Gin-catalyzed inversion occurs only rarely under native in vivo conditions. Inversion rates measured after a single round of lytic phage growth are about 10−6 per phage particle (53). On the other hand, induction of a Mu lysogen after long term growth gives rise to equal numbers of progeny containing their G segments in either orientation (54). This is advantageous to the phage since lytic infections will primarily result in progeny capable of attacking the same host, but phage induced from long term lysogenic growth will have a broad host range. Gin expression is limited by both low transcription and poor translation initiation signals (35, 55, 56). Transcription of gin is believed to initiate within the gix recombination site and thus be subject to autoregulation.
Whereas most DNA inversion reactions by serine recombinases involve single locus inversions, more complex systems have been reported (2, 57). The best analyzed is the Min system from p15B, a 94 kb plasmid from E. coli 15T− that exhibits considerable homology with phage P1 (58–60). There are six different 26 bp recombination sites within the 3.5 kb locus (Fig. 1D and 2). Restriction and sequence analyses are consistent with all being active for supporting inversion to generate 240 different sequence combinations. Transformation of individual recombinant plasmids containing the locus showed that an equilibrium population representing most or all of the 240 isomeric forms could be obtained after 40 generations of growth (60). The different rearrangements give rise to six different alternative 3′ ends of a tail fiber-like gene fused in-frame to a constant 5′ region.
The extended group of related phage, cryptic prophage, and plasmid serine DNA invertase genes that are associated with tail fiber gene expression typically share greater than 75% amino acid identity over the entire protein sequence (2). Hin is somewhat more distantly related, sharing 68% identity (82% similarity) with Gin (Fig. 3) and 66% identity (80% similarity) with Cin. Nevertheless, the phage DNA invertases can invert the H invertible segment as well as function on each other’s substrates (60–64). Kutsukake et al. reported that a serine DNA invertase in Salmonella Typhimurium called Fin that is associated with the phage P2-like Fels-2 prophage can function to promote flagellar phase variation at 1–3% of the hin+ rate, although it is only 63% identical to the Hin amino acid sequence (10).
Figure 3.
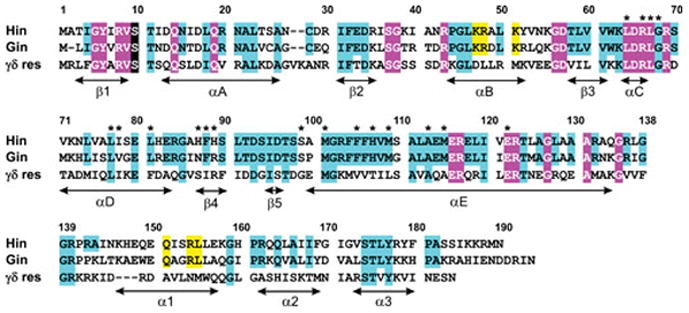
Amino acid sequence alignment of the Hin and Gin DNA invertases together with γδ resolvase. Highly conserved residues among all serine recombinases are highlighted in magenta. The active site serine residue is shaded black. Highly conserved residues among Fis/enhancer-dependent DNA invertases are in cyan; many of these are conserved among the small serine recombinases. Yellow highlighted residues within the α-helix B region are those that contact enhancer DNA and within α-helix 1 of the DNA binding domain (Hin residues 139–190) are those that interact with Fis. Residue numbering is according to Hin. Asterisks denote residues where single substitutions result in strong gain-of-function activities, often resulting in Fis/enhancer-independence (65, 109, 185). Secondary structure designations are from γδ resolvase (residues 1–138; PDB: 1GDT) and Hin (residues 139–190; PDB: 1IJW). An alignment that includes additional Fis/enhancer-dependent DNA invertases along with other small serine recombinases is given in ref. (2).
B.3 Specificity for DNA inversion by the Fis/enhancer-dependent serine invertase reactions
A hallmark of the Fis/enhancer-dependent DNA invertase reactions is that they only efficiently catalyze inversions between recombination sites located in cis. As discussed further below, the overall palindromic recombination sites have an orientation that is specified solely by the central non-palindromic two base pairs. In all cases, the active recombination sites are oriented in an inverted configuration with respect to each other as defined by this dinucleotide sequence. Where tested, the products of recombination reactions between sites oriented in a directly-repeated configuration are deletions, but the rate of this reaction is less than a few percent of the inversion reaction (53, 65–67). In the case of the min locus, deletions were detected but occurred at <0.1% the frequency of inversions (60). An extremely low rate of deletions is especially critical for maintaining the integrity of the min locus, which has five of its recombination sites in a directly repeated orientation. Likewise, unlike many other site-specific recombination systems, intermolecular reactions by serine DNA invertases occur at vanishingly low rates (53, 65, 66, 68). As elaborated below, the requirements for Fis, the enhancer, and DNA supercoiling combine to impart the strict directionality to the serine DNA invertase reactions.
B.3 Multiple inversions catalyzed by serine DNA recombinases in Bacteriodes
The abundant human intestinal symbiont Bacteriodes sp. are loaded with mobile DNA elements, and in particular, short DNA segments containing promoters that undergo reversible inversion (69, 70). The genome of Bacteriodes fragilis contains 30 potential DNA invertases; most are members of the tyrosine recombinase family, but three are serine recombinases. Unlike most site-specific recombination systems, individual recombinases in Bacteriodes promote inversions at many loci that are distributed throughout the chromosome. These inversion reactions are thought to enable the bacterium to continually adapt to the host environment and avoid immune surveillance, thereby enabling a long-term commensal relationship with their host.
A plasmid-encoded serine recombinase in B. fragilis called FinB (fragilis invertase B) catalyzes inversion of promoter regions upstream of seven polysaccharide biosynthesis genes (71, 72). Fin is quite similar to Hin (49% identical, 72% similar by Clustal O), and the sequences of the 32 bp inverted repeats flanking the promoter segments resemble the hix sequence. The same strain of B. fragilis also contains a 197 amino acid residue serine recombinase called Mpi (multiple promoter invertase) that promotes inversion of 13 different promoter elements that are responsible for expression of seven related capsular polysaccharide genes plus six other unidentified genes (73). Each of these invertible segments are flanked by related 20–22 bp recombination sites. Mpi is clearly a serine recombinase of similar overall structure as the invertase/resolvase fold but is less related to Hin in primary sequence (27% identical, 55% similar).
The Comstock group has also shown that the Gfi recombinase (glycoprotein family invertase) catalyzes inversions at promoters controlling seven different glycoprotein genes in Bacteroides distasonis (74). While these promoters are primarily maintained in the OFF orientation, all seven were found to invert at a low frequency under in vitro culturing conditions. Moreover, evidence for inversion into the ON orientation at some of the loci was obtained by PCR analysis of human stool samples, confirming glycoprotein phase variation in the organism’s native environment. Gfi is one of four serine recombinases present in B. distasonis.
The promoter-containing invertible segments in all the Bacteroides systems are extremely short with most being less than 225 bp and one being only 139 bp. Thus, it would not be unexpected if a host DNA bending protein facilitates assembly of recombination sites into synaptic complexes. It will be interesting to learn if a regulatory system analogous to the Fis/enhancer element is operating in these reactions and whether there are mechanisms to ensure that recombination only occurs between sites flanking a specific promoter to avoid large rearrangements of the chromosome.
C. Relationship of Fis/enhancer-dependent DNA invertases with other serine recombinases
The Fis/enhancer-dependent serine DNA invertase genes typically contain 185–200 residues that exhibit over 60% sequence identity over the entire protein, with many of the phage-associated DNA invertases being >75% identical to each other (Fig. 3) (2). Their domain organization consists of a ~100 amino acid residue catalytic domain connected to a long oligomerization helix followed by a 50–60 amino acid helix-turn-helix DNA binding domain (DBD) (Fig. 4) (1). It was first recognized in 1980 that the sequence of DNA invertases resembled the TnpR repressors/resolvases of transposons Tn3 and γδ (Tn1000) (75). The resolvases, which primarily catalyze deletion reactions, have the same protein architecture and share up to about 35% sequence identity (55% similarity) with DNA invertases over the entire polypeptide and 41% identity (62% similarity) over the catalytic domain + oligomerization helix (Fig. 3 and 4). These recombinases thus became the founding members of the DNA invertase/resolvase class, which together with more distantly related subgroups, constitute the serine recombinase superfamily (Fig. 4) (1, 76). Unlike DNA invertases, resolvases do not utilize Fis or a remote enhancer sequence. However, they do assemble elaborate supercoiling-dependent synaptic complexes, which employ additional resolvase subunits and sometimes DNA bending proteins like HU that perform critical DNA architectural and regulatory functions. They also share the same enzymology of DNA exchange (1, 3).
Figure 4.
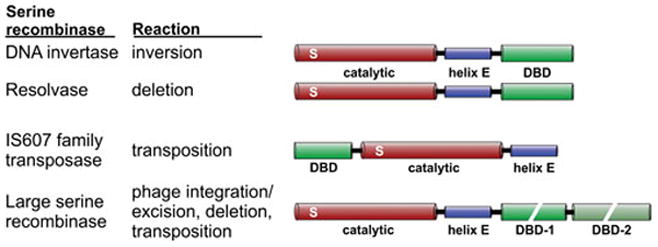
Serine recombinase subfamilies. The domain architectures of serine recombinase subfamily members denoting the ~100 amino acid residue catalytic domain containing the active site serine (S), the oligomerization helix E, and the DNA binding domain (DBD). The DNA binding regions of large serine recombinases can be quite large (300–450 residues) and consist of two discrete domains. The DNA invertases and resolvases are often grouped together as small serine recombinases.
The ‘small’ serine recombinases exhibiting the resolvase/invertase protein architecture cannot be easily classified into enzymes that are specific for promoting deletions or inversions based on their overall amino acid sequence. For example, the ISXc5 recombinase from Xanthomonas campestri catalyzes only deletions, yet is 55% identical to Hin and only 38% identical to γδ resolvase over the catalytic domain and oligomerization helix regions (77). Nevertheless, Fis/enhancer-dependent DNA invertases may be recognized by the presence of three conserved residues that contact Fis, which are located within helix 1 of their DNA binding domains, together with several basic residues in the helix B region of the catalytic domain, which contact the enhancer DNA (Fig. 3 and section D.6.5) (78). Some small serine recombinases have been shown to catalyze both DNA inversions and deletions. Examples include the β-recombinases (~32/34% identical to Hin/Gin, respectively) and Tn552 BinR (47/48% identical to Hin/Gin) (79–83). Neither of these utilize a Fis-like protein, as expected since they are active in gram (+) bacteria that do not contain Fis, but the β-recombinase requires a DNA bending protein related to HU (84, 85).
A large class of serine recombinases, which are known as ‘large serine recombinases’ and include the serine integrases, mediate a diversity of reactions in bacteria, including developmentally-controlled deletions and phage integration and excision from bacterial host chromosomes (Fig. 4) (76, 86, 87). These recombinases share the catalytic domain and oligomerization helix that is homologous with the resolvase/invertase subgroup but have a much larger 300–550 residue C-terminal domain. The C-terminal extension is folded into two subdomains that participate in DNA binding and contribute to synapsis (88–90). Many of the serine integrases have been shown to require recombination directionality factors or Xis-like proteins to promote the excision/deletion reaction (87). Some members of this class catalyze an excision-integration reaction that translocates the intervening DNA to a new location within the host DNA, and thus are considered transposases. Another class of serine recombinases, the IS607 class, also function as transposases (91). These are distinguished by their small winged-helix DNA binding domain being located N-terminal to the catalytic domain (Fig. 4) (92). Although less is currently known about the mechanistic and structural details of reactions catalyzed by members of these classes, the pathways and regulation of synaptic complex formation appear to be very different from those of the resolvases/invertase group. However, fundamental features regarding the enzymology of recombination, including the subunit rotation mechanism for DNA exchange, are believed to be conserved among all the serine recombinases.
D. Mechanism of Fis/enhancer-dependent serine DNA invertase reactions
D.1 General properties of Fis/enhancer-dependent DNA inversion reactions in vitro
Of the serine DNA invertase systems, the Hin- and Gin-catalyzed reactions have been studied most intensively in vitro. Reactions employing purified Hin or Gin and Fis proceed optimally at 37° in a buffered salt solution with a divalent cation like Mg2+ (23, 27, 47, 93, 94). The DNA substrate must be supercoiled, consistent with the sensitivity of the in vivo reaction to novobiocin (31). The Hin reaction also requires HU for efficient inversion on the wild-type substrate (discussed in section D.5) (23). Under standard inversion conditions, the in vitro Hin reaction is essentially dependent upon Fis. An extremely inefficient reaction occurs in the absence of Fis when up to 15% ethylene glycol or glycerol is added; this basal reaction without Fis is about 1% of the rate of the reaction with Fis. Gin-mediated DNA inversion is also strongly stimulated by Fis but may exhibit a higher Fis-independent activity than Hin (93, 94). A variety of RNA molecules such as rRNA or synthetic polycytidylic or polyuridylic acid or even long chain polyanions like polyglutamic acid were found to strongly stimulate the activity of partially purified Hin preparations, but the stimulatory effect is much less in reactions employing highly purified native or refolded Hin (23). The basis of the stimulation by RNA has not been determined; possibilities include stabilizing Hin or titrating an inhibitor.
Early preparations of Hin supported recombination in the presence of 10 mM EDTA (23, 30). However, highly purified native or refolded Hin preparations require a divalent cation or spermidine for complete inversion but not for the initial DNA cleavage step. Mg2+, Ca2+, or spermidine are similarly effective, but reactions with Mn2+ are poorer. Gin is reported to catalyze inversion in 10 mM EDTA (95) but is strongly stimulated by Mg2+ or Ca2+ and weakly by Mn2+ (94, 96, 97). The mechanism behind the requirement for divalent cations or polyamines, particularly for the DNA ligation step, is not known, but they are not believed to be directly functioning in DNA chemistry. Both Hin and Gin exhibit a robust substrate-specific supercoiled DNA relaxing activity that is also dependent on divalent cations or polyamines (30, 93, 98).
Hin and Gin promote multiple inversions on supercoiled plasmids to generate an equilibrium state where both DNA orientations are equally represented (23, 30, 93, 96, 97). Therefore, starting with one orientation the product population never exceeds 50% inversion because of the reverse reaction. Reactions with highly purified Hin are rapid, recombining the DNA substrate to near equilibrium within 1–2 minutes. Hin turns over between DNA substrate molecules very inefficiently but can catalyze multiple reactions utilizing different recombination sites on the same DNA molecule (R.C.J., unpublished). The inefficient turnover between substrates is at least partially due to the slow Hin-DNA dissociation rate (99). Deletion rates in vitro on substrates with directly-oriented recombination sites are typically <1% of inversion rates between sites in their native ‘inverted’ orientation (67), and intermolecular reactions are undetectable, making the specificity for the inversion reaction in vitro even greater than measured in vivo.
D.2 The DNA-cleaved invertasome intermediate
Hin generates inversion products without the appearance of readily detectable intermediates under standard reaction conditions. However, the combination of high concentrations of ethylene glycol (30% for Hin and up to 50% for Gin) and EDTA in place of Mg2+ stalls the reaction in an intermediate nucleoprotein complex where both recombination sites contain double strand cuts within their centers (100, 101). Formation of these ‘cleavage’ complexes is recognized by the appearance of the vector backbone and invertible segment following rapid quenching with a protein denaturant like SDS or HCl. Each of the four cleaved DNA ends contains a recombinase protomer covalently associated with the 5′ end and a two nucleotide protruding 3′ end terminating with a hydroxyl (100). The non-denatured cleavage complex represents a true reaction intermediate since it can be chased into ligated inversion products within seconds by adding back Mg2+ and diluting the ethylene glycol to ≤5%. Thus, DNA exchange occurs through double strand breaks over the central two base pairs of the 26 bp recombination sites (Fig. 2).
Unlike Hin, incubation of Gin with radiolabeled fragments generates a low level of site-specific DNA cleavages at the same respective locations within gix sites (98). Two dimensional thin layer chromatography of the covalent Gin-DNA complex after acid hydrolysis demonstrated that Gin was attached to the DNA 5′ end via a phosphoserine linkage, as observed earlier with γδ resolvase (102). Mutagenesis studies of Gin and Hin are consistent with the active site serine being at residue 9 in Gin and residue 10 in Hin, within one of the most conserved regions present in all serine recombinases (Fig. 3) (98, 103).
Electron microscopy of crosslinked Hin cleavage complexes revealed up to ~25% in an ‘invertasome’ structure containing the two hix sites looped onto the enhancer (Fig. 5) (104). Similar invertasome structures have also been observed from Gin reactions (105). Formation of Hin invertasomes required Hin, the two hix sites, Fis and the enhancer on a supercoiled DNA plasmid. Antibody probing confirmed that both Hin and Fis were present in the tripartite complex. Significantly, no such structures were observed from reactions starting with relaxed DNA. Moreover, single-looped molecules containing only one hix site associated with the enhancer were not observed, even when the enhancer was missing one of the two Fis binding sites (104).
Figure 5.
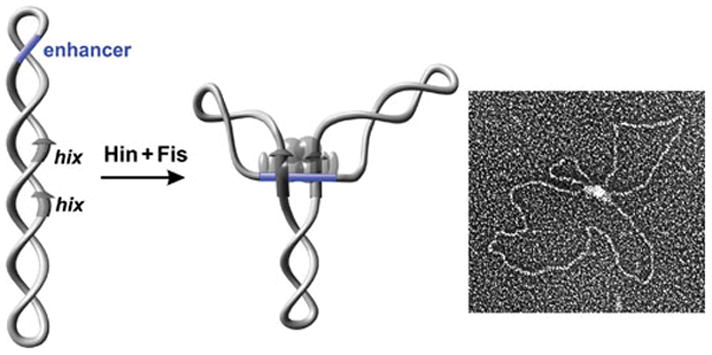
The Hin invertasome. Electron micrograph of an invertasome together with a schematic drawing of the structure. The invertasome structure was stabilized by crosslinking and supercoils removed prior to spreading onto the grid and low angle platinum shadowing (104). Hin subunits are rendered as translucent spheres; Fis subunits are rendered as ovals. Hix sites are depicted as arrows and the enhancer segment is blue.
Nucleoprotein structures assembled from complete reactions but consisting of only the two hix sites paired together were also observed by electron microscopy at high frequency after protein crosslinking (104). Most of these paired-hix structures probably represent complexes where the enhancer segment was released from an assembled invertasome during sample preparation because the majority of them contained Fis covalently crosslinked to the Hin complex by antibody probing. On the other hand, glutaraldehyde crosslinked paired-hix structures were also obtained in reactions without Fis or the enhancer. The formation of these complexes do not require DNA supercoiling and can even form with Hin mutants or disulfide-linked Hin dimers that cannot generate active synaptic complexes in the presence of Fis (106) (R.C.J. and co-workers, unpublished). Thus, the functional relevance of crosslinked paired-hix complexes formed by the wild-type enzyme without an operating Fis/enhancer system is questionable.
The imaging of invertasome intermediate complexes, combined with the requirements for their assembly, suggest a pathway for inversion in which the two recombination sites assemble into chemically-active synaptic complexes at the enhancer in a supercoiling-dependent reaction (Fig. 5 and 6). Stoichiometry calculations based on activity measurements or radiolabeling indicate that two dimers of Hin and two dimers of Fis are present in active synaptic complexes (104, 107). As discussed further below (section D.12), the supercoiling-directed assembly will orient the recombination sites in a configuration which specifies that the product of the DNA exchange reaction will be inversion of the intervening DNA.
Figure 6.
