Abstract
Saccharomyces cerevisiae selectively utilizes good nitrogen sources in preference to poor ones by down-regulating transcription of genes encoding proteins that transport and degrade poor nitrogen sources when excess nitrogen is available. This regulation is designated nitrogen catabolite repression (NCR). When cells are transferred from a good to a poor nitrogen source (glutamine to proline) or treated with rapamycin, an inhibitor of the protein kinases Tor1/2, Gln3 (NCR-sensitive transcription activator) moves from the cytoplasm into the nucleus. Gln3 re-accumulates in the cytoplasm when cells are returned to a good nitrogen source. However, Gln3 is not uniformly distributed in the cytoplasm. Such non-uniform distribution could result from a variety of interactions including association with a cytoplasmic vesicular system or components of the cytoskeleton. We used latrunculin, a drug that disrupts the actin cytoskeleton by inhibiting actin polymerization, to determine whether the actin cytoskeleton participates in intracellular Gln3 movement. Latrunculin-treatment prevents nuclear accumulation of Gln3 and NCR-sensitive transcription in cells transferred from ammonia to proline medium but does not prevent its accumulation in the cytoplasm of cells transferred from proline to glutamine medium. In contrast, rapamycin-induced nuclear accumulation of Gln3 is not demonstrably affected by latrunculin treatment. These data indicate the actin cytoskeleton is required for nuclear localization of Gln3 in response to limiting nitrogen but not rapamycin-treatment. Therefore, the actin cytoskeleton either participates in the response of Gln3 intracellular localization to nitrogen limitation before Tor1/2, or Tor1/2 inhibition only mimics the outcome of nitrogen limitation rather than directly regulating it.
A major mechanism to regulate transcription in eukaryotic cells is the intracellular partitioning of transcription factors. Saccharomyces cerevisiae GATA-factor (Gln3 and Gat1)-mediated transcription is an example of such regulation (1–9). One of the principle functions of Gln3 and Gat1, which bind to UASNTR elements containing the core sequence GATAA, is to activate transcription of genes whose products are required for the transport and catabolism of poor nitrogen sources (10–13). In the presence of a good nitrogen source (glutamine or ammonia in some strains), Gln3 is restricted to the cytoplasm in association with Ure2 (1–9, 14), a regulatory protein that also possesses characteristics expected of a glutathione S-transferase (15). In contrast, Gln3 accumulates in the nucleus and activates NCR1-sensitive transcription when cells are provided with a poor nitrogen source (proline) or when the supply of a good nitrogen source becomes limiting.
Although much has been reported about the mechanism by which Gln3 intracellular localization is regulated, the mechanistic details beyond observed correlations remain unclear. The most pertinent observations are as follows (1–9, 13, 16–31). (i) Gln3 and Ure2 form a complex that can be isolated in vitro. (ii) Gln3 accumulates in the nuclei of cells treated with the Tor1/2 inhibitor, rapamycin. Tor1/2 are protein kinases, the inactivation of which results in massive changes in cellular activity ranging from increased protein turnover, derepression of NCR-sensitive gene expression, and inhibition of protein synthesis initiation to cessation of actin cytoskeleton reorganization and cell division. (iii) The extent of Gln3 phosphorylation decreases in rapamycin-treated cells. (iv) Gln3 is not uniformly distributed within the cytoplasm of cells growing in the presence of a rich nitrogen source but, rather, appears to be distributed in the form of tubes and/or vesicles. (v) Tor1/2 have been found associated with multiple proteins, one of which is Lst8, known to function in a cytoplasmic vesicular system.
The vesicular/tubular appearance of the fluorescent Gln3 signal mentioned above might arise if Gln3 were associated with or contained in vesicles, such as the endoplasmic reticulum or an endosomal system (32–35). These systems participate in protein trafficking, endocytosis and other processes with which the actin cytoskeleton has been linked (36–40). In addition, in mammalian systems the actin cytoskeleton has been shown to function as a cytoplasmic anchor for transcription factors Nrf2 (41–43) and the dioxin receptor (44). In this role it is partially responsible for nucleocytoplasmic trafficking of these factors. Many of the studies that have identified roles of the actin cytoskeleton in yeast and other systems have made use of the drugs latrunculin A and B (45). Latrunculin binds to the actin monomer, inhibits its ability to polymerize, and thereby disrupts the actin cytoskeleton (46–48).
The above information motivated us to use latrunculin to determine whether the actin cytoskeleton plays a role in intracellular Gln3 localization. Our data show that an intact actin cytoskeleton is required for nuclear accumulation of Gln3 in response to nitrogen limitation but not for its cytoplasmic accumulation in response to nitrogen excess. It does not appear that Gln3 association with the endoplasmic reticulum is responsible for the actin cytoskeleton requirement of GATA-factor localization and transcription activity because co-localization between Gln3 and the endoplasmic reticulum marker protein, Kar2, is limited. However, the structures with which Gln3 appears to be associated are often seen in close proximity to the endoplasmic reticulum. Furthermore, Gln3 fluorescence localizes to tubular/vesicular structures that appear to be contiguous with the nucleus.
MATERIALS AND METHODS
Yeast Strains and Culture Conditions
S. cerevisiae strains TB123 (MATa, leu2-3,112, ura3-52, rme1, trp1, his4, GAL+, HMLa, GLN3-Myc13-KanMX) (1), DDY1495 (MATa, ACT1:HIS3, leu2-3,112, his3D200, tub2–201, ura3–52), and DDY1544 (MATa, act1-157:HIS3, leu2-3,112, hisD200, tub2–201, ura3-52) were grown in YNB (Difco) (without ammonium sulfate or amino acids), 2% glucose, 0.1% of the nitrogen source noted in the legends of Figs. 1–11 and supplemented with the appropriate auxotrophic requirements. Latrunculin B (Calbiochem) (7.5 or 15 mg/ml dissolved in ethanol) was added to a final concentration of 150 μM, rapamycin (Sigma) (1 mg/ml dissolved in 90% ethanol plus 10% Tween 20) was added to a final concentration of 200 ng/ml, and hydroxyurea was added to a final concentration of 200 mM. Cultures were grown to mid-log phase (A600 nm = 0.4–0.5, at 30 °C) at which time a sample was removed for microscopic assay. Latrunculin B or carrier (ethanol) was added to the remainder of the culture, and the cells were incubated for 30 min to permit latrunculin to act. Alternatively, hydroxyurea was added, and the cells were incubated for 120 min. At the end of this preincubation period the cells were collected by centrifugation, washed once with prewarmed, pre-aerated medium, and transferred to the indicated medium containing either carrier (ethanol), latrunculin, or hydroxyurea as noted in the legends of Figs. 1–11. Rapamycin was also added where indicated. Samples were collected at the indicated times for immunolocalization or RNA isolation.
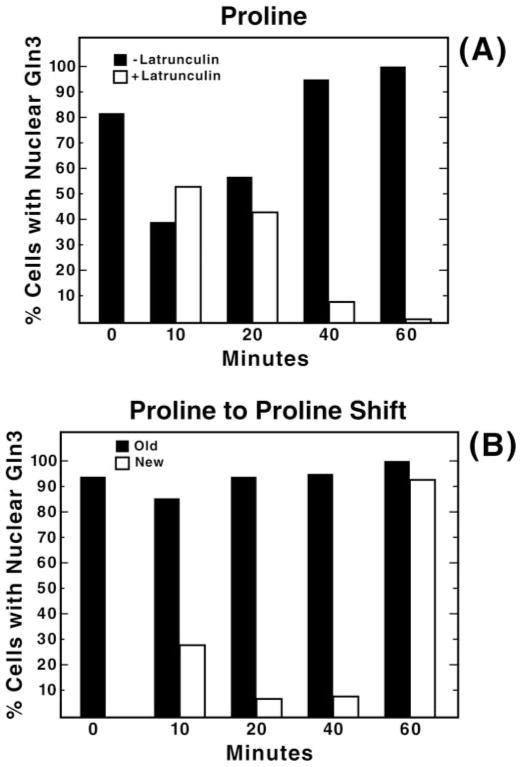
Fig. 1. Ethanol addition or transfer to fresh proline medium results in a transient disturbance in Gln3-Myc localization.
Yeast cultures (TB123) were grown to mid-log phase (A600 nm = 0.4–0.5) in YNB-proline medium. Panel A, aliquots were removed just before the addition of either 150 μM latrunculin B (open bars) or carrier, ethanol (filled bars) (0 min), and at the indicated times after the addition and processed for indirect immunofluorescence analysis. Panel B, TB123 cells, grown as in panel A, were sampled (0 min) and then divided into two portions. Cells in the first portion were harvested by centrifugation and resuspended again without a change of medium (filled bars). Cells in the second portion were similarly harvested but then resuspended in fresh prewarmed, pre-aerated proline medium identical to that used originally (open bars). Thereafter, at the times indicated both cultures were sampled for microscopic examination as in panel A.
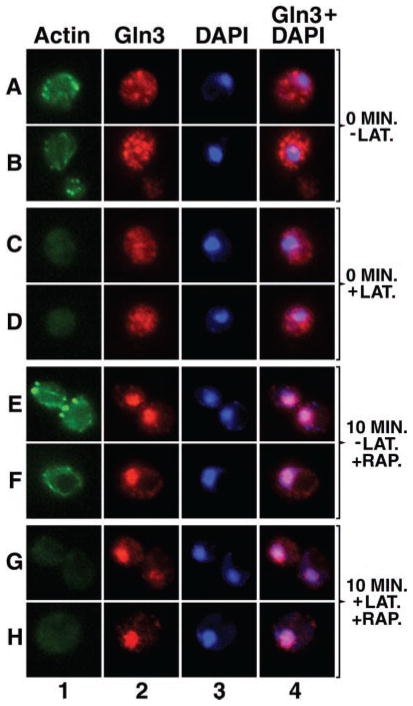
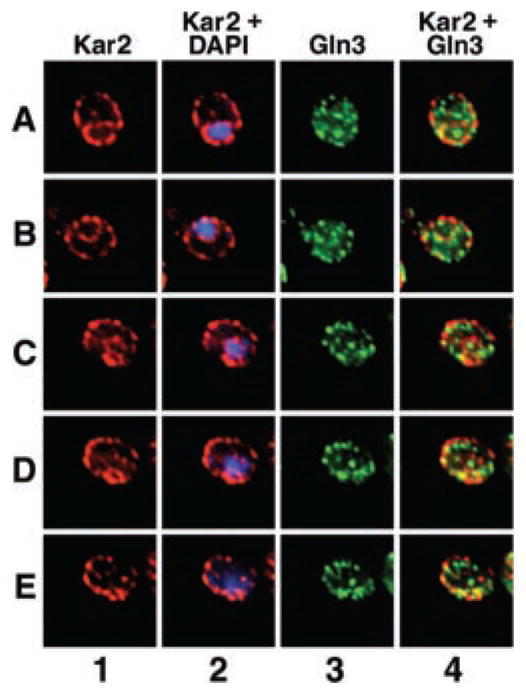
Fig. 11. Gln3 localizes to projections emanating from the nucleus during its exit from and entry into the nucleus.
Cultures (TB123) were either grown to mid-log in YNB-glutamine and transferred to proline medium for 30 min (rows A–E) or YNB-proline and transferred to glutamine for 1 min (row F); images in row F were taken from Cox et al. (7) for comparison. Cells were then processed for indirect immunofluorescence. The localization of Gln3-Myc13 and nuclei was determined by staining with monoclonal antibody 9E10 anti-Myc (columns 1 and 2) and DAPI (column 2, blue), respectively. Merged images (column 2) show the overlap (pink) of Gln3 and nuclei. Micrographs were imaged as described in Fig. 9.
Thirty minutes was chosen as the time for preincubation with latrunculin before the beginning of each experiment for two reasons. First, at this time the actin cytoskeleton was completely disrupted. In addition, we noticed in preliminary experiments that the addition of the carrier in which latrunculin was dissolved (ethanol) caused a modest and temporary disturbance of Gln3-Myc intracellular localization. The percentage of cells in which Gln3-Myc was localized to the nucleus transiently decreased by about 50%, then recovered by 20–40 min and remained constant thereafter at least up to 120 min, which is as long as we monitored the cells (Fig. 1A, filled bars, and data not shown). The decrease was somewhat variable and ranged from about 15 to 50%. Similar transient perturbation of Gln3-Myc localization also occurred when proline-grown cells were harvested, washed, and resuspended in fresh proline medium (Fig. 1B, open bars). Here, as in Fig. 1A, proper Gln3-Myc localization was recovered. This transient decrease in nuclear Gln3-Myc did not occur, however, if cells were pelleted by centrifugation and resuspended again without a change of medium, i.e. fresh prewarmed, pre-aerated medium did not replace the medium in which they were grown (Fig. 1B, filled bars). It also did not occur in a proline-grown ure2Δ mutant treated with ethanol. The physiological and biochemical bases of the transient decrease in nuclear Gln3-Myc reported above are not known.
Northern Blot Analysis
RNA was isolated by the phenol/freeze RNA method of Schmitt et al. (49). Northern blots were performed as described earlier (50, 60). The radioactive DNA probe for DAL80 was prepared as described earlier (15, 51). PC4 (a control used for sporulation assays whose expression mirrors ribosomal RNA concentration) was used as a control for RNA transfer and loading.
Immunofluorescence
Immunofluorescence staining was carried out as described by Cox et al. (7). Gln3-Myc and Kar2 were visualized using 9E10(c-myc) (Covance MMS-150P) monoclonal antibody at a dilution of 1:1000 or a Kar2 rabbit polyclonal antibody (52) at a dilution of 1:5000, respectively, as the primary antibody and either Alexa Fluor 488 or Alexa Fluor 594 goat anti-mouse IgG antibody (Molecular Probes) at a dilution of 1:200 or Alexa Fluor 594 goat anti-rabbit antibody at a dilution of 1:200, respectively, as secondary antibody. Actin was labeled using a 1:50 dilution of Alexa Fluor 488 phalloidin (Molecular Probes). Nuclei were stained with DAPI as described previously (7).
Immunofluorescence Microscopy
Cells were imaged using a Zeiss Axioplan 2 imaging microscope with a 100× Plan-Apochromat 1.40 oil objective. Images were acquired using a Zeiss Axio camera and AxioVision 3.0 (Zeiss) software. Images showing phalloidin staining of actin were taken at constant exposure. Images in Figs. 9–11 were deconvolved with AxioVision 3.0 (Zeiss) software using the constrained iterative algorithm.