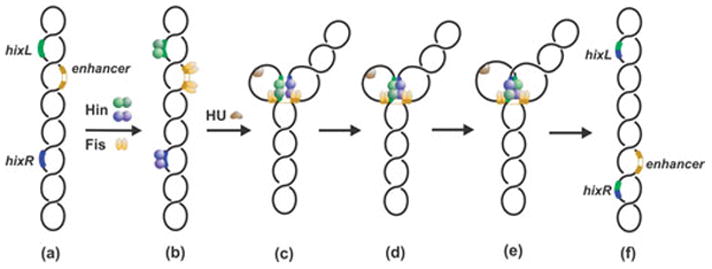
DNA inversion reaction pathway by Fis/enhancer-dependent serine invertases. In step (c) Hin dimers bound to hixL and hixR are associated with the Fis-bound enhancer at the base of a branch on supercoiled DNA. Formation of the Hin tetramer (c) generates an enzyme active for double strand cleavage and subunit rotation (d). Ligation and resolution of the complex (e) results in inversion of the DNA segment between recombination sites.
D.3 The serine DNA invertase dimer and recombination site binding
D.3.1 The serine invertase dimer
Hin chromatographs as a dimer by gel filtration in the presence of 0.5 – 1 M NaCl and 0.1% Triton X-100 or 4 mM CHAPS (108) (Y. Chang and R.C.J., unpublished). Dimeric interactions between subunits are highly sensitive to the zwitterionic detergent CHAPS; Hin behaves as a monomer in solution and cooperative DNA binding to hix is abrogated in the presence of high CHAPS concentrations (106, 108, 109). Gin is reported to chromatograph on Sephacryl S-200 as a monomer (94) but binds highly cooperatively as a dimer to gix sites (110). As noted above, Gin dimers exhibit weak DNA cleavage activity, but Hin dimers are chemically silent.
All evidence indicates that the dimeric structures of DNA invertases match very closely to those of resolvases (111, 112). A folded globular catalytic domain, which appears to be highly conserved among all serine recombinases, extends over the N-terminal ~100 amino acid residues (Fig. 3, 4, and 7). Within the catalytic domain are two particularly highly conserved regions, one around the active site serine (Hin residue 10) and the other around arginines at Hin residues 66 and 69 (Fig. 3), which are proximal to each other in the inactive dimer structures. Invariant arginines at Hin residues 8, 66, 69 are proposed to perform direct chemical roles in the cleavage-ligation reactions within the remodeled active tetramers of serine recombinases (113, 114). As expected, mutations of each of these arginines, along with Ser10, Arg43, and Asp65 which are also believed to participate in catalysis, abolish DNA cleavage by Hin (103, 109, 115, 116) (R.C.J., unpublished). Of particular significance is the fact that the serine hydroxyl nucleophiles from each subunit are separated by >35 Å and are not close to the scissile phosphate in the dimer structures of resolvase (111, 112). Thus, a major reorganization of the recombinase quaternary structure, together with adjustments surrounding the active site pockets, is required prior to any DNA chemistry.
Figure 7.
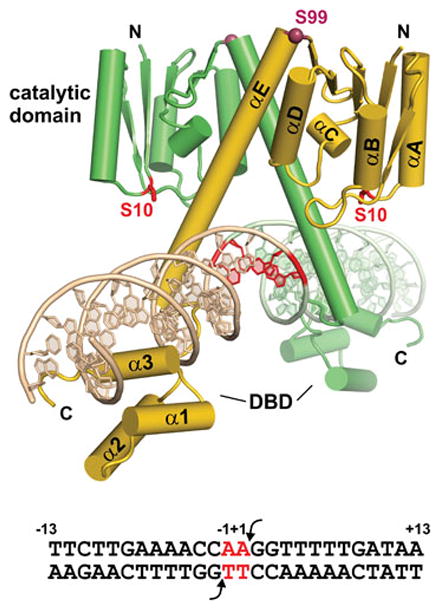
The serine DNA invertase dimer. Model of the Hin dimer bound to hixL derived from the catalytic domain and oligomerization helix E of γδ resolvase (PDB: 1GDT) linked to the DNA binding domain (DBD) of Hin (PDB: 1IJW) is shown. The Ser10 active site residue and core nucleotides where DNA exchange occurs are colored red. Hinge residue Ser99 (Cα) is rendered as a dark red sphere. The sequence of hixL showing the Hin cleavage sites (arrows) and core nucleotides (red) is given below.
The catalytic domain is followed by a long amphipathic α-helical region referred to as helix E from the γδ resolvase secondary structure designations (Fig. 3, 4, and 7). For Hin, modeling and experimental data are consistent with helix E extending from residues 100–134 (117, 118), though residues at the C-terminal end of helix E that are not in contact with the catalytic domain may be unstructured when not bound to DNA. Helix E residues between Met101 and Glu122 participate in most of the interactions between subunits in the dimer and undergo extensive remodeling to form much of the synaptic tetramer interface, including the connections between subunits of the newly formed rotating dimer and subunits across the rotating interface (discussed below). The ‘hinge’ residue connecting the catalytic domain to helix E, Ser99 in Hin and Ser97/98 in Gin, is believed to play a critical role in regulating the quaternary transition between the dimers and the tetramer (sections D.6.5 and D.13.3.5) (109, 119, 120). A four residue linker connects the oligomerization helix to the C-terminal DNA binding domain (DBD), which begins at Gly139 in Hin (121, 122).
D.3.2 The Hin DNA binding domain and recognition of the recombination site
X-ray crystal structures of the monomeric Hin DBD bound to the half-site that is identical between hixL and hixR reveal the details of sequence recognition of the recombination site (Fig. 8) (99, 122). At the N-terminal end, Gly139-Arg140 engage in extensive van der Waals interactions and make a series of hydrogen bonds with nucleotides along the floor of the minor groove of an A-tract sequence. The structure fits well with extensive mutagenesis and chemical probing studies which have shown that A/T or T/A base pairs, or more specifically the absence of a purine 2-amino group, at bp ±(5 and 6) are of critical importance (123, 124). Unlike most A-tract structures, however, the minor groove over this region is not highly compressed. Residues 146–180 fold into a helix-turn-helix (HTH) motif in which helix 1 and 2 are in a near antiparallel configuration, unlike classical phage repressor HTH structures but resembling homeodomains. However, the Hin recognition helix 3 is only 8 residues long, which is much shorter than homeodomains and more similar to repressors. The overall structure of the Hin DBD is similar to that of γδ and Sin resolvases, although the lengths of helix 1 differ (111, 112).
Figure 8.
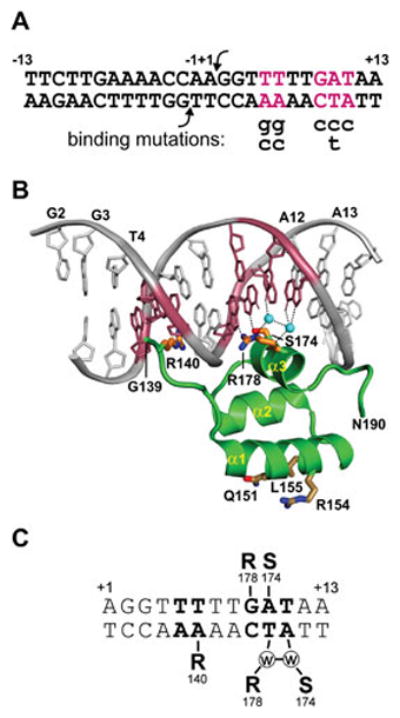
The Hin DNA binding domain bound to the conserved hix half-site located within the invertible segment. A. Sequence of hixL with the locations of severe DNA binding mutations (99, 123). B. Hin DBD – DNA complex (PDB: 1IJW). Side chains of residues Arg140, Ser174, and Arg178 along with the two ordered water molecules (cyan spheres) that make critical base contacts are shown. Conserved residues Gln151, Arg154, and Leu155 on α-helix 1 that contact Fis are also shown. C. Schematic representation of sequence-specific contacts, including water bridged hydrogen bonding.
Sequence recognition within the major groove involves direct contacts between only 2 amino acids on helix 3 and three base pairs at positions ±(9–11) (Fig. 8) (99, 122, 123). X-ray structures of the Hin DBD bound to the wild-type and four mutant hix sites reveal that in addition to direct base contacts by Arg178 and Ser174, two ordered water molecules engage in bridging contacts that are critical for hix binding. The importance of these waters is illustrated by a T/A to C/G substitution at position 11; this mutation decreases binding 25-fold but the only significant change in the structure is the loss of the two ordered water molecules (99).
D.3.3 Binding of the serine invertase dimer to the recombination site
The Hin dimer binds to the intact hix site with about 10-fold higher affinity than the isolated DNA binding domains (99, 121). In the case of Hin-hixR and Gin-gix where the sequence of one of the half sites is suboptimal, cooperative interactions are important for binding (108, 110). The C-terminal ends of the E helices traverse the minor groove on opposite sides of the DNA immediately adjacent to the two bp crossover region and are modeled to make a series of DNA contacts beginning with Hin Arg123 that are important for function (Fig. 7). Arg69 and Arg43 within the catalytic domain may indirectly influence DNA binding by the dimer, as evidenced by the ability of substitutions at these positions to suppress the effects of particular hix binding site mutations (116). The orientation of the recombination sites is determined solely by the identity of the nucleotides within the 2 bp crossover region (67). In native DNA invertase recombination sites, the sequence of the core nucleotides is never symmetrical (e.g., Fig. 2). Hin can efficiently bind and recombine hix sites containing different core sequences, including the symmetrical sequence A/T-T/A. Although a 2 bp core sequence separating the half sites is essential for recombination, Hin can still bind to a hix site with one or even three nucleotides introduced into the center (108, 125). Gel electrophoresis experiments imply that the DNA is bent within the Hin and Gin complexes (106, 110), but the DNA curvature in the Hin-hix complex is estimated to be much less than the ~60° bend angle present in the γδ resolvase-res site I crystals (111). Indeed, a recent low resolution X-ray structure of the Gin-gix complex models the DNA with only a small curvature (126).
D.4 The Fis/enhancer regulatory element
The most distinguishing feature of inversion reactions catalyzed by the serine DNA invertase family is the requirement for a remote recombinational enhancer and its binding partner Fis. Reactions performed without an enhancer but with the Fis protein exhibit very low inversion rates in vivo and in vitro (23, 27, 47). The ~1% inversion rate that occurs without an enhancer under standard in vitro conditions is dependent upon Fis binding to DNA and thus is believed to reflect the prolific non-specific DNA binding properties of Fis. Reactions without Fis are reduced >1000-fold, but as noted above, a detectable basal Fis-independent reaction occurs in vitro in the presence of ethylene glycol (25, 51). Enhancer sequences can be functionally exchanged between DNA inversion systems (48, 60).
D.4.1 Location of recombinational enhancers
The native distances from the center of the closest recombination site to the minimal boundary of the enhancer elements range from 89 bp (Gin) to 481 bp (Cin). As depicted in Fig. 1, the enhancer can be located inside the invertible segment (Hin) or outside the invertible segment (Gin and Cin), and the DNA specifying enhancer activity also functions as the coding sequence for residues 8/9–29/30 of the respective DNA invertase. The Hin enhancer has been shown to function effectively in either orientation and when located from 89 bp to over 4 kb from the nearest hix site, though a measureable decrease in inversion rates is evident when the enhancer is re-positioned many kb’s from a hix site (27). The hin enhancer does not function when positioned very close to a recombination site, and its activity varies with the helical repeat of supercoiled DNA over distances extending out to at least 150 bp, consistent with a physical interaction between the two sites through protein-mediated DNA looping (see section D.5) (28, 117).
Kanaar et al. reported a situation where the gin enhancer can function even though it is not strictly in cis with respect to the gix recombination site (127). Two multiply interlinked supercoiled DNA molecules containing the enhancer on one DNA circle and one or both gix sites on the second DNA circle support a remarkably robust recombination reaction. By contrast, similar experiments with singly catenated circles exhibit no activity, indicating that physical proximity of the three sites is insufficient for assembling an active recombination complex. Moreover, supplying the enhancer in trans on a short linear fragment at high concentrations is ineffective (101) (M. Haykinson and R.C.J., unpublished), unlike the enhancer-dependent Mu transposition reaction where similar experiments reveal enhancer activity (128). These properties of enhancers in the DNA inversion systems are consistent with the obligatory assembly of the tripartite invertasome at an interwound DNA branch on a plectonemically supercoiled DNA molecule.
D.4.2 Architecture of enhancers and Fis binding sites
The hin enhancer sequence is minimally contained within a 63 bp segment; an additional 3 bp of non-specific sequence on each end are required for Fis binding making the total length 69 bp (27, 129, 130). Deletion analysis of the cin enhancer gives similar boundaries (48). The Fis binding sites on each end of the enhancer are separated by 47 bp between their centers, which would position the Fis dimers on nearly opposite sides of the DNA helix separated by approximately 4.5 helical turns of DNA (Fig. 9C). Inactivation of either of the Fis binding sites abolishes all enhancer activity. The spacing between Fis binding sites is critical for activity (131) (Y. Chang, M.M. McLean, and R.C.J., unpublished). Addition of one bp is well tolerated, but a +2 insertion or −1 or −2 deletion severely decreases and a 5 bp insertion inactivates enhancer function. Ten bp insertions retain significant activity, but enhancers containing two or more DNA helical turns or a deletion of 10 bp are not functional. Recently, a modest effect of sequence in the segment between the Fis binding sites in the hin enhancer has been reported (78). The effect is localized to two A/T-rich patches separated by 10 bp where basic residues from the Hin catalytic domain interact (Fig. 9C and see below). The A/T-rich sequences, which face Hin in the invertasome structure, may form narrower and more negatively charged minor grooves and/or induce curvature into the DNA that facilitates assembly of the invertasome by enhancing Hin-enhancer and/or Fis-Hin interactions, respectively.
Figure 9.
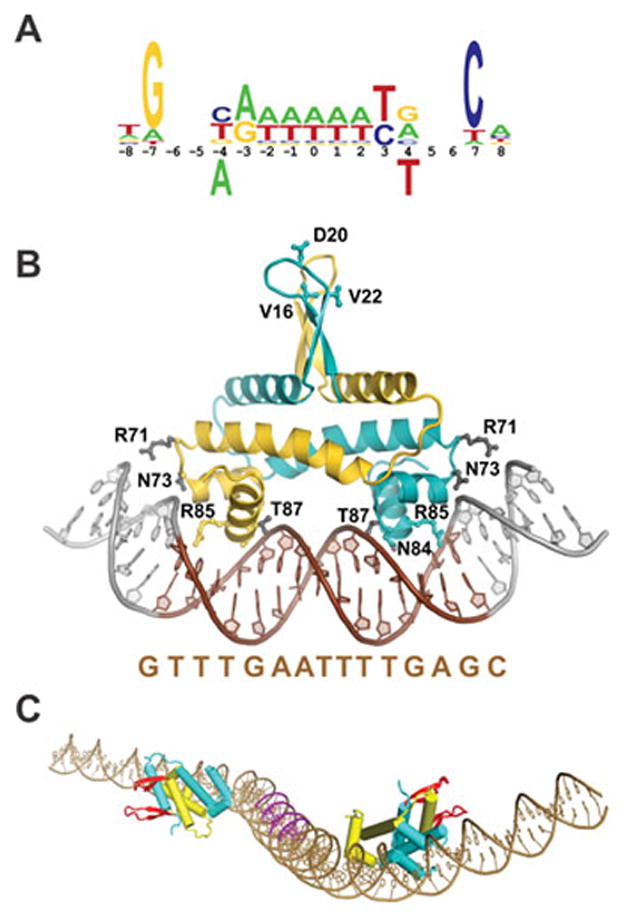
Fis and the recombinational enhancer. A. Fis binding motif derived from footprinting, mutagenesis, genome-wide ChIP, and X-ray crystallography (see ref. (130)). Bases below the numbering are strongly inhibitory for binding. B. Structure of the Fis dimer bound to a high affinity DNA segment (Fis residues 10–98; PDB: 3IV5); the sequence of the 15 bp core between ±7 (colored brown) is given below. Arg85 contacts the conserved guanines at the borders of the core sequence, Asn84 contacts the DNA backbone and often the base at ±4 and is responsible for the inhibitory effect of a thymine at this position (panel A). A subset of other important residues making DNA backbone contacts are colored grey. The Arg71 side chains, which are poorly resolved in most structures of DNA complexes, are shown oriented towards DNA. Bending of the flanking DNA segments varies depending on the DNA sequence. The triad of residues (Val16, Asp20, and Val22) near the tips of the mobile β-hairpin arms that contact DNA invertases are denoted for the cyan colored subunit. (C) Model of the hin enhancer. The two Fis dimers are docked onto the hin enhancer DNA sequence. The Fis β-hairpin arms are highlighted in red. The A/T-rich DNA segments contacted by the helix B regions of the DNA invertase tetramer in the invertasome are colored magenta (78).
DNA sequences of Fis binding sites are highly diverse but contain a 15 bp core “recognition” segment that usually has G/C and C/G base pairs at its boundaries combined with an A/T-rich region in the center (Fig. 9A) (49, 129, 132–134). High affinity Fis binding sites often contain pyrimidine-purine dinucleotide steps at the ±(3–4) positions, which correspond to locations of modest bending in Fis-DNA complexes (130). In all studied enhancers, one of the Fis binding sites (corresponding to site I or the hix proximal site in the hin enhancer) is considerably poorer than the other, in part due to a T/A instead of the preferred G/C bp on one end. This binding site overlaps the coding region for the highly conserved patch around the active site serine. The difference in binding sites appears irrelevant for enhancer function in vitro as a reconstructed enhancer containing two high affinity site II Fis binding sites functions nearly indistinguishably from the wild-type enhancer in the Hin system (29).
D.4.3 The Fis protein structure
Fis is a homodimer of 98 amino acid residue polypeptides. X-ray crystal structures reveal an ellipsoid core containing 4 α-helices beginning at Pro26 (Fig. 9B) (135, 136). Helices A and B from each subunit assemble into a four helix bundle and helices C and D of each subunit fold into a helix-turn-helix DNA binding unit stabilized by the C-terminal ends of helix B from the same polypeptide. A unique feature of the Fis dimer structure is that each DNA recognition helix D is separated by about 25 Å as opposed to the 32–34 Å separation that is typically found among helix-turn-helix DNA binding proteins. This separation is maintained in the DNA-bound complex and is accommodated by a severe narrowing of the central minor groove as described below. Another unusual feature of the Fis structure is the presence of mobile β-hairpin “arm” motifs at the N-terminal end of each subunit (residues 12–25) that protrude over 20 Å from the protein core (137). In many crystal forms, one or both of the β-hairpin arms are poorly resolved due to their mobility, but five different crystal forms, including Fis-DNA co-crystals, resolve most or all of the peptide chain of the β-hairpin arms (130, 137, 138) (S. Stella and R.C.J., unpublished). As expected, the arms are positioned somewhat differently in these crystals due to crystal lattice interactions. Although the two arms are well separated from each other in the crystal structures, direct disulfide linkages between residues in the two arms that are separated in crystals by up to ~20 Å rapidly form when they are substituted with cysteines (137). The residue-specific cysteine crosslinking data provides strong support for the structure of the β-hairpin arm motifs and for their mobility in solution.
Mutagenesis and protein crosslinking studies have mapped the sites of Fis-Hin interaction within the invertasome to be between residues near the tip of the β-hairpin arm of Fis and helix 1 of the Hin DBD (78, 137, 139, 140). Extensive mutagenesis and chemical modification studies over the Fis β-hairpin arm identified the critical residues for activation of Hin-catalyzed DNA inversion to be Val16, Asp20, and Val22 (Fig. 9B). The critical feature of residues 16 and 22 is their branched chain character; a subset of polar residues and even a leucine at residue 20 retain some activity (78). Loss of the Asn17 side chain, which stabilizes the conformation of the β-hairpin loop, also compromises Fis-activation of inversion (137). In addition, mutations at Fis residues Gln33 and Lys36 within helix A negatively impact Hin inversion and so may also directly or indirectly influence invertasome formation. Only one of the Fis arms on each dimer bound to the enhancer is required, as demonstrated by efficient DNA inversion promoted by heterodimers containing only one functional β-hairpin arm (103).
D.4.4 Fis binding to DNA and model of enhancer
Crystal structures of Fis bound to over 20 different DNA sequences with varying affinities have been determined (Fig. 9B) (130, 141) (S.P. Hancock, S. Stella, and R.C.J., unpublished). Fis covers a 21 bp DNA segment in the crystals, and most of the direct contacts over this region are to the backbone of the DNA. The most important base-specific contact is by Arg85 on helix D to the consensus guanines at the edges of the 15 bp core. The Asn84 side chain is also hydrogen bonded to a base in most structures, but its most significant role is to exclude a thymine at position ±3 due to a clash with the 5-methyl group (Fig. 9A and B). The DNA helix axis exhibits overall curvatures of around 65° in the various crystal structures with most of the bending being over the major groove interfaces. Solution biochemistry experiments imply variable amounts of additional DNA bending occur over the regions flanking the 15 bp core sequence; the sequence of the flanking DNA together with Arg71 on Fis modulate this dynamic bending (129, 142) (S. Hancock, S. Stella, and R.C.J., unpublished). The 5–7 bp A/T-rich segment at the center of the binding site is relatively straight but contains a highly compressed minor groove (130, 141). The narrow minor groove is critical for Fis binding because it enables the closely spaced recognition helices of the dimer to insert into the adjacent major grooves.
The Fis-DNA crystal structures enabled construction of a molecular model for the Fis-bound hin enhancer segment (Fig. 9C) (78). The DNA adopts an S-shaped structure with the β-hairpin arms from the two dimers oriented oppositely from one another. The S-shaped structure of the Fis-bound enhancer segment is supported by gel electrophoresis studies on spacing mutants (131, 142). As discussed below, the Fis-bound enhancer functions in the initial stages of the reaction as a scaffold to juxtapose serine invertase dimers for remodeling into the tetramer.
D.4.5 Functions of Fis in other DNA reactions and in chromosome compaction
Subsequent to the discovery of Fis as the factor required to activate DNA inversion reactions, Fis was shown to perform many other functions in the cell (24, 143). These include regulation of transcription and replication reactions, in addition to other specialized recombination reactions such as integration and excision of phage λ and Tn5 transposition. With respect to transcription, Fis can positively regulate promoter activity by interacting with RNA polymerase through its αCTD or σ subunits and negatively regulate transcription by competitively binding with RNAP or gene-specific activators (144). Transcriptome studies have reported significant changes in expression of over 20% of E. coli genes by the loss of Fis, but the correlation of Fis binding by genome-wide chromatin immunoprecipitation experiments and gene expression changes indicates that many of these effects are indirect (133, 134).
Fis is one of a small group of abundant nucleoid-associated proteins that also include HU, IHF, H-NS, and StpA. All of these proteins perform specific regulatory functions and are also thought to contribute to chromosome compaction by means of DNA bending and/or stabilization of DNA loops (145, 146). Single-DNA molecule experiments have demonstrated moderate global DNA compaction by Fis-induced bending, as predicted from the structural and biochemical data, plus robust DNA condensation activity through trapping of DNA loops (147, 148). Under rapid growth conditions, Fis is one of the most abundant DNA binding proteins in E. coli; however, cellular Fis levels are much lower under poor growth conditions and levels of Fis in stationary phase cells are very low (18, 32, 149). Genome-wide studies reveal that Fis binds singly or in clusters every 3–6 kb on average throughout the chromosome, and it has been proposed that interactions between tracts of bound Fis protein may contribute to the dynamic looped-domain structure of the bacterial nucleoid (133, 134). In addition to local effects on chromosome structure, Fis also has been reported to regulate expression of DNA gyrase and topoisomerase I and thereby modulate DNA supercoiling densities on a global scale (150, 151). Because assembly of active invertasomes is dependent upon supercoiling-induced DNA branching, these effects by Fis can also indirectly impact the efficiency of the inversion reaction in vivo.
Clear Fis homologs are found within many members of the gammaproteobacteria class. Members of the Enterobacteriales and related orders have conserved N-terminal β-hairpin arm sequences, but the sequence over this region becomes increasingly diverged and then partially or totally missing in Fis homologs from more distantly related gammaproteobacteria members. Distant Fis homologs are present in the betaproteobacteria class, but these are typically missing the N-terminal β-hairpin region that activates serine invertases.
D.5 DNA looping and the role of HU and other DNA bending proteins
Formation of invertasomes requires DNA looping between the recombination sites and the enhancer element, and in many systems, one of the DNA loops is relatively small. For the Hin reaction, the distance between the center of hixL and edge of the enhancer is 99 bp. DNA inversion in vitro is very inefficient on substrates with distances less than 87 bp, even in the presence of the DNA bending protein HU (28). On the other hand, DNA-cleaved invertasomes can assemble with as little as 56 bp between the two elements. The larger loop size required for the complete inversion reaction probably reflects the winding of DNA strands during the DNA exchange step (discussed below). With separations between 56 and 125 bp, the formation of DNA-cleaved invertasomes strongly correlates with the helical repeat of DNA, which was calculated from the activities of 38 different spacing substrates to be 11.2 bp/DNA turn (28). This value is similar to the helical repeat measured by others for supercoiled DNA (152, 153), and reflects the linking number deficit of supercoiled DNA relative to the 10.5 average for linear DNA.
Early in vitro Hin reactions with the wild-type substrate displayed up to a 10-fold stimulation of inversion rates by HU (23, 28), though more recent measurements employing different preparations of Hin exhibit less dependence. However, inversion reactions on substrates with shorter spacers are virtually dependent upon HU both in vitro and in vivo. Hin catalyzed inversions in hupAB+ cells can occur at low rates with hixL-enhancer segments as short as 56 bp, implying that DNA in vivo is more flexible than in a buffered salt solution with Mg2+ and HU in vitro (28). The additional flexibility in vivo may be in part mediated by spermidine, since in vitro reactions have shown that long chain polyamines like spermidine and spermine can enhance Hin invertasome assembly in the absence of HU. Nevertheless, the helical repeat derived from inversion activities of the spacer substrates in vivo also was calculated to be 11.2 bp/DNA turn, the same as determined in vitro in buffered salt solutions with Mg2+ and HU (28).
Hillyard et al. observed less than 4% Hin-mediated inversion events on the chromosome of hupAB mutant Salmonella Typhimurium strains as compared to the wild type (154). Using plasmid substrates, Haykinson et al. found that E. coli hupAB (HU deficient) mutants generated low but significant rates of Hin-catalyzed inversion with the wild-type hix-enhancer spacing but extremely low inversion was observed with shorter spacer lengths (28). As with in vitro reactions, plasmids with long spacers (e.g., 703 bp to the closest hix site) supported inversion rates in hupAB mutants that were less than 2-fold different than the wild-type parent. Wada et al reported a strong dependence on HU for inversions catalyzed in vivo by the Hin, Gin, and Pin DNA invertases (155). HU has been reported to not affect in vitro inversion rates by Gin, however, even though there is only 89 bp separation between the gixL site and the gin enhancer (26). As expected, HU does not have a significant effect on Cin-catalyzed DNA inversion because the enhancer is located nearly 500 bp from the closest cix site (50, 155).
The in vivo phenotypes of hupAB mutants and the inability of hupAB mutant extracts to complement HU-deficient inversion reactions in vitro, imply that HU is the primary protein in E. coli responsible for DNA looping activity in the Hin reaction. Surprisingly, mammalian nuclear extracts are more active than hupAB+ E. coli extracts for supplying activities that complement HU-deficient Hin-catalyzed inversion reactions (28). The proteins responsible are the abundant HMGB1 and HMGB2 chromatin-associated proteins (156). Likewise, the S. cerevisiae HMGB homologs Nhp6A and Nhp6B promote Hin invertasome assembly (157). Like HU, the non-specifically binding HMGB proteins have robust DNA bending activity, enabling ligation of DNA microcircles that are much smaller than the persistence length of DNA. Eukaryotic HMGB proteins can induce efficient formation of DNA microcircles down to 66 bp, one helical turn shorter than the smallest circle capable of ligation in the presence of HU. Likewise, HMGB proteins are more active than HU in promoting assembly of Hin invertasomes with short hix-enhancer segments and can enable invertasomes to be assembled on substrates containing hix-enhancer segments 10 bp shorter than possible by HU. Significant stimulation of invertasome assembly occurs with only a few molecules of HU or HMGB proteins per DNA substrate, although maximum amounts of invertasomes are assembled with sufficient protein to bind every 150–300 bp (28, 157).
D.6 Assembly and molecular structure of the invertasome
D.6.1 Overview of invertasome assembly pathway and DNA exchange
The critical regulatory step of the inversion reaction is the formation of the active tetramer at the enhancer. The overall pathway is summarized below and illustrated with molecular models in Fig. 10 (see also Fig. 6); experimental evidence for the individual steps is discussed in subsequent sections. The first step is the looping of recombinase dimers bound to the two recombination sites into the Fis-bound enhancer segment (Fig. 10A). Formation of this tripartite complex occurs at the base of a supercoiled DNA branch, which provides conformational energy to generate a specific geometry of crossing DNA strands (two negative nodes, see also Fig. 11 and below). The complex is stabilized by protein interactions between Fis dimers bound on each end of the enhancer and residues on helix 1 of one of the subunits on each recombinase dimer. The catalytic domains of each dimer are thereby positioned next to each other for isomerization into the active tetramer. The massive conformational rearrangement that generates the tetramer breaks most of the original contacts between subunits of the dimer, which are largely mediated by residues from the E helices that are oriented parallel to each other (Fig. 7 and 10A). Transition into the tetramer creates an extensive set of new contacts between synapsed subunits, which largely involve residues of the E helices that are oriented antiparallel to each other (Fig. 10B). During the final stage of the remodeling reaction, basic residues from the B helices on the catalytic domains of the same subunits clamp onto the enhancer DNA between the two Fis binding sites (Fig. 10C). During this stage, the subunits of the original dimer become separated from each other by a relatively flat and completely hydrophobic interface (illustrated in Fig. 15 below). This interface enables the top subunit pair as drawn in Fig. 10 D–F to rotate about the bottom subunit pair, which is fixed onto the enhancer through its connections with the Fis/enhancer segment. Upon full assembly of the tetramer, each subunit cleaves its respective DNA strand by forming the serine ester bond with the 5′ phosphodiester by a still rather mysterious process that probably involves additional coupled conformation changes. The rotation of one synapsed pair of subunits about the other translocates the covalently-linked DNA strands to the recombinant orientation. Reversal of the phosphoserine linkage through attack by the free 3′ OH end (ligation) restores the DNA phosphodiester backbone.
Figure 10.
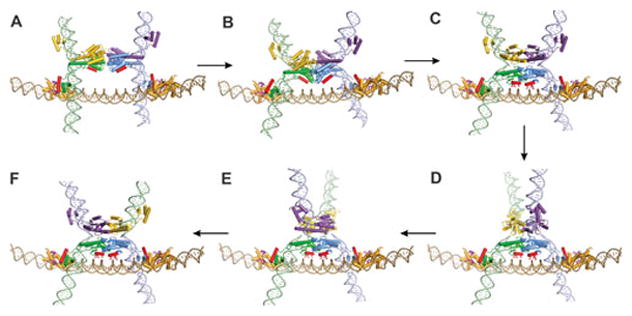
Assembly of the Hin invertasome and subunit rotation. A. Hix-Hin dimers associated at the Fis-bound enhancer. Fis dimers are gold with their β-hairpin arms colored magenta. Hin α-helix B and α-helix 1 are colored red. B. Pre-activated Hin tetramer (based on 3BVP) and C. post-cleavage tetramer (based on 1ZR4). Side chains of residues from helix B that contact enhancer DNA are denoted. D and E. Partial rotations (50° and 90°, respectively) of the top synaptic subunit pair and (F) complete subunit rotation to mediate the exchange of DNA strands. A movie depicting the assembly of the invertasome and DNA exchange by subunit rotation is provided in Video 2 of reference (78), from which these images are taken. Details of the models are described in ref. (78, 109, 117).
Figure 11.
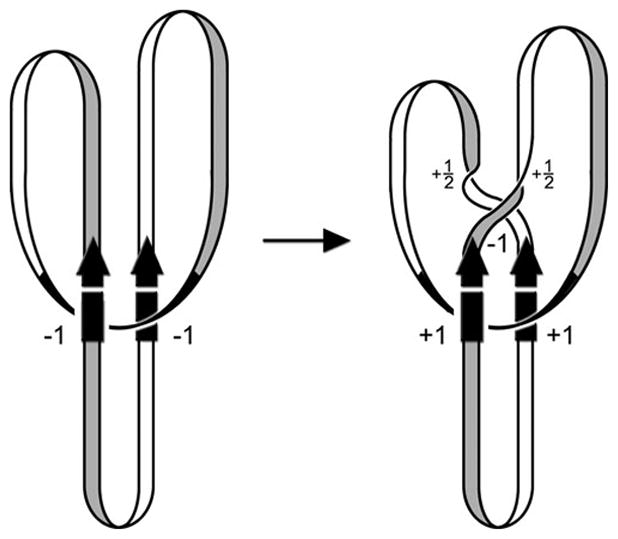
DNA topological changes during the inversion reaction. The starting complex (invertasome) between the two recombination sites (arrows) and the enhancer at the base of a plectonemic branch traps two negative DNA nodes (DNA depicted as a ribbon without supercoiling). Double strand cleavages and DNA exchange by the equivalent of a 180° clockwise rotation create a negative node but also introduce two half turns of helical twist that cancel the negative node. The recombinant configuration of DNA strands changes the trapped nodes to a positive sign resulting in an overall linking number change of +4. Node signs are determined by directionally tracing the entire path of the DNA molecule (193). By convention, a node is defined as negative when the DNA strand in front is pointed upwards and the strand underneath crosses in a rightward direction. A positive node is when the strand underneath crosses in a leftward direction. (Figure is modified from ref. (105)).
Figure 15.
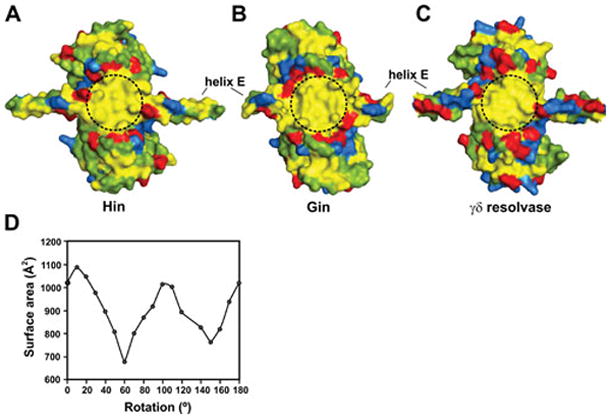
The subunit rotation interface. Surfaces of rotating subunit pairs from the (A) Hin model (residues 2–134 based on PDB: 2GM4), (B) Gin X-ray structure (residues 2–125, PDB: 3UJ3), and (C) γδ resolvase X-ray structure (residues 1–132, PDB: 2GM4) are shown after alignment. Hydrophobic residues are colored yellow, acidic residues are red, basic residues are blue, and polar residues are green. A 1.6 Å probe was used to render the surfaces. Dashed circles demarking the rotating interface have a diameter of ~20 Å. D. Surface area overlap calculated for different clockwise rotational conformers from the Hin model; γδ resolvase gives a very similar pattern (117, 119). Rotations of around 0–10° correspond to conformers where the DNA ends are in-line for ligation and conformers around 100° have the E-helices between dimer pairs in a parallel/antiparallel configuration. The Hin models are based on γδ resolvase structures (shown here based on 2GM4); comparison of subunit structures with those from resolvase tetramers (2GM4 or 1ZR4) give RMSD values of <0.7 Å over the peptide backbone (residues 1–120). Subunits from the Gin tetramer structure (3UJ3) exhibit RMSD values of 1.3 – 1.5 Å over the peptide backbone atoms from residues 3–120 and 1.1 – 1.4 Å over just the catalytic domains from the Hin models or γδ resolvase tetramers (1ZR4 or 2GM4). Much of the difference between Gin and resolvase structures or Hin models is over poorly resolved loops connecting β1 to αA and β2 to αB.
D.6.2 Fis-Hin contacts
The connections between the mobile β-hairpin arms of Fis and helix 1 of the Hin DBD were initially identified within DNA-cleaved invertasomes by site-directed protein-protein crosslinks utilizing reagents containing linker lengths as short as 4.4 Å (78). The crosslinkers targeted a cysteine introduced at Fis residue 19 or 21 between the critical Hin-activating residues Val16, Asp20, and Val22 within the β-hairpin loop (Fig. 9B) and either native lysines on helix 1 (Hin residues 146 or 158) or cysteines introduced at solvent-exposed residues between 146 and 151. Mutant analysis of residues on helix 1 then identified Arg154 and Leu155 as being most critical for activation by Fis, with contributions by Gln151 and possibly Lys158 (Fig. 8B). Notably, residues 151, 154, and 155 are completely conserved among members of the DNA invertase subfamily that are known to be regulated by Fis (Fig. 3). Fis-Hin crosslinking experiments in which individual Hin subunits were radiolabeled demonstrated that only the bottom subunits of the tetramer (colored blue and green in Fig. 10) are associated with Fis. Moreover, Fis could be crosslinked to inactive covalently-locked Hin dimers containing a disulfide bond across the dimer interface. Subsequent reduction of the disulfide bond enabled the Fis-linked Hin dimers to remodel into a tetramer that is active for DNA cleavage. This key result supports a model in which the Fis dimers initially contact inactive Hin dimers and maintain this contact during the dimer-tetramer remodeling reaction (Fig. 10 A–C). Hin tetramers crosslinked to Fis are also competent for DNA ligation, providing evidence that Hin normally remains associated with the Fis/enhancer element through all the chemical and mechanical steps of the reaction (78).
D.6.3 Structure of the invertase tetramer
Initial evidence for the quaternary arrangement of subunits in the Hin tetramer came from experiments employing a strong hyperactive Hin mutant H107Y that can assemble stable DNA-cleaved synaptic complexes on oligonucleotides substrates without the Fis/enhancer (158). Cysteines were substituted around the protein surface targeting specific interfaces, and site-directed crosslinking experiments employing bis-maleimide reagents with different spacer lengths were performed. Crosslinks between residues located proximal to the N-terminal end of the E helix (residues 94, 99, and 101) of subunits originating from different dimers implied that this region of the catalytic domain is within the synaptic interface of the tetramer and that the catalytic domains from the originating dimers are positioned orthogonally to each other. In this configuration the DNA crossover sites are far apart from each other on opposite sides of the synaptic tetramer. A similar overall architecture for synaptic tetramers of resolvase were also proposed based on molecular and biophysical data, including small angle scattering of X-rays and neutrons (159–161).
The Steitz laboratory has determined several X-ray crystal structures of reaction intermediates of γδ resolvase tetramers bound to DNA (PDB: 1ZR2, 1ZR4, and 2GM4; 3.9, 3.4, and 3.5 Å resolution, respectively) (119, 162). These were generated using hyperactive mutants of resolvase with the protein used for the 2GM4 crystals being a chimera containing Hin residues 94–103 substituted for the analogous residues of resolvase. This segment includes the ‘flexible joint’ between the catalytic domain and oligomerization helix E, in which a serine (Hin residue 99) replaces a glycine (resolvase residue 101). Each of the tetrameric structures represents a post cleavage complex where the 5′ ends of the cleaved DNA are covalently joined to each subunit at serine 10. Surprisingly, the cleaved DNA ends of each recombination site are separated by about 30 Å, nearly the distance between the two active site serines in the structure of the inactive resolvase dimer bound to DNA (PDB: 1GDT) (111). The post cleavage tetramers contain a flat and hydrophobic interface between synapsed subunit pairs that can support subunit rotation (discussed further in Section D.9).