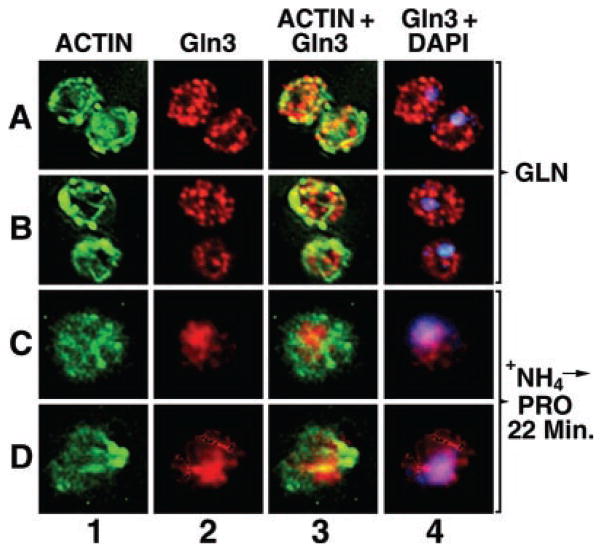
Fig. 9. Intracellular localization of Gln3 and actin.
Cultures (TB123) were grown to mid-log in 2% glucose, YNB-glutamine medium (rows A and B) or were transferred from glucose-ammonia to -proline medium, incubated at 30 °C for 22 min (rows C and D), and processed for indirect immunofluorescence. The localization of actin, Gln3-Myc13, and nuclei was determined by staining with phalloidin (columns 1 and 3, green), monoclonal antibody 9E10 anti-Myc (columns 2–4, red), and DAPI (column 4, blue), respectively. Merged images (column 3) show the overlap (column 3, yellow) of Gln3 and actin or Gln3 and DAPI-positive material (column 4, pink). Micrographs were imaged using a Zeiss Axioplan 2 imaging microscope. 0.4-μm sections were collected as a Z-stack, and one image from the center of that stack is shown. Images were deconvolved with AxioVision 3.0 software using the constrained iterative algorithm.
RESULTS
Nuclear Accumulation of Gln3 in Response to a Poor Nitrogen Source Requires the Actin Cytoskeleton
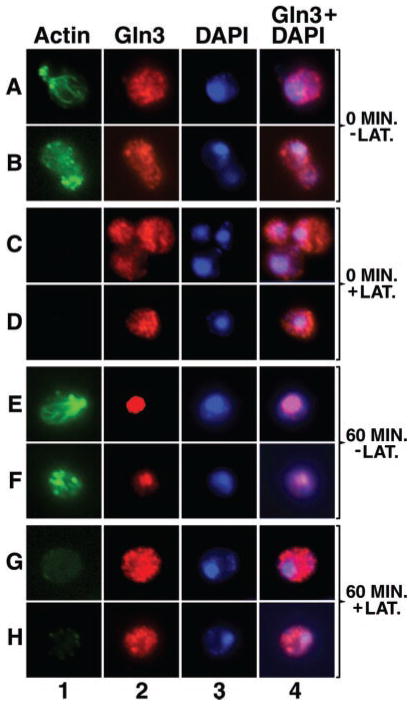
Gln3 moves from the cytoplasm into the nucleus when cells are transferred from a good to poor nitrogen source (ammonia or glutamine to proline medium). To determine whether an intact actin cytoskeleton is required for this process, we measured the effect of latrunculin treatment on intracellular Gln3 distribution in cells transferred from ammonia to proline medium. Actin patches and cables are clearly visible both before (0 min) and 60 min after transferring cells from ammonia to proline medium in the absence of latrunculin (Fig. 2, column 1, rows A, B, and E, F) but appear completely depolymerized in yeast cells preincubated 30 min with latrunculin (Fig. 2, column 1, rows C, D and G, H). Nuclear accumulation of Gln3 correlates with the presence of an intact actin cytoskeleton. At zero time, Gln3 is non-randomly distributed in the cytoplasm of both control and latrunculin-treated, ammonia-grown cells (Fig. 2, column 2, rows A, B and C, D). After 60 min in minimal-proline medium, Gln3 accumulated in the nuclei of untreated cells (Fig. 2, column 2, rows E and F), co-localizing with DAPI-stained material (Fig. 2, columns 2–4, rows E and F). In latrunculin-treated cells, little if any nuclear accumulation of Gln3 occurred after transfer to proline medium (Fig. 2, columns 2–4, rows G and H). In a few latrunculin-treated cells transferred to minimal-proline medium, faint actin clusters begin to reappear in long exposures of micrographs taken at the later time points; actin cables, however, do not reappear. This reappearance may derive from a previously noted characteristic of latrunculin B on mammalian cells, i.e. its inhibition of actin polymerization begins to reverse with time (47). This slight reappearance of actin clusters did not, however, affect Gln3 localization (data not shown).
Fig. 2. An intact actin cytoskeleton is required for nuclear localization of Gln3 when cells are transferred from a good nitrogen source to a poor one.
Yeast cultures (TB123) were grown to mid-log phase in 2% glucose, YNB (without amino acids or ammonium sulfate), and 0.1% ammonia sulfate. Cells were then pretreated with either 150 μM latrunculin B (+LAT.) or carrier, ethanol (−LAT.) for 30 min at 30 °C to permit latrunculin to act. At that time aliquots were removed just before transfer from ammonia to proline medium (0 min) and 60 min after transfer and processed for indirect immunofluorescence. Cells were stained to visualize actin (column 1), Gln3 (column 2), or the nucleus (column 3). Column 4 depicts merged images from columns 2 and 3.
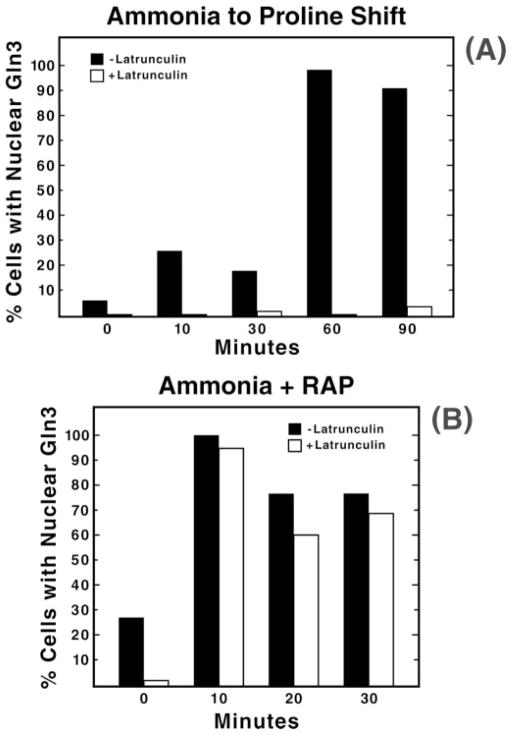
Previous Gln3 localization experiments demonstrate the value of quantitating time-dependent Gln3 partitioning among cellular compartments (7), and prompted us to perform such measurements here. Just before transfer from minimal ammonia to proline medium, Gln3 was nuclear in ~5% of the cells (Fig. 3A, closed bars). This value rose to about 25% at 10 min post-transfer, dropped slightly at 30 min, and reached nearly 100% at 60 min. In contrast, when latrunculin was present, Gln3 accumulated in the nuclei of no more than 5% of the cells (Fig. 3A, open bars). Therefore, nuclear accumulation of Gln3 requires an intact actin cytoskeleton. These data also demonstrate that nuclear accumulation of Gln3 is not simply delayed in latrunculin-treated cells.