A tetrameric crystal structure of a Gin hyperactive mutant containing the catalytic domains plus the first 26 residues of the E helices has also been reported (PDB: 3UJ3; 3.7 Å resolution) (120). Interestingly, this structure was crystallized in ethylene glycol that promotes stable tetramer formation in Hin. Each Gin subunit closely resembles γδ resolvase tetrameric subunits (RMSD over common backbone atoms = 1.3–1.5 Å; see Fig. 15 legend below). The synapsed subunit pairs are again separated by a flat and completely hydrophobic interface (see Fig. 15B below), but they are in a conformation that is equivalent to a 26° rotation of synapsed subunit pairs relative to the resolvase tetramer structures. A high resolution tetrameric crystal structure, also grown in ethylene glycol, of the Sin resolvase catalytic domains plus part of the E helices from a hyperactive mutant has also been determined (PDB: 3PKZ; 1.86 Å resolution) (114). The Sin tetramer exhibits yet a different rotational conformation, with the synapsed subunit pairs rotated 35–45° relative to those in the resolvase tetramers. Even though DNA is not present in the complex, the three Sin tetramer structures present in the asymmetric unit of the crystal currently provide the best view of the active site of a serine recombinase in a conformation poised for catalysis. The active site serines together with other key catalytic residues are organized around sulfate ions in the crystal, and docking of the bent uncleaved DNA of the γδ resolvase dimer structure positions the scissile phosphodiester bonds over the sulfates. Rice and co-workers suggest that this rotational state may represent the tetramer conformation that is active for cleavage and ligation (114).
D.6.4 Evidence for a structural intermediate in the formation of the active tetramer
Two additional X-ray structures provide evidence that the dimer-tetramer remodeling may occur through a defined intermediate step referred to here as a ‘pre-activated’ tetramer (Fig. 10B). One structure is a tetramer complex of the TP901 serine integrase catalytic domain plus N-terminal portion of its E helix (PDB: 3BVP; 2.1 Å resolution) (163). In the TP901 integrase structure, the four E helices are associated in a conformation similar to the tetrameric structures described above, but the catalytic domains are in an incompatible position for DNA chemistry and have not fully rotated about the hinge immediately prior to the E helices to form the flat interface between rotating dimers. A tetramer structure of another hyperactive γδ resolvase mutant (PDB: 2GM5; 2.1 Å resolution) appears to be a hybrid containing one dimer resembling the pre-activated TP901 conformation and one dimer matching that in the post cleavage resolvase tetramer (162). This asymmetric tetrameric structure may be relevant to the phenotypes of some mutant Hin proteins, particularly those with changes in the E helix region that exhibit uncoupling of double strand cleavages between the two hix sites (106, 109). Whereas wild-type Hin promotes near concerted double strand cuts at both hix sites, these mutants generate substantial amounts of double strand cuts at only one hix site. No Hin mutant, however, has been obtained that uncouples the cleavages of the two DNA strands at a single hix site to generate a single strand break; the highly concerted nature of the double strand cleavage reaction implies a coupled conformational change by the subunits of an original dimer. Site-directed crosslinks obtained from stable uncleaved mutant tetramers (section D.13) are consistent with a pre-activated tetramer structure (M.M. McLean, G. Dhar, and R.C.J., unpublished).
D.6.5 The dimer–tetramer remodeling reaction: role of the helix B region
Remodeling of inactive invertase dimers into the active tetramer is likely to be initiated by coincident scissor-like opening of the two juxtaposed dimer interfaces to expose their hydrophobic surfaces. During formation of the initial pre-activated tetramer, the E helices from synapsed subunits slide along each other over about 15 residues. In the later phase of assembly into the active tetramer, further rotation of the catalytic domains about the hinge located at Hin Ser99 (Gly101 in γδ resolvase) (109, 119, 162) associates the D helices of subunits that form rotating pairs (118) and positions Hin residues Lys47, Arg48, and Lys51 in the B helices of the two enhancer proximal subunits in contact with enhancer DNA between the bound Fis dimers (Fig. 10B–C) (78). Experimental evidence indicates that tetramers with rotating subunit pairs covalently linked across their D helices are competent for DNA cleavage, subunit rotation, and ligation, indicating that the DNA chemical and mechanical steps of the reaction occur within the fully assembled tetramer ((118) and M. M. McLean and R.C.J., unpublished data).
Attractive electrostatic forces between the basic residues in the B helices and the enhancer DNA are postulated to help drive the latter conformational changes to the active tetramer structure (78). In the initial pre-activated tetramer, the B helices from the enhancer proximal Hin subunits are modeled to be oriented at about a 50° angle with respect to the enhancer DNA (Fig. 10B). In the active tetramer, Lys47, Arg48, and Lys51 from both subunits are positioned against the enhancer DNA between the bound Fis dimers (Fig. 10C). Unlike other serine recombinases, all of the Fis/enhancer-dependent serine invertases have basic residues at these positions, although arginines or lysines are found at positions 48 and 51 (Fig. 3). Loss of one of the helix B basic residues has a relatively small effect on Hin-catalyzed inversion rates, but substitution of two of these residues with alanine reduces Fis-activated inversion rates ≥20-fold but does not affect Fis-independent inversion by Hin hyperactive mutants. The locations where the B helices contact enhancer DNA were mapped by coupling the chemical nuclease FeBABE to Hin residues near the C-terminal end of the B helix (78). The hydroxyl radicals cleaved within A/T-rich segments in the center of the enhancer where the DNA minor grooves face the Hin tetramer (Fig. 9C and 10C). Substitution of these segments with G/C-rich sequences has a demonstrable but small effect on inversion rates. The modest importance of base sequence, combined with the conservation of either arginine or lysine at the equivalent of residues 48 and 51 within Fis/enhancer-dependent invertases, suggests that nonspecific electrostatic forces are operating during this step.
D.7 DNA topological changes introduced by reactions initiated within the invertasome
DNA topological analyses of recombinant products generated by the Hin and Gin have provided crucial structural and mechanistic insights into the reaction. As discussed below, these studies established the starting geometry of DNA strands within the invertasome, provided the first experimental evidence for the subunit rotation model for DNA exchange, first revealed the role of the enhancer in regulating the processivity of the subunit rotation reaction, and identified the importance of DNA superstructure in mediating the bias for intramolecular DNA inversion over other recombinant products.
D.7.1 A single DNA inversion reaction results in a linking number change of +4
To determine the change in DNA linking number that accompanies a single DNA exchange reaction resulting in inversion, reactions were performed on unique negatively supercoiled plasmid topoisomers. High resolution electrophoresis in agarose gels containing chloroquine revealed that the overwhelming majority of inversion products are unknotted but have lost 4 negative supercoils. This was first demonstrated for Gin under standard inversion reaction conditions (164, 165) and was later shown for Hin under conditions ensuring only a single recombination reaction, which was necessary because of the robust DNA relaxing activity by Hin (166). The +4 linking number change was interpreted by Kanaar and Cozzarelli as having been generated by the following mechanism (Fig. 11) (105, 164). Two negative nodes (DNA crossings) are entrapped within the initiating recombination complex, consistent with the geometry of DNA strands at the base of a plectonemic supercoiled branch. DNA exchange by double strand breaks at the two recombination sites followed by a 180° clockwise rotation of each duplex above the crossing enhancer DNA and then ligation introduces a negative crossover node, but is also accompanied by a half turn of twist on each duplex thereby cancelling out the crossover node. However, because recombination switches the geometry of the entrapped nodes from −2 to +2, the linking number changes by +4. Thus the energetically favorable reaction, in which there is a net loss of 4 negative supercoils together with the release of twist, is the clockwise rotation of DNA strands. Because the recombinase subunits are covalently linked to the DNA ends by the serine phosphodiester bond, it was proposed that the subunits must also undergo the equivalent 180° clockwise rotation within the invertasome complex.
D.7.2 Additional DNA rotations generate specific DNA knots
DNA knot nodes in plasmid DNA can be resolved by agarose gel electrophoresis after nicking the DNA to remove supercoils. Under standard in vitro conditions Hin and Gin generate very few knotted products because most reactions ligate after a single 180° rotation (Fig. 12) (164, 167). Likewise, a comparison of knotted to inverted products of Hin-catalyzed reactions in vivo under conditions where topoisomerase IV, the major unknotting enzyme in E. coli, is blocked indicated that <5% of in vivo recombination reactions generate knots (166, 168). By contrast, Hin reactions performed on DNA substrates where one of the hix sites contains a mutation within the central 2 bp crossover region exhibit robust knotting. Reactions on these substrates are unable to ligate in the recombinant (inverted) orientation after a single 180° rotation because the 2 bp overlap region cannot base pair (Fig. 13) (166, 167). A second 180° rotation restores base pairing and enables ligation, but the two processive rotations generate a 3-noded knot (trefoil) with the invertible segment repositioned into the parental orientation (Fig. 12). Most of the products from a single reaction on a substrate with mutant core nucleotides are trefoils without inversion. However, occasionally the reaction fails to ligate after two rotations and undergoes a third rotation where it again cannot ligate over the mismatched crossover region and thus undergoes a fourth rotation that can support ligation back into the parental orientation; the four subunit rotations starting in the invertasome structure generate a 5-noded twist family knot (95, 101, 167). A distinguishing feature of Hin reactions on crossover mutant substrates is the absence of 4- and 6-noded twist knots (note that 6-noded compound knots can be generated from two independent reactions that each introduces 3 nodes). Reactions with wild-type recombination sites, however, do form twist knots with even numbers of nodes, albeit rarely. Knot formation in vitro by Gin has been most extensively studied in reactions employing low Mg2+ and low salt concentrations, conditions that inhibit ligation and destabilize the Fis/enhancer-bound invertasome complex (95). The stereostructures of knots generated by Hin and Gin have been determined by electron microscopy (95, 101, 167). In all cases for Hin and the vast majority of cases for Gin, the structures of the knots, including the chirality of the nodes, are compatible with only clockwise rotations starting with the −2 topology of DNA strands present in the starting invertasome. The structures of the DNA knots thus provide additional strong support for assembly of the invertasome at a DNA branch and the subunit rotation model for DNA exchange.
Figure 12.
DNA topological changes during processive recombination by serine DNA invertases. Initially, the recombination sites (green and blue) are assembled at the enhancer DNA (brown) in the invertasome structure. The initial assembly depicts short and long loops between the recombination sites and enhancer as found in the Hin and Gin systems. The first DNA exchange by the equivalent of 180° rotations of DNA strands covalently-linked to recombinase subunits generates inversion; these molecules have lost 4 negative supercoils (not shown) but no knot nodes are introduced. Subsequent processive rounds of DNA exchange generate knots of increasing complexity with the orientation of the invertible segment alternating between parental and inverted; these have all lost 2 negative supercoils. Multiple windings of DNA (processive DNA exchanges) are torsionally restricted as long as the enhancer remains associated with the recombination sites, but release of the enhancer removes this constraint. The structures of the knots, which have been confirmed by electron microscopy, are depicted below.
Figure 13.
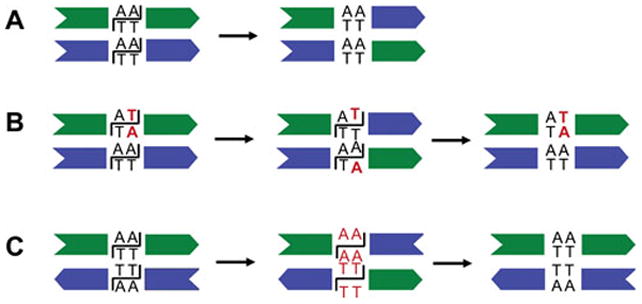
Synapsis and DNA exchange of recombination sites. A. Synapsis of wild-type recombination sites in a parallel configuration as in the standard invertasome complex. DNA cleavage followed by exchange and ligation inverts the intervening DNA segment. B. Synapsis where one recombination site contains a mutation (red) within the core nucleotides. DNA cleavage and a single exchange result in unpaired core nucleotides that cannot ligate. A second exchange is required for ligation, which reorients the intervening DNA segment into the starting (parental) configuration and generates a 3-noded knot (Fig. 12). C. Synapsis of wild-type recombination sites that are oriented in a directly-repeated configuration. Most of the time the sites align in an antiparallel configuration within an invertasome structure (see Fig. 16), and the core nucleotides cannot base pair after the first DNA exchange. Ligation can occur after a second exchange back into the parental orientation creating a 3-noded knot as in panel B.
Short DNA lengths between one of the recombination sites and the enhancer inhibit processive rotations because of torsional constraints introduced by the multiple DNA windings (Fig. 12). This has been demonstrated for Hin both in vitro and in vivo where substrates containing mutant crossover regions were shown to produce few knots when the separation between hixL and the enhancer is the wild-type length of 99 bp as compared to substrates with much longer spacings (166, 167). Reactions on the mutant substrates forming invertasomes with short loops accumulate significant amounts of broken DNA molecules because the mismatched DNA ends are held in a non-ligatible configuration after a single exchange (100). The low frequency knots that are generated under these conditions tend to be highly complex because of release of the Fis/enhancer from the invertasome complex with the consequent loss of torsional constraints (see section D.10). Knotted DNA molecules generated by at least 8 processive exchanges have been observed from Hin and Gin reactions (95, 167). It has been suggested that Gin can also recombine DNA via alternative pathways that are topologically distinct from the primary pathway, especially when the loop sizes are short (101).
D.8 Evidence for subunit rotation at the protein level
Time-resolved site-directed crosslinking experiments using Hin mutants with strategically placed cysteines have provided direct evidence for subunit rotation during the recombination reaction. Initially, these experiments employed tetramers assembled with the Hin-H107Y Fis/enhancer-independent mutant in covalently-linked cleavage complexes with radiolabeled linear DNA substrates of different lengths in order to tag individual subunits (158). Crosslinks formed within cleavage complexes assembled using proteins containing cysteines located shortly prior to (Hin residues 94 or 99) or within the N-terminal end of helix E (Hin residue 101) link trans-diagonally located subunits (Fig 14). Crosslinks formed early in the reaction are between subunits originating from synapsed dimers, but after a few seconds, subunits from the original dimers become crosslinked because of subunit translocation within the tetramer. The Hin tetramer models based on the resolvase post-cleavage structures suggest that even the starting configuration of subunits would require a small amount of rotational movement in order for the sulfhydryl groups to be within the 8 Å crosslinking distance for residues 94 (Fig. 14) and 99 (117).
Figure 14.
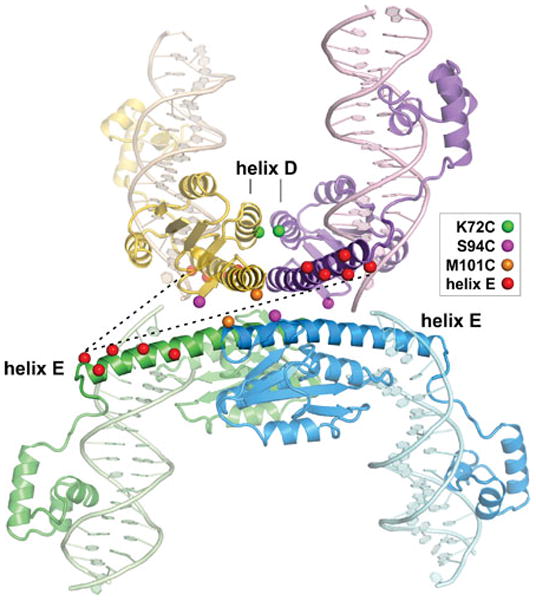
Site-directed crosslinking demonstrating subunit rotation within active Hin tetramers. Model of the Hin tetramer based on resolvase tetramer structures. View is looking into the rotation interface with residues converted to cysteine for site-directed crosslinking rendered as colored spheres (Cβ atom). Rotations refer to the top subunit pair (gold and purple) relative to a fixed bottom subunit pair. M101C residues (orange spheres) from the gold and blue trans-diagonally located subunits are within range for crosslinking with a bis-thio reactive reagent containing an 8 Å spacer but becomes optimally positioned for crosslinking after a ~20° clockwise (cw) rotation. Initial crosslinks at residue 101 are between gold and blue subunits, but with time, purple and blue crosslinked subunit products that are the result of subunit exchange become equally represented. Crosslinking between S94C residues (magenta spheres) requires either a counterclockwise (ccw) rotation of at least 20° (optimal between 40–70°) to link trans-diagonally subunits or a clockwise rotation of about 270° to link purple and blue subunits from an original dimer. Fis/enhancer-activated reactions on supercoiled DNA primarily form 94–94 crosslinks between purple and blue subunits (cw rotation), whereas crosslinks between gold and blue subunits (ccw rotation) are overrepresented initially in Fis/enhancer-independent reactions but then both products become equally represented. Red spheres mark residues within the C-terminal third of helix E that support robust crosslinking even though they can be up to 70 Å from their nearest partner; rotations of 90 ± 15° in the cw or ccw direction are required to generate crosslinks. Fis/enhancer-activated reactions on supercoiled DNA primarily form crosslinks between purple and green subunits (cw rotation) containing helix E cysteines, whereas Fis/enhancer-independent reactions without DNA supercoiling form crosslinks between gold and green subunits (ccw) or purple and green subunits (cw) with the same kinetics. Dashed lines highlight the positions of Q134C residues at the C-terminal ends of helix E. The kinetics of crosslinking between residues at positions 101, 94, and the C-terminal end of helix E provide strong support for bidirectional subunit rotation within Fis/enhancer-independent reactions and for primarily single round cw rotation in Fis/enhancer-dependent reactions on negatively supercoiled DNA. Crosslinks between helix D residues like K72C (green spheres) readily form upon full assembly of the tetramer and do not block subunit rotation or DNA ligation. Details are provided in refs. (117, 118, 158).
Strikingly, robust crosslinking also occurs between helically-phased cysteines on the C-terminal third of helix E (117, 118, 158). These residues are located up to 70 Å away from each other in the Hin tetramer model (Fig. 14). In order for the cysteines across the rotational interface to be in position for crosslinking, one pair of synapsed subunits would be required to rotate 90 ± 15° in either the clockwise or counterclockwise direction. Nevertheless, subunits from the original dimers (requires counterclockwise rotation) or from the synapsed dimers (requires clockwise rotation) crosslink within seconds of Hin cleaving both hix sites. Clockwise and counterclockwise rotation products exhibit nearly equivalent rates of crosslinking suggesting that both directions occur equally efficiently within the Hin tetramer complex on linear DNA. Bidirectional rotation within DNA invertase tetramers is also consistent with earlier topological analyses of reactions on relaxed plasmid DNA by a Fis-independent Gin mutants (169). Hin tetramers crosslinked between residues on the C-terminal end of the E helices are unable to support DNA ligation because the rotating subunit pairs are covalently locked about half way through a complete rotation leaving the DNA ends far apart from each other. As noted above, crosslinking between helix D residues 72 (Fig. 14) or 76 covalently links the synapsed rotating pairs of subunits with no effect on the subunit rotation and ligation steps (118).
Crosslinking reactions on supercoiled DNA using Fis/enhancer-dependent Hin recombinases generate a different kinetic profile than those with DNA fragments or relaxed plasmid DNA (117). The profiles are consistent with the vast majority of complexes undergoing a rapid but single clockwise translocation of subunits (Fig. 14). Most complexes remained in a singly rotated state for at least 20 min, even with mutant substrates where the crossover regions cannot base pair (Fig. 13B). Crosslinking reactions have also been performed on positively supercoiled DNA preparations. Although the Hin-catalyzed inversion reactions are less efficient than on negatively supercoiled DNA, a strong bias for rotation in the counterclockwise direction was observed. These results, combined with the evidence for equivalent rates of bidirectional rotations on linear fragments or relaxed plasmids, indicate that DNA supercoiling is determining the direction of subunit rotation in reactions initiated from invertasomes. As noted above, the clockwise direction of rotation is energetically favorable on native negatively supercoiled DNA because it is accompanied by a loss of supercoils and helical twist.