Fig. 3. Quantitation of Gln3 partitioning between the nucleus and cytoplasm in cells transferred from minimal-ammonia to -proline medium (panel A) or after treatment of ammonia-grown cells with rapamycin (RAP; panel B).
The experimental format used to transfer cells from glucose-ammonia to -proline medium was as described in Fig. 2. For rapamycin treatment, cultures (TB123) were grown to mid-log phase in 2% glucose, YNB (without amino acids or ammonium sulfate), and 0.1% ammonia sulfate and pretreated with 150 μM latrunculin B or carrier (ethanol) as described in Fig. 2. Aliquots were then removed just before the addition of 200 ng/ml rapamycin (0 min) and at the indicated times after the addition. Samples were processed for indirect immunofluorescence as in Fig. 2.
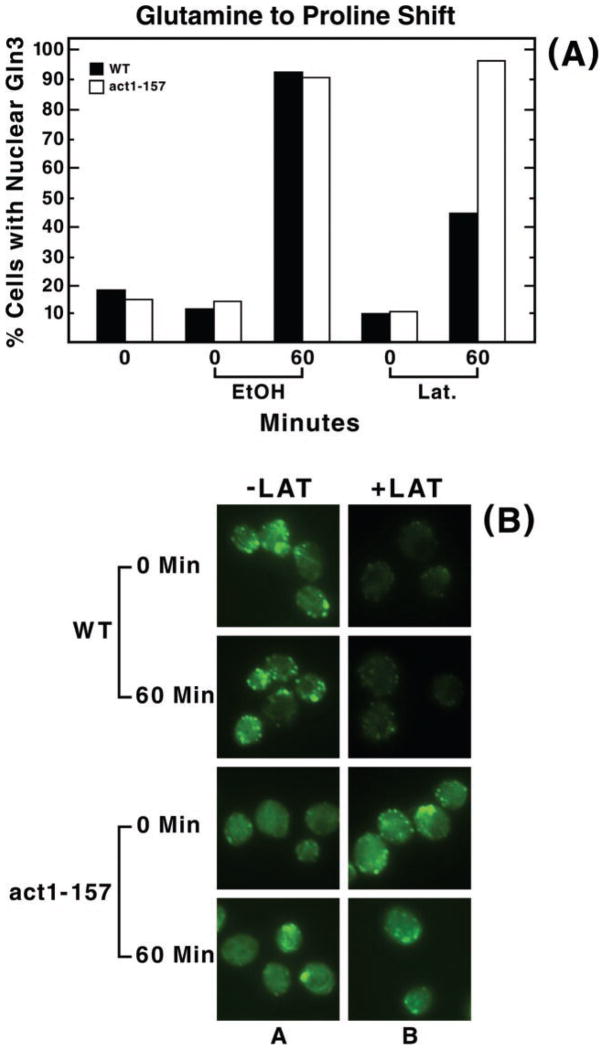
A potential problem interpreting drug-induced effects in whole cells is the possibility that the measured effect derives indirectly as a consequence of the drug action on a molecule or system other than the one being studied. Although latrunculin has not to our knowledge been reported to affect molecules other than actin, we tested this question more directly using act1-157 cells. This strain contains a site-specific mutation in the actin gene that confers resistance to latrunculin without substantially changing the structure of the actin cytoskeleton of the cell or its ability to undergo fluid-phase endocytosis (53). Therefore, latrunculin should not inhibit Gln3-Myc from accumulating in the nuclei of act1-157 cells transferred from glutamine to proline medium unless the drug acts by some mode other than binding to actin and preventing its polymerization. As shown in Fig. 4, latrunculin treatment neither demonstrably affected the appearance of the actin cytoskeleton visualized by phalloidin staining (panel B, bottom two rows) nor the percentage of act1-157 cells containing nuclear Gln3-Myc after transfer to proline medium (Fig. 4A, open bars). On the other hand, the wild type strain remained sensitive to latrunculin treatment. These data support the interpretation that latrunculin was acting specifically and that a functional actin cytoskeleton was required for nuclear entry of Gln3-Myc.
Fig. 4. act1-157, a yeast strain whose actin is resistant to latrunculin, shows no inhibition of Gln3 nuclear accumulation when cells are transferred from a good nitrogen source to a poor one after latrunculin treatment.
Yeast strains containing either wild type (WT) ACT1 (DDY 1495) (54) or the act1-157 mutation (DDY 1544) (53) were grown to mid-log in YNB-glutamine and sampled for microscopic examination (0 min, panel A). The cultures were then preincubated with latrunculin B as described in Fig. 2 for 30 min. Aliquots were then removed just before transfer from glutamine to proline medium (0 min, EtOH or Lat in panel A; 0 min, −LAT and +LAT in panel B) and 60 min after this transfer and processed for indirect immunofluorescence. Panel A, the percent of cells in which Gln3 is localized to the nucleus. Panel B, representative wild type and mutant cells treated with either 150 μM latrunculin or the carrier and stained with phalloidin to permit evaluation of latrunculin treatment and the act1-157 phenotype. The genetic background containing the act1-157 mutation is somewhat less latrunculin-sensitive than the one we used for the rest of our experiments. We did not, however, increase the latrunculin concentration in this control experiment because we preferred to maintain a uniform latrunculin concentration throughout all of the experiments reported.
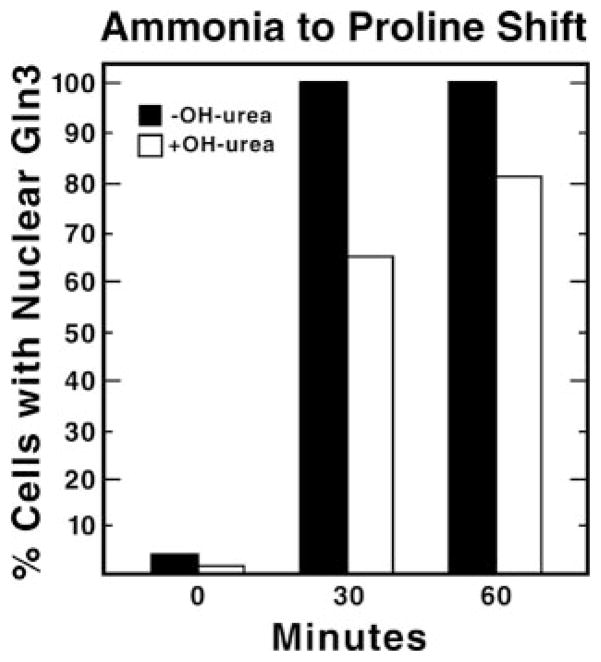
Another way in which latrunculin might inhibit nuclear accumulation of Gln3-Myc indirectly is if Gln3 nuclear transport requires that the cell be actively undergoing cell division. Actin disruption has been shown to cause cell cycle arrest at G2/M (19). More specifically, Belmont and co-workers (53, 54) show that in a culture treated with latrunculin A for 4 h, the number of large unbudded cells increased from 38 to 77%. To test the possibility that cell cycle arrest might inhibit nuclear accumulation of Gln3-Myc after transfer from a good to a poor nitrogen source, we compared Gln3-Myc nuclear localization in hydroxyurea pretreated and untreated cells transferred from ammonia to proline medium. Hydroxyurea arrests cells in S-phase by inhibiting ribonucleotide reductase, which in turn depletes cells of deoxyribonucleotides required for DNA synthesis (55, 56). As shown in Fig. 5, the number of both untreated and hydroxyurea-treated cells with nuclear Gln3-Myc increased from about 4 to 80% after transfer from ammonia to proline medium, arguing that cessation of the cell cycle does not interfere with nuclear accumulation of Gln3-Myc. Also consistent with this interpretation is the fact that the ability for nuclear accumulation of Gln3-Myc was lost in most latrunculin-treated cells at 10 min after transfer from ammonia to proline medium (Fig. 3). This represents only 40 min beyond the point where cells first came into contact with latrunculin, which would be insufficient time for all of the cells to arrest. Together, these data support the contention that latrunculin inhibition of Gln3-Myc accumulation in the nucleus occurs as a result of the drug effect upon actin and the actin cytoskeleton.
Fig. 5. Cell cycle arrest does not inhibit Gln3-Myc nuclear accumulation when cells are transferred from a good nitrogen source to a poor one.
A yeast culture (TB123) was grown to log phase (A600 nm = 0.2) in YNB-ammonia medium. Half of the culture was pretreated with 200 mM hydroxyurea for 2 h at 30 °C (+OH-urea). 70–80% of the cells were scored as large budded cells after this incubation, demonstrating that cell cycle arrest had been achieved. Aliquots were removed just before transfer to YNB-proline (0 min) and at 30 and 60min after transfer and processed for immunofluorescence.
NCR-sensitive, Gln3-mediated Transcription Is Inhibited in Latrunculin-treated Cells
Because latrunculin inhibited Gln3 accumulation in the nucleus, we measured NCR-sensitive transcription in cells transferred from ammonia to proline medium in the presence or absence of latrunculin. DAL80, a gene whose expression exhibits a typical NCR-sensitive profile, responds normally in an untreated culture after transfer to proline medium, i.e. low and high expression with good and poor nitrogen sources, respectively (Fig. 6, lanes A and B). Increased DAL80 expression did not occur, however, when the latrunculin-treated culture was similarly transferred (lanes C and D).
Fig. 6. Effect of latrunculin treatment on NCR-sensitive gene expression.
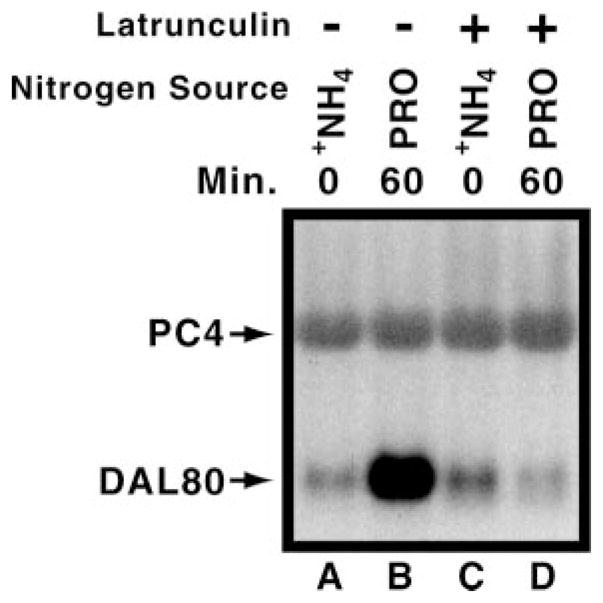
Wild type cultures (TB123) were treated as described in Fig. 2. However, in this case cell samples were harvested, and total RNA was isolated from each. A Northern blot measuring steady state DAL80 mRNA levels was then performed (9 μg of total RNA per lane). PC4 (a control used for sporulation assays whose expression mirrors ribosomal RNA concentration) was used as a control.
Cytoplasmic Accumulation of Gln3 Does Not Require an Intact Actin Cytoskeleton
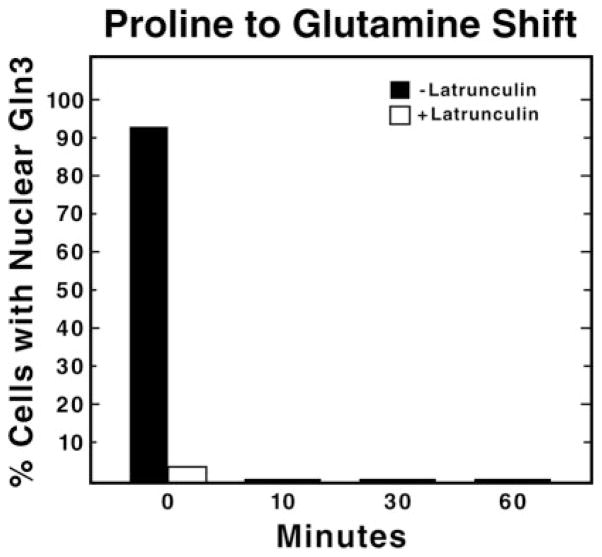
We used a similar experimental approach to query whether an intact actin cytoskeleton is required for Gln3 to leave the nucleus and accumulate in the cytoplasm after a transition from limiting to excess nitrogen, i.e. pretreating proline-grown cells with latrunculin and transferring them to glutamine medium. To our surprise, at the zero time point just before the transfer, Gln3 was already cytoplasmic in virtually all of the latrunculin-treated cells. (Fig. 7, open bars). In fact a similar result was seen at the 40- and 60-min time points in the control experiment depicted in Fig. 1A (open bars). The two experiments argue that (i) there is no demonstrable actin cytoskeleton requirement for Gln3 to leave the nucleus, and (ii) once Gln3 is cytoplasmic, it cannot re-enter the nucleus even when there is a cellular signal for it to do so (growth in proline) if latrunculin is present (Figs. 1A and 3A). These data imply that Gln3 may continuously cycle from the nucleus to the cytoplasm in proline-grown cells. However, we cannot exclude the possibility that an actin cytoskeleton is required to maintain Gln3 within the nucleus.
Fig. 7. An intact actin cytoskeleton is not required for the cytoplasmic accumulation of Gln3.
Cultures (TB123) were grown to mid-log in 2% glucose, YNB (without amino acids or ammonium sulfate), and 0.1% proline and treated with latrunculin B as described in Fig. 2. Aliquots were then removed just before transfer from proline to glutamine medium (0 min) and at the indicated times after transfer and processed for indirect immunofluorescence.
Effect of Latrunculin on Rapamycin-induced Nuclear Localization of Gln3
Rapamycin has been highly useful for investigating the molecular events associated with NCR-sensitive transcription (1–4, 6, 9). Therefore, we determined whether rapamycin-induced nuclear localization of Gln3 was sensitive to latrunculin as observed when transferring cells from excess to limiting nitrogen. In the absence of latrunculin, the actin cytoskeleton does not appear detectably different in rapamycin-treated and -untreated cells (Fig. 8, column 1, rows A, B and E, F). The only significant difference we noted was fewer cells in which a normal polarized distribution of actin clusters occurs, as reported by others (16, 19, 20) (Fig. 8, column 1, row E). In the presence of latrunculin, the actin cytoskeleton was largely absent in both rapamycin-treated and untreated cells (Fig. 8, column 1, rows C, D and G, H).