D.9 Structural nature of the rotation interface
The rotation interface separating the synapsed subunit pairs is almost exclusively composed of residues from the first half of the E helices. Hin and Gin share only about 25% sequence identity with γδ resolvase over this surface. Nevertheless, the molecular character of the rotating interfaces of Gin (120), Hin (117), and γδ resolvase (119, 162) are very similar (Fig. 15A–C). In each case the interface surface is circular (diameter 20–25 Å), relatively flat, exclusively aliphatic, and is surrounded by mostly acidic residues that are somewhat conserved between the invertases and resolvases. The interface surface areas of different rotational conformers vary with the greatest surface overlap where the cleaved DNA ends are linearly aligned (0–10° clockwise rotation) and where the E-helices between rotating pairs are aligned in a parallel/antiparallel configuration (~100° clockwise rotation) (Fig. 15D) (117, 119). The overall buried surface areas, combined with shape complementarity and van der Waals interactions between specific surface residues, may function as energetic barriers or pause sites in the rotation reaction. A pause in subunit rotation over the DNA-aligned conformer may facilitate the ligation step. Rotational pausing at helix E-aligned conformers may contribute to the robust crosslinking efficiencies observed between cysteines on the C-terminal third of the E helices (117, 118). Crosslinking reactions performed in high and low ethylene glycol concentrations suggest that high ethylene glycol favors Hin tetramers to be in the helix E-aligned rotational conformer over the DNA-aligned conformer, providing a partial explanation for why ligation is inhibited and the tetramer intermediate is stabilized by high ethylene glycol (117).
D.10 Factors regulating processivity of subunit exchange
The protein crosslinking experiments together with the topological changes in the recombinant products determined both in vitro and in vivo on wild-type substrates demonstrate that mechanisms exist to limit subunit rotations to a single exchange. Nevertheless, additional exchanges can occur with high efficiency under certain conditions, the most biologically important is when sites synapse in the incorrect orientation or pseudo-sites synapse that have different nucleotides at their crossover site (e.g., Fig. 13B and C). The identity of the 2 bp crossover sequence upon the first rotation is the only check after the initial DNA binding step to ensure that the reaction is between the correct DNA sites that are synapsed into the correct orientation. The ability to undergo additional forward subunit exchanges enables ligation back into the parental DNA sequence, thereby avoiding a chromosome break. Rare examples have been reported from in vivo reactions between pseudo-sites where ligation must have occurred over mismatched crossover regions to generate sequence conversions (170, 171).
One major mechanism inhibiting multiple rotations of subunits is the interaction between the Fis/enhancer element and the recombinase complex in the invertasome structure (101, 117, 166, 167). The short DNA loop that is trapped with the native Hin substrate inhibits the multiple DNA windings that occur coincidently with each subunit exchange (Fig. 12). Thus, formation of knotted products on substrates with sequence differences at the crossover dinucleotides requires an expanded length of DNA between the enhancer and hixL. Crosslinking reactions also demonstrate that the length of the DNA loop between the closest hix site and the enhancer controls the ability of the subunits to undergo multiple exchanges (117). Conditions that destabilize the connections between the Fis/enhancer element and recombinase complex once the active invertasome has formed result in processive subunit exchanges. These include: 1. mutations in the contact sites between Hin and Fis (78, 166), 2. mutations in the helix B region of Hin that contacts the enhancer DNA (78), and 3. positioning of the enhancer close to, but not in helical phase with, a hix site (117). In each case, reactions generate an increased frequency of higher-order knots, and where tested by crosslinking, generate a random mix of products that are indicative of multiple subunit exchanges even when the shortest loop size is ≤150 bp.
Even when the lengths of the DNA segments separating the enhancer from the recombination sites are sufficiently large such that multiple subunit rotations would not be expected to be torsionally constrained by DNA windings, most Hin reactions generate unknotted inversion products indicative of a single exchange (166, 167). For example, <3% of in vivo reactions on plasmid substrates in which the shortest loop size would be ~700 bp were estimated to have undergone multiple exchanges. Likewise, reactions on substrates with mutant crossover regions primarily ligate after 2 exchanges and only very rarely exceed 4 exchanges. These features suggest that additional mechanisms exist to restrict subunit rotations such that ligation will occur after exchange if the overlapping nucleotides can base pair.
Additional support for a mechanism limiting subunit rotations comes from single-DNA molecule experiments (B. Xiao, R.C.J., and J. F. Marko, unpublished). Reactions employing a Fis/enhancer-independent Hin mutant were performed between two linear DNA duplexes containing a hix site that were each attached to a glass surface on one end and a paramagnetic bead on the other. The DNA molecules were coiled about each other by winding of the beads by a magnet. Under conditions inhibiting ligation, the Hin mutant unwound the braided DNA molecules in discrete steps whereby unwinding events were separated by time periods of seconds to minutes. The profiles are unlike those observed in similar experiments employing the serine integrase Bxb1, where an apparently ungated reaction results in full relaxation of the braided DNA molecules (172). The single-DNA molecule experiments provide further support for the subunit rotation mechanism and strongly imply that an intrinsic feature of the Hin tetramer structure mediates pausing of the rotation reaction.
Rotational pausing over the surface overlap maxima between rotating subunit pairs where the DNA ends are linearly aligned for ligation is one mechanism that may contribute to gating of the subunit rotation reaction. Consistent with this idea, certain Hin mutations in large hydrophobic residues within the rotating interface are defective in ligation (Y. Chang and R.C.J, unpublished). Li et al. (119) have suggested that charge interactions between two arginines on the C-terminal end of the helix E on one subunit and two aspartic acids in the catalytic domain near the beginning of helix E on the subunit across the rotating interface may pause the rotation of γδ resolvase tetramers at the ligation-ready conformer. The Hin and Gin sequence is compatible with potential formation of one of these ion pairs, Hin residues Asp93-Arg123. The properties of Hin mutations at these residues imply that both play important roles early in the inversion reaction, but there is currently no evidence for a role of these residues in controlling subunit rotation (115, 118)(Y. Chang, S. Merickel. and R.C.J., unpublished).
D.11 DNA supercoiling and branching
DNA supercoiling plays an essential role in the Fis/enhancer-dependent reaction by localizing the three interacting sites at the base of a DNA branch with the correct −2 topology. Solution experiments and even assays employing proteins bound to relaxed DNA plasmids have failed to provide evidence for meaningful Fis-Hin interactions (104) (R.C.J., unpublished). Simulations by Vologodskii and coworkers predict that supercoiling increases the local concentration of 2 sites about 100-fold and three sites at a branch by more than 1000-fold over nonsupercoiled DNA (173). Thus, the conformational energy present in plectonemically supercoiled DNA is probably required to stabilize interactions between DNA-bound invertases and the Fis/enhancer element, which are otherwise too weak to form. The ability of hyperactive mutants of Hin and Gin to efficiently recombine DNA in Fis/enhancer-independent reactions without supercoiling indicates that supercoiling is not required for the protein conformational changes, chemical steps, or potential topological steps like DNA unwinding.
Electron microscopy and long-chain DNA modeling studies generally agree that naked supercoiled DNA is branched every 2 ±1 kb (173–176). The DNA duplexes within branched molecules are under constant thermal fluctuation and can readily undergo slithering motions to align distant sites (176–178). The extent of branching does not vary significantly over physiological levels of supercoiling and may actually decrease at high supercoiling densities (σ) (174, 175, 179). Formation of active invertasomes begins at σ ~ −0.025 and increases to up to σ ~ −0.06 (33) (I. Moskowitz and R.C.J., unpublished). This correlates with the reduction in the superhelix diameter over this range of σ (175); likewise, a decrease in the separation of DNA duplexes at a branch point with increasing σ would also be expected. Computer simulations give a superhelix diameter of about 100 Å at σ = −0.06 where invertasome assembly is most efficient (174). In the invertasome model, the separation between hix sites where they cross the enhancer is about 75 Å. Hin reactions employing mutant enhancers that contain an additional helical turn of DNA and thus would exhibit a separation between hix sites of around 110 Å remain remarkably efficient (131).
Once Hin dimers are localized together at the enhancer, it is possible that supercoiling contributes to remodeling into the active tetrameric structure utilizing energy in the underwound supercoiled DNA. Alternatively, energy associated with the mobility of the β-hairpin arms of Fis may assist in the dimer-tetramer remodeling. A prominent role for such forces, however, seem unlikely given that enhancers with additional 10 bp separation between Fis sites function within 30–50% the activity of wild-type enhancers (131). Moreover, Fis/enhancer activation proceeds efficiently when the mobility of the β-hairpin arms is predicted to be restricted due to disulfide crosslinks between arm residues 15 or 18 (137).
Invertasomes could assemble via several potential pathways. One is a near simultaneous collision of the three sites slithered into supercoiled DNA branch. A second pathway involves sequential looping of individual recombinase bound recombination sites into the Fis/enhancer. In vivo assays employing transcriptional repression reporters have been interpreted as providing evidence for single loops between a Fis binding site and a hix site, but recruitment of pseudo-hix sites in these experiments, which are observed in vitro, cannot be ruled out (115, 180). A third pathway posits an initial interaction between recombinase dimers bound to recombination sites in a manner that entraps a branch, followed by joining of the Fis/enhancer on the branched DNA to the paired-recombination site complex. In vivo data from transcriptional repression assays are also consistent with formation of paired-hix complexes (115, 180). In addition, glutaraldehyde readily crosslinks paired-hix complexes (104), but there is little current in vitro evidence that paired-hix complexes are precursors for invertasome formation and not the result of non-specific crosslinking.
DNA modeling studies by Halford and co-workers have concluded that there is only a 10-fold difference in the rates of 3-site as compared with 2-site synapses on supercoiled DNA. They modeled three sites on a 4.2 kb supercoiled DNA converging to a branch point within 0.1 – 1 sec (181). Tripartite Hin invertasomes have been measured to assemble within seconds under certain conditions (106). Taken together, the available experimental data combined with DNA modeling studies appears most compatible with a near 3-site collision at a supercoiled branch being the primary pathway for invertasome assembly.
D.12 The specificity for DNA inversion
The conformational energy of supercoiled DNA also mediates the large bias for promoting inversions over deletions between appropriately oriented recombination sites in cis. Formation of deletions by Hin is completely dependent upon Fis, but only about half of the Gin-catalyzed deletion reactions go through a Fis-dependent pathway (169). Significantly, all of the rare deletion products generated by Hin are catenated circles and in all analyzed cases were found to be linked only once (67). Alignment of directly-repeated recombination sites for deletion within an invertasome-like structure at a plectonemic branch demands that an additional loop must be introduced into the DNA (Fig. 16), which would be energetically costly. This alignment therefore occurs only rarely, but the product of a single clockwise rotation is the observed singly-interlinked catenane. The Hin-catalyzed deletion reaction is highly dependent upon HU, which fits with the additional DNA bending that is required to assemble the deletion complex.
Figure 16.
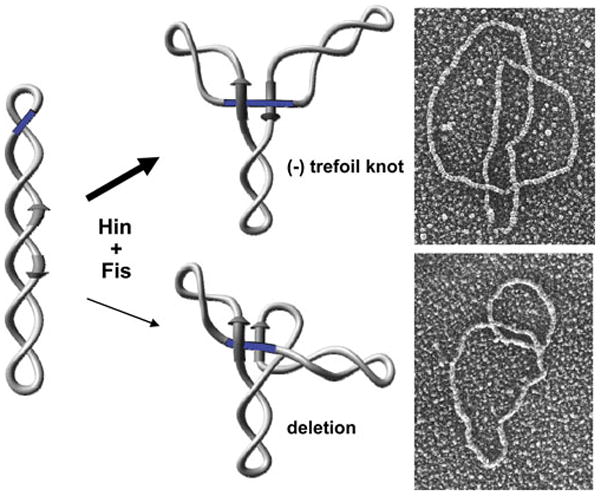
Products formed on substrates containing directly-repeated recombination sites. Most synaptic complexes assemble in an invertasome structure as diagrammed in the top pathway, but the recombination sites are in an antiparallel orientation. The unpaired core nucleotides after a single DNA exchange prevent ligation (Fig. 13C). Ligation after two exchanges results in a knot containing three negative nodes (Fig. 12), as shown in the electron micrograph of a trefoil generated by Hin. In the bottom pathway, which occurs rarely, the recombination sites assemble in parallel orientation that requires an additional DNA loop. This structure is energetically unfavorable on a negatively supercoiled substrate. A single DNA exchange results in a singly-linked catenated deletion product, as shown in the micrograph. The DNA molecules were coated with RecA protein to facilitate visualization of the DNA nodes.
Directly-repeated recombination sites overwhelmingly prefer to synapse in the standard invertasome configuration, but this generates an antiparallel orientation of sites (Fig. 16). An invertasome complex assembled with directly repeated recombination sites results in unpaired crossover nucleotides that cannot ligate after a single subunit rotation, and therefore must undergo an even number of rotations to enable ligation back into the parental orientation (Fig. 13C). In Hin- and Gin-catalyzed reactions both the robust rate of formation and structures of the knots produced by substrates with recombination sites oriented for deletion mimic the knotting behavior of substrates with mutant core sites in the standard inverted orientation (67, 95).
Further support for the supercoiling-directed alignment of recombination sites into active complexes comes from experiments employing sites in which the 2 bp crossover regions between recombining sites are internally symmetric (e.g., A/T-T/A). Recombination reactions between such sites overwhelmingly generate inversions and only rarely deletions, regardless of their relative orientations on the DNA molecule (67). These results confirm that site orientation is solely specified by the two nucleotides at the crossover region and that the configuration of sites that will generate inversion upon DNA exchange is preferentially assembled within the active synaptic complex (invertasome). Recombination site orientation by other serine recombinase reactions is also exclusively determined by the crossover dinucleotide sequence and tested only at the ligation step after DNA exchange (182–184).
D.13 Key residues controlling conformational changes: locations and properties of gain-of-function mutations that often lead to Fis/enhancer-independence
D.13.1 Identification and properties of Fis/enhancer-independent mutants
Gain-of-function mutants that exhibit altered regulatory properties have been isolated in the Hin, Gin and Cin serine invertases. The mutations identify important interactions within or between subunits that control the conformational rearrangements which occur during the dimer to tetramer remodeling reaction. As discussed above, strong Fis/enhancer-independent mutants of Hin and Gin have been invaluable for structural and mechanistic studies where regulatory controls imparted by the Fis/enhancer element and DNA supercoiling are unimportant. Moreover, comparison of the Fis/enhancer-dependent verses -independent reactions has provided valuable insights into the functions of the Fis/enhancer control element and DNA topological forces involved in regulating the specificity for inversion.
The first reported mutants that could function without the Fis/enhancer regulatory element were in Gin (H107Y, M115V, and weakly F105V) and Cin (H107Y and R72H) (Hin residue numbering is used throughout this discussion) (65, 169, 185). H107Y and M115V are also among the strongest Hin hyperactivating mutations, but changes in over 20 additional residues have been shown to confer altered regulatory properties (Fig. 3 and not shown) (106, 107, 109). Hin gain-of-function mutants have been isolated by genetic screens for mutants that (1) catalyze inversion in fis mutant hosts, (2) activate the cellular SOS DNA damage response, or (3) suppress inactive or weakly active Hin mutants, or by site-directed mutagenesis based on structure modeling or information from resolvase systems.
A hallmark of strong hyperactive mutants is their newfound ability to catalyze deletions and intermolecular fusions, in addition to promoting inversions, all without the requirement for Fis or the enhancer (65, 107, 169, 185). In vitro analyses also demonstrate that they no longer require DNA supercoiling and can synapse and recombine short oligonucleotide substrates containing their cognate recombination site sequence. They are further activated by the Fis/enhancer system provided the DNA is supercoiled, and under these conditions, recombination through an invertasome complex dominates (109, 169) (J. Heiss, Y. Chang, and R.C.J., unpublished).
D.13.2 DNA topological properties of Fis-independent reactions
Comprehensive topological studies on reactions catalyzed by the strong Fis-independent Gin-M115V and Hin-H107Y mutants have been performed with similar conclusions (101, 169) (S. Merickel, J. Heiss, and R.C.J., unpublished). Nearly all reactions on negatively supercoiled DNA in the presence of Fis initiate from a synaptic complex trapping two negative nodes indicative of the invertasome structure. However, unlike the wild-type enzymes, most reactions undergo multiple subunit rotations to generate a series of increasingly complex knotted products. The more processive nature of the subunit exchange reaction is probably due to the mutations stabilizing the active tetramer conformation (see below). In the absence of Fis, Gin-M115V and Hin-H107Y also preferentially promote recombination through a −2 synaptic complex to give a linking number change (ΔLK) of +4 after the first subunit exchange (169) and generate knotting profiles on substrates with wild type or mismatched recombination sites that reflect processive exchanges (101) (S. Merickel, J. Heiss, and R. Johnson, unpublished). However, unlike the wild-type enzymes, a diversity of other synaptic complex configurations also generates recombinant products. These include a substantial number of simple synapses between recombination sites on apposed supercoiled strands where no topological change occurs upon DNA exchange (ΔLK = 0 complex) and a lower number of complexes that trap from 3 to at least 12 (−) supercoiled nodes via random intramolecular collision pathways. Many of the diverse product structures generated by Gin-M115V have been confirmed by electron microscopy (101). Productive synapses on non-supercoiled substrates occur with equal efficiencies with recombination sites aligned in a parallel or antiparallel orientation (Fig. 13 A and C, respectively) (107, 169). Thus, in the absence of the Fis/enhancer system, the hyperactive mutants have lost the strict control of recombination site synapsis, and this enables all types of recombination products to occur, including those from intermolecular reactions. The overrepresentation of intramolecular recombination by synaptic complexes that trap two (−) nodes presumably reflects the propensity for branching within plectonemically supercoiled DNA and could also reflect the contribution of helix B – DNA interactions in promoting tetramer assembly at a branch.
D.13.3 Residues controlling conformational changes required for active tetramer assembly
The locations of regulatory mutations and the biochemical activities of the mutant proteins have identified key interactions between residues that control steps of the dimer-tetramer remodeling reaction. These include intrasubunit and intersubunit interactions that stabilize the dimer conformation, contacts that occur during the remodeling process, and contacts that are of particular importance in stabilizing the active tetramer conformation (109). Individual residues can participate in more than one step, and some of the hyperactivating mutations appear to gain new contacts that stabilize the pre-activated or activated tetramer. As might be expected, there is considerable overlap of positions where mutations generate hyperactive serine invertases and resolvases (186–189). A subset of the characterized DNA invertase hyperactivating mutations are discussed below focusing on those that have the largest effects and reveal particularly important features controlling the reaction. The proposed functions of the residues rely on structural models and morphed intermediates of Hin (78, 109), as discussed above, and the Gin tetramer structure (120).
D.13.3.1 Helix E residues involved in intermolecular interactions
Most of the strongest hyperactivating mutations involve residues on helix E, which undergoes extensive structural changes during the dimer-to-tetramer conversion. Whereas most mutations within helix E block steps after DNA binding, mutations in seven different residues can result in hyperactivating mutations. One of the strongest is H107Y that has been isolated in Hin, Gin, and Cin (65, 106, 185) and studied intensively in Hin (107, 118, 158). The histidine imidazole ring is predicted to be hydrogen bonded and in van der Waals contact with residues of the partner subunit on the surface of the dimer (Fig. 17A). The hyperactivating tyrosine side chain is predicted to clash or reorient into the solvent, consistent with the mild destabilization of the dimer interface by the mutant (109). Upon formation of the initial pre-activated tetramer, the mutant tyrosine rings between subunits of newly synapsed pairs are predicted to adopt a pi-stacked conformation (Fig. 17B), thereby stabilizing the tetrameric synaptic complex as experimentally observed. A glutamine to arginine substitution at the analogous position of the Sin recombinase also leads to a strong hyperactivating mutation, and a crystal structure of this mutant shows the arginine side chains from subunits of each rotating pair in a pi-stacked configuration (114). In the low resolution Gin tetramer X-ray structure, the native imidazole side chain is modeled to be packed against the helix E backbone (120). It seems likely that the larger tyrosine, with its greater pi-stacking potential (190, 191), adopts the alternate conformation. A phenylalanine at this position confers a milder hyperactive phenotype, but all other substitutions tested inactivate the invertase (65, 106, 185, 192).
Figure 17.
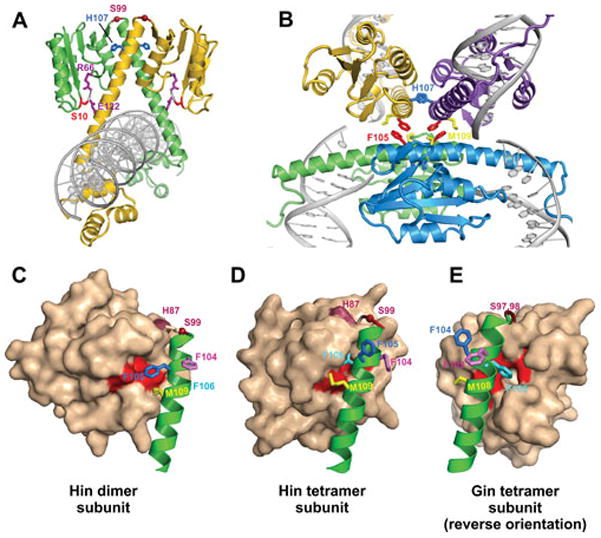
Residues regulating conformational transitions. A. Hin dimer model depicting some of the residues where mutations have been isolated that control the dimer-tetramer transition. B. Hin tetramer model (based on PDB: 2GM4) highlighting helix E residues Phe105 and Met109, whose side chains are located within the rotation interface and His107, whose side chain is predicted to stabilize the synaptic interface. C. Subunit from the Hin dimer model showing helix E residues Phe104, Phe105, Phe106 (behind helix E), and Met109. The surface of the catalytic domain is also shown highlighting the pocket organized by residues surrounding Phe88. Residues around the Phe88 pocket where mutations lead to Fis/enhancer-independence are colored red. D. Subunit from the Hin tetramer model rendered similarly as in panel C. Note that the side chains of Phe105 and Met109 have rotated away from the catalytic domain and are now within the rotation interface (panel B). Phe104 rotates away from the partner dimer subunit and becomes associated with the synaptic and rotation interfaces. Certain mutations at His87 (colored maroon) lead to strong Fis/enhancer-independence, and different substitutions of the hinge residue Ser99 generate various phenotypes (see text). E. Subunit from the Gin tetramer X-ray structure (PDB: 3UJ3) in the reverse orientation as shown in panel D. Numbering of residues in Gin is one less than Hin.
Methionine 115 to valine creates another very robust Fis/enhancer-independent mutant that has been studied extensively (65, 101, 109, 120, 169). During formation of the tetramer, the Met115 side chain is modeled to move out of a cleft in the dimer partner subunit, slide along the dimer interface, and insert into a new pocket organized by residues from the N-terminal end of helix E of the synaptic subunit partner to both stabilize the synaptic subunit pair and contribute to the rotating interface of the tetramer (109, 120) (see Movie 2 in ref. (109)). Hin mutations within residues of the dimer partner cleft (V71 and Leu64) and synaptic subunit pocket (M101) also lead to hyperactivity (109). In Hin, the M115V mutation strongly destabilizes dimeric interactions and results in efficient formation of DNA-cleaved tetrameric synaptic complexes. In contrast, M115I generates tetrameric synaptic complexes that are inefficiently cleaved and less able to support complete recombination without Fis/enhancer-activation. The mutant isoleucine side chain may impede latter rearrangements to the fully active tetramer conformation or alter the tetramer structure for the cleavage step.
D.13.3.2 The helix E phenylalanine triad and Phe88 catalytic domain pocket
An invariant triad of phenylalanines between residues 104–106 on helix E of serine invertases play a critical role in controlling the conformational changes required for formation of the active tetramer (109). In the dimer, Phe105 is inserted into a hydrophobic pocket of the catalytic domain of its own subunit (Fig. 17C). This pocket is organized by Phe88 and a series of surrounding hydrophobic residues; Hin hyperactivating mutations have been isolated in six of these, and in all cases, are changed to a smaller hydrophobic side chain. During tetramer formation, the Phe105 side chain rotates out and the Phe106 side chain inserts into this pocket (Fig. 17D and E) (see Movie 3 in ref. (109)). The Phe105 ring is modeled to become stacked on the Phe105 ring across the rotating interface from the diagonally-oriented subunit (Fig. 17B). In the Gin tetramer, which is rotated by 26°, the Phe105 rings have separated (120). Substitutions at Phe105 in Hin generate a wide range of phenotypes ranging from inactivity to strong hyperactivity (Y. Chang and R. Johnson, unpublished). Most of the active mutants have defects in ligation, consistent with a prominent role of Phe105 in stabilizing the DNA-aligned rotational conformer. All Phe106 substitutions are defective for steps subsequent to DNA binding, except for weak inversion activity by tyrosine (192) (Y. Chang and R. Johnson, unpublished). The Phe104 side chain ring switches from the dimer interface to the synaptic and rotating interfaces and all substitutions of this residue tested are also severely defective in post DNA binding steps (106, 192).
D.13.3.3 Met109 on helix E
Met109 on helix E is an invariant residue among serine invertases that is predicted to also engage in intrasubunit van der Waals contacts with its catalytic domain in the dimer (Fig. 17C). During tetramer formation, the Met109 side chain is repositioned into the rotating interface (Fig. 17B, D, and E). Most substitutions at residue 109 in Hin severely interfere with inversion, but a large subset generate cleaved DNA molecules, even in the absence of Fis, that are competent for subunit rotation but defective in ligation (106) (Y. Chang and R.C.J., unpublished). The Fis-independent cleavage activities of Met109 mutations can be rationalized by destabilization of the dimer, whereas the ligation defects presumably reflect perturbations of the rotation interface by the mutant side chains.
D.13.3.4 Additional regulatory dimeric contacts: residues Ser10, Arg66, and Glu122
Hin gain-of-function mutants point to another set of interactions between conserved residues that appear to stabilize the inactive dimer conformation. Mutations at Glu122 on helix E and at Arg66 within the active site region lead to efficient tetramer formation without Fis (109). The side chains of Glu122, Arg66, and the active site Ser10 have the potential to engage in hydrogen bonding between dimer subunits that would need to be broken very early during the remodeling process (Fig. 17A). One side of the γδ resolvase dimer crystal structure has the equivalent glutamic acid on helix E hydrogen bonded to the active site serine (111), and mutations of this glutamic acid also exhibit altered regulatory properties (119, 160, 162, 186, 189). Arg66 is believed to play a chemical role in the DNA cleavage/ligation reaction, consistent with the synaptic tetramers formed by Hin-R66D being inactive for DNA cleavage (109, 113, 114). The phenotypes of different substitutions at Hin residue 122 range from no activity after dimer DNA binding to efficient formation of uncleaved or cleaved tetramers. The diversity can be rationalized in light of the large number of transient interactions residue 122 is predicted to make during the transition to the tetramer.
D.13.3.5 The hinge residue Ser99
Serine 99 at the junction between the catalytic domain and helix E is modeled to be at the position where the pivoting and rotation occurs between these structural units during the dimer-tetramer remodeling (Fig. 17A, C–E) (109, 119). In some serine invertases, this residue is a threonine, and in γδ/Tn3 resolvase it is a glycine, but a serine (along with an aspartic acid to tyrosine change at the next residue) encourages formation of resolvase tetramers (189). Serine 99 has unfavorable peptide backbone angles in the Hin dimer models and thus may facilitate remodeling to the tetramer where the backbone angles are in the normal range. Consistent with this idea, Hin-S99G remains as an inactive dimer even under Fis/enhancer stimulation (109). On the other hand, Hin Ser99 mutants with lysine and arginine substitutions generate small numbers of uncleaved synaptic tetramers in reactions without Fis that exhibit slightly altered electrophoretic migrations from DNA-cleaved tetramers. These mutations synergize with other hyperactivating mutations to promote robust synaptic tetramer formation, but the resulting complexes remain blocked for DNA cleavage. These properties suggest that certain substitutions at position 99 facilitate early steps of tetramer formation, but cannot complete assembly of catalytically-competent tetramers in the absence of Fis.
Acknowledgments
I thank John Marko (Northwestern University) for discussions and comments on the manuscript. I am very grateful to the present and past members of my laboratory for their many contributions to work presented in this chapter. Research on recombination systems in my laboratory has been supported by the National Institutes of Health (GM038509).
References
- 1.Grindley ND, Whiteson KL, Rice PA. Mechanisms of site-specific recombination. Annu Rev Biochem. 2006;75:567–605. doi: 10.1146/annurev.biochem.73.011303.073908. [DOI] [PubMed] [Google Scholar]
- 2.Johnson RC. Bacterial site-specific DNA inversion systems. In: Craig NL, Craigie R, Gellert M, Lambowitz AM, editors. Mobile DNA II. ASM Press; Washington, D.C: 2002. pp. 230–271. [Google Scholar]
- 3.Rice PA. Title to be added. In: Craig NL, editor. Mobile DNA III. ASM Press; Washington, D.C: 2014. in press. [Google Scholar]
- 4.Stark WM. The serine recombinases. In: Craig NL, editor. Mobile DNA III. ASM Press; Washington, D.C: 2014. in press. [Google Scholar]
- 5.Andrewes FW. Studies in group-agglutination - The Salmonella group and its antigenic structure. J Pathology and Bacteriology. 1922;25:505–521. [Google Scholar]
- 6.Iino T. Genetics and chemistry of bacterial flagella. Bacteriol Rev. 1969;33:454–475. doi: 10.1128/br.33.4.454-475.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lederberg J, Edwards P. Serotypic recombination in Salmonella. J Immunol. 1953;71:323–340. [PubMed] [Google Scholar]
- 8.Lederberg J, Iino T. Phase variation in Salmonella. Genetics. 1956;41:743–757. doi: 10.1093/genetics/41.5.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stocker BAD. Measurement of the rate of mutation of flagellar antigenic phase in Salmonella typhimurium. J Hyg. 1949;47:398–413. doi: 10.1017/s002217240001473x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kutsukake K, Nakashima H, Tominaga A, Abo T. Two DNA invertases contribute to flagellar phase variation in Salmonella enterica serovar Typhimurium strain LT2. J Bacteriol. 2006;188:950–957. doi: 10.1128/JB.188.3.950-957.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gillen KL, Hughes KT. Negative regulatory loci coupling flagellin synthesis to flagellar assembly in Salmonella typhimurium. J Bacteriol. 1991;173:2301–2310. doi: 10.1128/jb.173.7.2301-2310.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ikeda JS, Schmitt CK, Darnell SC, Watson PR, Bispham J, Wallis TS, Weinstein DL, Metcalf ES, Adams P, O’Connor CD, O’Brien AD. Flagellar phase variation of Salmonella enterica serovar Typhimurium contributes to virulence in the murine typhoid infection model but does not influence Salmonella-induced enteropathogenesis. Infect Immun. 2001;69:3021–3030. doi: 10.1128/IAI.69.5.3021-3030.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zieg J, Silverman M, Hilmen M, Simon M. Recombinational switch for gene expression. Science. 1977;196:170–172. doi: 10.1126/science.322276. [DOI] [PubMed] [Google Scholar]
- 14.Zieg J, Hilmen M, Simon M. Regulation of gene expression by site-specific inversion. Cell. 1978;15:237–244. doi: 10.1016/0092-8674(78)90098-3. [DOI] [PubMed] [Google Scholar]
- 15.Zieg J, Simon M. Analysis of the nucleotide sequence of an invertible controlling element. Proc Natl Acad Sci USA. 1980;77:4196–4200. doi: 10.1073/pnas.77.7.4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Silverman M, Simon M. Phase variation: genetic analysis of switching mutants. Cell. 1980;19:845–854. doi: 10.1016/0092-8674(80)90075-6. [DOI] [PubMed] [Google Scholar]
- 17.Silverman M, Zieg J, Mandel G, Simon M. Analysis of the functional components of the phase variation system. Cold Spring Harb Symp Quant Biol. 1981;45:17–26. doi: 10.1101/sqb.1981.045.01.005. [DOI] [PubMed] [Google Scholar]
- 18.Osuna R, Lienau D, Hughes KT, Johnson RC. Sequence, regulation, and functions of fis in Salmonella typhimurium. J Bacteriol. 1995;177:2021–2032. doi: 10.1128/jb.177.8.2021-2032.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Silverman M, Zieg J, Simon M. Flagellar-phase variation: isolation of the rh1 gene. J Bacteriol. 1979;137:517–523. doi: 10.1128/jb.137.1.517-523.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aldridge PD, Wu C, Gnerer J, Karlinsey JE, Hughes KT, Sachs MS. Regulatory protein that inhibits both synthesis and use of the target protein controls flagellar phase variation in Salmonella enterica. Proc Natl Acad Sci USA. 2006;103:11340–11345. doi: 10.1073/pnas.0602127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamamoto S, Kutsukake K. FljA-mediated posttranscriptional control of phase 1 flagellin expression in flagellar phase variation of Salmonella enterica serovar Typhimurium. J Bacteriol. 2006;188:958–967. doi: 10.1128/JB.188.3.958-967.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bonifield HR, Hughes KT. Flagellar phase variation in Salmonella enterica is mediated by a posttranscriptional control mechanism. J Bacteriol. 2003;185:3567–3574. doi: 10.1128/JB.185.12.3567-3574.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson RC, Bruist MF, Simon MI. Host protein requirements for in vitro site-specific DNA inversion. Cell. 1986;46:531–539. doi: 10.1016/0092-8674(86)90878-0. [DOI] [PubMed] [Google Scholar]
- 24.Finkel SE, Johnson RC. The Fis protein: it’s not just for DNA inversion anymore. Mol Microbiol. 1992;6:3257–3265. doi: 10.1111/j.1365-2958.1992.tb02193.x. [DOI] [PubMed] [Google Scholar]
- 25.Johnson RC, Ball CA, Pfeffer D, Simon MI. Isolation of the gene encoding the Hin recombinational enhancer binding protein. Proc Natl Acad Sci USA. 1988;85:3484–3488. doi: 10.1073/pnas.85.10.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koch C, Kahmann R. Purification and properties of the Escherichia coli host factor required for inversion of the G segment in bacteriophage Mu. J Biol Chem. 1986;261:15673–15678. [PubMed] [Google Scholar]
- 27.Johnson RC, Simon MI. Hin-mediated site-specific recombination requires two 26 bp recombination sites and a 60 bp recombinational enhancer. Cell. 1985;41:781–791. doi: 10.1016/s0092-8674(85)80059-3. [DOI] [PubMed] [Google Scholar]
- 28.Haykinson MJ, Johnson RC. DNA looping and the helical repeat in vitro and in vivo: effect of HU protein and enhancer location on Hin invertasome assembly. EMBO J. 1993;12:2503–2512. doi: 10.1002/j.1460-2075.1993.tb05905.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bruist MF, Glasgow AC, Johnson RC, Simon MI. Fis binding to the recombinational enhancer of the Hin DNA inversion system. Genes Dev. 1987;1:762–772. doi: 10.1101/gad.1.8.762. [DOI] [PubMed] [Google Scholar]
- 30.Johnson RC, Bruist MB, Glaccum MB, Simon MI. In vitro analysis of Hin-mediated site-specific recombination. Cold Spring Harb Symp Quant Biol. 1984;49:751–760. doi: 10.1101/sqb.1984.049.01.085. [DOI] [PubMed] [Google Scholar]
- 31.Bruist MF, Simon MI. Phase variation and the Hin protein: in vivo activity measurements, protein overproduction, and purification. J Bacteriol. 1984;159:71–79. doi: 10.1128/jb.159.1.71-79.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ball CA, Osuna R, Ferguson KC, Johnson RC. Dramatic changes in Fis levels upon nutrient upshift in Escherichia coli. J Bacteriol. 1992;174:8043–8056. doi: 10.1128/jb.174.24.8043-8056.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lim HM, Simon MI. The role of negative supercoiling in Hin-mediated site-specific recombination. J Biol Chem. 1992;267:11176–11182. [PubMed] [Google Scholar]
- 34.Ó Cróinín T, Carroll RK, Kelly A, Dorman CJ. Roles for DNA supercoiling and the Fis protein in modulating expression of virulence genes during intracellular growth of Salmonella enterica serovar Typhimurium. Mol Microbiol. 2006;62:869–982. doi: 10.1111/j.1365-2958.2006.05416.x. [DOI] [PubMed] [Google Scholar]
- 35.Koch C, Mertens G, Rudt F, Kahmann R, Kanaar R, Plasterk RH, van de Putte P, Sandulache R, Kamp D. The invertible G segment. In: Symonds N, Toussaint A, van de Putte P, Howe MM, editors. Phage Mu. Cold Spring Harbor Laboratory; New York: Cold Spring Harbor: 1987. pp. 75–91. 0 ed. [Google Scholar]
- 36.Hiestand-Nauer R, Iida S. Sequence of the site-specific recombinase gene Cin and of its substrates serving in the inversion of the C segment of bacteriophage P1. EMBO J. 1983;2:1733–1740. doi: 10.1002/j.1460-2075.1983.tb01650.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chow LT, Bukhari AI. The invertible DNA segments of coliphages Mu and P1 are identical. Virology. 1976;74:242–248. doi: 10.1016/0042-6822(76)90148-3. [DOI] [PubMed] [Google Scholar]
- 38.Toussaint A, Lefebvre N, Scott JR, Cowan JA, de Bruijn F, Bukhari AI. Relationships between temperate phages Mu and P1. Virology. 1978;89:146–161. doi: 10.1016/0042-6822(78)90048-x. [DOI] [PubMed] [Google Scholar]
- 39.Grundy FJ, Howe MM. Involvement of the invertible G segment in bacteriophage Mu tail fiber biosynthesis. Virology. 1984;134:296–317. doi: 10.1016/0042-6822(84)90299-x. [DOI] [PubMed] [Google Scholar]
- 40.Howe MM, Schumm JW, Taylor AL. The S and U genes of bacteriophage Mu are located in the invertible G segment of Mu DNA. Virology. 1979;92:108–124. doi: 10.1016/0042-6822(79)90218-6. [DOI] [PubMed] [Google Scholar]
- 41.Giphart-Gassler M, Plasterk RH, van de Putte P. G inversion in bacteriophage Mu: a novel way of gene splicing. Nature. 1982;297:339–342. doi: 10.1038/297339a0. [DOI] [PubMed] [Google Scholar]
- 42.Kamp D, Kahmann R, Zipser D, Broker TR, Chow LT. Inversion of the G DNA segment of phage Mu controls phage infectivity. Nature. 1978;271:577–580. doi: 10.1038/271577a0. [DOI] [PubMed] [Google Scholar]
- 43.van de Putte P, Cramer S, Giphart-Gassler M. Invertible DNA determines host specificity of bacteriophage Mu. Nature. 1980;286:218–222. doi: 10.1038/286218a0. [DOI] [PubMed] [Google Scholar]
- 44.Plasterk RH, Brinkman A, van de Putte P. DNA inversions in the chromosome of Escherichia coli and in bacteriophage Mu: relationship to other site-specific recombination systems. Proc Natl Acad Sci USA. 1983;80:5355–5358. doi: 10.1073/pnas.80.17.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Iida S, Meyer J, Kennedy KE, Arber W. A site-specific, conservative recombination system carried by bacteriophage P1. Mapping the recombinase gene cin and the cross-over sites cix for the inversion of the C segment. EMBO J. 1982;1:1445–1453. doi: 10.1002/j.1460-2075.1982.tb01336.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Iida S, Huber H, Hiestand-Nauer R, Meyer J, Bickle TA, Arber W. The bacteriophage P1 site-specific recombinase Cin: recombination events and DNA recognition sequences. Cold Spring Harb Symp Quant Biol. 1984;49:769–777. doi: 10.1101/sqb.1984.049.01.087. [DOI] [PubMed] [Google Scholar]