Fig. 8. An intact actin cytoskeleton is not required for the nuclear localization of Gln3 after rapamycin treatment.
Cultures (TB123) were grown to mid-log in 2% glucose, YNB (without amino acids or ammonium sulfate), and 0.1% ammonium sulfate and pretreated with either 150 μM latrunculin B (+LAT) or the carrier, ethanol (−LAT) as described in Fig. 2. Aliquots were then removed just before the addition of 200 ng/ml rapamycin (0 min) and at the indicated times after the addition. Samples were processed for indirect immunofluorescence as in Fig. 2.
In the absence of latrunculin, Gln3 localized to the nuclei of nearly all cells within 10 min of rapamycin treatment (Fig. 8, column 2, rows E, F and Fig. 3B, closed bars). In contrast with data in Figs. 2 and 3A, Gln3 accumulated in the nuclei of most rapamycin-treated cells pretreated with latrunculin (Fig. 8, column 2, rows G and H and Fig. 3B, open bars). Therefore, rapamycin-induced nuclear localization of Gln3 responds differently to latrunculin treatment than does Gln3 localization after a transition from excess to limiting nitrogen.
Gln3-Myc Does Not Appear to Co-localize with Actin
In mammalian systems, where NrF2 and the dioxin receptor are directly regulated by the actin cytoskeleton, there is significant co-localization of these proteins with actin (41, 42, 44). Although Gln3 appears to be sequestered in the cytoplasm by Ure2 rather than by actin, the actin cytoskeleton requirement seen in Figs. 2 and 3A prompted us to determine whether Gln3 and actin co-localized. Two conditions were assayed; (i) cells growing logarithmically in minimal glutamine medium where actin would not apparently be needed, i.e. no requirement could be demonstrated, and (ii) after transfer of cells from ammonia to proline medium, where the actin cytoskeleton is required for Gln3 nuclear accumulation. In the latter case we selected a time after the transfer at which some but not all Gln3 was nuclear, thereby hopefully increasing the probability that Gln3 molecules in the process of being accumulated in the nucleus would be assayed. Most polymerized actin and Gln3 do not co-localize (Fig. 9). There are instances in which the signals co-localize or appear to overlap. However, when two molecules each occupy a large fraction of the cell volume, as occurs with Gln3 and actin (Fig. 9, columns 1 and 2), they may artifactually appear to co-localize. Therefore, a conclusion of partial co-localization must be viewed with a degree of skepticism.
Gln3-Myc Lies Close to the Endoplasmic Reticulum but Does Not Appear to Co-localize with It
Non-uniform cytoplasmic distribution of Gln3 is reminiscent of results observed with proteins that localize to the endoplasmic reticulum, prompting determination of whether cytoplasmic Gln3 co-localized with the endoplasmic reticulum marker, Kar2 (32, 57, 58). Proteins localized to the endoplasmic reticulum appear as punctate foci situated around the periphery of the cell and circling the nucleus (Fig. 10, column 1). Gln3-Myc appears similarly punctate (Fig. 10, column 3), but it does not co-localize with Kar2 fluorescence except occasionally near the nucleus (Fig. 10, column 4). However, Gln3 and Kar2 foci are often situated very close to one another, even alternating around the cell periphery (Fig. 10, column 4).
Fig. 10. Intracellular localization of Gln3 and Kar2 in cells provided with glutamine as sole nitrogen source.
Cultures (TB123) were grown to mid-log in YNB-glutamine medium and processed for indirect immunofluorescence. Localization of Gln3-Myc13, Kar2, and the nuclei were determined by staining with monoclonal antibody 9E10 anti-Myc (columns 3 and 4, green), rabbit polyclonal antibody anti-Kar2 (columns 1, 2, and 4, red), and DAPI (column 2, blue), respectively. Merged images (columns 2 and 4) show the overlap of Kar2 and DAPI-stained material and Gln3 and Kar2 (yellow), respectively. Micrographs were imaged using a Zeiss Axioplan 2 imaging microscope. 0.4-μm sections were collected as a Z-stack, and one image from the center of that stack is shown in rows A and B. Images in rows C–E show three contiguous sections from a single Z-stack. Images were deconvolved with AxioVision 3.0 software using the constrained iterative algorithm.
Gln3 Localizes to Projections Emanating from the Nucleus
Gln3 quickly exits from the nuclei of cells transferred from proline to glutamine medium. However, shortly after transfer, i.e. in cells where Gln3 is still mostly nuclear, projections with which Gln3 is associated emanate from the nucleus (Fig. 11, row F, taken from Cox et al. (7)). This prompted us to determine whether Gln3 behaved similarly when it was being accumulated within the nucleus. As before, we selected cells in which some, but not all Gln3 was situated in the nucleus, thereby limiting the amount of fluorescence in the cytoplasm. As shown in Fig. 11, rows A–E, similar projections are clearly visible in cells immediately after transfer from glutamine to proline medium. Although the identity of these Gln3-containing projections is not presently known, their presence is consistent with the suggestions that (i) Gln3 movement into and out of the nucleus does not occur uniformly across its surface and (ii) a vesicular system may play a role in Gln3 entry into and exit from the nucleus.
DISCUSSION
Data presented in this work demonstrate NCR-sensitive gene expression and Gln3 accumulation in the nuclei of cells transferred from conditions of nitrogen excess to limitation is almost completely inhibited by latrunculin, suggesting a strong requirement for an intact actin cytoskeleton. In contrast, nuclear accumulation of Gln3 in response to rapamycin-treatment did not exhibit this requirement, i.e. latrunculin does not inhibit rapamycin-induced Gln3 accumulation in the nucleus.
There are at least two ways of interpreting these observations. In the first instance, the data are those expected if the requirement for an actin cytoskeleton occurs before the participation of Tor1/2 in the response of Gln3 intracellular localization to nitrogen limitation. This explanation would in turn argue that polymerized actin is required for transmitting the signal indicating nitrogen limitation or the loss of a signal indicating nitrogen excess to Tor1/2. Here, it is unlikely that the process requiring polymerized actin directly involves the Gln3 molecule itself. Alternatively, the pathways through which intracellular Gln3 localization responds to nitrogen limitation and rapamycin inhibition of Tor1/2 differ from one another. In this regard, data described above may represent another in a growing number of instances in which the results observed in response to rapamycin-mediated inhibition of Tor1/2 are not congruent with those observed after the onset of nitrogen limitation (13, 59). Although the latter interpretation would conceptually permit the actin cytoskeleton-dependent process to be situated close to Gln3 in the regulatory pathway, the limited co-localization of Gln3 and actin we observed does not rigorously demonstrate a stable interaction between the majority of cytoplasmic Gln3 and the actin cytoskeleton. At the same time it is important to note that transient direct or indirect interactions between Gln3 and actin would not likely be detected by the methods we used.
One of the observations we made, if it did not derive from an artifact, may provide some insight into the kinetics of intracellular Gln3 movement. We observed that when cells provided with proline as sole nitrogen source are treated with latrunculin, Gln3 leaves the nucleus in the absence of a cellular signal indicating nitrogen excess (Fig. 1, open bars). Although there is a modest, transient release of Gln3 from nuclei that derives from the ethanol used to dissolve latrunculin, this release does not account for the effects observed with latrunculin treatment because untreated cells fully recover, i.e. Gln3 is nuclear in 95–98% of them. In contrast, the fraction of latrunculin-treated cells in which Gln3 remains nuclear continues to decrease until Gln3 is cytoplasmic in all but 1–3% of them. We can envision at least two ways of interpreting this observation; (i) an intact actin cytoskeleton is required for Gln3 not only to move into the nucleus but also to be maintained there and, (ii) Gln3 continuously cycles into and out of the nucleus, i.e. it is in formal terms not unlike the dynamic assembly and disassembly of the actin cytoskeleton itself. By the latter interpretation, Gln3 accumulates in the nucleus because its rate of entry is greater than the combined rates of the processes in which Gln3 participates within the nucleus and then exits from it. Latrunculin disrupts Gln3 cycling by preventing it from moving into the nucleus, thus causing it to aberrantly collect in the cytoplasm of cells provided with a poor nitrogen source. Two significant consequences derive from such potential cycling of Gln3 into and out of the nucleus even during nitrogen limitation as follows. (i) The sensitivity and resolution of the regulatory system to changes in the nitrogen supply is much greater than if Gln3 remained constantly in the nucleus until cells encountered conditions of nitrogen excess. (ii) The response time to changing conditions is much shorter because entry is slowed or disrupted depending upon the quality of the nitrogen source rather than development, transmission, and transduction of a second signal being required to bring about Gln3 exit from the nucleus.
Finally, our experiments demonstrate two significant characteristics of the cytoplasmic distribution of Gln3. First, Gln3-Myc does not appear to co-localize with a marker of the endoplasmic reticulum, but it does often appear in close proximity to this vesicular system and at times co-localizes with it in areas near the nucleus. In fact, we frequently observe Gln3 and endoplasmic reticulum marker Kar2 alternating around the periphery of the cell. Whether this organizational feature of Gln3 and the endoplasmic reticulum possesses physiological significance is uncertain. Second, Gln3 appears to be localized with tubular/vesicular structures as it enters and exits from the nucleus rather than being uniformly distributed around it. These structures appear to extend out from the nucleus itself. Such observations would be those expected if a tubular/vesicular system participated in the movement of Gln3 between the cytoplasmic and nuclear compartments. Clearly, much remains to be learned about the molecular mechanisms that regulate Gln3 intracellular localization and movement. However, these and earlier data increasingly point toward the participation of an intracellular trafficking system in this process.
Acknowledgments
We thank Dr. Randy Schekman for providing Kar2 specific antibody, Dr. Michael Hall for wild type strain TB123, Dr. David Drubin for the act1-157 mutant, Dr. John Cox for helpful suggestions, Tim Higgins for preparing the artwork, and the University of Tennessee Yeast Group for suggestions in improving the manuscript.
Footnotes
This work was supported by National Institutes of Health Grant GM-35642.
The abbreviations used are: NCR, nitrogen catabolite repression; DAPI, 4,6-diamidino-2-phenylindole; YNB, yeast nitrogen base.
References
- 1.Beck T, Hall MN. Nature. 1999;402:689–692. doi: 10.1038/45287. [DOI] [PubMed] [Google Scholar]
- 2.Hardwick JS, Kuruvilla FG, Tong JK, Shamji AF, Schreiber SL. Proc Natl Acad Sci U S A. 1999;96:14866–14870. doi: 10.1073/pnas.96.26.14866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cardenas ME, Cutler NS, Lorenz MC, Di Como CJ, Heitman J. Genes Dev. 1999;13:3271–3279. doi: 10.1101/gad.13.24.3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bertram PG, Choi JH, Carvalho J, Ai W, Zeng C, Chan TF, Zheng XFS. J Biol Chem. 2000;275:35727–35733. doi: 10.1074/jbc.M004235200. [DOI] [PubMed] [Google Scholar]
- 5.Cox KH, Rai R, Distler M, Daugherty JR, Coffman JA, Cooper TG. J Biol Chem. 2000;275:17611–17618. doi: 10.1074/jbc.M001648200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shamji AF, Kuruvilla FG, Schreiber SL. Curr Biol. 2000;10:1574–1581. doi: 10.1016/s0960-9822(00)00866-6. [DOI] [PubMed] [Google Scholar]
- 7.Cox KH, Tate JJ, Cooper TG. J Biol Chem. 2002;277:37559–37566. doi: 10.1074/jbc.M204879200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carvalho J, Zheng XFS. J Biol Chem. 2003;278:16878–16886. doi: 10.1074/jbc.M300429200. [DOI] [PubMed] [Google Scholar]
- 9.Carvalho J, Bertram PG, Wente SR, Zheng XFS. J Biol Chem. 2001;276:25359–25365. doi: 10.1074/jbc.M103050200. [DOI] [PubMed] [Google Scholar]
- 10.Cooper TG. In: Mycota III. Marzluf G, Bambrl R, editors. Springer-Verlag; Berlin: 1996. pp. 139–169. [Google Scholar]
- 11.Hoffman-Bang J. Mol Biotechnol. 1999;12:35–73. doi: 10.1385/MB:12:1:35. [DOI] [PubMed] [Google Scholar]
- 12.ter Schure EG, van Riel NA, Verrips CT. FEMS Microbiol Rev. 2000;24:67–83. doi: 10.1111/j.1574-6976.2000.tb00533.x. [DOI] [PubMed] [Google Scholar]
- 13.Cooper TG. FEMS Microbiol Rev. 2002;26:223–238. doi: 10.1111/j.1574-6976.2002.tb00612.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kulkarni AA, Abul-Hamd AT, Rai R, El Berry H, Cooper TG. J Biol Chem. 2001;276:32136–32144. doi: 10.1074/jbc.M104580200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rai R, Tate JJ, Cooper TG. J Biol Chem. 2003;278:12826–12833. doi: 10.1074/jbc.M212186200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Torres J, Di Como CJ, Herrero E, De La Torre-Ruiz MA. J Biol Chem. 2002;277:43495–43504. doi: 10.1074/jbc.M205408200. [DOI] [PubMed] [Google Scholar]
- 17.Crespo JL, Daicho K, Ushimaru T, Hall MN. J Biol Chem. 2001;276:34441–34444. doi: 10.1074/jbc.M103601200. [DOI] [PubMed] [Google Scholar]