- 47.Kahmann R, Rudt F, Koch C, Mertens G. G inversion in bacteriophage Mu DNA is stimulated by a site within the invertase gene and a host factor. Cell. 1985;41:771–780. doi: 10.1016/s0092-8674(85)80058-1. [DOI] [PubMed] [Google Scholar]
- 48.Huber HE, Iida S, Arber W, Bickle TA. Site-specific DNA inversion is enhanced by a DNA sequence element in cis. Proc Nat Acad Sci USA. 1985;82:3776–3780. doi: 10.1073/pnas.82.11.3776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hubner P, Arber W. Mutational analysis of a prokaryotic recombinational enhancer element with two functions. EMBO J. 1989;8:577–585. doi: 10.1002/j.1460-2075.1989.tb03412.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Haffter P, Bickle TA. Purification and DNA-binding properties of FIS and Cin, two proteins required for the bacteriophage P1 site-specific recombination system, cin. J Mol Biol. 1987;198:579–587. doi: 10.1016/0022-2836(87)90201-4. [DOI] [PubMed] [Google Scholar]
- 51.Koch C, Vandekerckhove J, Kahmann R. Escherichia coli host factor for site-specific DNA inversion: cloning and characterization of the fis gene. Proc Natl Acad Sci USA. 1988;85:4237–4241. doi: 10.1073/pnas.85.12.4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kanaar R, van Hal JP, van de Putte P. The recombinational enhancer for DNA inversion functions independent of its orientation as a consequence of dyad symmetry in the Fis-DNA complex. Nucleic Acids Res. 1989;17:6043–6053. doi: 10.1093/nar/17.15.6043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Plasterk RH, Ilmer TA, Van de Putte P. Site-specific recombination by Gin of bacteriophage Mu: inversions and deletions. Virology. 1983;127:24–36. doi: 10.1016/0042-6822(83)90367-7. [DOI] [PubMed] [Google Scholar]
- 54.Symonds N, Coelho A. Role of the G segment in the growth of phage Mu. Nature. 1978;271:573–574. doi: 10.1038/271573a0. [DOI] [PubMed] [Google Scholar]
- 55.Kahmann R, Rudt F, Mertens G. Substrate and enzyme requirements for in vitro site-specific recombination in bacteriophage Mu. Cold Spring Harb Symp Quant Biol. 1984;49:285–294. doi: 10.1101/sqb.1984.049.01.034. [DOI] [PubMed] [Google Scholar]
- 56.Plasterk RH, van de Putte P. Inversion of DNA in vivo and in vitro by Gin and Pin proteins. Cold Spring Harb Symp Quant Biol. 1984;49:295–300. doi: 10.1101/sqb.1984.049.01.035. [DOI] [PubMed] [Google Scholar]
- 57.Komano T. Shufflons: multiple inversion systems and integrons. Annu Rev Genet. 1999;33:171–191. doi: 10.1146/annurev.genet.33.1.171. [DOI] [PubMed] [Google Scholar]
- 58.Iida S, Sandmeier H, Hubner P, Hiestand-Nauer R, Schneitz K, Arber W. The Min DNA inversion enzyme of plasmid p15B of Escherichia coli 15T-: a new member of the Din family of site-specific recombinases. Mol Microbiol. 1990;4:991–997. doi: 10.1111/j.1365-2958.1990.tb00671.x. [DOI] [PubMed] [Google Scholar]
- 59.Sandmeier H, Iida S, Hubner P, Hiestand-Nauer R, Arber W. Gene organization in the multiple DNA inversion region min of plasmid p15B of E. coli 15T-: assemblage of a variable gene. Nucleic Acids Res. 1991;19:5831–5838. doi: 10.1093/nar/19.21.5831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sandmeier H, Iida S, Meyer J, Hiestand-Nauer R, Arber W. Site-specific DNA recombination system Min of plasmid p15B: a cluster of overlapping invertible DNA segments. Proc Natl Acad Sci USA. 1990;87:1109–1113. doi: 10.1073/pnas.87.3.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kamp D, Kahmann R. The relationship of two invertible segments in bacteriophage Mu and Salmonella typhimurium DNA. Mol Gen Genet. 1981;184:564–566. doi: 10.1007/BF00352543. [DOI] [PubMed] [Google Scholar]
- 62.Kutsukake K, Iino T. Inversions of specific DNA segments in flagellar phase variation of Salmonella and inversion systems of bacteriophages P1 and Mu. Proc Natl Acad Sci USA. 1980;77:7338–7341. doi: 10.1073/pnas.77.12.7338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kutsukake K, Nakao T, Iino T. A gene for DNA invertase and an invertible DNA in Escherichia coli K-12. Gene. 1985;34:343–350. doi: 10.1016/0378-1119(85)90143-x. [DOI] [PubMed] [Google Scholar]
- 64.van de Putte P, Plasterk R, Kuijpers A. A Mu Gin complementing function and an invertible DNA region in Escherichia coli K-12 are situated on the genetic element e14. J Bacteriol. 1984;158:517–522. doi: 10.1128/jb.158.2.517-522.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Klippel A, Cloppenborg K, Kahmann R. Isolation and characterization of unusual Gin mutants. EMBO J. 1988;7:3983–3989. doi: 10.1002/j.1460-2075.1988.tb03286.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Scott TN, Simon MI. Genetic analysis of the mechanism of the Salmonella phase variation site specific recombination system. Mol Gen Genet. 1982;188:313–321. doi: 10.1007/BF00332694. [DOI] [PubMed] [Google Scholar]
- 67.Moskowitz IP, Heichman KA, Johnson RC. Alignment of recombination sites in Hin-mediated site-specific DNA recombination. Genes Dev. 1991;5:1635–1645. doi: 10.1101/gad.5.9.1635. [DOI] [PubMed] [Google Scholar]
- 68.Kennedy KE, Iida S, Meyer J, Stalhammar-Carlemalm M, Hiestand-Nauer R, Arber W. Genome fusion mediated by the site specific DNA inversion system of bacteriophage P1. Mol Gen Genet. 1983;189:413–421. doi: 10.1007/BF00325903. [DOI] [PubMed] [Google Scholar]
- 69.Cerdeno-Tarraga AM, Patrick S, Crossman LC, Blakely G, Abratt V, Lennard N, Poxton I, Duerden B, Harris B, Quail MA, Barron A, Clark L, Corton C, Doggett J, Holden MT, Larke N, Line A, Lord A, Norbertczak H, Ormond D, Price C, Rabbinowitsch E, Woodward J, Barrell B, Parkhill J. Extensive DNA inversions in the B. fragilis genome control variable gene expression. Science. 2005;307:1463–1465. doi: 10.1126/science.1107008. [DOI] [PubMed] [Google Scholar]
- 70.Kuwahara T, Yamashita A, Hirakawa H, Nakayama H, Toh H, Okada N, Kuhara S, Hattori M, Hayashi T, Ohnishi Y. Genomic analysis of Bacteroides fragilis reveals extensive DNA inversions regulating cell surface adaptation. Proc Natl Acad Sci USA. 2004;101:14919–14924. doi: 10.1073/pnas.0404172101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Patrick S, Parkhill J, McCoy LJ, Lennard N, Larkin MJ, Collins M, Sczaniecka M, Blakely G. Multiple inverted DNA repeats of Bacteroides fragilis that control polysaccharide antigenic variation are similar to the hin region inverted repeats of Salmonella typhimurium. Microbiology. 2003;149:915–924. doi: 10.1099/mic.0.26166-0. [DOI] [PubMed] [Google Scholar]
- 72.Krinos CM, Coyne MJ, Weinacht KG, Tzianabos AO, Kasper DL, Comstock LE. Extensive surface diversity of a commensal microorganism by multiple DNA inversions. Nature. 2001;414:555–558. doi: 10.1038/35107092. [DOI] [PubMed] [Google Scholar]
- 73.Coyne MJ, Weinacht KG, Krinos CM, Comstock LE. Mpi recombinase globally modulates the surface architecture of a human commensal bacterium. Proc Natl Acad Sci USA. 2003;100:10446–10451. doi: 10.1073/pnas.1832655100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fletcher CM, Coyne MJ, Bentley DL, Villa OF, Comstock LE. Phase-variable expression of a family of glycoproteins imparts a dynamic surface to a symbiont in its human intestinal ecosystem. Proc Natl Acad Sci USA. 2007;104:2413–2418. doi: 10.1073/pnas.0608797104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Simon M, Zieg J, Silverman M, Mandel G, Doolittle R. Phase variation: evolution of a controlling element. Science. 1980;209:1370–1374. doi: 10.1126/science.6251543. [DOI] [PubMed] [Google Scholar]
- 76.Smith M, Thorpe H. Diversity in the serine recombinases. Mol Micro. 2002;44:299–307. doi: 10.1046/j.1365-2958.2002.02891.x. [DOI] [PubMed] [Google Scholar]
- 77.Liu CC, Huhne R, Tu J, Lorbach E, Droge P. The resolvase encoded by Xanthomonas campestris transposable element ISXc5 constitutes a new subfamily closely related to DNA invertases. Genes Cells. 1998;3:221–233. doi: 10.1046/j.1365-2443.1998.00182.x. [DOI] [PubMed] [Google Scholar]
- 78.McLean MM, Chang Y, Dhar G, Heiss JK, Johnson RC. Multiple interfaces between a serine recombinase and an enhancer control site-specific DNA inversion. Elife. 2013;2:e01211. doi: 10.7554/eLife.01211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Canosa I, Lurz R, Rojo F, Alonso JC. beta Recombinase catalyzes inversion and resolution between two inversely oriented six sites on a supercoiled DNA substrate and only inversion on relaxed or linear substrates. J Biol Chem. 1998;273:13886–13891. doi: 10.1074/jbc.273.22.13886. [DOI] [PubMed] [Google Scholar]
- 80.Janniere L, McGovern S, Pujol C, Petit MA, Ehrlich SD. In vivo analysis of the plasmid pAM beta 1 resolution system. Nucleic Acids Res. 1996;24:3431–3436. doi: 10.1093/nar/24.17.3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rojo F, Alonso JC. The beta recombinase from the Streptococcal plasmid pSM19035 represses its own transcription by holding the RNA polymerase at the promoter region. Nucleic Acids Res. 1994;22:1855–1860. doi: 10.1093/nar/22.10.1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rowland SJ, Dyke KG. Characterization of the staphylococcal beta-lactamase transposon Tn552. EMBO J. 1989;8:2761–2773. doi: 10.1002/j.1460-2075.1989.tb08418.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rowland SJ, Dyke KG. Tn552, a novel transposable element from Staphylococcus aureus. Mol Microbiol. 1990;4:961–975. doi: 10.1111/j.1365-2958.1990.tb00669.x. [DOI] [PubMed] [Google Scholar]
- 84.Alonso JC, Gutierrez C, Rojo F. The role of chromatin-associated protein Hbsu in beta-mediated DNA recombination is to facilitate the joining of distant recombination sites. Mol Microbiol. 1995;18:471–478. doi: 10.1111/j.1365-2958.1995.mmi_18030471.x. [DOI] [PubMed] [Google Scholar]
- 85.Alonso JC, Weise F, Rojo F. The Bacillus subtilis histone-like protein Hbsu is required for DNA resolution and DNA inversion mediated by the beta recombinase of plasmid pSM19035. J Biol Chem. 1995;270:2938–2945. doi: 10.1074/jbc.270.7.2938. [DOI] [PubMed] [Google Scholar]
- 86.Smith MC, Brown WR, McEwan AR, Rowley PA. Site-specific recombination by phiC31 integrase and other large serine recombinases. Biochem Soc Trans. 2010;38:388–394. doi: 10.1042/BST0380388. [DOI] [PubMed] [Google Scholar]
- 87.Smith MC. Title to be added. In: Craig NL, editor. Mobile DNA III. ASM Press; Washington, D.C: 2014. in press. [Google Scholar]
- 88.Van Duyne GD, Rutherford K. Large serine recombinase domain structure and attachment site binding. Crit Rev Biochem Mol Biol. 2013;48:476–491. doi: 10.3109/10409238.2013.831807. [DOI] [PubMed] [Google Scholar]
- 89.Rutherford K, Yuan P, Perry K, Sharp R, Van Duyne GD. Attachment site recognition and regulation of directionality by the serine integrases. Nucleic Acids Res. 2013;41:8341–8356. doi: 10.1093/nar/gkt580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rutherford K, Van Duyne GD. The ins and outs of serine integrase site-specific recombination. Curr Opin Struct Biol. 2014;24:125–131. doi: 10.1016/j.sbi.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kersulyte D, Mukhopadhyay AK, Shirai M, Nakazawa T, Berg DE. Functional organization and insertion specificity of IS607, a chimeric element of Helicobacter pylori. J Bacteriol. 2000;182:5300–5308. doi: 10.1128/jb.182.19.5300-5308.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Boocock MR, Rice PA. A proposed mechanism for IS607-family serine transposases. Mob DNA. 2013;4:24. doi: 10.1186/1759-8753-4-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kanaar R, van de Putte P, Cozzarelli NR. Purification of the Gin recombination protein of Escherichia coli phage Mu and its host factor. Biochim Biophys Acta. 1986;866:170–177. doi: 10.1016/0167-4781(86)90114-4. [DOI] [PubMed] [Google Scholar]
- 94.Mertens G, Fuss H, Kahmann R. Purification and properties of the DNA invertase Gin encoded by bacteriophage Mu. J Biol Chem. 1986;261:15668–15672. [PubMed] [Google Scholar]
- 95.Kanaar R, Klippel A, Shekhtman E, Dungan JM, Kahmann R, Cozzarelli NR. Processive recombination by the phage Mu Gin system: implications for the mechanisms of DNA strand exchange, DNA site alignment, and enhancer action. Cell. 1990;62:353–366. doi: 10.1016/0092-8674(90)90372-l. [DOI] [PubMed] [Google Scholar]
- 96.Plasterk RH, Kanaar R, van de Putte P. A genetic switch in vitro: DNA inversion by Gin protein of phage Mu. Proc Natl Acad Sci USA. 1984;81:2689–2692. doi: 10.1073/pnas.81.9.2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mertens G, Hoffmann A, Blocker H, Frank R, Kahmann R. Gin-mediated site-specific recombination in bacteriophage Mu DNA: overproduction of the protein and inversion in vitro. EMBO J. 1984;3:2415–2421. doi: 10.1002/j.1460-2075.1984.tb02148.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Klippel A, Mertens G, Patschinsky T, Kahmann R. The DNA invertase Gin of phage Mu: formation of a covalent complex with DNA via a phosphoserine at amino acid position 9. EMBO J. 1988;7:1229–1237. doi: 10.1002/j.1460-2075.1988.tb02935.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chiu TK, Sohn C, Dickerson RE, Johnson RC. Testing water-mediated DNA recognition by the Hin recombinase. EMBO J. 2002;21:801–814. doi: 10.1093/emboj/21.4.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Johnson RC, Bruist MF. Intermediates in Hin-mediated DNA inversion: a role for Fis and the recombinational enhancer in the strand exchange reaction. EMBO J. 1989;8:1581–1590. doi: 10.1002/j.1460-2075.1989.tb03542.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Crisona NJ, Kanaar R, Gonzalez TN, Zechiedrich EL, Klippel A, Cozzarelli NR. Processive recombination by wild-type Gin and an enhancer-independent mutant. Insight into the mechanisms of recombination selectivity and strand exchange. J Mol Biol. 1994;243:437–457. doi: 10.1006/jmbi.1994.1671. [DOI] [PubMed] [Google Scholar]
- 102.Reed RR, Moser CD. Resolvase-mediated recombination intermediates contain a serine residue covalently linked to DNA. Cold Spring Harbor Symp Quant Biol. 1984;49:245–249. doi: 10.1101/sqb.1984.049.01.028. [DOI] [PubMed] [Google Scholar]
- 103.Merickel SK, Haykinson MJ, Johnson RC. Communication between Hin recombinase and Fis regulatory subunits during coordinate activation of Hin-catalyzed site-specific DNA inversion. Genes and Dev. 1998;12:2803–2816. doi: 10.1101/gad.12.17.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Heichman KA, Johnson RC. The Hin invertasome: protein-mediated joining of distant recombination sites at the enhancer. Science. 1990;249:511–517. doi: 10.1126/science.2166334. [DOI] [PubMed] [Google Scholar]
- 105.Kanaar R, Cozzarelli NR. Roles of supercoiled DNA structure in DNA transactions. Cur Opin Struct Biol. 1992;2:369–379. [Google Scholar]
- 106.Haykinson MJ, Johnson LM, Soong J, Johnson RC. The Hin dimer interface is critical for Fis-mediated activation of the catalytic steps of site-specific DNA inversion. Curr Biol. 1996;6:163–177. doi: 10.1016/s0960-9822(02)00449-9. [DOI] [PubMed] [Google Scholar]
- 107.Sanders ER, Johnson RC. Stepwise dissection of the Hin-catalyzed recombination reaction from synapsis to resolution. J Mol Biol. 2004;340:753–766. doi: 10.1016/j.jmb.2004.05.027. [DOI] [PubMed] [Google Scholar]
- 108.Glasgow AC, Bruist MF, Simon MI. DNA-binding properties of the Hin recombinase. J Biol Chem. 1989;264:10072–10082. [PubMed] [Google Scholar]
- 109.Heiss JK, Sanders ER, Johnson RC. Intrasubunit and intersubunit interactions controlling assembly of active synaptic complexes during Hin-catalyzed DNA recombination. J Mol Biol. 2011;411:744–764. doi: 10.1016/j.jmb.2011.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Mertens G, Klippel A, Fuss H, Blocker H, Frank R, Kahmann R. Site-specific recombination in bacteriophage Mu: characterization of binding sites for the DNA invertase Gin. EMBO J. 1988;7:1219–1227. doi: 10.1002/j.1460-2075.1988.tb02934.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yang W, Steitz TA. Crystal structure of the site-specific recombinase gammadelta resolvase complexed with a 34 bp cleavage site. Cell. 1995;82:193–207. doi: 10.1016/0092-8674(95)90307-0. [DOI] [PubMed] [Google Scholar]
- 112.Mouw KW, Rowland SJ, Gajjar MM, Boocock MR, Stark WM, Rice PA. Architecture of a serine recombinase-DNA regulatory complex. Mol Cell. 2008;30:145–155. doi: 10.1016/j.molcel.2008.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Olorunniji FJ, Stark WM. The catalytic residues of Tn3 resolvase. Nucleic Acids Res. 2009;37:7590–7602. doi: 10.1093/nar/gkp797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Keenholtz RA, Rowland SJ, Boocock MR, Stark WM, Rice PA. Structural basis for catalytic activation of a serine recombinase. Structure. 2011;19:799–809. doi: 10.1016/j.str.2011.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Nanassy OZ, Hughes KT. In vivo identification of intermediate stages of the DNA inversion reaction catalyzed by the Salmonella Hin recombinase. Genetics. 1998;149:1649–1663. doi: 10.1093/genetics/149.4.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Adams CW, Nanassy O, Johnson RC, Hughes KT. Role of arginine-43 and arginine-69 of the Hin recombinase catalytic domain in the binding of Hin to the hix DNA recombination sites. Mol Microbiol. 1997;24:1235–1247. doi: 10.1046/j.1365-2958.1997.4141789.x. [DOI] [PubMed] [Google Scholar]
- 117.Dhar G, Heiss JK, Johnson RC. Mechanical constraints on Hin subunit rotation imposed by the Fis/enhancer system and DNA supercoiling during site-specific recombination. Mol Cell. 2009;34:746–759. doi: 10.1016/j.molcel.2009.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Dhar G, McLean MM, Heiss JK, Johnson RC. The Hin recombinase assembles a tetrameric protein swivel that exchanges DNA strands. Nucleic Acids Res. 2009;37:4743–4756. doi: 10.1093/nar/gkp466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Li W, Kamtekar S, Xiong Y, Sarkis GJ, Grindley ND, Steitz TA. Structure of a synaptic gamma delta resolvase tetramer covalently linked to two cleaved DNAs. Science. 2005;309:1210–1215. doi: 10.1126/science.1112064. [DOI] [PubMed] [Google Scholar]