- 18.Schmidt A, Beck T, Koller A, Kunz J, Hall MN. EMBO J. 1998;17:6924–6931. doi: 10.1093/emboj/17.23.6924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Helliwell SB, Howald I, Barbet N, Hall MN. Genetics. 1998;148:99–112. doi: 10.1093/genetics/148.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmidt A, Kunz J, Hall MN. Proc Natl Acad Sci U S A. 1996;93:13780–13785. doi: 10.1073/pnas.93.24.13780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barbet NC, Schneider U, Helliwell SB, Stansfield I, Tuite MF, Hall MN. Mol Biol Cell. 1996;7:25–42. doi: 10.1091/mbc.7.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kunz J, Henriquez R, Schneider U, Deuter-Reinhard M, Movva NR, Hall MN. Cell. 1993;73:585–596. doi: 10.1016/0092-8674(93)90144-f. [DOI] [PubMed] [Google Scholar]
- 23.Heitman J, Movva NR, Hall MN. Science. 1991;253:905–909. doi: 10.1126/science.1715094. [DOI] [PubMed] [Google Scholar]
- 24.Noda T, Ohsumi Y. J Biol Chem. 1998;273:3963–3966. doi: 10.1074/jbc.273.7.3963. [DOI] [PubMed] [Google Scholar]
- 25.Thomas G, Hall MN. Curr Opin Cell Biol. 1997;9:782–787. doi: 10.1016/s0955-0674(97)80078-6. [DOI] [PubMed] [Google Scholar]
- 26.Zheng XFS, Schreiber SL. Proc Natl Acad Sci U S A. 1997;94:3070–3075. doi: 10.1073/pnas.94.7.3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zheng XFS, Florentino D, Chen J, Crabtree GR, Schreiber SL. Cell. 1995;82:121–130. doi: 10.1016/0092-8674(95)90058-6. [DOI] [PubMed] [Google Scholar]
- 28.Liu Z, Sekito T, Epstein CB, Butow RA. EMBO J. 2001;20:7209–7219. doi: 10.1093/emboj/20.24.7209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Loewith R, Jacinto E, Wullschleger S, Lorberg A, Crespo JL, Bonenfant D, Oppliger W, Jenoe P, Hall MN. Mol Cell. 2002;10:457–468. doi: 10.1016/s1097-2765(02)00636-6. [DOI] [PubMed] [Google Scholar]
- 30.Chen EJ, Kaiser CA. J Cell Biol. 2003;161:333–347. doi: 10.1083/jcb.200210141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wedaman KP, Reinke A, Anderson S, Yates J, McCaffery JM, Powers T. Mol Biol Cell. 2003;14:1204–1220. doi: 10.1091/mbc.E02-09-0609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Preuss D, Mulholland J, Kaiser CA, Orlean P, Albright C, Rose MD, Robbins PW, Botstein D. Yeast. 1991;7:891–911. doi: 10.1002/yea.320070902. [DOI] [PubMed] [Google Scholar]
- 33.Vida TA, Emr SD. J Cell Biol. 1995;128:779–792. doi: 10.1083/jcb.128.5.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kunz J, Schneider U, Howald I, Schmidt A, Hall MN. J Biol Chem. 2000;275:37011–37020. doi: 10.1074/jbc.M007296200. [DOI] [PubMed] [Google Scholar]
- 35.Cardenas ME, Heitman J. EMBO J. 1995;14:5892–5907. doi: 10.1002/j.1460-2075.1995.tb00277.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ayscough KR, Stryker J, Pokala N, Sanders M, Crews P, Drubin DG. J Cell Biol. 1997;137:399–416. doi: 10.1083/jcb.137.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ayscough KR. Curr Biol. 2000;10:1587–1590. doi: 10.1016/s0960-9822(00)00859-9. [DOI] [PubMed] [Google Scholar]
- 38.Kaminska J, Gajewska B, Hopper AK, Zoladek T. Mol Cell Biol. 2002;22:6946–6948. doi: 10.1128/MCB.22.20.6946-6958.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dewar H, Warren DT, Gardiner FC, Gourlay CG, Satish N, Richardson MR, Andrews PD, Ayscough KR. Mol Biol Cell. 2002;13:3646–3661. doi: 10.1091/mbc.E02-05-0262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eitzen G, Wang L, Thorngren N, Wickner W. J Cell Biol. 2002;158:669–679. doi: 10.1083/jcb.200204089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kang KW, Lee SJ, Park JW, Kim SG. Mol Pharmacol. 2002;62:1001–1010. doi: 10.1124/mol.62.5.1001. [DOI] [PubMed] [Google Scholar]
- 42.Zipper LM, Mulcahy RT. J Biol Chem. 2002;277:36544–36552. doi: 10.1074/jbc.M206530200. [DOI] [PubMed] [Google Scholar]
- 43.Itoh K, Wakabayashi N, Katoh Y, Ishii T, Igarashi K, Engel JD, Yamamoto M. Genes Dev. 1999;13:76–86. doi: 10.1101/gad.13.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Berg P, Pongratz I. J Biol Chem. 2002;277:32310–32319. doi: 10.1074/jbc.M203351200. [DOI] [PubMed] [Google Scholar]
- 45.Spector I, Shochet NR, Kashman Y, Groweiss A. Science. 1983;219:493–495. doi: 10.1126/science.6681676. [DOI] [PubMed] [Google Scholar]
- 46.Coue M, Brenner SL, Spector I, Korn ED. FEBS Lett. 1987;213:316–318. doi: 10.1016/0014-5793(87)81513-2. [DOI] [PubMed] [Google Scholar]
- 47.Spector I, Shochet NR, Blasberger D, Kashman Y. Cell Motil Cytoskeleton. 1989;13:127–144. doi: 10.1002/cm.970130302. [DOI] [PubMed] [Google Scholar]
- 48.Morton WM, Ayscough K, McLaughlin PJ. Nat Cell Biol. 2000;2:376–378. doi: 10.1038/35014075. [DOI] [PubMed] [Google Scholar]
- 49.Schmitt ME, Brown TA, Trumpower BL. Nucleic Acids Res. 1990;18:3091–3092. doi: 10.1093/nar/18.10.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tate JJ, Cox KH, Rai R, Cooper TG. J Biol Chem. 2002;277:20477–20482. doi: 10.1074/jbc.M200962200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tate JJ, Cooper TG. J Biol Chem. 2003;278:36924–36933. doi: 10.1074/jbc.M301829200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lyman SK, Schekman R. Cell. 1997;88:85–96. doi: 10.1016/s0092-8674(00)81861-9. [DOI] [PubMed] [Google Scholar]
- 53.Belmont LD, Patterson GML, Drubin DG. 1999;112:1325–1336. doi: 10.1242/jcs.112.9.1325. [DOI] [PubMed] [Google Scholar]
- 54.Belmont LD, Drubin DG. J Cell Biol. 1998;142:1289–1299. doi: 10.1083/jcb.142.5.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Di Como CJ, Arndt KT. Genes Dev. 1996;10:1904–1916. doi: 10.1101/gad.10.15.1904. [DOI] [PubMed] [Google Scholar]
- 56.Garber PM, Rine J. Genetics. 2002;161:521–534. doi: 10.1093/genetics/161.2.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rose MD, Misra LM, Vogel JP. Cell. 1989;57:1211–1221. doi: 10.1016/0092-8674(89)90058-5. Correction (1990) Cell 58, 801. [DOI] [PubMed] [Google Scholar]
- 58.Fehrenbacher KL, Davis D, Wu M, Boldogh I, Pon LA. Mol Biol Cell. 2002;13:854–865. doi: 10.1091/mbc.01-04-0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cox KH, Kulkarni A, Tate JJ, Cooper TG. J Biol Chem. 2004;279:10270–10278. doi: 10.1074/jbc.M312023200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cox KH, Pinchak AB, Cooper TG. Yeast. 1999;15:703–713. doi: 10.1002/(SICI)1097-0061(19990615)15:8<703::AID-YEA413>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]