- 120.Ritacco CJ, Kamtekar S, Wang J, Steitz TA. Crystal structure of an intermediate of rotating dimers within the synaptic tetramer of the G-segment invertase. Nucleic Acids Res. 2013;41:2673–2682. doi: 10.1093/nar/gks1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Bruist MF, Horvath SJ, Hood LE, Steitz TA, Simon MI. Synthesis of a site-specific DNA-binding peptide. Science. 1987;235:777–780. doi: 10.1126/science.3027895. [DOI] [PubMed] [Google Scholar]
- 122.Feng JA, Johnson RC, Dickerson RE. Hin recombinase bound to DNA: the origin of specificity in major and minor groove interactions. Science. 1994;263:348–355. doi: 10.1126/science.8278807. [DOI] [PubMed] [Google Scholar]
- 123.Hughes KT, Gaines PC, Karlinsey JE, Vinayak R, Simon MI. Sequence-specific interaction of the Salmonella Hin recombinase in both major and minor grooves of DNA. EMBO J. 1992;11:2695–2705. doi: 10.1002/j.1460-2075.1992.tb05335.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Sluka JP, Horvath SJ, Glasgow AC, Simon MI, Dervan PB. Importance of minor-groove contacts for recognition of DNA by the binding domain of Hin recombinase. Biochemistry. 1990;29:6551–6561. doi: 10.1021/bi00480a002. [DOI] [PubMed] [Google Scholar]
- 125.Hughes KT, Youderian P, Simon MI. Phase variation in Salmonella: analysis of Hin recombinase and hix recombination site interaction in vivo. Genes Dev. 1988;2:937–948. doi: 10.1101/gad.2.8.937. [DOI] [PubMed] [Google Scholar]
- 126.Ritacco CJ, Steitz TA, Wang J. Exploiting large non-isomorphous differences for phase determination of a G-segment invertase-DNA complex. Acta Crystallogr D Biol Crystallogr. 2014;70:685–693. doi: 10.1107/S1399004713032392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kanaar R, van de Putte P, Cozzarelli NR. Gin-mediated recombination of catenated and knotted DNA substrates: implications for the mechanism of interaction between cis-acting sites. Cell. 1989;58:147–159. doi: 10.1016/0092-8674(89)90411-x. [DOI] [PubMed] [Google Scholar]
- 128.Surette MG, Chaconas G. The Mu transpositional enhancer can function in trans: requirement of the enhancer for synapsis but not strand cleavage. Cell. 1992;68:1101–1108. doi: 10.1016/0092-8674(92)90081-m. [DOI] [PubMed] [Google Scholar]
- 129.Pan CQ, Finkel SE, Cramton SE, Feng JA, Sigman DS, Johnson RC. Variable structures of Fis-DNA complexes determined by flanking DNA-protein contacts. J Mol Biol. 1996;264:675–695. doi: 10.1006/jmbi.1996.0669. [DOI] [PubMed] [Google Scholar]
- 130.Stella S, Cascio D, Johnson RC. The shape of the DNA minor groove directs binding by the DNA-bending protein Fis. Genes Dev. 2010;24:814–826. doi: 10.1101/gad.1900610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Johnson RC, Glasgow AC, Simon MI. Spatial relationship of the Fis binding sites for Hin recombinational enhancer activity. Nature. 1987;329:462–465. doi: 10.1038/329462a0. [DOI] [PubMed] [Google Scholar]
- 132.Shao Y, Feldman-Cohen LS, Osuna R. Functional characterization of the Escherichia coli Fis-DNA binding sequence. J Mol Biol. 2008;376:771–785. doi: 10.1016/j.jmb.2007.11.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kahramanoglou C, Seshasayee AS, Prieto AI, Ibberson D, Schmidt S, Zimmermann J, Benes V, Fraser GM, Luscombe NM. Direct and indirect effects of H-NS and Fis on global gene expression control in Escherichia coli. Nucleic Acids Res. 2011;39:2073–2091. doi: 10.1093/nar/gkq934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Cho BK, Knight EM, Barrett CL, Palsson BO. Genome-wide analysis of Fis binding in Escherichia coli indicates a causative role for A-/AT-tracts. Genome Res. 2008;18:900–910. doi: 10.1101/gr.070276.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Yuan HS, Finkel SE, Feng JA, Kaczor-Grzeskowiak M, Johnson RC, Dickerson RE. The molecular structure of wild-type and a mutant Fis protein: relationship between mutational changes and recombinational enhancer function or DNA binding. Proc Natl Acad Sci USA. 1991;88:9558–9562. doi: 10.1073/pnas.88.21.9558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kostrewa D, Granzin J, Koch C, Choe HW, Raghunathan S, Wolf W, Labahn J, Kahmann R, Saenger W. Three-dimensional structure of the E. coli DNA-binding protein FIS. Nature. 1991;349:178–180. doi: 10.1038/349178a0. [DOI] [PubMed] [Google Scholar]
- 137.Safo MK, Yang WZ, Corselli L, Cramton SE, Yuan HS, Johnson RC. The transactivation region of the Fis protein that controls site-specific DNA inversion contains extended mobile beta-hairpin arms. EMBO J. 1997;16:6860–6873. doi: 10.1093/emboj/16.22.6860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Cheng YS, Yang WZ, Johnson RC, Yuan HS. Structural analysis of the transcriptional activation region on Fis: crystal structures of six Fis mutants with different activation properties. J Mol Biol. 2000;302:1139–1151. doi: 10.1006/jmbi.2000.4123. [DOI] [PubMed] [Google Scholar]
- 139.Osuna R, Finkel SE, Johnson RC. Identification of two functional regions in Fis: the N-terminus is required to promote Hin-mediated DNA inversion but not lambda excision. EMBO J. 1991;10:1593–1603. doi: 10.1002/j.1460-2075.1991.tb07680.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Koch C, Ninnemann O, Fuss H, Kahmann R. The N-terminal part of the E. coli DNA binding protein FIS is essential for stimulating site-specific DNA inversion but is not required for specific DNA binding. Nucleic Acids Res. 1991;19:5915–5922. doi: 10.1093/nar/19.21.5915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Hancock SP, Ghane T, Cascio D, Rohs R, Di Felice R, Johnson RC. Control of DNA minor groove width and Fis protein binding by the purine 2-amino group. Nucleic Acids Res. 2013;41:6750–6760. doi: 10.1093/nar/gkt357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Perkins-Balding D, Dias DP, Glasgow AC. Location, degree, and direction of DNA bending associated with the Hin recombinational enhancer sequence and Fis-enhancer complex. J Bacteriol. 1997;179:4747–4753. doi: 10.1128/jb.179.15.4747-4753.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Johnson RC, Johnson LM, Schmidt JW, Gardner JF. Major nucleoid proteins in the structure and function of the Escherichia coli chromosome. In: Higgins NP, editor. The Bacterial Chromosome. ASM Press; Washington, D.C: 2005. pp. 65–132. [Google Scholar]
- 144.Browning DF, Grainger DC, Busby SJ. Effects of nucleoid-associated proteins on bacterial chromosome structure and gene expression. Curr Opin Microbiol. 2010;13:773–780. doi: 10.1016/j.mib.2010.09.013. [DOI] [PubMed] [Google Scholar]
- 145.Luijsterburg MS, Noom MC, Wuite GJ, Dame RT. The architectural role of nucleoid-associated proteins in the organization of bacterial chromatin: A molecular perspective. J Struct Biol. 2006;156:262–272. doi: 10.1016/j.jsb.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 146.Dillon SC, Dorman CJ. Bacterial nucleoid-associated proteins, nucleoid structure and gene expression. Nat Rev Microbiol. 2010;8:185–195. doi: 10.1038/nrmicro2261. [DOI] [PubMed] [Google Scholar]
- 147.Skoko D, Yan J, Johnson RC, Marko JF. Low-force DNA condensation and discontinuous high-force decondensation reveal a loop-stabilizing function of the protein Fis. Phys Rev Lett. 2005;95:208101. doi: 10.1103/PhysRevLett.95.208101. [DOI] [PubMed] [Google Scholar]
- 148.Skoko D, Yoo D, Bai H, Schnurr B, Yan J, McLeod SM, Marko JF, Johnson RC. Mechanism of chromosome compaction and looping by the Escherichia coli nucleoid protein Fis. J Mol Biol. 2006;364:777–798. doi: 10.1016/j.jmb.2006.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Ishihama A, Kori A, Koshio E, Yamada K, Maeda H, Shimada T, Makinoshima H, Iwata A, Fujita N. Intracellular concentrations of 65 species of transcription factors with known regulatory functions in Escherichia coli. J Bacteriol. 2014;196:2718–2727. doi: 10.1128/JB.01579-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Schneider R, Travers A, Kutateladze T, Muskhelishvili G. A DNA architectural protein couples cellular physiology and DNA topology in Escherichia coli. Mol Microbiol. 1999;34:953–964. doi: 10.1046/j.1365-2958.1999.01656.x. [DOI] [PubMed] [Google Scholar]
- 151.Weinstein-Fischer D, Elgrably-Weiss M, Altuvia S. Escherichia coli response to hydrogen peroxide: a role for DNA supercoiling, topoisomerase I and Fis. Mol Microbiol. 2000;35:1413–1420. doi: 10.1046/j.1365-2958.2000.01805.x. [DOI] [PubMed] [Google Scholar]
- 152.Richardson SM, Boles TC, Cozzarelli NR. The helical repeat of underwound DNA in solution. Nucleic Acids Res. 1988;16:6607–6616. doi: 10.1093/nar/16.14.6607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Bellomy GR, Record MTJ. Stable DNA loops in vivo and in vitro: Roles in gene regulation at a distance and in biophysical characterization of DNA. Prog Nucleic Acid Res Mol Biol. 1990;39:81–127. doi: 10.1016/s0079-6603(08)60624-8. [DOI] [PubMed] [Google Scholar]
- 154.Hillyard DR, Edlund M, Hughes KT, Marsh M, Higgins NP. Subunit-specific phenotypes of Salmonella typhimurium HU mutants. J Bacteriol. 1990;172:5402–5407. doi: 10.1128/jb.172.9.5402-5407.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Wada M, Kutsukake K, Komano T, Imamoto F, Kano Y. Participation of the hup gene product in site-specific DNA inversion in Escherichia coli. Gene. 1989;76:345–352. doi: 10.1016/0378-1119(89)90174-1. [DOI] [PubMed] [Google Scholar]
- 156.Paull TT, Haykinson MJ, Johnson RC. The nonspecific DNA-binding and -bending proteins HMG1 and HMG2 promote the assembly of complex nucleoprotein structures. Genes Dev. 1993;7:1521–1534. doi: 10.1101/gad.7.8.1521. [DOI] [PubMed] [Google Scholar]
- 157.Paull TT, Johnson RC. DNA looping by Saccharomyces cerevisiae high mobility group proteins NHP6A/B. Consequences for nucleoprotein complex assembly and chromatin condensation. J Biol Chem. 1995;270:8744–8754. doi: 10.1074/jbc.270.15.8744. [DOI] [PubMed] [Google Scholar]
- 158.Dhar G, Sanders ER, Johnson RC. Architecture of the Hin synaptic complex during recombination: the recombinase subunits translocate with the DNA strands. Cell. 2004;119:33–45. doi: 10.1016/j.cell.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 159.Leschziner AE, Grindley NDF. The architecture of the gamma delta resolvase crossover site synaptic complex revealed by using constrained DNA substrates. Mol Cell. 2003;12:775–781. doi: 10.1016/s1097-2765(03)00351-4. [DOI] [PubMed] [Google Scholar]
- 160.Sarkis GJ, Murley LL, Leschziner AE, Boocock MR, Stark WM, Grindley ND. A model for the gamma delta resolvase synaptic complex. Mol Cell. 2001;8:623–631. doi: 10.1016/s1097-2765(01)00334-3. [DOI] [PubMed] [Google Scholar]
- 161.Nollmann M, He J, Byron O, Stark WM. Solution structure of the Tn3 resolvase-crossover site synaptic complex. Mol Cell. 2004;16:127–137. doi: 10.1016/j.molcel.2004.09.027. [DOI] [PubMed] [Google Scholar]
- 162.Kamtekar S, Ho RS, Cocco MJ, Li W, Wenwieser SV, Boocock MR, Grindley NDF, Steitz TA. Implications of structures of synaptic tetramers of gamma delta resolvase for the mechanism of recombination. Proc Natl Acad Sci USA. 2006;103:10642–10647. doi: 10.1073/pnas.0604062103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Yuan P, Gupta K, Van Duyne GD. Tetrameric structure of a serine integrase catalytic domain. Structure. 2008;16:1275–1286. doi: 10.1016/j.str.2008.04.018. [DOI] [PubMed] [Google Scholar]
- 164.Kanaar R, van de Putte P, Cozzarelli NR. Gin-mediated DNA inversion: product structure and the mechanism of strand exchange. Proc Natl Acad Sci USA. 1988;85:752–756. doi: 10.1073/pnas.85.3.752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Kahmann R, Mertens G, Klippel A, Brauer B, Rudt R, Koch C. The mechanism of G inversion. In: McMacken R, Kelly TJ, editors. DNA Replication and Recombination, UCLA Symposium on Molecular and Cellular Biology. Vol. 47. Alan R. Liss, Inc; New York, N.Y: 1987. pp. 681–689. [Google Scholar]
- 166.Merickel SK, Johnson RC. Topological analysis of Hin-catalysed DNA recombination in vivo and in vitro. Mol Micro. 2004;51:1143–1154. doi: 10.1046/j.1365-2958.2003.03890.x. [DOI] [PubMed] [Google Scholar]
- 167.Heichman KA, Moskowitz IP, Johnson RC. Configuration of DNA strands and mechanism of strand exchange in the Hin invertasome as revealed by analysis of recombinant knots. Genes Dev. 1991;5:1622–1634. doi: 10.1101/gad.5.9.1622. [DOI] [PubMed] [Google Scholar]
- 168.Deibler RW, Rahmati S, Zechiedrich EL. Topoisomerase IV, alone, unknots DNA in E. coli. Genes Dev. 2001;15:748–761. doi: 10.1101/gad.872301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Klippel A, Kanaar R, Kahmann R, Cozzarelli NR. Analysis of strand exchange and DNA binding of enhancer-independent Gin recombinase mutants. EMBO J. 1993;12:1047–1057. doi: 10.1002/j.1460-2075.1993.tb05746.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Iida S, Hiestand-Nauer R. Localized conversion at the crossover sequences in the site-specific DNA inversion system of bacteriophage P1. Cell. 1986;45:71–79. doi: 10.1016/0092-8674(86)90539-8. [DOI] [PubMed] [Google Scholar]
- 171.Iida S, Hiestand-Nauer R. Role of the central dinucleotide at the crossover sites for the selection of quasi sites in DNA inversion mediated by the site-specific Cin recombinase of phage P1. Mol Gen Genet. 1987;208:464–468. doi: 10.1007/BF00328140. [DOI] [PubMed] [Google Scholar]
- 172.Bai H, Sun M, Ghosh P, Hatfull GF, Grindley ND, Marko JF. Single-molecule analysis reveals the molecular bearing mechanism of DNA strand exchange by a serine recombinase. Proc Natl Acad Sci USA. 2011;108:7419–7424. doi: 10.1073/pnas.1018436108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Vologodskii A, Cozzarelli NR. Effect of supercoiling on the juxtaposition and relative orientation of DNA sites. Biophys J. 1996;70:2548–2556. doi: 10.1016/S0006-3495(96)79826-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Vologodskii AV, Levene SD, Klenin KV, Frank-Kamenetskii M, Cozzarelli NR. Conformational and thermodynamic properties of supercoiled DNA. J Mol Biol. 1992;227:1224–1243. doi: 10.1016/0022-2836(92)90533-p. [DOI] [PubMed] [Google Scholar]
- 175.Boles TC, White JH, Cozzarelli NR. Structure of plectonemically supercoiled DNA. J Mol Biol. 1990;213:931–951. doi: 10.1016/S0022-2836(05)80272-4. [DOI] [PubMed] [Google Scholar]
- 176.Marko JF. The internal ‘slithering’ dynamics of supercoiled DNA. Physica A. 1997;244:263–277. [Google Scholar]
- 177.Oram M, Marko JF, Halford SE. Communications between distant sites on supercoiled DNA from non-exponential kinetics for DNA synapsis by resolvase. J Mol Biol. 1997;270:396–412. doi: 10.1006/jmbi.1997.1109. [DOI] [PubMed] [Google Scholar]
- 178.Marko JF. Short note on the scaling behavior of communication by ‘slithering’ on a supercoiled DNA. Physica A. 2001;296:289–292. [Google Scholar]
- 179.Vologodskii AV, Cozzarelli NR. Conformational and thermodynamic properties of supercoiled DNA. Annu Rev Biophys Biomol Struct. 1994;23:609–643. doi: 10.1146/annurev.bb.23.060194.003141. [DOI] [PubMed] [Google Scholar]
- 180.Lee SY, Lee HJ, Lee H, Kim S, Cho EH, Lim HM. In vivo assay of protein-protein interactions in Hin-mediated DNA inversion. J Bacteriol. 1998;180:5954–5960. doi: 10.1128/jb.180.22.5954-5960.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Sessions RB, Oram M, Szczelkun MD, Halford SE. Random walk models for DNA synapsis by resolvase. J Mol Biol. 1997;270:413–425. doi: 10.1006/jmbi.1997.1108. [DOI] [PubMed] [Google Scholar]
- 182.Stark WM, Grindley NDF, Hatfull GF, Boocock MR. Resolvase-catalysed reactions between res sites differing in the central dinucleotide of subsite I. EMBO J. 1991;10:3541–3548. doi: 10.1002/j.1460-2075.1991.tb04918.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Ghosh P, Kim AI, Hatfull GF. The orientation of mycobacteriophage Bxb1 integration is solely dependent on the central dinucleotide of attP and attB. Mol Cell. 2003;12:1101–1111. doi: 10.1016/s1097-2765(03)00444-1. [DOI] [PubMed] [Google Scholar]
- 184.Mandali S, Dhar G, Avliyakulov NK, Haykinson MJ, Johnson RC. The site-specific integration reaction of Listeria phage A118 integrase, a serine recombinase. Mobile DNA. 2013;4:2. doi: 10.1186/1759-8753-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Haffter P, Bickle TA. Enhancer-independent mutants of the Cin recombinase have a relaxed topological specificity. EMBO J. 1988;7:3991–3996. doi: 10.1002/j.1460-2075.1988.tb03287.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Burke ME, Arnold PH, He J, Wenwieser SV, Rowland SJ, Boocock MR, Stark WM. Activating mutations of Tn3 resolvase marking interfaces important in recombination catalysis and its regulation. Mol Microbiol. 2004;51:937–948. doi: 10.1046/j.1365-2958.2003.03831.x. [DOI] [PubMed] [Google Scholar]
- 187.Rowland SJ, Boocock MR, McPherson AL, Mouw KW, Rice PA, Stark WM. Regulatory mutations in Sin recombinase support a structure-based model of the synaptosome. Mol Microbiol. 2009;74:282–298. doi: 10.1111/j.1365-2958.2009.06756.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Arnold PH, Blake DG, Grindley NDF, Boocock MR, Stark WM. Mutants of Tn3 resolvase which do not require accessory binding sites for recombination activity. EMBO J. 1999;18:1407–1414. doi: 10.1093/emboj/18.5.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Olorunniji FJ, He J, Wenwieser SV, Boocock MR, Stark WM. Synapsis and catalysis by activated Tn3 resolvase mutants. Nucleic Acids Res. 2008;36:7181–7191. doi: 10.1093/nar/gkn885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.McGaughey GB, Gagne M, Rappe AK. pi-Stacking interactions. Alive and well in proteins. J Biol Chem. 1998;273:15458–15463. doi: 10.1074/jbc.273.25.15458. [DOI] [PubMed] [Google Scholar]
- 191.Brocchieri L, Karlin S. Geometry of interplanar residue contacts in protein structures. Proc Natl Acad Sci USA. 1994;91:9297–9301. doi: 10.1073/pnas.91.20.9297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Lee HJ, Lee SY, Lee HM, Lim HM. Effects of dimer interface mutations in Hin recombinase on DNA binding and recombination. Mol Genet Genomics. 2001;266:598–607. doi: 10.1007/s004380100576. [DOI] [PubMed] [Google Scholar]
- 193.Cozzarelli NR, Krasnow MA, Gerrard SP, White JH. A topological treatment of recombination and topoisomerases. Cold Spring Harb Symp Quant Biol. 1984;49:383–400. doi: 10.1101/sqb.1984.049.01.045. [DOI] [PubMed] [Google Scholar]


